Abstract
The diagnosis of conventional and oncocytic poorly differentiated thyroid carcinomas is difficult. The aim of this study was to characterize their largely unknown miRNA expression profile and to compare it to well differentiated thyroid tumors as well as to identify miRNAs which could potentially serve as diagnostic and prognostic markers.
A total of 14 poorly differentiated, 13 oncocytic poorly differentiated, 72 well differentiated thyroid carcinomas and 8 normal thyroid specimens were studied for expression of 768 miRNAs using PCR-Microarrays.
MiRNA expression was different between poorly differentiated and oncocytic poorly differentiated thyroid carcinomas demonstrating individual clusters on the clustering analysis. Both tumor types showed upregulation of miR-125a-5p, -15a-3p, -182, -183-3p, -222, -222-5p and downregulation of miR-130b, -139-5p, -150, -193a-5p, -219-5p, -23b, -451, -455-3p and of miR-886-3p as compared to normal thyroid tissue. In addition, the oncocytic poorly differentiated thyroid carcinomas demonstrated upregulation of miR-221 and miR-885-5p. The difference in expression was also observed between miRNA expression in poorly differentiated and well differentiated tumors. The CHAID algorithm allowed to separate poorly differentiated from well differentiated thyroid carcinomas with a 73–79% accuracy using miR-23b and miR-150 as a separator. Kaplan-Meier and multivariate analysis showed a significant association with tumor relapses (for miR-23b) and with tumor specific death (for miR-150) in poorly differentiated and oncocytic poorly differentiated thyroid carcinomas.
MiRNA expression is different in conventional and oncocytic poorly differentiated thyroid carcinomas in comparison to well differentiated thyroid cancers and can be used for discrimination between these tumor types. The newly identified deregulated miRNAs (miR-150, miR-23b) bear the potential to be used in a clinical setting delivering prognostic and diagnostic information.
Keywords: thyroid cancer, poorly differentiated carcinoma carcinoma, follicular thyroid carcinoma, papillary carcinoma, miRNA profiling, diagnosis, prognosis
INTRODUCTION
Poorly differentiated thyroid carcinomas of conventional type (PD) and oncocytic (Hürthle) type (oPD) are biologically situated between well differentiated papillary (PTC) and follicular (FTC) thyroid carcinomas on the one hand and anaplastic thyroid carcinomas (ATC) on the other. In contrast to the latter which belongs to the group of the most lethal human neoplasms, PTC and FTC have an excellent prognosis (DeLellis, et al. 2004).
These neoplasms are known to be particularly difficult to diagnose and different diagnostic criteria for these entities have been used in the past, with some considering more the pattern of growth (solid, trabecular, insular), and others emphasizing high-grade features such as necrosis, atypia or a high mitotic index (Carcangiu, et al. 1984; Sakamoto, et al. 1983). The entity was finally recognized by the WHO in 2004, however, the criteria were not well defined (DeLellis et al. 2004). A consensus meeting was held in 2007 and a diagnostic algorithm was developed by different experts in thyroid pathology throughout the world (Volante, et al. 2007). PDs can be admixed with well differentiated thyroid tumors like PTCs or FTCs but even a small PD component determines the patient outcome (Dettmer, et al. 2011). Oncocytic PD were originally not included in the consensus proposal, however it was shown that proposed criteria were also applicable to them, demonstrating an even worse outcome for oncocytic PD compared to conventional PD (Dettmer, et al. 2012).
The use of immunohistochemical markers like Galectin-3 or HBME-1 are only of limited use because signs of malignancy are easily identifiable (Tallini 2011; Volante and Papotti 2010; Volante, et al. 2008). Thus so far, this diagnosis relies on hematoxylin-eosin (H&E) stained slides.
The molecular background of these tumors is only partially understood so far. Specific mutations for this tumor type have not been described. RET/PTC rearrangements and PAX8/PPARγ rearrangements appear not to play a role in PD or oPD (Soares, et al. 2011). Characteristic mutational profiles like BRAF and RAS mutations in PTC have not been described so far, although at least a subset of these tumors seem to originate from classic PTC, showing these mutations (Ricarte-Filho, et al. 2009; Volante, et al. 2009). TP53 mutations can be detected more frequently in these tumors than in well differentiated carcinomas but less frequently than in ATC (Soares et al. 2011). To facilitate and to confirm the diagnosis of PD and oPD carcinomas, reliable molecular tests would be beneficial, ideally combined with prognostic information about tumor behavior.
MicroRNAs (miRNAs) are a class of non-coding RNAs that has been discovered about twenty years ago (Lee, et al. 1993). However it took ten more years until the scientific community recognized their important role in practically all cell processes (Bartel 2004). It is known today, that they are also involved in human cancer (Bartel 2004; Keutgen, et al. 2012; Leone, et al. 2012; Nikiforova, et al. 2008). These tiny regulators can function as oncogenes or tumor suppressor genes by regulating the expression of target genes through loss or gain of miRNA functions (Galasso, et al. 2012). MicroRNA expression signatures have been identified in various human solid malignancies and in thyroid carcinomas (Dettmer, et al. 2013a; Galasso et al. 2012; He, et al. 2005; Nikiforova et al. 2008). A range of miRNAs (miR-155, miR-21, miR-31, miR-146b, miR-221, miR-222) are known to be deregulated in papillary thyroid carcinomas (Chen, et al. 2008; Nikiforova et al. 2008; Schwertheim, et al. 2009; Tetzlaff, et al. 2007; Yip, et al. 2011). They have been proven to be a valuable diagnostic tool in fine needle aspiration biopsies (FNAB) and surgical specimens and were also able to predict patient outcome (Chen et al. 2008; Menon and Khan 2009; Nikiforova, et al. 2009; Yip et al. 2011). However, the information in the literature on PD is very limited and absent for oPD (Nikiforova et al. 2008; Schwertheim et al. 2009).
The aim of this study was to analyze a large set of PD and oPD carcinomas and to establish the miRNA profile of PD and oPD on a large scale, covering the expression of almost 800 different miRNAs and compare it to well differentiated thyroid carcinomas. Further, we investigated whether PD and oPD had distinct miRNA signatures, as was demonstrated recently for FTC, oncocytic FTC (oFTC) and FVPTC (Dettmer et al. 2013a; Dettmer, et al. 2013b). Finally, we evaluated the clinical relevance of deregulated novel candidate miRNAs and assessed their prognostic value.
MATERIAL AND METHODS
Thyroid samples
The study population was enriched with patients having an adverse clinical outcome (ACO) as described elsewhere (Dettmer et al. 2011). This approach increases tremendously the statistical power if one want to assess the factors which may be responsible for an adverse outcome. Nevertheless, one has to bear in mind that this patient collective does not reflect the normal population in a pathology department. ACO was defined when a patient had a least one of the following features: local relapse after first radioiodine therapy, distant metastases or tumor associated death.
In total, we identified 99 thyroid carcinomas with an ACO and used 128 age-, stage- and gender-matched cases as controls. Of those 227 tumors, 64 with an ACO and 35 controls underwent miRNA expression analysis. In total, 107 thyroid neoplastic and non-neoplastic samples were analyzed including 27 poorly differentiated thyroid carcinomas (14 PD and 13oPD), 27 PTC and 17 FVPTC (follicular variant of PTC), 16 follicular thyroid tumors (FTC) and 12 oncocytic FTC (oFTC) and 8 normal thyroid tissues. Patient characteristics are summarized in table 1. All samples of this retrospective study were formalin-fixed paraffin embedded (FFPE) tissues. FFPE tissues were received from the University Hospital Zurich and surrounding pathology institutes, approved by the Cantonal Research Ethics Board (STV 28-2006). The study was conducted according to the REMARK guidelines (McShane, et al. 2005).
Table 1.
Patient characteristics
| tumor type | PTC ACO |
PTC CG |
p-value | FVPTC ACO |
FVPTC CG |
p-value | FTC ACO |
FTC CG |
p-value | oFTC ACO |
oFTC CG |
p-value | PD | oPD | p-value | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | male | 7 | 2 | 0.75 | 4 | 3 | 0.77 | 0 | 2 | 0.27 | 1 | 5 | 0.21 | 4 | 6 | 0.34 |
| female | 13 | 5 | 5 | 5 | 4 | 10 | 3 | 3 | 10 | 7 | ||||||
| Age (mean in years ± Std. Error) | 47.4 ± 3.6 | 48.9 ± 5.5 | 49.2 ± 5.9 | 52.8 ± 6.7 | 42.5 ± 4.6 | 42.7 ± 5.1 | 76.5 ± 0.9 | 60.9 ± 2.7 | 65.4 ± 10.4 | 71.5±9.2 | ||||||
| Tumor stage | pT1 | 2 | 2 | 0.5 | 0 | 1 | 0.18 | 0 | 0 | 0.007 | 0 | 0 | 0.59 | 0 | 0 | 0.098 |
| pT2 | 3 | 0 | 0 | 0 | 3 | 0 | 1 | 1 | 0 | 3 | ||||||
| pT3 | 15 | 5 | 8 | 7 | 1 | 12 | 3 | 7 | 14 | 10 | ||||||
| pT4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| t overall survival (OS) (months ± Std. Error) | 76.9 ± 11.7 | 67.9 ± 10.6 | 73.7 ± 15.7 | 89.0 ± 17.5 | 83.4 ± 22.1 | 114.9 ± 16.3 | 114.8 ± 35.9 | 116.6 ± 19.7 | 70.0 ± 39.6 | 63.4 ± 57.5 | ||||||
| t tumor specific survival (TSS) (months ± Std. Error) | 76.9 ± 11.7 | 67.9 ± 10.6 | 73.7 ± 15.7 | 89.0 ± 17.5 | 83.4 ± 22.1 | 114.9 ± 16.3 | 114.8 ± 35.9 | 119.8 ± 22.4 | 70.0 ± 39.6 | 63.4 ± 57.5 | ||||||
| t relapse free survival (RFS (months ± Std. Error) | 11.0 ± 1.8 | 67.9 ± 10.6 | 26.3 ± 13.3 | 89.1 ± 17.6 | 6.5 ± 1.3 | 114.9 ± 16.3 | 52 ± 34.6 | 119.9 ± 22.4 | 15.8 ± 18.2 | 20.3 ± 22.7 |
PTC = Papillary Thyroid Carcinoma; FVPTC = Follicular Variant of Papillary Thyroid Carcinoma; FTC min. inv. = Minimally Invasive Thyroid Carcinoma; PD = Poorly differentiated Thyroid Carcinoma; oFTC min. inv. = Oncocytic Minimally Invasive Thyroid Carcinoma; oPD = Oncocytic Poorly differentiated Thyroid Carcinoma. ACO = Adverse Clinical Outcome; CG = Control Group
All tumors were classified according to widely accepted diagnostic histologic criteria (DeLellis et al. 2004; Volante et al. 2007). Six 15 μm thick FFPE tissue samples per case were microdissected prior to molecular analysis and it was ensured that representative tumor material was used for RNA extraction. The examiner was blinded to the clinical outcome and the histological diagnosis.
RNA Isolation
Total RNA was extracted from FFPE tissue samples with the RecoverAll kit (Ambion, Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. RNA quantity was assessed with a Spectrophotometer (NanoDrop 1000, Thermo Scientific, USA). MiRNA quality was assessed by amplification of small nucleolar RNA RNU44.
miRNA Expression Analysis
Quantitation of mature miRNA expression levels in thyroid tumors and normal thyroid tissue was performed using TaqMan® Human Microarray Assays v3 (Applied Biosystems, Life Technologies, Carlsbad, CA) which is designed to detect 768 human miRNAs. The array was investigated on an ABI 7900 platform (Applied Biosystems, Life Technologies, Carlsbad, CA). Briefly 150 ng of total RNA was reverse transcribed using a high-capacity cDNA archive kit (Applied Biosystems, Life Technologies, Carlsbad, CA) followed by a preamplification and amplification on ABI 7900 Real-Time PCR System (Applied Biosystems, Life Technologies, Carlsbad, CA). Endogenous controls RNU44 and U6 snRNA (Applied Biosystems, Life Technologies, Carlsbad, CA) were used for the normalization of RNA input and non-human miRNA ath-miR159a was used as negative control.
miRNA expression levels were calculated by relative quantitation using DataAssist v3.0 software (Applied Biosystems, Life Technologies, Carlsbad, CA) and the fold-expression changes were determined by 2−ΔΔCT method as compared to normal thyroid tissue(Livak and Schmittgen 2001). The maximum allowed Ct value for calculations was 37. Outliers among replicates were excluded and p-values were adjusted using Benjamini-Hochberg false discovery rate. The data is presented as the fold change of miRNA expression in tumors relatively to normal thyroid tissues after normalization to endogenous controls RNU44 and U6 snRNA.
Patient follow-up
Complete follow-up data were collected using chart reports and the cancer registery of the canton Zuerich and recorded as overall survival (OS), tumour-specific survival (TSS) and relapse-free survival (RFS) as previously described (Dettmer et al. 2011).
Statistical analysis
DataAssist v3.0 software (Applied Biosystems, Life Technologies, Carlsbad, CA) was used to calculate agglomerative hierarchical clustering and RQ Plots between thyroid specimens. Assessment of the sample distribution (Kolmogorov-Smirnov-test), descriptive statistics, Chi-squared Automatic Interaction Detection (CHAID), Kaplan Meier survival analysis (log rank test) and Cox regression analysis were performed with SPSS 21 (IBM, Armonk, USA).
RESULTS
miRNA expression profiles of PD and oPD
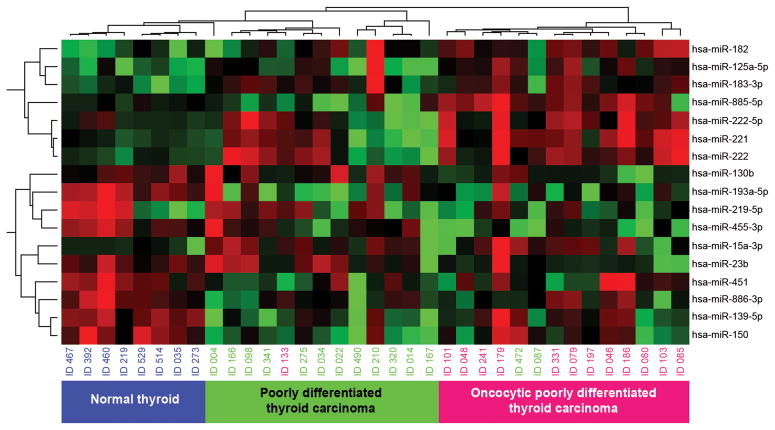
Seventeen miRNAs showed significant deregulation in PD and oPD as compared to normal thyroid tissue. Both, PD and oPDs showed upregulation of miR-125a-5p, miR-15a-3p, miR-182, miR-183-3p, miR-222, miR-222-5p and downregulation of miR-130b, -139-5p, -150, -193a-5p, -219-5p, -23b, -451, -455-3p and of miR-886-3p (Table 2). The most upregulated miRNAs were miR-183-3p (7 fold) in PD and miR-221 and miR-885-5p in oPD. The most downregulated miRNA in both tumor types was miR-219-5p demonstrating 30 fold downregulation in PD and 160 fold in oPD as compared to normal tissue (Table 2). The unsupervised hierarchical clustering analysis of miRNA expression showed separate clusters for PD, oPD and for normal thyroid tissue (Fig. 1).
Table 2.
Fold change of miRNAs as compared to normal thyroid tissue.
| Assay | PD origin PTC (RQ) |
PD origin PTC (P- Value) |
onc PD (RQ) |
onc PD (P- Value) |
FTC min inv ACO (RQ) |
FTC min inv ACO (P- Value) |
FTC min inv CG (RQ) |
FTC min inv CG (P- Value) |
onc FTC min inv ACO (RQ) |
onc FTC min inv ACO (P- Value) |
onc FTC min inv CG (RQ) |
onc FTC min inv CG (P- Value) |
FVPTC ACO (RQ) |
FVPTC ACO (P- Value) |
FVPTC CG (RQ) |
FVPTC CG (P- Value) |
classic PTC ACO (RQ) |
classic PTC ACO (P- Value) |
classic PTC CG (RQ) |
classic PTC CG (P- Value) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-125a-5p | 1.5102 | 0.4881 | 2.4358 | 0.0187 | 1.2707 | 0.4421 | 1.6433 | 0.2316 | 4.0741 | 0.3049 | 4.7852 | 0.0075 | 2.5696 | 0.2219 | 1.7166 | 0.066 | 1.4341 | 0.3937 | 2.1557 | 0.5639 |
| hsa-miR-130b | 0.8801 | 0.9389 | 0.1602 | 0.0187 | 1.783 | 0.6458 | 2.5355 | 0.2316 | 1.731 | 0.7234 | 0.3676 | 0.0926 | 0.4902 | 0.2219 | 2.3146 | 0.3188 | 0.6463 | 0.3937 | 0.7208 | 0.6894 |
| hsa-miR-139-5p | 0.1707 | 0.0201 | 0.3858 | 0.1434 | 0.677 | 0.4421 | 0.388 | 0.0238 | 0.536 | 0.3049 | 0.7897 | 0.507 | 0.6616 | 0.2219 | 0.3725 | 0.066 | 0.2414 | 0.0066 | 0.405 | 0.0453 |
| hsa-miR-150 | 0.1761 | 0.0504 | 0.2722 | 0.0947 | 0.354 | 0.2963 | 0.1426 | 0.103 | 0.4136 | 0.3049 | 0.2988 | 0.134 | 0.6563 | 0.4591 | 0.2031 | 0.1116 | 0.7581 | 0.5937 | 1.3036 | 0.6661 |
| hsa-miR-15a-3p | 5.1826 | 0.0504 | 4.6616 | 0.1309 | 5.1007 | 0.2963 | 3.3332 | 0.1252 | 8.4616 | 0.4806 | 4.7142 | 0.1786 | 5.0306 | 0.1008 | 3.7938 | 0.1657 | 4.9066 | 0.0628 | 8.3654 | 0.5639 |
| hsa-miR-182 | 4.1321 | 0.3134 | 15.7878 | 0.0102 | 3.0254 | 0.4586 | 5.8046 | 0.103 | 10.0218 | 0.3049 | 17.0327 | 0.1449 | 4.4518 | 0.2219 | 1.7252 | 0.5809 | 2.4388 | 0.0383 | 3.2818 | 0.6282 |
| hsa-miR-183-3p | 7.529 | 0.099 | 13.6166 | 0.0102 | 3.388 | 0.4118 | 10.2013 | 0.0027 | 10.4589 | 0.3049 | 29.1113 | 0.0667 | 2.4758 | 0.2219 | 8.1253 | 0.1401 | 1.5139 | 0.5937 | 3.393 | 0.5639 |
| hsa-miR-193a-5p | 0.1959 | 0.0504 | 0.2206 | 0.0105 | 0.3138 | 0.2352 | 0.2039 | 0.0053 | 0.4504 | 0.3049 | 0.288 | 0.0075 | 0.5907 | 0.2125 | 0.4214 | 0.066 | 0.2925 | 0.0066 | 0.4258 | 0.0639 |
| hsa-miR-219-5p | 0.0301 | 0.0504 | 0.006 | 0.009 | 0.0132 | 0.2963 | 0.0415 | 0.231 | 0.1188 | 0.3049 | 0.0102 | 0.1786 | 0.1312 | 0.2219 | 0.0422 | 0.178 | 0.0093 | --- | 0.1217 | 0.4679 |
| hsa-miR-221 | 0.9105 | 0.9788 | 12.2539 | 0.0081 | 4.3356 | 0.4118 | 5.1579 | 0.1045 | 17.4066 | 0.3049 | 29.5232 | 0.0075 | 13.3133 | 0.0649 | 19.4575 | 0.066 | 8.8605 | 0.0692 | 8.4463 | 0.0453 |
| hsa-miR-222 | 1.6391 | 0.8249 | 9.7331 | 0.0081 | 4.8395 | 0.2963 | 4.9973 | 0.2857 | 7.3821 | 0.3049 | 7.0607 | 0.0084 | 27.2159 | 0.1829 | 11.7024 | 0.0853 | 13.9184 | 0.001 | 19.3691 | 0.4679 |
| hsa-miR-222-5p | 1.2917 | 0.9389 | 7.2611 | 0.0491 | 1.2929 | 0.6677 | 1.5184 | 0.5663 | 4.754 | 0.4118 | 5.5375 | 0.0192 | 2.8774 | 0.1599 | 11.8716 | 0.264 | 1.4406 | 0.5676 | 1.4444 | 0.6379 |
| hsa-miR-23b | 0.7987 | 0.8369 | 0.1962 | 0.0466 | 0.9723 | 0.9397 | 1.4223 | 0.3509 | 1.0668 | 0.9409 | 0.745 | 0.5174 | 1.3934 | 0.4802 | 2.2949 | 0.1401 | 0.7094 | 0.5228 | 0.8915 | 0.804 |
| hsa-miR-451 | 0.2865 | 0.0504 | 0.4047 | 0.2194 | 0.3729 | 0.2963 | 0.4661 | 0.1612 | 0.4923 | 0.3049 | 0.2964 | 0.0447 | 0.3157 | 0.1008 | 0.3498 | 0.089 | 0.1935 | 0.0292 | 0.2783 | 0.1313 |
| hsa-miR-455-3p | 0.406 | 0.4881 | 0.1474 | 0.0454 | 0.2969 | 0.2963 | 0.141 | 0.103 | 1.2864 | 0.8904 | 0.0566 | 0.0447 | 0.4076 | 0.2219 | 0.2612 | 0.1116 | 0.3434 | 0.1663 | 0.7686 | 0.6661 |
| hsa-miR-885-5p | 1.917 | 0.7069 | 42.8892 | 0.0204 | 0.2762 | 0.2963 | 1.1838 | 0.739 | 105.7278 | 0.575 | 520.8887 | 0.3223 | 0.7637 | 0.5749 | 0.8291 | 0.6616 | 15.8382 | 0.4348 | 4.2795 | 0.6155 |
| hsa-miR-886-3p | 0.1247 | 0.0504 | 0.2513 | 0.1482 | 0.3488 | 0.3146 | 0.4431 | 0.2978 | 0.3415 | 0.3049 | 0.779 | 0.6609 | 0.5886 | 0.4591 | 0.1852 | 0.1401 | 0.3833 | 0.3051 | 0.5471 | 0.5639 |
P-values are indicated in relation to normal thyroid tissue. PTC = Papillary Thyroid Carcinoma; FVPTC = Follicular Variant of Papillary Thyroid Carcinoma; FTC min. inv. = Minimally Invasive Thyroid Carcinoma; PD = Poorly differentiated Thyroid Carcinoma; oFTC min. inv. = Oncocytic Minimally Invasive Thyroid Carcinoma; oPD = Oncocytic Poorly differentiated Thyroid Carcinoma. ACO = Adverse Clinical Outcome; CG = Control Group
Figure 1.
Unsupervised hierarchical clustering analysis (Pearson correlation, complete linkage) of poorly differentiated (PD) and oncocytic poorly differentiated (oPD) thyroid carcinomas and normal thyroid tissue based on their miRNA expression. PD (green), oPD (pink), and normal thyroid tissue (blue) form three distinct clusters.
Progression of deregulated miRNAs from well differentiated to poorly differentiated tumors
Next, we compared expression of these 17 deregulated miRNAs in PD tumors with their expression in well differentiated thyroid carcinomas (Table 2). Several miRNAs (miR-221, -222) that were upregulated in PTC and FVPTC showed loss of expression in PD (p<0.05). Other miRNAs (miR-15a-3p and miR-183-3p (p<0.05)) were upregulated in PD tumors as compared to well differentiated tumors.
When oPD cancers were compared to well differentiated oncocytic tumors, a number of miRNAs (miR-125a-5p, -183-3p, -219-5p, -221 and miR-885-5p) showed loss of expression (Table 2). In contrast, miR-222 was twice more highly expressed in oPD as compared to oFTC.
We also evaluated the possibility of discriminating PD and oPD from well differentiated thyroid tumors using the Chi-squared Automatic Interaction Detection (CHAID) algorithm. It revealed a 73.3% overall accuracy for the separation between FTC and PD and a 75% accuracy for oFTC and oPD using miR-23b as a separator with a cut-off of 0.5 fold. MiR-150 (cut-off of 0.2 fold) was able to separate PTC from PD with an accuracy of 79.3% using CHAID.
Deregulated miRNAs and patient survival
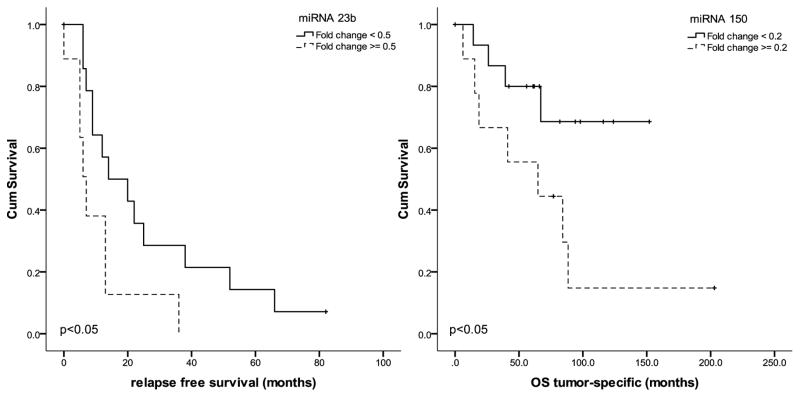
One significant deregulated miRNA in PD and oPD was able to predict a decreased relapse free survival (RFS) in these tumors: miR-23b (Fig. 2). This miRNA has been confirmed as the only independent predictor of tumor relapse in a multivariate Cox regression analysis including patient age, tumor stage and gender-matched tumors. The hazard ratio was Exp(B)=2.62; 95% CI: 1.01 – 6.77). MiR-150 demonstrates a significant decreased tumor specific survival (TSS) in a Kaplan Meier analysis (Fig. 2), which could also be confirmed in a multivariate Cox regression analysis including age, stage and gender-matched tumors for TSS with patients having a 5.03 fold increased (95% CI: 1.29 – 19.69) risk of a fatal outcome (table 3). Of note, tumor necrosis and or an increased mitotic index were not able to identify further patients with an adverse outcome in PD patients.
Figure 2.
Kaplan Meier survival analysis including poorly differentiated (PD) and oncocytic poorly differentiated (oPD) thyroid carcinomas: Relapse free survival is significantly reduced for miR-23b. Tumor specific survival is reduced for miR-150. Tests are log rank tests.
Table 3.
Clinicopathological characteristics and expression of miR-150 and miR-23b. Although there were initially 27 PD and oPD, the assay for miR-150 failed in one sample and for miR-23b in 4 samples, reflecting the difficulties with downregulated markers.
| MiRNA (fold change) | miR-150 <0.2 |
miR-150 ≥0.2 |
P-value | |
|---|---|---|---|---|
| Age (years) | 68.8±9.7 | 67.1±11.2 | 0.774 | |
| pT | pT 1–2 | 1 | 2 | 0.446 |
| pT 3–4 | 14 | 9 | ||
| gender | male | 7 | 3 | 0.082 |
| female | 8 | 8 | ||
| MiRNA (fold change) | miR-23b <0.5 |
miR-23b ≥0.5 |
||
| Age (years) | 69.6±9.6 | 64.7±12.5 | 0.615 | |
| pT | pT 1–2 | 2 | 1 | 0.584 |
| pT 3–4 | 12 | 8 | ||
| gender | male | 6 | 2 | 0.024 |
| female | 8 | 7 | ||
DISCUSSION
We analyzed a large series of poorly differentiated thyroid carcinomas for the complete miRNA profile of the Sanger Database v16 generating more than 80,000 TaqMan PCR-based data points and compared it with the miRNA profile of well differentiated tumors. All PD tumors were diagnosed according to widely accepted criteria and the Turin proposal (DeLellis et al. 2004; Volante et al. 2007) ensuring diagnostic accuracy and comparability of these data to other studies.
MiRNAs have drawn increasing attention in recent years. Their deregulation has been shown in many different human tumors including thyroid neoplasms and they have been increasingly used as diagnostic and prognostic markers by our group and others (Dettmer et al. 2013a; Lu, et al. 2005; Nikiforova et al. 2008; Sheu, et al. 2010).
miRNA expression profiles of PD and oPD
So far, there is only very limited information available on miRNA profiles in PD carcinomas and to our knowledge no information on oPD (Nikiforova et al. 2008; Schwertheim et al. 2009). The first study on miRNA profiles in PD was done by our group and out of ten reported miRNAs at that time, we were able to confirm seven in the present study including miR-183, -221 and -222 (Nikiforova et al. 2008). Four more miRNAs were concordantly expressed in our present study as well (miR-129, -146b, -181a and -339). However they were not found to be significantly deregulated in this present work and therefore were excluded from further analysis. Out of the remaining three miRNAs, miR-181b was not included on the present array, miR-187 was not amplified, while miR-213 was the only one not being concordantly expressed. The other study on PD miRNA profiles was carried out by Schwertheim and colleagues (Schwertheim et al. 2009). They reported ten miRNAs and we are currently able to confirm eight of them, miR-222 is discordantly expressed and does not reach statistical significance in their study and one (miR-181b) was not on our array.
As facts accumulate, we know that tumors originating from the same cell of origin can present with distinct miRNA profiles (Dettmer et al. 2013a; Dettmer et al. 2013b; Nikiforova et al. 2008). Although there were a few outliers, the unsupervised hierarchical clustering analysis including significant deregulated miRNAs in PD and oPDs separates these tumors into two distinct groups, suggesting that this also might be true for these types of thyroid carcinomas.
Progression of deregulated miRNAs from well differentiated to poorly differentiated tumors
Several miRNAs are increasingly deregulated between well differentiated and poorly differentiated tumors. Among them miR-221 and miR-222 have been well described in PTC for a long time (Nikiforova et al. 2008). They are both known to negatively regulate p27 and an absent p27 expression can be observed in various aggressive human neoplasms as well (Visone, et al. 2007). Interestingly, the latter two miRNAs are less deregulated in PD compared to PTC, suggesting that this pathway is not driving tumor progression in PD. We were unable to find significant correlations when looking into survival data between miR-221 and miR-222 and PD or oPD, which is consistent with this observation. In contrast, miR-183-3p is upregulated in PD as compared to well differentiated PTC or FVPTC and is also involved in decreased patient survival, supporting the progression model from well differentiated to PD carcinomas. Interestingly, miR-183 has been shown to be also upregulated in aggressive prostate cancer where it targets SMAD4 and DKK3 in the WNT/β-catenin pathway (Ueno, et al. 2013).
MiR-222 is more highly expressed in oPD than in oFTC while other miRNAs are lost in the more aggressive tumors like miR-139-5p, -219-5p, and -23b, further supporting the progression model. MiR-139-5p is lost in aggressive breast carcinomas, targeting Ras and PI3K members as well as NFκB (REF KRISHNAN) while miR-23b is known to be suppressed by c-Myc (Gao, et al. 2009). All those pathways and genes are known to be involved in thyroid tumorigenesis for a long time (Nikiforov 2009; Yamashita, et al. 1986).
Diagnostic use
Several miRNAs may be used diagnostically to distinguish between PTC/FVTC/FTC and PD. The most promising candidates would be the ones which are significantly deregulated between the entities like miR-150, -183-3p, -222 and miR-222. The separation between FTC and PD and between oFTC and oPD can be particularly difficult. While feeding the CHAID-Algorithm with these four biomarkers, an overall accuracy of about 75% could be achieved for both clinically relevant questions with the use of only one miRNA: miR-23b. The accuracy increased to almost 80% when using miR-150 as a separator between PTC and PD. These miRNAs may help to diagnose difficult cases in the future and to stratify patients appropriately. One limitation of the study is the fact that most of these markers are downregulated. If one tests for their expression and gets a negative result, it is difficult to know whether the tested marker is strongly downregulated and therefore not detectable by PCR or whether the assay did not work properly.
Biological implications of reported miRNAs and patient survival
A very important aspect of the present work is not only to show the actual miRNA profile of the tumors but also to provide information about the clinical consequences of their deregulation. A subset of the significantly deregulated miRNAs has an impact on patient survival and can predict tumor relapse and survival even in tumors with such an adverse outcome. These miRNAs are known to play important roles in other malignancies such as esophageal squamous cell carcinoma (Yokobori, et al. 2013) for miR-150 or prostate cancer for miR-23b (Gao et al. 2009). Tumor necrosis, an increased mitotic index or convoluted nuclei, normally very strong indicators of an adverse outcome, were not able to further sub-stratify patients in the PD groups which is not surprising, because these features are part of the Turin criteria which define these tumors (Volante et al. 2007).
So far, miRNAs in thyroid cancer are used as diagnostic and predictive tools only. Nevertheless, miRNA based therapies have been shown to be an effective instrument in hepatocellular carcinoma and may hopefully be one day also available in thyroid cancer (Kota, et al. 2009).
Conclusions
This is the first comprehensive miRNA profile for PD and oPD in the literature. We compared the expression of the most deregulated miRNAs in PD and oPD to their expression in the most common thyroid carcinomas in PTC, FVPTC, FTC and oFTC, aggressive and non-aggressive. A subset of seventeen miRNAs is able to separate PD from oPD and normal thyroid tissue, suggesting that these are in fact two distinct entities. Two of those markers, miR-150 and miR-23b bear the potential to provide prognostic and diagnostic evidence at the same time.
Acknowledgments
This work was supported by the National Institute of Health grant R01 CA88041 (Y.E. N.) and by the Fondation pour la recherche Nuovo-Soldati, the Gertrud-Hagmann-Stiftung für Malignomforschung and the Research Support Foundation (M.D.). We thank Martina Storz for technical assistance and Dr. Inti Zlobec for reviewing the manuscript.
Footnotes
Conflict of interest statement: None declared.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Carcangiu ML, Zampi G, Rosai J. Poorly differentiated (“insular”) thyroid carcinoma. A reinterpretation of Langhans’ “wuchernde Struma”. Am J Surg Pathol. 1984;8:655–668. doi: 10.1097/00000478-198409000-00005. [DOI] [PubMed] [Google Scholar]
- Chen YT, Kitabayashi N, Zhou XK, Fahey TJ, 3rd, Scognamiglio T. MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Mod Pathol. 2008;21:1139–1146. doi: 10.1038/modpathol.2008.105. [DOI] [PubMed] [Google Scholar]
- DeLellis R, Lloyd R, Heitz P, Eng C. Pathology and genetics of tumours of endocrine organs. Lyon: IARC Press; 2004. World Health Organization classification of tumours. [Google Scholar]
- Dettmer M, Schmitt A, Steinert H, Haldemann A, Meili A, Moch H, Komminoth P, Perren A. Poorly differentiated thyroid carcinomas: how much poorly differentiated is needed? Am J Surg Pathol. 2011;35:1866–1872. doi: 10.1097/PAS.0b013e31822cf962. [DOI] [PubMed] [Google Scholar]
- Dettmer M, Schmitt A, Steinert H, Moch H, Komminoth P, Perren A. Poorly differentiated oncocytic thyroid carcinoma--diagnostic implications and outcome. Histopathology. 2012;60:1045–1051. doi: 10.1111/j.1365-2559.2012.04188.x. [DOI] [PubMed] [Google Scholar]
- Dettmer M, Vogetseder A, Durso MB, Moch H, Komminoth P, Perren A, Nikiforov YE, Nikiforova MN. MicroRNA Expression Array Identifies Novel Diagnostic Markers for Conventional and Oncocytic Follicular Thyroid Carcinomas. Journal of Clinical Endocrinology & Metabolism. 2013a;98:E1–E7. doi: 10.1210/jc.2012-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer MS, Perren A, Moch H, Komminoth P, Nikiforov YE, Nikiforova MN. Comprehensive microRNA expression profiling identifies novel markers in follicular variant of papillary thyroid carcinoma. Thyroid. 2013b doi: 10.1089/thy.2012.0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galasso M, Sandhu SK, Volinia S. MicroRNA expression signatures in solid malignancies. Cancer J. 2012;18:238–243. doi: 10.1097/PPO.0b013e318258b5f4. [DOI] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keutgen XM, Filicori F, Crowley MJ, Wang Y, Scognamiglio T, Hoda R, Buitrago D, Cooper D, Zeiger MA, Zarnegar R, et al. A panel of four miRNAs accurately differentiates malignant from benign indeterminate thyroid lesions on fine needle aspiration. Clin Cancer Res. 2012;18:2032–2038. doi: 10.1158/1078-0432.CCR-11-2487. [DOI] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Leone V, D’Angelo D, Pallante P, Croce CM, Fusco A. Thyrotropin Regulates Thyroid Cell Proliferation by Up-Regulating miR-23b and miR-29b that Target SMAD3. J Clin Endocrinol Metab. 2012 doi: 10.1210/jc.2012-1349. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumour MARKer prognostic studies (REMARK) Eur J Cancer. 2005;41:1690–1696. doi: 10.1016/j.ejca.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Menon MP, Khan A. Micro-RNAs in Thyroid neoplasms:molecular, diagnostic and therapeutic implications. J Clin Pathol. 2009 doi: 10.1136/jcp.2008.063909. [DOI] [PubMed] [Google Scholar]
- Nikiforov YEB, Paul W, Thompson Lester D, editors. Diagnostic Pathology and Molecular Genetics of the Thyroid. Lippincott Williams & Wilkins (LWW); 2009. [Google Scholar]
- Nikiforova MN, Chiosea SI, Nikiforov YE. MicroRNA Expression Profiles in Thyroid Tumors. Endocr Pathol. 2009;20:85–91. doi: 10.1007/s12022-009-9069-z. [DOI] [PubMed] [Google Scholar]
- Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricarte-Filho JC, Ryder M, Chitale DA, Rivera M, Heguy A, Ladanyi M, Janakiraman M, Solit D, Knauf JA, Tuttle RM, et al. Mutational profile of advanced primary and metastatic radioactive iodine-refractory thyroid cancers reveals distinct pathogenetic roles for BRAF, PIK3CA, and AKT1. Cancer Res. 2009;69:4885–4893. doi: 10.1158/0008-5472.CAN-09-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Kasai N, Sugano H. Poorly differentiated carcinoma of the thyroid. A clinicopathologic entity for a high-risk group of papillary and follicular carcinomas. Cancer. 1983;52:1849–1855. doi: 10.1002/1097-0142(19831115)52:10<1849::aid-cncr2820521015>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Schwertheim S, Sheu SY, Worm K, Grabellus F, Schmid KW. Analysis of deregulated miRNAs is helpful to distinguish poorly differentiated thyroid carcinoma from papillary thyroid carcinoma. Horm Metab Res. 2009;41:475–481. doi: 10.1055/s-0029-1215593. [DOI] [PubMed] [Google Scholar]
- Sheu SY, Grabellus F, Schwertheim S, Worm K, Broecker-Preuss M, Schmid KW. Differential miRNA expression profiles in variants of papillary thyroid carcinoma and encapsulated follicular thyroid tumours. Br J Cancer. 2010;102:376–382. doi: 10.1038/sj.bjc.6605493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P, Lima J, Preto A, Castro P, Vinagre J, Celestino R, Couto JP, Prazeres H, Eloy C, Maximo V, et al. Genetic alterations in poorly differentiated and undifferentiated thyroid carcinomas. Curr Genomics. 2011;12:609–617. doi: 10.2174/138920211798120853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallini G. Poorly differentiated thyroid carcinoma. Are we there yet? Endocr Pathol. 2011;22:190–194. doi: 10.1007/s12022-011-9176-5. [DOI] [PubMed] [Google Scholar]
- Tetzlaff MT, Liu A, Xu X, Master SR, Baldwin DA, Tobias JW, Livolsi VA, Baloch ZW. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocr Pathol. 2007;18:163–173. doi: 10.1007/s12022-007-0023-7. [DOI] [PubMed] [Google Scholar]
- Ueno K, Hirata H, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. microRNA-183 is an oncogene targeting Dkk-3 and SMAD4 in prostate cancer. Br J Cancer. 2013;108:1659–1667. doi: 10.1038/bjc.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- Volante M, Collini P, Nikiforov YE, Sakamoto A, Kakudo K, Katoh R, Lloyd RV, LiVolsi VA, Papotti M, Sobrinho-Simoes M, et al. Poorly differentiated thyroid carcinoma: the Turin proposal for the use of uniform diagnostic criteria and an algorithmic diagnostic approach. Am J Surg Pathol. 2007;31:1256–1264. doi: 10.1097/PAS.0b013e3180309e6a. [DOI] [PubMed] [Google Scholar]
- Volante M, Papotti M. Poorly differentiated thyroid carcinoma: 5 years after the 2004 WHO classification of endocrine tumours. Endocr Pathol. 2010;21:1–6. doi: 10.1007/s12022-009-9100-4. [DOI] [PubMed] [Google Scholar]
- Volante M, Rapa I, Gandhi M, Bussolati G, Giachino D, Papotti M, Nikiforov YE. RAS mutations are the predominant molecular alteration in poorly differentiated thyroid carcinomas and bear prognostic impact. J Clin Endocrinol Metab. 2009;94:4735–4741. doi: 10.1210/jc.2009-1233. [DOI] [PubMed] [Google Scholar]
- Volante M, Rapa I, Papotti M. Poorly differentiated thyroid carcinoma: diagnostic features and controversial issues. Endocr Pathol. 2008;19:150–155. doi: 10.1007/s12022-008-9040-4. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Ong J, Fagin JA, Melmed S. Expression of the myc cellular proto-oncogene in human thyroid tissue. J Clin Endocrinol Metab. 1986;63:1170–1173. doi: 10.1210/jcem-63-5-1170. [DOI] [PubMed] [Google Scholar]
- Yip L, Kelly L, Shuai Y, Armstrong MJ, Nikiforov YE, Carty SE, Nikiforova MN. MicroRNA Signature Distinguishes the Degree of Aggressiveness of Papillary Thyroid Carcinoma. Ann Surg Oncol. 2011;18:2035–2041. doi: 10.1245/s10434-011-1733-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori T, Suzuki S, Tanaka N, Inose T, Sohda M, Sano A, Sakai M, Nakajima M, Miyazaki T, Kato H, et al. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013;104:48–54. doi: 10.1111/cas.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]