Abstract
This study aimed to determine whether exposure of the oocyte and/or embryo to maternal undernutrition results in the later programming of insulin action in the liver and factors regulating gluconeogenesis. To do this, we collect livers from singleton and twin fetal sheep that were exposed to periconceptional (PCUN; −60 to 7 days) or preimplantation (PIUN; 0–7 days) undernutrition at 136–138 days of gestation (term = 150 days). The mRNA and protein abundance of insulin signaling and gluconeogenic factors were then quantified using qRT-PCR and Western blotting, respectively, and global microRNA expression was quantified using deep sequencing methodology. We found that hepatic PEPCK-C mRNA (P < 0.01) and protein abundance and the protein abundance of IRS-1 (P < 0.01), p110β (P < 0.05), PTEN (P < 0.05), CREB (P < 0.01), and pCREB (Ser133; P < 0.05) were decreased in the PCUN and PIUN singletons. In contrast, hepatic protein abundance of IRS-1 (P < 0.01), p85 (P < 0.01), p110β (P < 0.001), PTEN (P < 0.01), Akt2 (P < 0.01), p-Akt (Ser473; P < 0.01), and p-FOXO-1 (Thr24) (P < 0.01) was increased in twins. There was a decrease in PEPCK-C mRNA (P < 0.01) but, paradoxically, an increase in PEPCK-C protein (P < 0.001) in twins. Both PCUN and PIUN altered the hepatic expression of 23 specific microRNAs. We propose that the differential impact of maternal undernutrition in the presence of one or two embryos on mRNAs and proteins involved in the insulin signaling and gluconeogenesis is explained by changes in the expression of a suite of specific candidate microRNAs.
Keywords: pregnancy, nutrition, fetus, epigenetic
it has been demonstrated in a range of epidemiological and experimental studies that exposure of the oocyte, embryo, or fetus to a range of environmental stressors, including poor maternal nutrition, results in poor metabolic and cardiovascular outcomes in postnatal life (8, 9, 13, 22, 25, 41, 42, 53, 56). In sheep, maternal undernutrition from 60 days before to 30 days after conception resulted in an impairment of the insulin and glucose responses to a glucose tolerance test at 10 mo after birth (49). The effects of exposure to maternal undernutrition during early gestation were also more pronounced in singleton than in twin offspring. However, it is not known whether exposure to maternal undernutrition limited to around the time of conception alone is sufficient to program changes in the insulin-signaling pathway in tissues of metabolic importance such as the liver or whether there is a differential impact of maternal undernutrition in the periconceptional period in singletons and twins.
Insulin acts through the insulin receptor (IR), which is stabilized by caveolin-1 (Cav-1), resulting in a series of activations by phosphorylation of the insulin receptor substrate-1 (IRS-1) or -2 (IRS-2), phosphatidylinositol 3-kinase (PI3K), and conversion of phosphatidylinositol 4,5-bisphosphate (PIP2) to phosphatidylinositol 3,4,5-trisphosphate (PIP3). Conversion of PIP2 to PIP3 is negatively regulated by phosphatase and tensin homolog (PTEN) (48). PIP3 phosphorylates protein kinase B (PKB/Akt), which may result in the phosphorylation of the transcription factor Forkhead box protein O1 (FOXO1) and the inhibition of nuclear export of FOXO1 protein, thus inhibiting the role of FOXO1 in the stimulation of the mRNA expression of phosphoenolpyruvate carboxykinase cytosolic isoform (PEPCK-C) (4). When the liver is relatively resistant to the actions of insulin, the mRNA expression of PEPCK-C is not suppressed by insulin, thus resulting in an inadequate suppression of hepatic glucose output that may in turn lead to hyperglycemia.
The expression of PEPCK-C mRNA before and after birth is also stimulated by glucocorticoids, which promote gluconeogenesis (4, 11, 45, 47). Glucocorticoid action in tissues is mediated by glucocorticoid receptor (GR) and the activity of the intracellular 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD-1), which converts cortisone to cortisol (51). PEPCK-C mRNA expression is also regulated by the binding of transcription factors, including cAMP response element-binding protein (CREB), hepatocyte nuclear factor 4α (HNF4α), and CCAAT enhancer-binding protein-α and -β (C/EBPα and C/EBPβ) on the promoter region of the gene encode for PEPCK-C (PCK1) (4, 51). Additionally, methylation of CpG sites in the promoter region can inhibit the binding of transcription factors (40). Therefore, methylation of the PEPCK-C (PCK1) gene promoter can contribute to the regulation of PEPCK-C mRNA expression.
The rate of gluconeogenesis is also controlled by the activity of the constitutively expressed PEPCK mitochondrial isoform (PEPCK-M), which maintains basal gluconeogenic capacity (31, 40). Furthermore, cellular energy homeostasis in the liver is maintained by passive glucose transport, which occurs primarily through the activity of the insulin-independent glucose transporter 1 (GLUT1) during fetal life (17, 52). It is not known, however, whether there are specific effects of maternal undernutrition on the oocyte and/or embryo on the mRNA expression or protein abundance of factors regulating gluconeogenesis within the liver.
MicroRNAs (miRs) are small (∼22 nucleotides) species of noncoding RNA that act as posttranscriptional regulators, and their action requires a perfect or near-perfect binding of the “seed” sequence of the miR to the 3′-untranslated Region (3′-UTR) of the target transcript (3, 14). microRNAs play an essential role in the maintenance of insulin signaling and glucose homeostasis and dysregulation of miRs; e.g., miR-103, miR-107, miR-29a/b/c, and the let-7 family in liver, muscle, and fat are associated with features of the metabolic syndrome, obesity, and type 2 diabetes in a range of experimental models (12, 18, 20, 23, 24, 30, 39, 50, 57).
The period around the time of conception (oocyte maturation and preimplantation period) is a critical window during which existing epigenetic marks are erased and reestablished (46). There is also evidence to suggest that incomplete erasure of parental epigenetic marks and the environmental influence on the reestablishment of the epigenetic marks in the offspring may underlie the mechanisms of developmental programming of metabolic diseases in adult life (6). It is not known, however, whether there are specific effects of maternal undernutrition in different time windows during this period that result in the programmed changes of miRs or the methylation of genes and the programming of specific metabolic phenotypes. Therefore, we have investigated the separate effects of maternal undernutrition in the periconceptional period (PCUN; for ≥2 mo before and 1 wk after conception) or preimplantation period (PIUN; for 1 wk after conception) on the mRNA expression and protein abundance of the insulin-signaling molecules and on factors regulating gluconeogenesis in the liver of the fetal sheep in singleton and twin pregnancies. In addition, we have determined the impact of PCUN or PIUN on the methylation level at three CpG sites within the proximal PCK1 gene promoter region and on the expression of hepatic miRs using next generation small-RNA sequencing.
We hypothesize that the preimplantation period is a critical period during which nutritional restriction may result in changes in the abundance of key factors regulating hepatic insulin signaling and gluconeogenesis in fetal life, which may predispose to the development of insulin resistance and glucose intolerance in later life. Additionally, we propose that these effects will be greater in twin fetuses.
MATERIALS AND METHODS
All procedures were approved by the University of Adelaide Animal Ethics Committee and by the Primary Industries and Resources South Australia Animal Ethics Committee.
Nutritional Management
South Australian Merino ewes were fed a diet that provided 100% of nutritional requirements and consisted of lucerne chaff and pellets containing cereal hay, lucerne hay, barley, oats, almond shells, lupins, oat bran, lime, and molasses (Johnsons & Sons, Kapunda, South Australia, Australia). Eighty percent of the total energy requirements were obtained from the lucerne chaff (8.3 MJ/kg metabolizable energy, 193 g/kg of crude protein, and 85% dry matter) and 20% of the energy requirements from the pellet mixture (8.0 MJ/kg metabolizable energy, 110 g/kg of crude protein, and 90% dry matter). At the end of an acclimatization period, ewes were randomly assigned to a feeding regime: 1) control (C; n = 12), 100% of nutritional requirements from ∼60 days prior to mating until 6 days after mating; 2) PCUN (n = 13), with 70% of the control allowance from ∼60 days prior to mating until 6 days after mating, with all dietary components reduced by an equal amount; and 3) PIUN (n = 9), with 70% of the control diet from mating until 6 days after mating. All dietary components were reduced by an equal amount.
From 7 days after conception, all ewes were fed 100% of requirements.
Mating and Pregnancy
Ewes were released in a group to a pen every evening at 1600 with rams of proven fertility that were fitted with harnesses and crayons. Ewes were individually penned the following morning at 0800, and the occurrence of mating was confirmed by the presence of a crayon mark on the ewe's rump. The day of mating was defined as day 0, and fetal number was established by ultrasound between 40 and 80 days of gestation.
All ewes (n = 34) were euthanized humanely with an overdose of sodium pentobarbitone between 136 and 138 days of gestation. Fetuses (singleton: C, n = 6; PCUN, n = 8; PIUN, n = 3; twin: C, n = 11; PCUN, n = 8; PIUN, n = 11) were weighed and euthanized, and liver samples (dorsal lobe) were collected and snap-frozen in liquid nitrogen.
Quantification of mRNA Expression
RNA was extracted from ∼50 mg of liver tissue using Trizol reagent (Invitrogen, Groningen, The Netherlands) from singleton (C, n = 6; PCUN, n = 8; PIUN, n = 3) and twin fetuses (C, n = 11; PCUN, n = 8; PIUN, n = 11). RNA was purified using the RNeasy Mini Kit (Qiagen, Basel, Switzerland). The quality and concentration of the RNA were determined by measuring absorbance at 260 and 280 nm, and RNA integrity was confirmed by agarose gel electrophoresis. Complementary DNA (cDNA) was synthezised using the purified RNA and superscript 3 reverse transcriptase (Invitrogen) with random hexamers.
The relative expression of mRNA transcripts of the insulin receptors A (IRA) and B (IRB), IRS-1, IRS-2, PI3K regulatory subunit (p85), PI3K catalytic subunit (p110β), GLUT1, GLUT2, FOXO-1, CREB, HNF4α, C/EBPα, C/EBPβ, GR, 11β-HSD1, PEPCK-M, PEPCK-C, and the housekeeper gene cyclophilin was measured by quantitative real-time PCR (qRT-PCR) using the SYBR Green system in an ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA).
Primer sequences were validated for use in sheep in this (Table 1) or prior studies (7, 26, 32). Each amplicon was sequenced to ensure the authenticity of the DNA product, and a dissociation melt curve analysis was performed after each run to demonstrate amplicon homogeneity. Each qRT-PCR reaction well contained 5 μl of SYBR Green Master Mix (Applied Biosystems); 2 μl of primer (forward and reverse), 2 μl of molecular grade H2O, and 1 μl of cDNA (50 ng/μl). Controls for each sample containing no cDNA were also used to confirm absence of DNA contamination. The cycling conditions consisted of 40 cycles of 95°C for 15 min and 60°C for 1 min.
Table 1.
Primer sequences for quantitative RT-PCR
| Gene Name | Sequence | Accession No. |
|---|---|---|
| Cyclophilin | F: 5′CCTGCTTTCACAGAATAATTCCA3′ | BC105173 |
| R: 5′CATTTGCCATGGACAAGATGCCA3′ | ||
| IRS-1 | F: 5′TATGGCAAGCTTTGGACAAACGGG3′ | XM_581382.3 |
| R: 5′ACAAGAGTTTGCCACTACCGCTCT3′ | ||
| IRS-2 | F: 5′GTTCCAAGCTGTCCATGGAG3′ | NM_003749 |
| R: 5′CTCATGAGCACGTACTGGTC3′ | ||
| p85 | F: 5′GAAATTGAACGAGTGGCTGGGCAA3′ | M61745.1 |
| R: 5′TTCGGTTGCTGCTTCCAACATTCC3′ | ||
| p110β | F: 5′AATGCTTACCGTGAAGCCCTCTCT3′ | NM_001206047.1 |
| R: 5′ATCAAGCGCAGCATTTGGAGTGTC3′ | ||
| GLUT1 | F: 5′TAACCGCAACGAGGAGAACC3′ | U89029 |
| R: 5′CCACGCAGCTTCTTCAGCA3′ | ||
| GLUT2 | F: 5′ATGGCACATCCTGCTTGGTTTGTC3′ | AJ318925.1 |
| R: 5′TAGCTGGCATTGGTGAAGAGCTGA3′ | ||
| FOXO-1 | F: 5′ACACCTTTACAAGTGCCTCTG3′ | XM_342244 |
| R: 5′TAGCCATTGCAGCTGCTCAC3′ | ||
| CREB | F: 5′CCAAGGAGGAGCAATACAAC3′ | AF006042.1 |
| R: 5′GACACTCTCGTGCTGCTTC3′ | ||
| HNF4α | F: 5′GATGAGTTGGTGCTGCCCTTTCAA3′ | NM_001205153.1 |
| R: 5′ACTGGCGGTCGTTGATGTAGTCTT3′ | ||
| C/EBPα | F: 5′TGGCCGACCTGTTCCAACACA3′ | AY621546.1 |
| R: 5′TAGTCAAAGTCGTTGCCGCCTCCT3′ | ||
| C/EBPβ | F: 5′CACAGCGACGAGTGCAAGATCC3′ | AY371068.1 |
| R: 5′CTTGAACAAGTTCCGCAGGGTG3′ | ||
| PEPCK-M | F: 5′TACGAGGCCTTCAACTGGCGT3′ | Y11484 |
| R: 5′AGATCCAAGGCGCCTTCCTTA3′ | ||
| PEPCK-C | F: 5′AAC TC CGGTTCTGCACTCCA3′ | BC112664.1 |
| R: 5′GGT CGTGCATGATGACTTTGC3′ |
F, forward; R, reverse; IRS-1 and -2, insulin receptor substrate-1 and -2, respectively; GLUT1 and -2, glucose transporter 1 and 2, respectively; FOXO-1, forkhead box protein O1; CREB, cAMP response element-binding protein; HNF4α, hepatocyte nuclear factor 4α; C/EBPα and -β, CCAAT enhancer-binding protein-α and -β, respectively; PEPCK-M, phosphoenolpyruvate carboxykinase mitochondrial isoform; PEPCK-C, phosphoenolpyruvate carboxykinase cytosolic isoform.
The expression of each mRNA transcript was measured, and expression relative to cyclophilin (which did not differ between groups) was calculated using the comparative threshold cycle (CT) method (Q-gene qRT-PCR analysis software).
Quantification of Protein Abundance
Protein abundance was determined using Western blotting. Briefly, liver samples (∼100 mg) from singleton (C, n = 4; PCUN, n = 4; PIUN, n = 3) and twin fetuses (C, n = 4; PCUN, n = 4; PIUN, n = 5) were homogenized in 1 ml of lysis buffer (50 mM Tris·HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 1 mM Na3VO4, 30 mM NaF, 10 mM Na4P2O7, 10 mM EDTA, and 1 protease inhibitor tablet) and centrifuged at 12,000 g at 4°C for 15 min to remove insoluble material. Protein content of the clarified extracts was quantified using bicinchoninic acid protein assay. Prior to Western blot analysis, samples (10 μg protein) were subjected to SDS-PAGE and stained with Coomassie blue reagent (Thermo Fisher Scientific, Rockford, IL) to ensure equal loading of the proteins. Equal volumes and concentrations of protein were subjected to SDS-PAGE. The proteins were transferred onto a PolyScreen polyvinylidene difluoride hybridization transfer membrane (Perkin-Elmer, Waltham, MA) using a semidry blotter (Hoefer, Holliston, MA). The membranes were blocked with 5% BSA in Tris-buffered saline with 0.1% Tween-20 (TBS-T) at room temperature for 1 h and then incubated overnight with primary antibody against IRβ, GLUT1, HNF4α, PEPCK-C (Santa Cruz Biotechnology, Santa Cruz, CA), Cav-1, PTEN, Akt1, Akt2, p-Akt (Ser473), FOXO-1, p-FOXO-1 (Thr24), CREB, p-CREB (Ser133) (Cell Signaling, Danvers, MA), IRS-1, p85, GLUT2 (Merck Millipore, Billerica, MA), and p110β (Epitomics, Burlingame, CA). Membranes were washed and bound antibody was detected using anti-rabbit, anti-mouse (Cell Signaling Technology), or anti-goat (Merck Millipore) horseradish peroxidase-conjugated secondary IgG antibodies at room temperature for 1 h. Enhanced chemiluminescence reagents SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) and ImageQuant LAS 4000 (GE Healthcare, Rydalmere, NSW, Australia) were used to detect the protein-antibody complexes. AlphaEaseFC (Alpha Innotech, Santa Clara, CA) was utilized to quantify specific bands of the target proteins.
Measurement of PEPCK-C (PCK1) Gene Promoter Methylation
DNA methylation within the PCK1 promoter was analyzed by combined bisulphite restriction assay (COBRA) (54, 56). Briefly, DNA was extracted from ∼30 mg of liver samples from singleton (C, n = 4; PCUN, n = 6; PIUN, n = 3) and twin fetuses (C, n = 7; PCUN, n = 6; PIUN, n = 6). DNA (∼2 μg) was subjected to bisulphite conversion (Epitect; Qiagen). The PCR was performed on 100 ng of bisulphite-converted DNA using primers (forward: 5′ TAAAGGTTTGTTATGGTTGGTTTAG 3′; reverse: 3′ CTAACCTTTAAATTCCAAAAAAA 5′) and conditions that amplified methylated and unmethylated templates with no bias. Amplicons were measured covering three CpG sites at −49, −58, and −88, where +1 denotes the PCK1 gene start site in the bovine sequence. COBRA was performed using restriction endonucleases that cleave only those amplicons derived from methylated templates. The PCK1 amplicons were digested with 20 U of either Taq I (Thermofisher Scientific) or Dpn II (New England Biolabs, Ipswich, MA) for 2 h at 37°C, followed by deactivation step at 65°C for 20 min or TaiI (Thermofisher Scientific) for 2 h at 37°C, followed by deactivation step by adding 20 mM of 5 M EDTA, pH 8.0, which digests methylated templates at −49, −58, and −88, respectively. The intensity of the cut and uncut fragments was quantified using an Experion Automated Electrophoresis System (Bio-Rad, Hercules, CA).
Micro RNA Expression
MicroRNA (miR) data were obtained using the next generation small-RNA sequencing, as described previously (19). Briefly, total RNA was prepared from ∼25 mg of liver samples from singleton (C, n = 3; PCUN, n = 3; PIUN, n = 3) and twin fetuses (C, n = 3; PCUN, n = 3; PIUN, n = 3) using TRIzol reagent. Libraries were created with the NEBnext small RNA kit and sequenced using a SOLiD 5500 next generation sequencer (Applied Biosystems). Reads were mapped using Applied Biosystems “Lifescope” software (default settings) against human hg19 and cow UMD3.1 genomes (sheep genome is poorly annotated for miRs). Lifescope software uses a pipeline that parses the reads over miRBase first to capture miRs and prior to parsing the remaining reads over the rest of the genome. Reads that were not captured by the human miRBasev18 reference set were discarded, which is the definitive catalog of precursor and mature miRs. Precursor miRs were defined by the genomic coordinates as defined by miRBase version 18, and processed miR counts were counted from reads that started within 3 nt of the sequence defined by miRBase.
Statistical Analyses
mRNA expression, protein abundance, and PCK1 methylation.
All data are presented as means ± SE. All data were analyzed using the Statistical Package for the Social Sciences Software (SPSS, Chicago, IL). We used the density test of normality using STATA software and found that the data sets were normally distributed. Two-way analysis of variance (ANOVA) was used to determine the effects of maternal nutritional treatment (PCUN and PIUN) and fetal number (singleton or twin) on mRNA expression, protein abundance, and methylation levels of the PCK1 promoter in the liver. When there was no interaction between the effects of nutritional treatment and fetal number, the data from singleton and twin fetuses were pooled for presentation of the effects of nutritional treatment. When there was an interaction between the effects of nutritional treatment and fetal number, data from singletons and twins were split, and the effects of nutritional treatment were then determined using a one-way ANOVA. The Duncans post hoc test was used to determine the level of significant difference in mean values between nutritional treatment groups. A probability level of 5% (P < 0.05) was taken as significant.
miR expression.
A threshold for a fold difference of expression of miRs between the PCUN or PIUN treatment groups relative to controls was set at >1.5 or <0.67 with a threshold of >100 reads/million or at >1.2 or <0.83 with a threshold of >10,000 reads/million, where the relative standard deviation was <50% among animals within a treatment group. Selected miRs based on these criteria from data mapped to the human miRBase were then cross-checked with the corresponding miRs mapped to the bovine miRBase. MicroRNAs were then selected as high-confidence “candidates.”
Proteins within signaling pathways targeted by the candidate miRs were analyzed using DIANA-mirPath (Targetscan 5) (36). The correlation of the expression of all candidate miRs with protein abundance was determined using linear regression analysis (SPSS). Candidate miRs were also analyzed using Targetscan to identify 8mer, 7mer-m8, or 7mer-1A match between the seed sequence of the candidate miRs within the 3′-UTR of the putative mRNA targets within the insulin-signaling pathway conserved across species.
RESULTS
Hepatic mRNA Expression and Protein Abundance Of Insulin-Signaling Molecules
There was no difference in hepatic mRNA expression of IRA, IRB, IRS-1, IRS-2, p85, p110β, FOXO-1, GLUT1, or GLUT-2 in either PCUN or PIUN fetal sheep compared with controls in singletons or twins. The hepatic abundance of IRβ, Cav-1, Akt1, GLUT1, and GLUT2 proteins was also not different between treatment groups in singletons or twins.
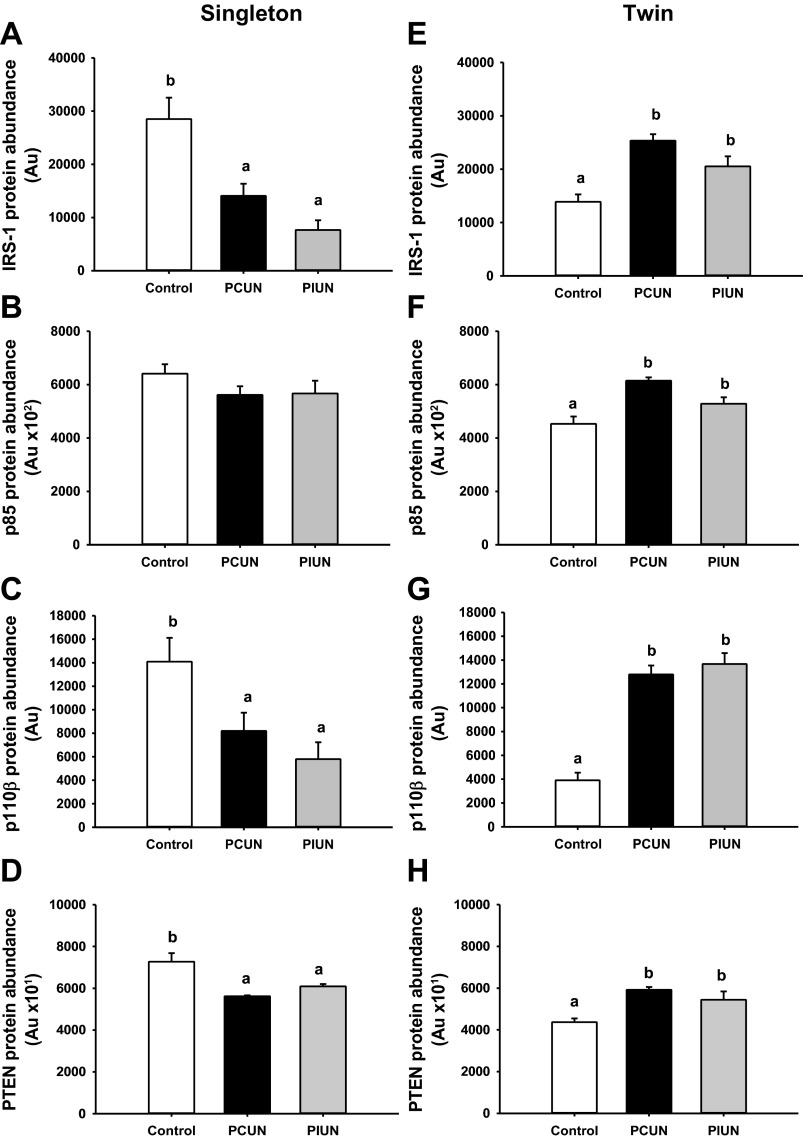
The protein abundance of IRS-1 (P < 0.05), p85 (P < 0.01), p110β (P < 0.01), PTEN (P < 0.01) (Fig. 1), Akt2 (P < 0.05), and phosphorylated FOXO-1 (Thr24; P < 0.05) (Fig. 2), as well as CREB (P < 0.05) (Fig. 3) and PEPCK-C (P < 0.01) (Fig. 4), was lower in the control twins than in control singletons.
Fig. 1.
Protein abundance of insulin receptor substrate (IRS-1), phosphatidylinositol 3-kinase (PI3K; p85), PI3K (p110β), and phosphatase and tensin homolog (PTEN) in singleton and twin fetuses in late gestation. Abundance of IRS-1, p110β, and PTEN protein was lower in singletons (A, C, and D), and abundance of IRS-1, p85, p110β, and PTEN protein was higher in twins (E, F, G, and H) in the periconceptional undernutrition (PCUN) and preimplantation undernutrition (PIUN) groups compared with controls. p85 protein abundance was not different in singletons in either treatment group (B). Different letters denote significant differences between treatment groups.
Fig. 2.
Protein abundance of Akt2, phosphorylated Akt at Ser473, forkhead box protein O1 (FOXO-1), and phosphorylated FOXO-1 at Thr24 in singleton and twin fetuses in late gestation. A: Akt2 protein abundance tended to be lower (P = 0.06) in the PCUN and PIUN groups compared with controls in singletons. B, C, and D: the protein abundances of phosphorylated Akt (Ser473), FOXO-1, and phosphorylated FOXO-1 (Thr24) were not different in either treatment group in singletons. E, F, and H: the protein abundances of Akt2, phosphorylated Akt (Ser473), and phosphorylated FOXO-1 (Thr24) were higher in the PCUN and PIUN groups compared with controls in twins. G: FOXO-1 protein abundance was decreased only in PIUN group compared with controls in twins. Different letters denote significant differences between treatment groups.
Fig. 3.
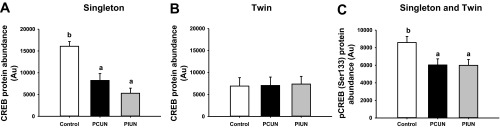
Protein abundance of cAMP response element-binding protein (CREB) and phosphorylated CREB at Ser133 in singleton and twin fetuses in late gestation. A and B: CREB protein abundance was lower in the PCUN and PIUN groups compared with controls in singletons (A), but there was no difference in either treatment groups in twins (B). C: the protein abundance of phosphorylated CREB (Ser133) was lower in the PCUN and PIUN groups compared with controls in singletons and twins. Different letters denote significant differences between treatment groups.
Fig. 4.
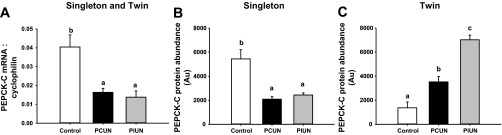
mRNA expression and protein abundance of phosphoenolpyruvate carboxykinase cytosolic isoform (PEPCK-C) in singleton and twin fetuses in late gestation. A: PEPCK-C mRNA expression was lower in the PCUN and PIUN groups compared with controls in singletons and twins. B and C: protein abundance of PEPCK-C was lower in singletons (B) and higher in twins (C) in the PCUN and PIUN groups compared with controls. Different letters denote significant differences between treatment groups.
Singletons.
The protein abundance of IRS-1 (P < 0.01), p110β (P < 0.05), and PTEN (P < 0.05), but not p85, was lower in the liver of the PCUN and PIUN fetuses compared with controls (Fig. 1). There was also a trend toward a decrease in hepatic Akt2 protein abundance (P < 0.06) in the PCUN and PIUN groups compared with controls. There was no difference, however, in the protein abundance of total phosphorylated Akt (Ser473), FOXO-1, or phosphorylated FOXO-1 (Thr24) between treatment groups (Fig. 2).
Twins.
Hepatic protein abundance of IRS-1 (P < 0.01), p85 (P < 0.01), p110β (P < 0.001), PTEN (P < 0.01), Akt2 (P < 0.01), and total phosphorylated Akt (Ser473) (P < 0.01) was higher in the PCUN and PIUN groups compared with controls (Figs. 1 and 2). The hepatic protein abundance of FOXO-1 was lower (P < 0.01) in the PIUN group, whereas the phosphorylated FOXO-1 (Thr24) was higher (P < 0.01) in both the PCUN and PIUN groups compared with controls (Fig. 2).
Factors Regulating PEPCK-C Expression
Expression of C/EBPβ, but not C/EBPα mRNA, was lower (P < 0.01) in the liver of the PCUN group compared with controls in both singletons and twins. There was no effect of either PCUN or PIUN on HNF4α mRNA and protein or on CREB, GR, and 11β-HSD1 mRNA expression in either singletons or twins. The hepatic protein abundance of CREB was lower (P < 0.01) in PCUN and PIUN singletons, but not twins, compared with controls (Fig. 3). The phosphorylation of CREB (Ser133) was lower (P < 0.05), however, in both PCUN and PIUN singletons and twins compared with controls (Fig. 3).
PEPCK-M mRNA Expression, PEPCK-C mRNA Expression and Protein Abundance, and PCK1 Promoter Methylation
There was no effect of either nutritional treatment or fetal number on PEPCK-M mRNA expression. However, PEPCK-C mRNA expression was lower (P < 0.01) in PCUN and PIUN singletons and twins compared with controls (Fig. 4). The hepatic abundance of PEPCK-C protein was lower in singletons (P < 0.01) and higher in twins (P < 0.001) in the PCUN and PIUN groups compared with controls (Fig. 4). However, there was no effect of nutritional treatment on the level of methylation at PCK1 promoter sites [methylation level for the TaiI and Taq I site (−49 and −88) was 3–6% and for the Dpn II site (−58) was 6–10% across treatment groups in singletons and twins; data not shown].
Hepatic Expression of miRs
The expression of 23 miRs changed in either the PCUN or PIUN groups relative to controls (Table 2).
Table 2.
Effects of PCUN and PIUN on the expression of candidate miRs in the liver in singleton and twin fetal sheep in late gestation
| Singletons |
Twins |
Predicted Target Protein |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| microRNA | Control [mean expression (reads/million)] | PCUN (fold change relative to controls) | PIUN (fold change relative to controls) | Control [mean expression (reads/million)] | PCUN (fold change relative to controls) | PIUN (fold change relative to controls) | Protein | nt match | No. of matches |
| hsa-miR-142-5p (−1 isomir) | 195 | ×1.60↑ | ×1.99↑ | 250 | ×0.63↓ | ×0.60↓ | IRS-2 | 7mer-1A | 1 |
| hsa-miR-493-3p | 280 | ×1.66↑ | 436 | p85 | 7mer-1A | 1 | |||
| hsa-miR-146b-5p | 990 | ×1.87↑ | 1,061 | ||||||
| hsa-miR-339-5p | 10,779 | ×0.80↓ | ×0.80↓ | 7,818 | |||||
| hsa-miR-148a-3p | 11,494 | ×0.76↓ | 11,592 | ×0.71↓ | |||||
| hsa-miR-19a-3p | 13,193 | ×0.72↓ | 8,729 | ||||||
| hsa-miR-19b-3p | 28,477 | ×0.66↓ | 18,837 | ||||||
| hsa-miR-30a-5p (+2 isomir) | 41,451 | ×0.76↓ | 36,325 | FOXO1 | 7mer-m8 | 1 | |||
| Cav-1 | 7mer-1A | 1 | |||||||
| hsa-miR-30e-5p (−4 isomir) | 25,165 | ×0.80↓ | 21,641 | ||||||
| hsa-miR-122-5p | 117,180 | 140,513 | ×1.37↑ | ×0.77↓ | |||||
| hsa-let-7a-5p | 3,304 | 5,709 | ×0.57↓ | ×0.57↓ | IRS-2 | 8mer | 1 | ||
| IR | 8mer | 1 | |||||||
| Akt2 | 8mer | 1 | |||||||
| hsa-let-7b-5p | 1,966 | 2,742 | ×0.61↓ | ×0.61↓ | IRS-2 | 8mer | 1 | ||
| IR | 8mer | 1 | |||||||
| Akt2 | 8mer | 1 | |||||||
| hsa-miR-130a-3p | 139 | 155 | ×0.63↓ | ×0.66↓ | |||||
| hsa-miR-16-2-3p (−4 isomir) | 207 | 200 | ×0.40↓ | ||||||
| hsa-miR-34c-5p | 215 | 242 | ×0.57↓ | ||||||
| hsa-miR-106b-5p | 8,734 | 6,137 | ×1.68↑ | ||||||
| hsa-let-7 g-5p (+1 isomir) | 5,260 | 6,624 | ×0.62↓ | ||||||
| hsa-miR-335-5p | 6,079 | 6,865 | ×0.67↓ | ||||||
| hsa-miR-379-3p (+3 isomir) | 101 | 150 | ×0.61↓ | ||||||
| hsa-miR-369-3p | 483 | 518 | ×0.64↓ | ||||||
| hsa-miR-34a-5p | 595 | 560 | ×0.67↓ | ||||||
| hsa-miR-382-5p | 147 | 154 | ×0.64↓ | ||||||
| hsa-miR-126-5p | 11,447 | 10,243 | ×0.77↓ | ||||||
PCUN, periconceptional undernutrition; PIUN, preimplantation undernutrition; miR, microRNA; hsa denotes that data were mapped to human miRBase using the Applied Biosystems “Lifescope” software, representing sheep miR orthologs. ↑>1.5 with threshold of >100 reads/million or >1.2 with threshold of >10,000 reads/million; RSD <50%. ↓<0.67 with threshold of >100 reads/million or <0.83 with threshold of >10,000 reads/million; RSD <50%.
Singletons.
Hepatic expression of miR-142-5p was higher in the PCUN and PIUN groups, whereas the expression of miR-146b-5p was higher in the PCUN group and the expression of miR-493-3p higher in the PIUN group relative to controls (Table 2). Hepatic expression of miR-339-5p was lower in the PCUN and PIUN groups, and the expression of miR-148a-3p, miR-19a-3p, miR-19b-3p, miR-30a-5p, and miR-30e-5p in the PIUN group was also lower relative to controls (Table 2).
Twins.
In the PIUN group, hepatic expression of miR-106b-5p was higher, whereas the expression of miR-122-5p was higher in the PCUN but lower in the PIUN group relative to controls (Table 2).
Hepatic expression of miR-142-5p, let-7a-5p, let-7b-5p, and miR-130a-3p was lower in the PCUN and PIUN groups, whereas the expression of miR-16-2-3p and miR-34c-5p was lower in the PCUN group only (Table 2). Expression of miR-148a-3p, let-7g-5p, miR-335-5p, miR-379-3p, miR-369-3p, miR-34a-5p, miR-382-5p, and miR-126-5p was also lower in the PIUN group only (Table 2).
Relationship Between miR Expression and the Abundance of the Insulin-Signaling Proteins and PEPCK-C
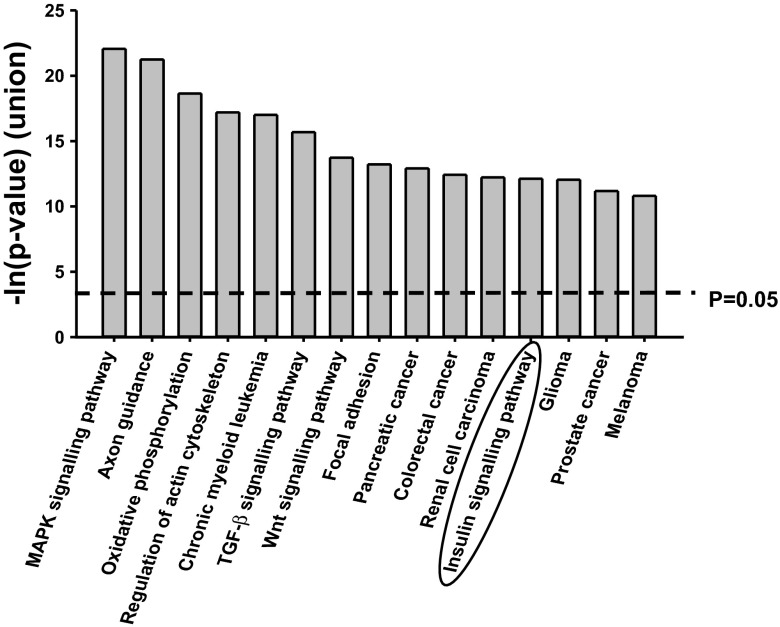
DIANA-mirPath software revealed that 23 miRs were associated significantly with multiple pathways, including the insulin-signaling pathway (Fig. 5). The −1 isomir of miR-142-5p was predicted to regulate IRS-2, miR-493-3p was predicted to regulate p85, the +2 isomir of miR-30a-5p was predicted to regulate FOXO-1 and Cav-1, and the let-7 family (seed sequence conserved between members) was predicted to regulate IRS-2, IR, and Akt2 (Table 2).
Fig. 5.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with the 23 candidate microRNAs (miRs) defined by target scan multiple miR analysis. The 23 candidate miRs found to have altered expression in singleton and twin fetuses in late gestation were associated with MAPK signaling, axon guidance, oxidative phosphorylation, regulation of actin cytoskeleton, TGFβ signaling, Wnt signaling, focal adhesion, a number of cancers, and, more importantly, insulin signaling.
There was an inverse relationship between the expression of miR-130a-3p and IRS-1 protein abundance (P < 0.01, r2 = 0.47) and between let-7a-5p expression and Akt2 protein abundance (P < 0.01, r2 = 0.50) in singletons and twins (Table 3). In twins only, there was an inverse relationship between miR-379-3p (P < 0.01, r2 = 0.81) and miR-369-3p (P < 0.05, r2 = 0.62) expression and Akt2 protein abundance (Table 3). Additionally, there was an inverse relationship between miR-34c-5p and p85 protein abundance (P < 0.01, r2 = 0.77) and between miR-106b-5p expression and FOXO-1 protein abundance (P < 0.05, r2 = 0.55) in twins only (Table 3).
Table 3.
Relationship between the expression of candidate miRs and the protein abundance of the insulin-signaling molecules in fetal liver in late gestation
| Correlations |
||||||
|---|---|---|---|---|---|---|
| microRNA | IRS-1 | Akt2 | PI3K (p85) | FOXO-1 | nt Match | No. of Match |
| hsa-miR-130a-3p | y = −106.59× +31,438; P < 0.01, r2 = 0.47, singleton and twin | NA | NA | |||
| hsa-let-7a-5p | y = −3.12× +30,068; P < 0.01, r2 = 0.50, singleton and twin | 8mer | 1 | |||
| hsa-miR-379-3p (+3 isomir) | y = −32.49× +5,844; P < 0.01, r2 = 0.81, twin only | NA | NA | |||
| hsa-miR-369-3p | y = −6.62× +4,677; P < 0.05, r2 = 0.62, twin only | NA | NA | |||
| hsa-miR-34c-5p | y = −1,108.8× +737,410; P < 0.01, r2 = 0.77, twin only | NA | NA | |||
| hsa-miR-106b-5p | y = −0.97× +16,046; P < 0.05, r2 = 0.55, twin only | NA | NA | |||
PI3K, phosphatidylinositol 3-kinase.
Interestingly, there was a positive relationship between miR-339-5p expression and PI3K (p110β) (P < 0.05, r2 = 0.36), PTEN (P < 0.05, r2 = 0.31), Akt2 (P < 0.01, r2 = 0.42), and PEPCK-C (P < 0.01, r2 = 0.53) protein abundance in singletons and twins (data not shown). There was also a positive relationship between miR-19a-3p (P < 0.01, r2 = 0.77), miR-19b-3p (P < 0.01, r2 = 0.70), and miR-335-3p (P < 0.01, r2 = 0.73) expression and IRS-1 protein abundance in singletons only (data not shown).
DISCUSSION
The “thrifty phenotype” hypothesis postulated that poor maternal nutrition before or during gestation resulted in an increased risk of metabolic, endocrine, and cardiovascular disease in adulthood (2, 16, 28, 29). There have been no studies to date, however, on the effects of maternal undernutrition during the period around the time of conception on the factors that regulate hepatic glucose metabolism. In this study, we have demonstrated that maternal undernutrition during the periconceptional and preimplantation periods resulted in significant changes in the hepatic protein abundance of the insulin-signaling molecules CREB and PEPCK-C and that these effects were different in singleton and twin pregnancies (Fig. 6). We have also demonstrated for the first time that exposure to maternal undernutrition around the time of conception results in changes in the expression of specific miRs that may play a role in the programming of insulin signaling and hepatic gluconeogenesis.
Fig. 6.
A summary diagram that shows the impact of PCUN and PIUN on hepatic insulin-signaling molecules and gluconeogenic factors in singleton and twin fetuses.
Impact of PCUN and PIUN on Insulin Signaling and Gluconeogenic Pathways in the Fetal Liver
Singletons.
In singleton fetuses, the hepatic protein abundance of IRS-1, p110β, and PTEN was lower in each of the PCUN and PIUN groups. The lower PTEN protein abundance may explain the lack of change in Akt2 or FOXO-1 phosphorylation in the PCUN and PIUN groups despite the lower protein abundance of p110β. Therefore, it appears that maternal undernutrition in the first week of pregnancy alone is sufficient to program a downregulation of the hepatic insulin-signaling pathway. These changes in the hepatic insulin-signaling pathway may contribute to the development of insulin resistance and glucose intolerance that have been observed in offspring after exposure to maternal undernutrition in the first 4 days after conception in the rodent (21) and in the postnatal animal after exposure to maternal undernutrition in the first 30 days after conception in the sheep (38). Interestingly, it has also been shown in the Dutch Winter Hunger Famine study that exposure to famine during early gestation also resulted in an increased risk of insulin resistance and glucose intolerance in adult life (41).
Paradoxically, however, whereas the hepatic protein abundance of insulin-signaling molecules was lower in the singleton fetuses in the PCUN and PIUN groups, PEPCK-C mRNA expression and protein abundance was also lower in these groups compared with controls. This finding is in contrast to previous experimental studies where exposure to a maternal low-protein diet during the preimplantation period in rats (21), chronic fetal hypoglycemia and hypoinsulinemia during midgestation in sheep (33), and moderate maternal undernutrition during mid- and late gestation in the baboon (34) resulted in an increase in hepatic PEPCK-C mRNA expression in late fetal life. In these prior studies, it is likely that maternal undernutrition also resulted in increased fetal cortisol concentrations to result in the programming of an increase in expression of gluconeogenic enzymes (10, 11, 35).
The protein abundance of CREB and its phosphorylated form was lower in PCUN and PIUN singletons. Thus a decrease in substrate supply to the embryo during the first week after conception may program a decrease in CREB protein abundance, and this may then contribute to the paradoxical decrease in hepatic PEPCK-C mRNA expression. Previously, it has been shown that phosphorylation of CREB (Ser133) is positively associated with hepatic PEPCK-C mRNA expression in the absence of changes in Akt, FOXO-1, HNF4α, or C/EBPβ (43). C/EBPα mRNA expression was also lower in PCUN fetuses, which may contribute to the lower PEPCK-C mRNA expression in these fetuses.
Twins.
A novel finding in this study was that exposure to maternal PCUN or PIUN had different effects in twins compared with singletons (Fig. 6). In the twin, exposure to PCUN or PIUN resulted in a higher rather than lower protein abundance of IRS-1, p85, p110β, PTEN, Akt2, phosphorylated Akt, and phosphorylated FOXO-1 in the fetal liver compared with controls. Although hepatic PTEN protein abundance was higher in the PCUN and PIUN twins, the protein abundance of phosphorylated Akt was also higher in these groups. Consequently, phosphorylated FOXO-1 was higher in the PCUN and PIUN groups, which may contribute to the lower PEPCK-C mRNA expression present in the twin. However, whereas PEPCK-C mRNA expression was lower, the protein abundance of PEPCK-C was higher in the liver of the twin fetuses in the PCUN and PIUN groups. Thus, although an enhanced intrahepatic insulin-signaling response may downregulate PEPCK-C mRNA expression in the PCUN or PIUN twin, there may be other factors that upregulate the protein abundance of the gluconeogenic enzyme and contribute to the emergence of glucose intolerance.
The hepatic protein abundance of IRS-1, p85, p110β, PTEN, Akt2 and phosphorylated FOXO-1, as well as CREB and PEPCK-C, was lower than in the control twin than in the control singleton and similar to the level present in the PCUN and PIUN singleton fetuses. The changes that occur in insulin signaling in the liver of the control twin may be programmed through similar epigenetic mechanisms as those present in the PCUN and PIUN singleton fetus, perhaps in anticipation of the limitation in fetal substrate supply that is characteristic of a normal twin pregnancy. We note that the upregulation of hepatic insulin signaling in the twin fetus after exposure to PCUN or PIUN results in a level of insulin signaling similar to that in the control singleton. This relative increase in hepatic insulin signaling in the PCUN and PIUN twins may be a response to maintain hepatic substrate utilization to meet metabolic demand in the face of an anticipated further decrease in fetal substrate supply.
Impact of PCUN and PIUN on miRs
We have shown for the first time that there is an impact of maternal undernutrition in the periconceptional and preimplantation periods on the expression specific miRs in the fetal liver and that the impact on the specific miRs is different in the twin and singleton fetus. In singletons, the expression of miR-142-5p, miR-493-3p, and miR-146b-5p was higher in the PCUN and/or PIUN groups, and in contrast, the expression of candidate miRs was generally lower in the PCUN and/or PIUN twin relative to controls. Additionally, the expression of six candidate miRs was inversely correlated with the protein abundance of the key insulin-signaling and gluconeogenic molecules, and the expression of four candidate miRs was positively correlated with the protein abundance of these molecules. A positive relationship suggests that these miRs may act indirectly to inhibit factors that regulate the translation or degradation of the target transcript or protein. Among the 10 candidate miRs, the expression of miR-130a, miR-19a, miR-19b, miR-335, miR-34c, miR-379, and let-7 family has been shown to be altered in states of insulin resistance, glucose intolerance, and/or type 2 diabetes in adult life (12, 15). This corresponds with our finding that insulin signaling is one of the top 15 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with the 23 candidate miRs collectively, and target scan software predicts that the −1 isomir of miR-142-5p regulates IRS-2, miR-493-3p regulates p85, and the +2 isomir of miR-30a-5p regulates FOXO-1 and Cav-1, and the let-7 family was predicted to regulate IRS-2 and IR. We also found that the let-7a seed sequence had a highly conserved 8mer match within the 3′-UTR of Akt2 and that the expression of let-7a was decreased in the PCUN and PIUN twins, in which Akt2 is more abundant. This suggests that PCUN and PIUN in twin fetuses recruit let-7a to modulate the translation and thus protein abundance of Akt2. Interestingly, although the level of expression of the candidate miRs was not different in the fetal liver of control twins and control singletons, PCUN and PIUN recruited a different “suite” of miRs in singleton and twin fetuses. Furthermore, there were a larger number of candidate miRs recruited by PIUN compared with PCUN, suggesting that a mismatch between pre- and postconception nutritional levels may be important in the programming of miRs in early prenatal life.
The expression of a number of miRs was correlated with the protein abundance of insulin signaling or gluconeogenic factors in singletons or in twins only. It has been shown that an mRNA transcript can have multiple miR target binding sites within its 3′-UTR (3). Therefore, the regulation of a transcript and the subsequent protein abundance may depend on the expression level of a different number of miRs in the singleton and twin.
Although not all of the KEGG pathways found to be associated with the 23 miRs using DIANA-mirPath analysis will be relevant to hepatic function and metabolism, dysregulation of expression of miR-19b, miR-106b, and miR-130a is associated with the MAPK/ERK tumor-promoting pathway (44). miR-106-25 is associated with TGFβ tumor suppressor signaling pathway in the liver (37), and miR-122 is associated with hepatitis-C (27) and nonalcoholic steatohepatitis (5), which is one of the risk factors for hepatocellular carcinoma (1). Furthermore, plasma levels of miR-122 and miR-34a were higher in patients with nonalcoholic fatty liver disease (55). These findings suggest that exposure to maternal undernutrition in early prenatal life may have an impact on hepatic growth and physiology beyond programming of the insulin-signaling and gluconeogenic pathways.
Summary
We have demonstrated that maternal undernutrition during the periconceptional and/or preimplantation periods results in a lower protein abundance of hepatic insulin-signaling molecules and a paradoxical downregulation in the mRNA expression and protein abundance of PEPCK-C in the singleton fetus. We propose that this paradox is explained by the separate programming of a decrease in protein abundance of the hepatic transcription factor CREB and phosphorylated CREB. In contrast, exposure to maternal PCUN and PIUN results in an increase in hepatic protein abundance of insulin-signaling molecules, a decrease in PEPCK-C mRNA expression, and an increase in PEPCK protein abundance in the twin fetus. We propose that these changes and the differential impact of maternal undernutrition in the presence of one or two embryos are explained by the impact of the hormonal and nutritional environment in early pregnancy on the expression of a suite of specific candidate miRs that regulate insulin action in the hepatocyte. The candidate miRs identified in this study that may be important regulators of the hepatic insulin response include miR-130a, miR-19a, miR-19b, miR-335, miR-34c, miR-369, miR-379, miR-106b, and let-7a.
The current study has identified specific changes in miRs and metabolic pathways in the fetal liver after exposure to maternal undernutrition in the periconceptional and preimplantation periods. These findings highlight the biological importance of the early nutritional environment and the adaptations of the early embryo to that environment for later metabolic health. It is clear that a deeper understanding of the impact of the in vivo or ex vivo nutritional environment on the embryo and on the programming of insulin-signaling pathways in tissues of metabolic importance in the offspring is required. Furthermore, this study clearly highlights the importance of good nutrition in young women of reproductive age around the time of conception for the metabolic health of the next generation.
GRANTS
This study was supported by funding from the Australian Research Council (I. C. McMillen, C. T. Roberts, and S. K. Walker) and the National Health and Medical Research Council of Australia (I. C. McMillen). C. T. Roberts is supported by a National Health and Medical Research Council Senior Research Fellowship (APP1020749). J. L. Morrison was supported by a South Australian Cardiovascular Research Network Fellowship (CR10A4988).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
S.L., J.L.M., C.M.S., D.T.H., S.E.O., S.M.M., C.T.R., and I.C.M. conception and design of research; S.L., J.L.M., O.W.-W., S.M.M., D.O.K., and S.K.W. performed experiments; S.L., O.W.-W., C.M.S., D.T.H., S.Z., and I.C.M. analyzed data; S.L., J.L.M., C.M.S., D.T.H., S.E.O., S.Z., D.O.K., and I.C.M. interpreted results of experiments; S.L. prepared figures; S.L. drafted manuscript; S.L., J.L.M., O.W.-W., C.M.S., D.T.H., S.E.O., S.Z., S.M.M., D.O.K., S.K.W., C.T.R., and I.C.M. edited and revised manuscript; S.L., J.L.M., O.W.-W., C.M.S., D.T.H., S.E.O., S.Z., S.M.M., D.O.K., S.K.W., C.T.R., and I.C.M. approved final version of manuscript.
ACKNOWLEDGMENTS
We gratefully acknowledge the research assistance provided by Anne Jurisevic, Laura O'Carroll, and Andrew Snell during the course of this study.
REFERENCES
- 1.Baffy G, Brunt EM, Caldwell SH. Hepatocellular carcinoma in non-alcoholic fatty liver disease: an emerging menace. J Hepatol 56: 1384–1391, 2012 [DOI] [PubMed] [Google Scholar]
- 2.Barker DJ. The fetal and infant origins of adult disease. BMJ 301: 1111, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Chakravarty K, Cassuto H, Reshef L, Hanson RW. Factors that control the tissue-specific transcription of the gene for phosphoenolpyruvate carboxykinase-C. Crit Rev Biochem Mol Biol 40: 129–154, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic microRNA expression. Hepatology 48: 1810–1820, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong S, Whitelaw E. Epigenetic germline inheritance. Curr Opin Genet Dev 14: 692–696, 2004 [DOI] [PubMed] [Google Scholar]
- 7.Dodic M, Hantzis V, Duncan J, Rees S, Koukoulas I, Johnson K, Wintour EM, Moritz K. Programming effects of short prenatal exposure to cortisol. FASEB J 16: 1017–1026, 2002 [DOI] [PubMed] [Google Scholar]
- 8.Edwards LJ, Bryce AE, Coulter CL, McMillen IC. Maternal undernutrition throughout pregnancy increases adrenocorticotrophin receptor and steroidogenic acute regulatory protein gene expression in the adrenal gland of twin fetal sheep during late gestation. Mol Cell Endocrinol 196: 1–10, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Edwards LJ, McMillen IC. Periconceptional nutrition programs development of the cardiovascular system in the fetal sheep. Am J Physiol Regul Integr Comp Physiol 283: R669–R679, 2002 [DOI] [PubMed] [Google Scholar]
- 10.Edwards LJ, Symonds ME, Warnes KE, Owens JA, Butler TG, Jurisevic A, McMillen IC. Responses of the fetal pituitary-adrenal axis to acute and chronic hypoglycemia during late gestation in the sheep. Endocrinology 142: 1778–1785, 2001 [DOI] [PubMed] [Google Scholar]
- 11.Fowden AL, Mijovic J, Silver M. The effects of cortisol on hepatic and renal gluconeogenic enzyme activities in the sheep fetus during late gestation. J Endocrinol 137: 213–222, 1993 [DOI] [PubMed] [Google Scholar]
- 12.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA 108: 21075–21080, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner DS, Pearce S, Dandrea J, Walker R, Ramsay MM, Stephenson T, Symonds ME. Peri-implantation undernutrition programs blunted angiotensin II evoked baroreflex responses in young adult sheep. Hypertension 43: 1290–1296, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guay C, Roggli E, Nesca V, Jacovetti C, Regazzi R. Diabetes mellitus, a microRNA-related disease? Transl Res 157: 253–264, 2011 [DOI] [PubMed] [Google Scholar]
- 16.Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia 35: 595–601, 1992 [DOI] [PubMed] [Google Scholar]
- 17.Hay WW., Jr Placental transport of nutrients to the fetus. Horm Res 42: 215–222, 1994 [DOI] [PubMed] [Google Scholar]
- 18.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol 21: 2785–2794, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Humphreys DT, Hynes CJ, Patel HR, Wei GH, Cannon L, Fatkin D, Suter CM, Clancy JL, Preiss T. Complexity of murine cardiomyocyte miRNA biogenesis, sequence variant expression and function. PLoS One 7: e30933, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornfeld JW, Baitzel C, Könner AC, Nicholls HT, Vogt MC, Herrmanns K, Scheja L, Haumaitre C, Wolf AM, Knippschild U, Seibler J, Cereghini S, Heeren J, Stoffel M, Brüning JC. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature 494: 111–115, 2013 [DOI] [PubMed] [Google Scholar]
- 21.Kwong WY, Miller DJ, Wilkins AP, Dear MS, Wright JN, Osmond C, Zhang J, Fleming TP. Maternal low protein diet restricted to the preimplantation period induces a gender-specific change on hepatic gene expression in rat fetuses. Mol Reprod Dev 74: 52–60, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Kwong WY, Wild AE, Roberts P, Willis AC, Fleming TP. Maternal undernutrition during the preimplantation period of rat development causes blastocyst abnormalities and programming of postnatal hypertension. Development 127: 4195–4202, 2000 [DOI] [PubMed] [Google Scholar]
- 23.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Ling HY, Hu B, Hu XB, Zhong J, Feng SD, Qin L, Liu G, Wen GB, Liao D, F. MiRNA-21 reverses high glucose and high insulin induced insulin resistance in 3T3-L1 adipocytes through targeting phosphatase and tensin homologue. Exp Clin Endocrinol Diabetes 120: 553–559, 2012 [DOI] [PubMed] [Google Scholar]
- 25.MacLaughlin SM, Walker SK, Kleemann DO, Sibbons JP, Tosh DN, Gentili S, Coulter CL, McMillen IC. Impact of periconceptional undernutrition on adrenal growth and adrenal insulin-like growth factor and steroidogenic enzyme expression in the sheep fetus during early pregnancy. Endocrinology 148: 1911–1920, 2007 [DOI] [PubMed] [Google Scholar]
- 26.MacLaughlin SM, Walker SK, Kleemann DO, Tosh DN, McMillen IC. Periconceptional undernutrition and being a twin each alter kidney development in the sheep fetus during early gestation. Am J Physiol Regul Integr Comp Physiol 298: R692–R699, 2010 [DOI] [PubMed] [Google Scholar]
- 27.Marquez RT, Bandyopadhyay S, Wendlandt EB, Keck K, Hoffer BA, Icardi MS, Christensen RN, Schmidt WN, McCaffrey AP. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest 90: 1727–1736, 2010 [DOI] [PubMed] [Google Scholar]
- 28.McMillen IC, MacLaughlin SM, Muhlhausler BS, Gentili S, Duffield JL, Morrison JL. Developmental origins of adult health and disease: the role of periconceptional and foetal nutrition. Basic Clin Pharmacol Toxicol 102: 82–89, 2008 [DOI] [PubMed] [Google Scholar]
- 29.McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev 85: 571–633, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Meerson A, Traurig M, Ossowski V, Fleming JM, Mullins M, Baier LJ. Human adipose microRNA-221 is upregulated in obesity and affects fat metabolism downstream of leptin and TNF-α. Diabetologia 56: 1971–1979, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modaressi S, Brechtel K, Christ B, Jungermann K. Human mitochondrial phosphoenolpyruvate carboxykinase 2 gene. Structure, chromosomal localization and tissue-specific expression. Biochem J 333: 359–366, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muhlhausler BS, Duffield JA, Ozanne SE, Pilgrim C, Turner N, Morrison JL, McMillen IC. The transition from fetal growth restriction to accelerated postnatal growth: a potential role for insulin signalling in skeletal muscle. J Physiol 587 4199–4211, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narkewicz MR, Carver TD, Hay WWJ. Induction of cytosolic phosphoenolpyruvate carboxykinase in the ovine fetal liver by chronic fetal hypoglycemia and hypoinsulinemia. Pediatr Res 33: 493–496, 1993 [DOI] [PubMed] [Google Scholar]
- 34.Nijland MJ, Mitsuya K, Li C, Ford S, McDonald TJ, Nathanielsz PW, Cox LA. Epigenetic modification of fetal baboon hepatic phosphoenolpyruvate carboxykinase following exposure to moderately reduced nutrient availability. J Physiol 588: 1349–1359, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyirenda MJ, Lindsay RS, Kenyon CJ, Burchell A, Seckl JR. Glucocorticoid exposure in late gestation permanently programs rat hepatic phosphoenolpyruvate carboxykinase and glucocorticoid receptor expression and causes glucose intolerance in adult offspring. J Clin Invest 101: 2174–2181, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics 25: 1991–1993, 2009 [DOI] [PubMed] [Google Scholar]
- 37.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17–92 clusters in the control of transforming growth factor-beta signaling. Cancer Res 68: 8191–8194, 2008 [DOI] [PubMed] [Google Scholar]
- 38.Poore KR, Cleal JK, Newman JP, Boullin JP, Noakes DE, Hanson MA, Green LR. Nutritional challenges during development induce sex-specific changes in glucose homeostasis in the adult sheep. Am J Physiol Endocrinol Metab 292: E32–E39, 2007 [DOI] [PubMed] [Google Scholar]
- 39.Ramírez CM, Goedeke L, Rotllan N, Yoon JH, Cirera-Salinas D, Mattison JA, Suárez Y, de Cabo R, Gorospe M, Fernández-Hernando C. MicroRNA 33 regulates glucose metabolism. Mol Cell Biol 33: 2891–2902, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Razin A, Kantor B. DNA methylation in epigenetic control of gene expression. In: Epigenetics and Chromatin, edited by Jeanteur P. Heidelberg, Germany: Springer-Berlin, 2005, p. 151–167. [DOI] [PubMed] [Google Scholar]
- 41.Roseboom T, de Rooij S, Painter R. The Dutch famine and its long-term consequences for adult health. Early Hum Dev 82: 485–491, 2006 [DOI] [PubMed] [Google Scholar]
- 42.Roseboom TJ, van der Meulen JH, Ravelli AC, Osmond C, Barker DJ, Bleker OP. Effects of prenatal exposure to the Dutch famine on adult disease in later life: an overview. Twin Res 4: 293–298, 2001 [DOI] [PubMed] [Google Scholar]
- 43.Rozance PJ, Limesand SW, Barry JS, Brown LD, Thorn SR, LoTurco D, Regnault TR, Friedman JE, Hay WW., Jr Chronic late-gestation hypoglycemia upregulates hepatic PEPCK associated with increased PGC1α mRNA and phosphorylated CREB in fetal sheep. Am J Physiol Endocrinol Metab 294: E365–E370, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sand M, Skrygan M, Sand D, Georgas D, Hahn SA, Gambichler T, Altmeyer P, Bechara FG. Expression of microRNAs in basal cell carcinoma. Br J Dermatol 167: 847–855, 2012 [DOI] [PubMed] [Google Scholar]
- 45.Sasaki K, Cripe TP, Koch SR, Andreone TL, Petersen DD, Beale EG, Granner DK. Multihormonal regulation of phosphoenolpyruvate carboxykinase gene transcription. The dominant role of insulin. J Biol Chem 259: 15242–15251, 1984 [PubMed] [Google Scholar]
- 46.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484: 339–344, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Y, Liu S, Ferguson S, Wang L, Klepcyk P, Yun JS, Friedman JE. Phosphoenolpyruvate carboxykinase over expression selectively attenuates insulin signaling and hepatic insulin sensitivity in transgenic mice. J Biol Chem 277: 23301–23307, 2002 [DOI] [PubMed] [Google Scholar]
- 48.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol 7: 85–96, 2006 [DOI] [PubMed] [Google Scholar]
- 49.Todd SE, Oliver MH, Jaquiery AL, Bloomfield FH, Harding JE. Periconceptional undernutrition of ewes impairs glucose tolerance in their adult offspring. Pediatr Res 65: 409–413, 2009 [DOI] [PubMed] [Google Scholar]
- 50.Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474: 649–653, 2011 [DOI] [PubMed] [Google Scholar]
- 51.Walker BR. Extra-adrenal regeneration of glucocorticoids by 11b-hydroxysteroid dehydrogenase type 1: physiological regulator and pharmacological target for energy partitioning. Proc Nutr Soc 66: 1–8, 2007 [DOI] [PubMed] [Google Scholar]
- 52.Watford M, Hod Y, Chiao YB, Utter MF, Hanson RW. The unique role of the kidney in gluconeogenesis in the chicken. The significance of a cytosolic form of phosphoenolpyruvate carboxykinase. J Biol Chem 256: 10023–10027, 1981 [PubMed] [Google Scholar]
- 53.Watkins AJ, Wilkins A, Cunningham C, Perry VH, Seet MJ, Osmond C, Eckert JJ, Torrens C, Cagampang FR, Cleal J, Gray WP, Hanson MA, Fleming TP. Low protein diet fed exclusively during mouse oocyte maturation leads to behavioural and cardiovascular abnormalities in offspring. J Physiol 586: 2231–2244, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res 25: 2532–2534, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamada H, Suzuki K, Ichino N, Ando Y, Sawada A, Osakabe K, Sugimoto K, Ohashi K, Teradaira R, Inoue T, Hamajima N, Hashimoto S. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta 424: 99–103, 2013 [DOI] [PubMed] [Google Scholar]
- 56.Zhang S, Rattanatray L, MacLaughlin SM, Cropley JE, Suter CM, Molloy L, Kleemann D, Walker SK, Muhlhausler BS, Morrison JL, McMillen IC. Periconceptional undernutrition in normal and overweight ewes leads to increased adrenal growth and epigenetic changes in adrenal IGF2/H19 gene in offspring. FASEB J 24: 2772–2782, 2010 [DOI] [PubMed] [Google Scholar]
- 57.Zhou B, Li C, Qi W, Zhang Y, Zhang F, Wu JX, Hu YN, Wu DM, Liu Y, Yan TT, Jing Q, Liu MF, Zhai QW. Downregulation of miR-181a upregulates sirtuin-1 (SIRT1) and improves hepatic insulin sensitivity. Diabetologia 55: 2032–2043, 2012 [DOI] [PubMed] [Google Scholar]