Abstract
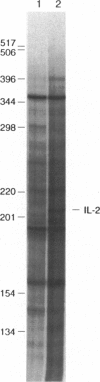
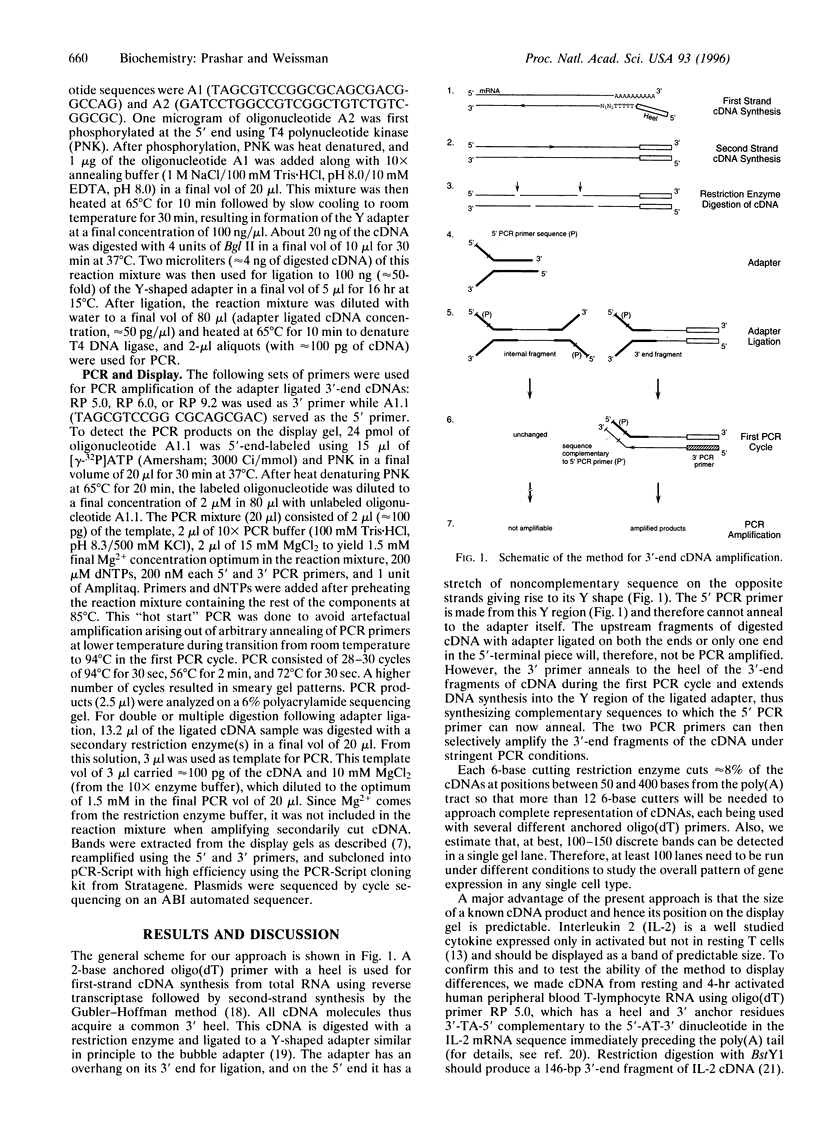
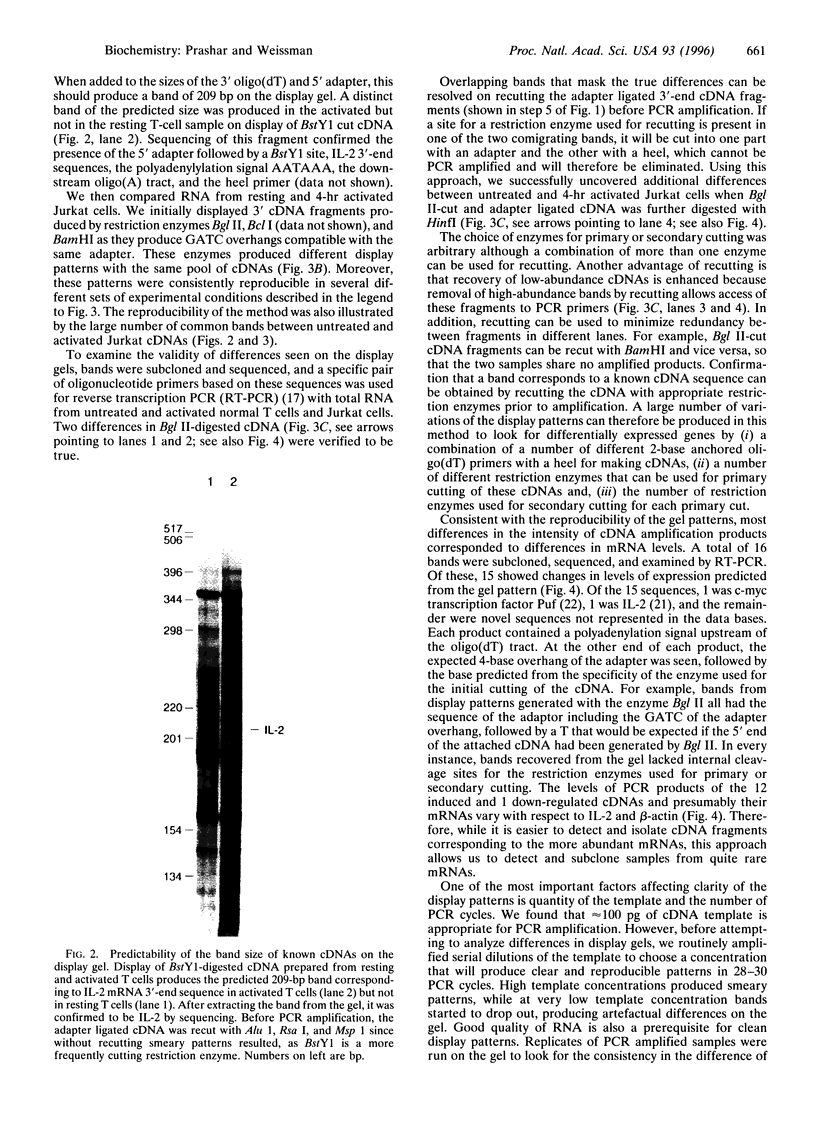
We have developed an approach to study changes in gene expression by selective PCR amplification and display of 3' end restriction fragments of double-stranded cDNAs. This method produces highly consistent and reproducible patterns, can detect almost all mRNAs in a sample, and can resolve hidden differences such as bands that differ in their sequence but comigrate on a gel. Bands corresponding to known cDNAs move to predictable positions on the gel, making this a powerful approach to correlate gel patterns with cDNA data bases. Applying this method, we have examined differences in gene expression patterns during T-cell activation. Of a total of 700 bands that were evaluated in this study, as many as 3-4% represented mRNAs that are upregulated, while approximately 2% were down-regulated within 4 hr of activation of Jurkat T cells. These and other results suggest that this approach is suitable for the systematic, expeditious, and nearly exhaustive elucidation of subtle changes in the patterns of gene expression in cells with altered physiologic states.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhargava A. K., Li Z., Weissman S. M. Differential expression of four members of the POU family of proteins in activated and phorbol 12-myristate 13-acetate-treated Jurkat T cells. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):10260–10264. doi: 10.1073/pnas.90.21.10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. C., Arya S. K., Wong-Staal F., Matsumoto-Kobayashi M., Kay R. M., Kaufman R. J., Brown E. L., Shoemaker C., Copeland T., Oroszlan S. Human T-cell growth factor: partial amino acid sequence, cDNA cloning, and organization and expression in normal and leukemic cells. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2543–2547. doi: 10.1073/pnas.81.8.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hedrick S. M., Cohen D. I., Nielsen E. A., Davis M. M. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984 Mar 8;308(5955):149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- Ito T., Kito K., Adati N., Mitsui Y., Hagiwara H., Sakaki Y. Fluorescent differential display: arbitrarily primed RT-PCR fingerprinting on an automated DNA sequencer. FEBS Lett. 1994 Sep 5;351(2):231–236. doi: 10.1016/0014-5793(94)00867-1. [DOI] [PubMed] [Google Scholar]
- Ivanova N. B., Belyavsky A. V. Identification of differentially expressed genes by restriction endonuclease-based gene expression fingerprinting. Nucleic Acids Res. 1995 Aug 11;23(15):2954–2958. doi: 10.1093/nar/23.15.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K., Siebenlist U. Immediate-early genes induced by antigen receptor stimulation. Curr Opin Immunol. 1995 Jun;7(3):327–332. doi: 10.1016/0952-7915(95)80106-5. [DOI] [PubMed] [Google Scholar]
- Koyama T., Hall L. R., Haser W. G., Tonegawa S., Saito H. Structure of a cytotoxic T-lymphocyte-specific gene shows a strong homology to fibrinogen beta and gamma chains. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1609–1613. doi: 10.1073/pnas.84.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Sehajpal P. K., Khanna A., Vlassara H., Cerami A., Stenzel K. H., Suthanthiran M. Differential regulation of transforming growth factor beta and interleukin 2 genes in human T cells: demonstration by usage of novel competitor DNA constructs in the quantitative polymerase chain reaction. J Exp Med. 1991 Nov 1;174(5):1259–1262. doi: 10.1084/jem.174.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Pardee A. B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992 Aug 14;257(5072):967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Liang P., Pardee A. B. Recent advances in differential display. Curr Opin Immunol. 1995 Apr;7(2):274–280. doi: 10.1016/0952-7915(95)80015-8. [DOI] [PubMed] [Google Scholar]
- Liang P., Zhu W., Zhang X., Guo Z., O'Connell R. P., Averboukh L., Wang F., Pardee A. B. Differential display using one-base anchored oligo-dT primers. Nucleic Acids Res. 1994 Dec 25;22(25):5763–5764. doi: 10.1093/nar/22.25.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfioletti G., Ruaro M. E., Del Sal G., Philipson L., Schneider C. A growth arrest-specific (gas) gene codes for a membrane protein. Mol Cell Biol. 1990 Jun;10(6):2924–2930. doi: 10.1128/mcb.10.6.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Mathieu-Daude F., Welsh J. RNA fingerprinting and differential display using arbitrarily primed PCR. Trends Genet. 1995 Jun;11(6):242–246. doi: 10.1016/s0168-9525(00)89058-7. [DOI] [PubMed] [Google Scholar]
- Mou L., Miller H., Li J., Wang E., Chalifour L. Improvements to the differential display method for gene analysis. Biochem Biophys Res Commun. 1994 Mar 15;199(2):564–569. doi: 10.1006/bbrc.1994.1265. [DOI] [PubMed] [Google Scholar]
- Paillard F., Vaquero C. Down-regulation of lck mRNA by T cell activation involves transcriptional and post-transcriptional mechanisms. Nucleic Acids Res. 1991 Sep 11;19(17):4655–4661. doi: 10.1093/nar/19.17.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel E. H., Berberich S. J., Flint S. J., Ferrone C. A. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science. 1993 Jul 23;261(5120):478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- Riley J., Butler R., Ogilvie D., Finniear R., Jenner D., Powell S., Anand R., Smith J. C., Markham A. F. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990 May 25;18(10):2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., King R. M., Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988 Sep 9;54(6):787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- Sokolov B. P., Prockop D. J. A rapid and simple PCR-based method for isolation of cDNAs from differentially expressed genes. Nucleic Acids Res. 1994 Sep 25;22(19):4009–4015. doi: 10.1093/nar/22.19.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman K. S., Northrop J. P., Verweij C. L., Crabtree G. R. Transmission of signals from the T lymphocyte antigen receptor to the genes responsible for cell proliferation and immune function: the missing link. Annu Rev Immunol. 1990;8:421–452. doi: 10.1146/annurev.iy.08.040190.002225. [DOI] [PubMed] [Google Scholar]
- Welcher A. A., Torres A. R., Ward D. C. Selective enrichment of specific DNA, cDNA and RNA sequences using biotinylated probes, avidin and copper-chelate agarose. Nucleic Acids Res. 1986 Dec 22;14(24):10027–10044. doi: 10.1093/nar/14.24.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Ooi S. L., Pardee A. B. New primer strategy improves precision of differential display. Biotechniques. 1995 May;18(5):842-6, 848, 850. [PubMed] [Google Scholar]
- Zipfel P. F., Irving S. G., Kelly K., Siebenlist U. Complexity of the primary genetic response to mitogenic activation of human T cells. Mol Cell Biol. 1989 Mar;9(3):1041–1048. doi: 10.1128/mcb.9.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]