ABSTRACT
Antibiotic usage is the most commonly cited risk factor for hospital-acquired Clostridium difficile infections (CDI). The increased risk is due to disruption of the indigenous microbiome and a subsequent decrease in colonization resistance by the perturbed bacterial community; however, the specific changes in the microbiome that lead to increased risk are poorly understood. We developed statistical models that incorporated microbiome data with clinical and demographic data to better understand why individuals develop CDI. The 16S rRNA genes were sequenced from the feces of 338 individuals, including cases, diarrheal controls, and nondiarrheal controls. We modeled CDI and diarrheal status using multiple clinical variables, including age, antibiotic use, antacid use, and other known risk factors using logit regression. This base model was compared to models that incorporated microbiome data, using diversity metrics, community types, or specific bacterial populations, to identify characteristics of the microbiome associated with CDI susceptibility or resistance. The addition of microbiome data significantly improved our ability to distinguish CDI status when comparing cases or diarrheal controls to nondiarrheal controls. However, only when we assigned samples to community types was it possible to differentiate cases from diarrheal controls. Several bacterial species within the Ruminococcaceae, Lachnospiraceae, Bacteroides, and Porphyromonadaceae were largely absent in cases and highly associated with nondiarrheal controls. The improved discriminatory ability of our microbiome-based models confirms the theory that factors affecting the microbiome influence CDI.
IMPORTANCE
The gut microbiome, composed of the trillions of bacteria residing in the gastrointestinal tract, is responsible for a number of critical functions within the host. These include digestion, immune system stimulation, and colonization resistance. The microbiome’s role in colonization resistance, which is the ability to prevent and limit pathogen colonization and growth, is key for protection against Clostridium difficile infections. However, the bacteria that are important for colonization resistance have not yet been elucidated. Using statistical modeling techniques and different representations of the microbiome, we demonstrated that several community types and the loss of several bacterial populations, including Bacteroides, Lachnospiraceae, and Ruminococcaceae, are associated with CDI. Our results emphasize the importance of considering the microbiome in mediating colonization resistance and may also direct the design of future multispecies probiotic therapies.
INTRODUCTION
Since the discovery of penicillin in 1928, antibiotics have revolutionized health care, saving patients from life-threatening infections, such as bacteremia, bacterial meningitis, tuberculosis, and pneumonia. It has recently been estimated that over 250 million courses of antibiotics are prescribed to outpatients in the United States annually (1). However, besides eradicating the pathogen of interest, antibiotics disturb members of the indigenous bacterial community of the gastrointestinal tract, i.e., the gut microbiome. In hospitals, this disruption may result in Clostridium difficile infection (CDI), the leading nosocomial infectious disease in the United States. Cases of CDI have more than doubled since 2001, with over 300,000 new diagnoses in 2009 (2).
An intact microbiome is crucial for its role in providing resistance to C. difficile colonization. Antibiotic use, proton pump inhibitors, and advancing age, which are all known to influence the composition of the gut microbiome, are all risk factors for CDI (3–8). However, no one has developed a set of microbiome-based biomarkers for CDI to complement these risk factors. Therefore, characterizing the differences in the microbiomes of individuals with and without CDI is essential for understanding the changes within the microbiome associated with CDI. This knowledge would also potentially lead to novel targeted therapies, as the typical treatments of metronidazole and vancomycin feed the cycle of disrupting the gut microbiome. For the estimated 25% of cases of CDI-characterized recurrent infection, the most effective treatment has been fecal microbial transplant (FMT), which has a 92% success rate in limiting further recurrence (9). The remarkable success of FMT, which restores the normal microbiome, underscores the importance of understanding the role of the microbiome in providing colonization resistance.
Current microbiome-related studies use three general methods to characterize differences in the microbiomes between groups of individuals. First, the microbial community composition of an individual can be distilled into a single parameter (i.e., alpha diversity) to describe the community in terms of richness or diversity. For instance, it has been shown that individuals with diarrhea tend to have a less diverse community composition than healthy individuals (10, 11). Unfortunately, such results do not lend themselves to subsequent mechanistic investigations and do not provide a therapeutic avenue, since it is difficult to predict whether an antibiotic will increase or decrease an individual’s diversity. Second, cross-community comparisons (i.e., beta diversity) have been made to relate the similarity of microbial communities between individuals (e.g., the UniFrac method and Bray-Curtis index). In humans, these metrics are useful in tracking an individual’s recovery from antibiotic therapy (6); however, they have had limited use in discriminating between treatment groups (see, e.g., reference 12). Using beta diversity metrics, it is again difficult to develop a mechanistic understanding of the relationship between the community structure and disease or to provide a therapeutic avenue to change a community structure. Finally, in an approach similar to that of genome-wide association studies, comparisons of the relative abundances of individual bacterial populations can be made between groups of individuals (10, 13). This approach does not account for the possibility that mixtures of populations can be protective or causative or that different mixtures can have the same phenotype in different individuals. Here we propose a comprehensive modeling approach that incorporates clinical metadata to identify collections of bacteria that can be associated with health and disease. A similar approach was recently used to model the microbiome to identify microbiome signatures of psoriasis; however, clinical data for the subjects was not included (14).
To better understand how clinical and microbiome-based factors are associated with CDI, we characterized the gut microbiomes of hospitalized individuals with and without CDI who developed diarrhea and of healthy individuals from the broader community. We used clinical and microbiome data to generate models of CDI status in order to differentiate between the three groups of subjects. Addition of microbiome data to clinically based models for CDI significantly improved the ability to differentiate these patient groups. Using these models as tools, we identified bacteria with potential roles in the resistance to C. difficile colonization, while controlling for clinically relevant risk factors.
RESULTS
Patient sampling and base model framework.
Fecal samples were collected from 338 individuals. Within this collection, 183 diarrheal stool samples were acquired from inpatients at the University of Michigan Hospital, including subjects both with CDI (n = 94) and without CDI (n = 89). These samples were tested as positive or negative for C. difficile by the clinical microbiology lab at the hospital and subsequently confirmed through PCR using C. difficile-specific 16S rRNA primers (see Materials and Methods). The remaining 155 nondiarrheal control stool samples were collected from individuals in the surrounding community. We collected a broad set of clinical data, including risk factors for development of CDI from the subjects’ questionnaire responses and their medical records (Table 1). As expected, antibiotic usage was more prevalent in CDI cases than in individuals in either of the control groups (P < 0.001). Fluoroquinolones represented the most-used at-risk antibiotics in hospitalized patients, followed by amoxicillin and cephalosporins. Interestingly, individuals that were CDI positive were more likely to have lived with a health care worker. Together, these clinical data represented the framework for our base model.
TABLE 1 .
Demographic information for subjects in each experimental group
| Characteristic | Value for: |
P value | ||
|---|---|---|---|---|
| Cases (n = 94) | Diarrheal controls (n = 89) |
Nondiarrheal controls (n = 155) |
||
| Sex, n (%) | ||||
| Females | 53 (56.4) | 49 (55.1) | 102 (65.8) | |
| Males | 41 (43.6) | 40 (44.9) | 53 (34.2) | 0.166 |
| Age (yr) | ||||
| Mean (SD) | 55.9 (18.3) | 58.7 (14.9) | 52.2 (21.5) | 0.034 |
| Range | 18-89 | 18-85 | 19-88 | |
| Race, n (%) | 0.712 | |||
| White | 84 (89.4) | 76 (85.4) | 129 (83.2) | |
| Black | 7 (7.4) | 9 (10.1) | 16 (10.3) | |
| Other/unknown | 3 (3.2) | 4 (4.5) | 10 (6.5) | |
| Wt, mean no. of lbs (SD) | 169.9 (56.9) | 177.9 (54.5) | 171.5 (47.3) | 0.549 |
| Vegetarian, n (%) | 2 (2.1) | 5 (5.6) | 8 (5.2) | 0.435 |
| Drug use, n (%) | ||||
| Antibiotics (<3 mo) | 72 (76.6) | 56 (62.9) | 21 (13.5) | <0.001 |
| Fluoroquinolone | 21 (22.3) | 17 (19.1) | 4 (2.6) | <0.001 |
| Amoxicillin | 10 (10.6) | 6 (6.7) | 7 (4.5) | 0.182 |
| Cephalosporin | 11 (11.7) | 3 (3.4) | 3 (1.9) | 0.004 |
| Clindamycin | 2 (2.1) | 0 | 5 (3.2) | 0.297 |
| Ampicillin | 1 (1.1) | 0 | 0 | 0.541 |
| Other factors, n (%) | ||||
| Antacid use for <30 days | 20 (21.3) | 20 (22.5) | 11 (7.1) | 0.001 |
| Surgery within the previous 6 mo | 48 (51.1) | 38 (42.7) | 14 (9.0) | <0.001 |
| History of C. difficile | 1 (1.1) | 2 (2.2) | 4 (2.6) | 0.793 |
| Residing with person with CDI | 1 (1.1) | 2 (2.2) | 1 (0.6) | 0.593 |
| Residing with health care worker | 25 (26.6) | 13 (14.6) | 17 (11.0) | 0.005 |
We used age, gender, race, antibiotic use, antacid use, a vegetarian diet, surgery within the past 6 months, a history of CDI, residence with another person who had CDI, and residence with another person who works in a health care setting as explanatory variables for three logit models. For each model, we evaluated the ability of these variables to discriminate a group’s classification using the area under the receiver operator characteristic (ROC) curve (AUC) (15). These curves look at the true-positivity rate, i.e., sensitivity, as it relates to the false-positivity rate, i.e., 1 minus the specificity. Using the full collection of explanatory variables, we were interested in three comparisons. The first comparison differentiated between the cases and the nondiarrheal controls (AUC = 0.891). The second differentiated between the cases and diarrheal controls (AUC = 0.659). The third differentiated between the diarrheal and nondiarrheal controls (AUC = 0.849). The base model for cases and diarrheal controls was the only comparison that was not significantly different than an empty model (i.e., a model without independent variables; P = 0.189). The AUC and 95% confidence intervals for all models are listed in Table S1 in the supplemental material. These three models served as the base for our development of other logit models that incorporated microbiome-based data.
Incorporation of diversity measures into logit models.
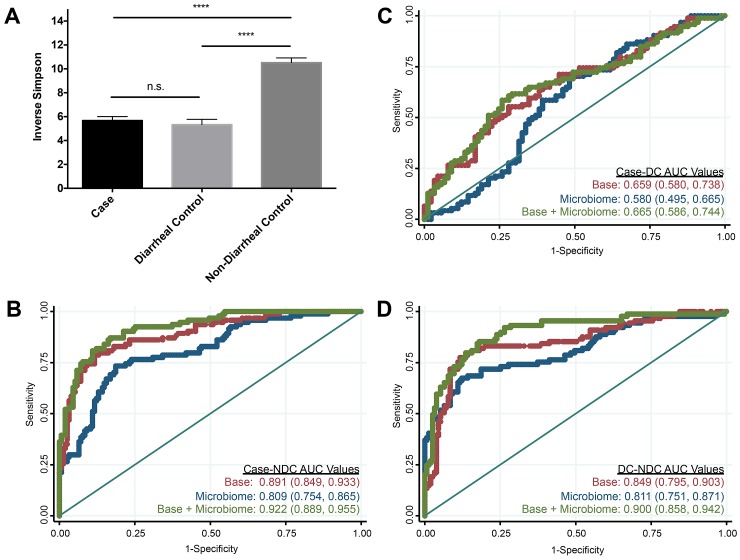
We first looked at the overall microbiome structural differences among the individuals in our study (see Fig. S1 in the supplemental material). There were apparent structural differences between nondiarrheal control samples and hospital-acquired samples (cases and diarrheal controls). These differences were statistically significant by analysis of molecular variance (AMOVA) (P < 0.001). The structures of cases and diarrheal controls were additionally significantly different from one another by AMOVA (P = 0.02), although to a lesser degree. In order to identify the differences between these experimental groups, we first looked at their levels of overall bacterial diversity. Previous studies have shown that the bacterial diversity of subjects with initial and recurrent CDI is markedly lower than that of healthy subjects (10, 11). Measuring diversity using the inverse Simpson index, we found that hospital-acquired samples had a 2-fold-lower diversity than those of the nondiarrheal controls but were not significantly different from each other (Fig. 1A). A model based on the inverse Simpson index alone significantly differentiated nondiarrheal controls from either cases or diarrheal controls (Fig. 1B to D), although this model performed no better than the base model alone. When we incorporated the inverse Simpson index into our base models, differentiation of cases from nondiarrheal controls was significantly improved (AUC = 0.922, P = 0.0072), and differentiation of diarrheal controls from nondiarrheal controls was also significantly improved (AUC = 0.900, P = 0.0009). Cases and diarrheal controls were indistinguishable when we used models that incorporated the inverse Simpson index. We performed the same analysis using the Shannon diversity index and observed similar results. These results indicate that although low diversity was a characteristic of CDI-positive subjects, subjects with diarrhea had lower diversity than healthy outpatients.
FIG 1 .
Bacterial diversity distinguishes between subjects with and without diarrhea. (A) Alpha diversity was measured using the inverse Simpson index. Statistical analysis was performed using Dunn’s multiple-comparison test. ****, P < 0.0001; n.s., no significance. Error bars represent ± the standard errors of the means (SEM). (B to D) ROC curves and AUC values with 95% confidence intervals in parentheses for each model comparing cases and nondiarrheal controls (NDC) (B), cases and diarrheal controls (DC) (C), and diarrheal controls and nondiarrheal controls (D). Red represents the base model, blue represents the inverse Simpson model, and green represents the base plus inverse Simpson model. The straight line represents the null model.
Incorporation of bacterial community types into logit models.
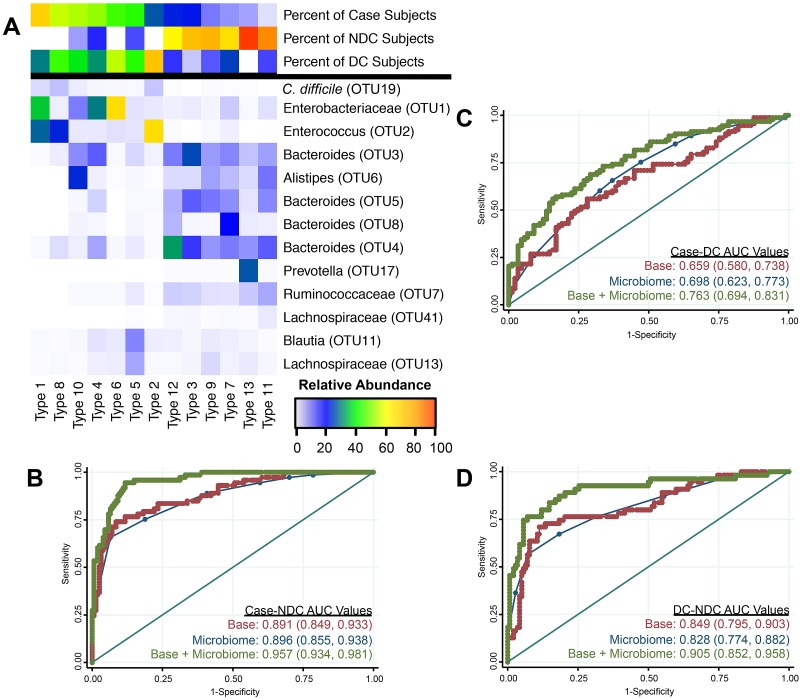
Next, we sought to determine whether a subject’s overall community composition differentiated CDI status from diarrheal status. We assigned the samples to a specified number of clusters (k = 2 to 15) based on their similarity to other samples after removing the C. difficile operational taxonomic unit, 19 (OTU 19). We selected 13 as the appropriate number of clusters (i.e., community types), as this resulted in the optimal AUC for the base model when we incorporated the subject’s community type. These community types varied in CDI prevalence among individuals within each type (Fig. 2A, percent of case subjects). Results from models using community types alone were similar to results from the base models (Fig. 2B to D). When the community type assignments were added to the base models, the AUCs significantly improved relative to those of the base models in all comparisons (Table S1). These results indicate that specific community types differentiated CDI status and suggest that certain community types may be more susceptible to colonization by C. difficile.
FIG 2 .
Specific community structure types significantly differentiate all population comparisons. (A) Heat map showing the structural differences between each community type. The top 3 rows show the percentages of individuals classified as a case, diarrheal control, or nondiarrheal control across each type. The remaining rows show the relative abundances for OTUs identified using feature selection through randomForest analysis. Although the relative abundance of C. difficile OTU 19 is shown, it was not considered in the formation of these community types. Types are ordered by decreasing percentage of case individuals. (B to D) ROC curves and AUC values with 95% confidence intervals in parentheses for each model comparing cases and nondiarrheal controls (B), cases and diarrheal controls (C), and diarrheal controls and nondiarrheal controls (D). Red represents the base model, blue represents the community types model, and green represents the base plus community type model. The straight line represents the null model.
To determine the taxonomic composition of each of these community types, we used the randomForest feature selection algorithm to identify those taxa that were indicators for the different community types (Fig. 2A). There were 6 community types that were less prevalent among case individuals or diarrheal controls than among nondiarrheal controls (i.e., types 12, 3, 9, 7, 13, and 11). These 6 types had higher relative abundances of OTUs belonging to the Bacteroides genus (OTUs 3, 4, 5, and 8), Alistipes genus (OTU 6), Prevotella genus (OTU 17), and the Ruminococcaceae (OTU 7) than the other community types, while the remaining 7 types (i.e., 1, 8, 10, 4, 6, 5, and 2) were enriched in Enterobacteriaceae (OTU 1), Enterococcus species (OTU 2), Blautia species (OTU 11), and Lachnospiraceae (OTU 13). There were 6 community types that were less prevalent among case individuals than among the diarrheal controls (i.e., types 6, 5, 2, 9, 7, and 11). These types could be further subdivided by the overall percentage of diarrheal controls within each type. Types 6, 5, and 2 had a high percentage of diarrheal controls, while types 9, 7, and 11 had a low percentage of diarrheal controls (Table S2). Types 6, 5, and 2 were also low in nondiarrheal controls and lacked several OTUs found primarily in that group (Fig. 2A). Furthermore, type 2 or 6 was highly enriched in either Enterococcus species or Enterobacteriaceae, respectively. Types 9, 7, and 11 also were found in a high proportion of nondiarrheal controls and were more abundant in OTUs found predominantly in the nondiarrheal control group. These group-specific taxonomic features that we have identified may be involved in susceptibility or resistance to CDI.
Incorporation of specific bacterial populations into logit models.
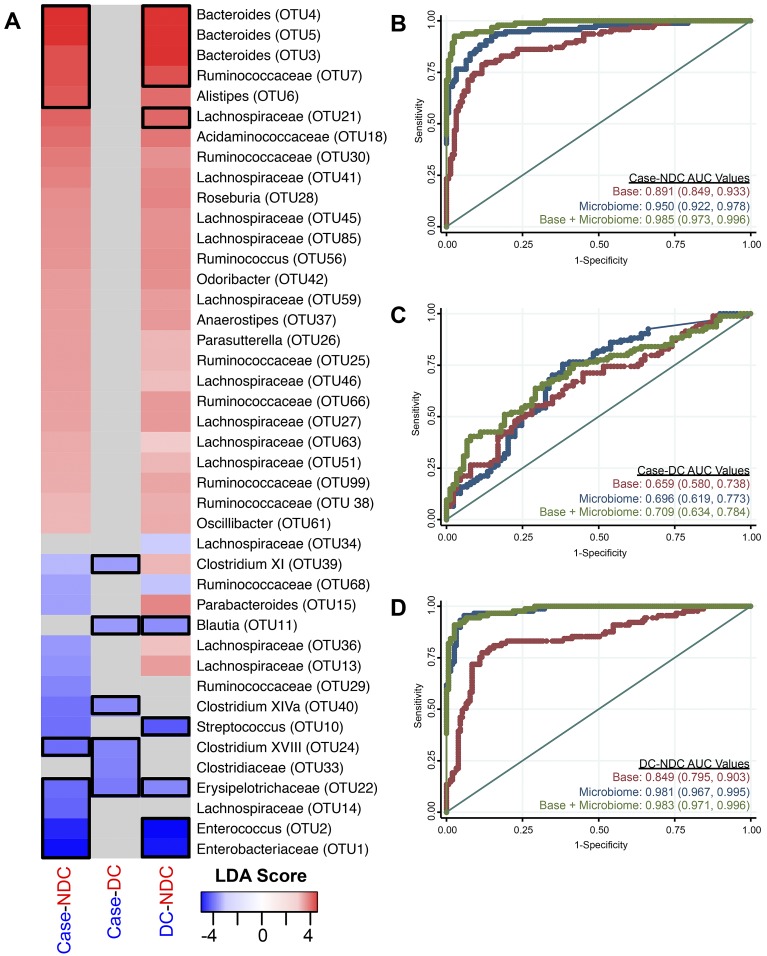
Having established that incorporation of a subject’s community type could better reflect their CDI or diarrheal status than a diversity index, we attempted to determine whether more-specific components of those community types could improve our models. To accomplish this, we first identified those bacterial populations that were differentially represented in each of the three comparisons using the linear discriminant analysis (LDA) effect size (LEfSe) algorithm (13); these analyses excluded C. difficile (OTU 19). Briefly, LEfSe uses (i) the Kruskal-Wallis rank sum test to identify taxonomic features that characterize the differences between our study groups and (ii) linear discriminant analysis to evaluate the effect size of each feature. Within each comparison, the OTUs with the largest effect size in each group and comparison were included in the respective base model (Fig. 3A).
FIG 3 .
Specific bacterial populations clearly differentiate non-diarrheal controls from subjects with CDI and non-CDI-associated diarrhea. (A) LEfSe was used to compare cases and nondiarrheal controls, cases and diarrheal controls, and diarrheal controls and nondiarrheal controls. Only the LDA scores of significant OTUs are shown. Gray boxes indicate that values were not significant for the given comparison. The number following the bacterial name indicates the OTU number. Black boxes show the OTUs at the 0.25-percentile cutoff for each group that were chosen for inclusion in the microbiome models. DC, diarrheal control; NDC, nondiarrheal control. (B to D) ROC curves and AUC values with 95% confidence intervals in parentheses for each model comparing cases and nondiarrheal controls (B), cases and diarrheal controls (C), and diarrheal controls and nondiarrheal controls (D). Red represents the base model, blue represents the specific OTUs model, and green represents the base plus specific-OTU model. The straight line represents the null model.
Within the case versus nondiarrheal control comparison, 13 OTUs were significantly enriched in cases and 26 OTUs were significantly enriched in the nondiarrheal controls (Fig. 3A). We selected 5 OTUs that were enriched in the cases and 5 that were enriched in the nondiarrheal controls (based on the most extreme differences) and included their relative abundances as independent variables in logit regression. The relative abundances of these specific OTUs discriminated quite well between cases and nondiarrheal controls (AUC = 0.950) (Table S1). Additionally, the combined base-microbiome model significantly outperformed the base model (AUC = 0.985, P < 0.0001). Subjects having CDI were significantly more likely to harbor Enterococcus species (OTU 2), Lachnospiraceae (OTU 14), and Erysipelotrichaceae (OTU 22) and significantly less likely to harbor Bacteroides species (OTU 5) than nondiarrheal controls. These results confirm the differences that were observed between the case and nondiarrheal control enriched community types and highlight the populations that had the greatest contribution to the model.
In the comparison of cases and diarrheal controls, no OTUs were significantly enriched in the diarrheal controls over the cases; however, we identified 6 OTUs that were significantly enriched in the cases (Fig. 3A). The relative abundances of these OTUs, when combined in a logit model (Fig. 3C), did not significantly distinguish cases and diarrheal controls (AUC = 0.696, P = 0.0934). Furthermore, the base plus microbiome model was not significantly different from the base clinical model (AUC = 0.709, P = 0.0652). Unlike with the bacterial community type analysis, we were unable to identify specific structural differences that could distinguish between cases and diarrheal controls in this model. These results confirm that overall microbiome structure was more discriminatory for patients with non-C. difficile-associated and C. difficile-associated diarrhea.
Finally, in the comparison of diarrheal controls and nondiarrheal controls, we identified 30 OTUs that were enriched in the nondiarrheal controls and 7 OTUs that were enriched in the diarrheal controls (Fig. 3A). Individuals with non-C. difficile-associated diarrhea were more likely to have higher relative abundances of Enterobacteriaceae (OTU 1), Enterococcus species (OTU 2), Erysipelotrichaceae (OTU 22), Streptococcus species (OTU 10), and Blautia species (OTU 11). The nondiarrheal controls were more likely to have higher levels of several Bacteroides, Lachnospiraceae, and Ruminococcaceae OTUs. These taxa are commonly associated with a healthy microbiome. We used the 5 most enriched OTUs in each of the diarrheal control and nondiarrheal control groups to create a logit model to differentiate between the two (Fig. 3D). These OTUs significantly differentiated the two control groups (AUC = 0.981). The inclusion of both clinical data and these OTUs provided considerable discrimination between the two groups compared with the base model alone (AUC = 0.983; P < 0.0001). These results indicate that there were significant changes to the microbiome when individuals had diarrhea.
DISCUSSION
We found distinct differences in the microbiomes of people with and without CDI as well as with and without diarrhea. We developed classification models to differentiate whether individuals had C. difficile infection or non-C. difficile-associated diarrhea based on clinical and microbiome data. The microbiome was incorporated into these models using three approaches: diversity indices, community types, and defined bacterial subsets. These approaches of representing the microbiome allowed us to describe the communities at various levels of resolution. When differentiating between the cases and diarrheal controls, incorporation of community types provided the only significant improvement in detection between these two groups of patients. The inverse Simpson index and the utilization of specific OTUs did not differentiate these two groups. For the comparisons of nondiarrheal controls to cases or diarrheal controls, inclusion of the microbiome data significantly enhanced our ability to differentiate the groups regardless of the approach we used to represent the microbiome. The highest AUCs were observed when we differentiated between hospitalized patients (either cases or diarrheal controls) and community residents (nondiarrheal controls), with AUCs consistently greater than 0.9 when both clinical and microbiome data were considered. Specifically, representing the microbiome using specific sets of OTUs was the best approach for differentiating between hospitalized patients and community subjects. These findings stress the importance of not just one individual bacterial population or one metric of the community (e.g., diversity) but rather collections of bacterial populations or overall community types in detecting disease state. They also suggest the presence of gut dysbiosis in patients with diarrhea. These results demonstrate that knowledge of bacterial communities, not just single species, and in combination with clinical factors may be beneficial in generating epidemiological models of disease.
It was notable that, among our three comparisons, the cases and diarrheal controls were the most similar. First, the base model to differentiate the two groups was unable to perform significantly better than a null model (P = 0.19). Second, the inverse Simpson diversity index revealed similar levels in both groups (P = 0.85). Third, many community types characterized by high numbers of cases were also more likely to contain diarrheal controls than nondiarrheal controls, which tended to cluster separately from both diarrheal groups. Finally, our OTU-based analysis did not identify any OTUs as being significantly enriched in the diarrheal controls relative to the cases. Because of the similarity in community structures and in clinical risk factors for CDI, results suggest that many of the diarrheal control subjects may actually be susceptible to CDI and have not yet been exposed to C. difficile. This implies that any perturbation resulting in diarrhea may also contribute to CDI. This hypothesis is particularly relevant within a hospital setting, where C. difficile spores are abundant and where there are numerous potential causes of diarrhea, including antibiotics (independent of CDI), infection, chemotherapy, and dietary changes.
Our models that compared cases to nondiarrheal controls showed that Bacteroides species, Lachnospiraceae, and Ruminococcaceae were enriched in controls and that Enterococcus species, Enterobacteriaceae, Erysipelotrichaceae, and some Lachnospiraceae were enriched in cases. These results confirm those of previous related studies (10, 16–18). Members of the Lachnospiraceae and Ruminococcaceae are the primary butyrate-producing bacteria in the human gastrointestinal tract. Butyrate has been associated with inhibition of C. difficile growth in vitro (19), inflammation suppression, and the health of colonic cells. Thus, butyrate as well as other short-chain fatty acids may represent one mechanism of colonization resistance. Comparison of the sequences within our Bacteroides OTU (OTU 5), which was enriched in our nondiarrheal controls, to sequences in an annotated 16S rRNA gene database showed that they were highly similar to Bacteroides uniformis and Bacteroides acidifaciens. B. uniformis was previously shown to ameliorate metabolic dysfunction caused by diet-induced obesity via changes in metabolic and immune responses (20). Because obesity is a risk factor for CDI (21), it is possible that B. uniformis also provides protection against infection by C. difficile. B. acidifaciens was demonstrated to increase IgA+ B cells in the large intestine (22), which may also limit the growth of gastrointestinal pathogens such as C. difficile. Overall, this shift in community structure is thought to be associated with a change in colonization resistance. Murine models of CDI have shown that similar changes in community structure render normally resistant mice sensitive to colonization by C. difficile (23, 24). Similarly, a mixture of 6 bacterial species that included a member of the Bacteroides genus and a member of the Lachnospiraceae, both of which were found to be significantly enriched in our nondiarrheal control population, was sufficient to clear C. difficile in a murine model of recurrent CDI (17).
Microbiome analyses have revealed that bacterial populations are patchy across individuals and that there is no core microbiome (12). This hinders one’s ability to consistently associate specific bacterial populations with disease. Instead, others have developed the concept of communities or enterotypes (25–27). Although the biological interpretation of these clusters is controversial, our study demonstrates that categorizing individuals into community types or utilizing subsets of the bacterial community improves our ability to identify individuals that belong to specific disease states. Similar approaches have been used to associate specific community types with the composition of one’s diet, obesity, inflammatory bowel disease, Crohn’s disease, Trichomonas vaginalis infection, vaginal pH, and ethnicity (25, 27–32); however, these studies have not combined the subject’s clinical information and community type to evaluate disease state.
The models evaluated in this study reflect bacterial communities at a specific point in time for these three patient groups. Thus, we are limited in our ability to assess the contribution of the microbiome toward risk or prevention of CDI. We also cannot determine the length of time that cases were colonized by C. difficile prior to sample collection. The aim of this investigation was not to enhance CDI diagnostics but to use a model-based framework to characterize features of the microbiome that are associated with CDI and health. Our approach suggests that knowledge of an individual’s microbiome composition is useful in distinguishing disease from health. However, prospective studies are needed to validate microbiome-based biomarkers of CDI risk. Identification of such risk factors will be possible only if samples are collected before the development of CDI. Furthermore, previous modeling has shown that albumin levels, white blood cell counts, creatinine levels, age, and increased leukocyte count can be used to predict CDI severity and mortality (4, 9, 33). It is possible that the incorporation of microbiome data can also be used to improve predictions of disease outcome. However, in the current investigation, we did not collect sufficient CDI severity data to address this possibility. As we have demonstrated in this study, there are distinct microbiome signatures that are associated with CDI. Understanding which community-wide changes are responsible for the loss in resistance to colonization leading to CDI is critical for future risk models and therapeutics.
MATERIALS AND METHODS
Sample collection and definitions.
This study was approved by the University of Michigan Institutional Review Board. The inpatient samples were collected from October 2010 to January 2012 at the University of Michigan Hospital, Ann Arbor, MI. All enrollees granted patient consent. Inpatient subjects were not pregnant, they were suspected of having an initial episode of CDI (not recurrent CDI), and their stool sample was diarrheal. Within 24 h of stool collection, these specimens were screened for C. difficile using the C.Diff Quik Chek Complete assay (Techlab, Blacksburg, VA). This rapid membrane enzyme immunoassay tests for the presence of both the C. difficile antigen glutamate dehydrogenase (GDH) and the C. difficile toxin proteins A and B. If this test resulted in a positive or negative result for both GDH and toxin proteins, the sample was classified as a case or as a diarrheal control, respectively. If the test was positive only for GDH, a PCR screen for the C. difficile tcdB gene, which encodes the toxin B protein, was performed (34). To confirm the results of the clinical lab, we additionally performed PCR on all inpatient samples using C. difficile-specific 16S rRNA gene primers as described elsewhere (35). Nondiarrheal, C. difficile-negative samples were collected between January 2011 and January 2012 from individuals residing in the area surrounding Ann Arbor, MI. Subjects were excluded if they had had any signs of diarrhea in the previous 7 days or were pregnant. Once enrolled, individuals collected a stool sample using the provided home stool specimen kit.
DNA sequencing and curation.
Total bacterial DNA was extracted from each stool sample using the PowerSoil-htp 96-well soil DNA isolation kit (MO Bio Laboratories, Carlsbad, CA) on an EpMotion 5075 liquid-handling workstation (Eppendorf, Hamburg, Germany). The V35 region of the 16S rRNA gene was amplified and sequenced using the 454 GS FLX pyrosequencing platform and curated using mothur as previously described (36, 37). We sequenced and processed a mock community in parallel with the samples sequenced for this study (37). The observed error rate among the mock community samples was 0.009%. Sequences were clustered into operational taxonomic units (OTUs) using a 3% distance cutoff (38). Taxonomic assignments were determined by using a naive Bayesian classifier with the Ribosomal Database Project (RDP) training set (version 9) with an 80% bootstrap confidence threshold. To mitigate against the effects of uneven sampling, all samples were rarefied to 1,450 sequences per sample (37). Among the samples with more than 1,450 sequences, the number of sequences per sample varied from 1,450 to 17,120, with a mean of 6,091 sequences/sample, a median of 5,986, and a median absolute deviation of 1,296. The OTU corresponding to C. difficile (OUT 19) was identified by checking the representative sequence against the NCBI nucleotide database with BLASTn. All 16S rRNA gene sequence data and the associated MIMARKS table are available at http://www.mothur.org/CDI_MicrobiomeModeling.
Statistical analyses.
Initial statistical analyses were conducted to assess differences among the three study groups (C. difficile cases, diarrheal controls, and nondiarrheal controls). For continuous variables (e.g., age and weight), one-way analysis of variance was utilized. For categorical variables, Pearson’s chi-square test or Fisher’s exact test was performed when expected cell frequencies were less than or equal to 5. The principal intent of the analyses was to assess whether the addition of microbiome data added to case differentiation, and as such, nested logit models were constructed with clinical data, with and without the incorporation of microbiome data. We utilized three approaches to capture the biodiversity of the gut microbiome. First, the inverse Simpson index was calculated for each sample and treated as a continuous variable in the models (39). Second, we assigned each sample to a different community type and used the type as a categorical variable in the model. These community types were identified by partitioning around medoids (PAM) of a Jensen-Shannon divergence distance matrix calculated from the microbiome data (27). The randomForest package in R (http://cran.r-project.org/), with the number of trees set to 1,000, was used to differentiate the composition of each cluster. Third, we built models using the relative abundances of a subset of the OTUs observed across the individuals. These OTUs were selected using LEfSe based on the comparisons of cases versus diarrheal controls, cases versus nondiarrheal controls, and diarrheal controls versus nondiarrheal controls (13). OTUs demonstrating the greatest differences (at a 0.25-percentile cutoff at both ends) were used as continuous variables in our logit models. The 0.25-percentile cutoff was selected to restrict the number of significant OTUs in order to build the models and avoid overfitting. Differences between nested models were compared using the test for the equality of ROC areas (15). Analyses were conducted in Stata/MP 12.1 and R version 3.0.1.
SUPPLEMENTAL MATERIAL
Overall structural differences between experimental groups. This nonmetric multidimensional scaling (NMDS) plot was generated from a distance matrix of Yue and Clayton measure of dissimilarity (θYC) values, which assess structural differences between samples (40). Gray represents nondiarrheal control samples, blue represents diarrheal control samples, and red represents case samples. Download
AUC comparisons of all models. The AUC values and 95% confidence intervals (CI) were calculated for each model in our study. Comparisons were made either between the listed model and an empty model (*) or between the combined base and microbiome model and the base model (**), as indicated in the text. Comparisons that were significant are shown as bold P values.
Breakdown of the individuals in each community type. All 338 subjects in the study were sorted into 13 community types using PAM (see Materials and Methods). These 13 community types were ordered by the percentage of case individuals belonging to each type. The numbers of diarrheal control, nondiarrheal control, and case subjects belonging to each community type are shown.
ACKNOWLEDGMENTS
This work was supported by several grants from the National Institutes of Health (1R01GM099514 to P.D.S., R01HG005975 to P.D.S., U19AI090871 to D.M.A., V.B.Y., and P.D.S., and P30DK034933 to P.D.S. and V.B.Y.). The funding agency had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Citation Schubert AM, Rogers MAM, Ring C, Mogle J, Petrosino JP, Young VB, Aronoff DM, Schloss PD. 2014. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio 5(3):e01021-14. doi:10.1128/mBio.01021-14.
REFERENCES
- 1. Hicks LA, Taylor TH, Hunkler RJ. 2013. U.S. outpatient antibiotic prescribing, 2010. N. Engl. J. Med. 368:1461–1462. 10.1056/NEJMc1212055 [DOI] [PubMed] [Google Scholar]
- 2. Lucado J, Gould C, Elixhauser A. 2012. Clostridium difficile infections (CDI) in hospital stays, 2009. Healthcare Cost and Utilization Project, Agency for Healthcare Research and Quality, Rockville, MD: http://www.hcup-us.ahrq.gov/reports/statbriefs/sb124.jsp [PubMed] [Google Scholar]
- 3. Bassetti M, Villa G, Pecori D, Arzese A, Wilcox M. 2012. Epidemiology, diagnosis and treatment of Clostridium difficile infection. Expert Rev. Anti Infect. Ther. 10:1405–1423. 10.1586/eri.12.135 [DOI] [PubMed] [Google Scholar]
- 4. Walk ST, Micic D, Jain R, Lo ES, Trivedi I, Liu EW, Almassalha LM, Ewing SA, Ring C, Galecki AT, Rogers MA, Washer L, Newton DW, Malani PN, Young VB, Aronoff DM. 2012. Clostridium difficile ribotype does not predict severe infection. Clin. Infect. Dis. 55:1661–1668. 10.1093/cid/cis786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yakob L, Riley TV, Paterson DL, Clements AC. 2013. Clostridium difficile exposure as an insidious source of infection in healthcare settings: an epidemiological model. BMC Infect. Dis. 13:376. 10.1186/1471-2334-13-376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U. S. A. 108:4554–4561. 10.1073/pnas.1000087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amir I, Konikoff FM, Oppenheim M, Gophna U, Half EE. 20 September 2013. Gastric microbiota is altered in oesophagitis and Barrett’s oesophagus and further modified by proton pump inhibitors. Environ. Microbiol. 10.1111/1462-2920.12285 [DOI] [PubMed] [Google Scholar]
- 8. Rea MC, O’Sullivan O, Shanahan F, O’Toole PW, Stanton C, Ross RP, Hill C. 2012. Clostridium difficile carriage in elderly subjects and associated changes in the intestinal microbiota. J. Clin. Microbiol. 50:867–875. 10.1128/JCM.05176-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shivashankar R, Khanna S, Kammer PP, Harmsen WS, Zinsmeister AR, Baddour LM, Pardi DS. 2013. Clinical factors associated with development of severe-complicated Clostridium difficile infection. Clin. Gastroenterol. Hepatol. 11:1466–1471. 10.1016/j.cgh.2013.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, Wang GP. 2013. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 51:2884–2892. 10.1128/JCM.00845-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197:435–438. 10.1086/525047 [DOI] [PubMed] [Google Scholar]
- 12. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Statnikov A, Alekseyenko AV, Li Z, Henaff M, Perez-Perez GI, Blaser MJ, Aliferis CF. 2013. Microbiomic signatures of psoriasis: feasibility and methodology comparison. Sci. Rep. 3:2620. 10.1038/srep02620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeLong ER, DeLong DM, Clarke-Pearson DL. 1988. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837–845 [PubMed] [Google Scholar]
- 16. Manges AR, Labbe A, Loo VG, Atherton JK, Behr MA, Masson L, Tellis PA, Brousseau R. 2010. Comparative metagenomic study of alterations to the intestinal microbiota and risk of nosocomial Clostridium difficile-associated disease. J. Infect. Dis. 202:1877–1884. 10.1086/657319 [DOI] [PubMed] [Google Scholar]
- 17. Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, Deakin LJ, Pickard DJ, Duncan SH, Flint HJ, Clark TG, Parkhill J, Dougan G. 2012. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8:e1002995. 10.1371/journal.ppat.1002995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vincent C, Stephens DA, Loo VG, Edens TJ, Behr MA, Dewar K, Manges AR. 2013. Reductions in intestinal Clostridiales precede the development of nosocomial Clostridium difficile infection. Microbiome 1:18. 10.1186/2049-2618-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rolfe RD. 1984. Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect. Immun. 45:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gauffin Cano P, Santacruz A, Moya Á, Sanz Y. 2012. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS One 7:e41079. 10.1371/journal.pone.0041079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bishara J, Farah R, Mograbi J, Khalaila W, Abu-Elheja O, Mahamid M, Nseir W. 2013. Obesity as a risk factor for Clostridium difficile infection. Clin. Infect. Dis. 57:489–493. 10.1093/cid/cit280 [DOI] [PubMed] [Google Scholar]
- 22. Yanagibashi T, Hosono A, Oyama A, Tsuda M, Suzuki A, Hachimura S, Takahashi Y, Momose Y, Itoh K, Hirayama K, Takahashi K, Kaminogawa S. 2013. IgA production in the large intestine is modulated by a different mechanism than in the small intestine: Bacteroides acidifaciens promotes IgA production in the large intestine by inducing germinal center formation and increasing the number of IgA+ B cells. Immunobiology 218:645–651. 10.1016/j.imbio.2012.07.033 [DOI] [PubMed] [Google Scholar]
- 23. Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. 2012. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 80:62–73. 10.1128/IAI.05496-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. 2011. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile infection. Gut Microbes 2:145–158. 10.4161/gmic.2.3.16333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holmes I, Harris K, Quince C. 2012. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One 7:e30126. 10.1371/journal.pone.0030126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. 2013. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput. Biol. 9:e1002863. 10.1371/journal.pcbi.1002863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, MetaHIT Consortium. Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M’rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Quince C, Lundin EE, Andreasson AN, Greco D, Rafter J, Talley NJ, Agreus L, Andersson AF, Engstrand L, D'Amato M. 2013. The impact of Crohn’s disease genes on healthy human gut microbiota: a pilot study. Gut 62:952–954. 10.1136/gutjnl-2012-304214 [DOI] [PubMed] [Google Scholar]
- 30. Brotman RM, Bradford LL, Conrad M, Gajer P, Ault K, Peralta L, Forney LJ, Carlton JM, Abdo Z, Ravel J. 2012. Association between Trichomonas vaginalis and vaginal bacterial community composition among reproductive-age women. Sex. Transm. Dis. 39:807–812. 10.1097/OLQ.0b013e3182631c79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gajer P, Brotman RM, Bai G, Sakamoto J, Schütte UM, Zhong X, Koenig SS, Fu L, Ma ZS, Zhou X, Abdo Z, Forney LJ, Ravel J. 2012. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 4:132ra152. 10.1126/scitranslmed.3003605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4680–4687. 10.1073/pnas.1002611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Butt E, Foster JA, Keedwell E, Bell JE, Titball RW, Bhangu A, Michell SL, Sheridan R. 2013. Derivation and validation of a simple, accurate and robust prediction rule for risk of mortality in patients with Clostridium difficile infection. BMC Infect. Dis. 13:316. 10.1186/1471-2334-13-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Agaronov M, Karak SG, Maldonado Y, Tetreault J, Aslanzadeh J. 2012. Comparison of GeneXpert PCR to BD GeneOhm for detecting C. difficile toxin gene in GDH positive toxin negative samples. Ann. Clin. Lab. Sci. 42:397–400 http://www.annclinlabsci.org/content/42/4/397.long [PubMed] [Google Scholar]
- 35. Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. 2004. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 97:1166–1177. 10.1111/j.1365-2672.2004.02409.x [DOI] [PubMed] [Google Scholar]
- 36. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. 10.1371/journal.pone.0027310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schloss PD, Westcott SL. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 77:3219–3226. 10.1128/AEM.02810-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Magurran AE. 1988. Ecological diversity and its measurement, vol 534 Princeton University Press, Princeton, NJ. [Google Scholar]
- 40. Yue JC, Clayton MK. 2005. A similarity measure based on species proportions. Commun. Statist. Theory Methods 34:2123–2131. 10.1080/STA-200066418 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overall structural differences between experimental groups. This nonmetric multidimensional scaling (NMDS) plot was generated from a distance matrix of Yue and Clayton measure of dissimilarity (θYC) values, which assess structural differences between samples (40). Gray represents nondiarrheal control samples, blue represents diarrheal control samples, and red represents case samples. Download
AUC comparisons of all models. The AUC values and 95% confidence intervals (CI) were calculated for each model in our study. Comparisons were made either between the listed model and an empty model (*) or between the combined base and microbiome model and the base model (**), as indicated in the text. Comparisons that were significant are shown as bold P values.
Breakdown of the individuals in each community type. All 338 subjects in the study were sorted into 13 community types using PAM (see Materials and Methods). These 13 community types were ordered by the percentage of case individuals belonging to each type. The numbers of diarrheal control, nondiarrheal control, and case subjects belonging to each community type are shown.