ABSTRACT
The Trojan horse Escherichia coli antibiotic microcin C (McC) consists of a heptapeptide attached to adenosine through a phosphoramidate linkage. McC is synthesized by the MccB enzyme, which terminally adenylates the ribosomally synthesized heptapeptide precursor MccA. The peptide part is responsible for McC uptake; it is degraded inside the cell to release a toxic nonhydrolyzable aspartyl-adenylate. Bionformatic analysis reveals that diverse bacterial genomes encoding mccB homologues also contain adjacent short open reading frames that may encode MccA-like adenylation substrates. Using chemically synthesized predicted peptide substrates and recombinant cognate MccB protein homologs, adenylated products were obtained in vitro for predicted MccA peptide-MccB enzyme pairs from Helicobacter pylori, Streptococcus thermophilus, Lactococcus johnsonii, Bartonella washoensis, Yersinia pseudotuberculosis, and Synechococcus sp. Some adenylated products were shown to inhibit the growth of E. coli by targeting aspartyl-tRNA synthetase, the target of McC.
IMPORTANCE
Our results prove that McC-like adenylated peptides are widespread and are encoded by both Gram-negative and Gram-positive bacteria and by cyanobacteria, opening ways for analyses of physiological functions of these compounds and for creation of microcin C-like antibiotics targeting various bacteria.
INTRODUCTION
The antibiotic microcin C (McC) (Fig. 1A) is produced by Escherichia coli strains harboring a plasmid-borne mcc operon (1, 2). McC consists of a heptapeptide whose C terminus is covalently linked through a phosphoramidate bond to adenosine; the phosphoramidate linker is esterified with an aminopropyl moiety (3). McC is active against E. coli strains that lack the mcc operon as well as other enteric bacteria (1, 2). McC penetrates the inner membrane of E. coli cells through the YejABEF transporter (4). In the cytoplasm of sensitive cells, the N-terminal formyl group of McC is removed by the peptide deformylase, followed by removal of N-terminal methionine by methionine aminopeptidase and degradation of the rest of the peptide by nonspecific aminopeptidases (5). The last remaining amino acid of McC peptide is an aspartate. Thus, following proteolytic processing, a modified nonhydrolyzable aspartyl-adenylate, referred to as processed McC, is released. Processed McC is a potent inhibitor of aspartyl-tRNA synthetase (AspRS) (6), an enzyme essential for protein synthesis.
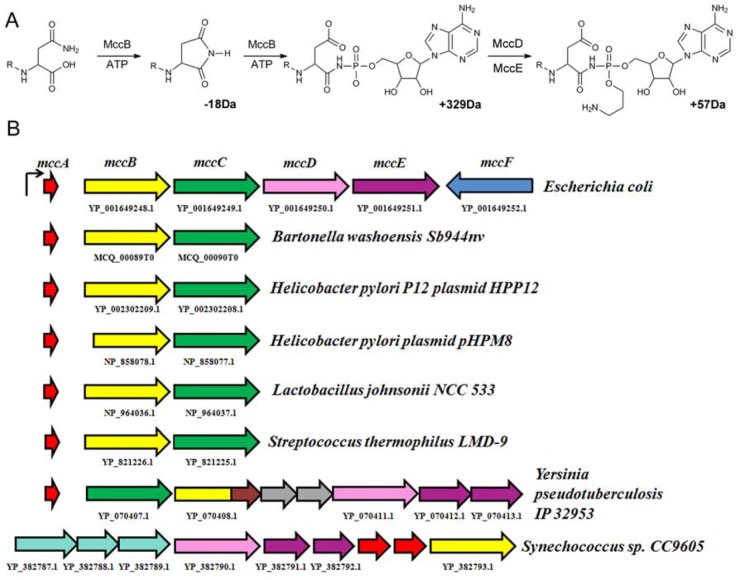
FIG 1 .
E. coli McC and homologous operons encoded by other bacteria. (A) Schematic representation of E. coli McC synthesis from the MccA peptide. Enzymes and cofactors involved are listed above and below the arrows; R represents N-terminal residues of MccA (formyl-MRTGNA). Numbers (in daltons [Da]) indicate differences in molecular masses between the products and the substrates of individual reaction steps. (B) At the top, the mcc operon of E. coli is shown. Arrows indicate individual genes (mccA, red; mccB, yellow; mccC, green; mccD, pink; mccE, purple; mccF, blue). Homologous operons from indicated sources are shown under the arrows. Open reading frames coding for predicted MccA-like peptides are not drawn to scale. Open reading frames indicated as gray arrows in the Y. pseudotuberculosis mcc operon correspond to yqcI and ycgG gene homologs. The part of the mccB homolog indicated in brown encodes a methylase domain. For the Synechococcus sp. CC9605 mcc operon, the three open reading frames indicated in cyan encode ABC-type transporters.
While intact McC inhibits the growth of sensitive E. coli cells at low micromolar concentrations, processed McC does not affect cell growth, even at millimolar concentrations (6). The peptide chain therefore promotes active uptake of toxic processed McC, which itself is unable to penetrate cells, via the YejABEF transporter. The biological function of YejABEF (other than McC transport) is unknown.
The McC peptide is encoded by a seven-codon-long mccA gene (7). During biosynthesis of the antibiotic, the MccA peptide (amino acid sequence MRTGNAN) is C-terminally adenylated by the MccB synthetase. The reaction proceeds through two rounds of adenylation and results in a conversion of terminal asparagine residue of the MccA peptide to aspartate (8, 9). The product of MccA adenylation is additionally decorated by an aminopropyl moiety in a reaction that proceeds through a mechanism yet to be defined and requires the products of mccD and mccE genes (10). The resulting mature McC is exported from the producing cells by MccC, an MFS-type transporter (11–13). Self-immunity to processed McC that accumulates in the cytoplasm of the producing cell is determined by the C-terminal domain of MccE, a GNAT (Gcn5-related acetyl transferase) family acetyltransferase (14), and MccF, an S66 family protease (15). The mccF gene is transcribed separately from the mccABCDE operon and is not strictly required for McC production and/or immunity of the producing cell (11–13).
The product of MccB-catalyzed adenylation of MccA, a peptide adenylate with a molecular mass of 1,119 Da (or 1,091 Da for a compound without an N-terminal formyl group) is biologically active, albeit its activity is significantly less than that of mature McC (10). Thus, the mccD and mccE genes, necessary for the additional aminopropyl decoration, are not strictly required for biological function. Therefore, a three-gene arrangement (mccABC) may fulfill the minimal requirements for biosynthesis and export of an McC-like molecule. Earlier, we analyzed bacterial DNA sequences for the presence of adjacent genes coding for proteins similar to E. coli MccB and MccC and found such pairs in Helicobacter pylori plasmids pHPM8 and HPP12 and in the genomes of Streptococcus thermophilus LMD-9, Lactococcus johnsonii NCC 533, and Bartonella washoensis Sb944nv (16). No other adjacent mcc-like genes were found in these organisms (Fig. 1B). Manual analysis of regions upstream of mccB homologs from these organisms identified putative seven-codon-long open reading frames (ORFs) whose products contained a C-terminal asparagine (16) (Fig. 1B and Table 1). The products of these open reading frames differ from E. coli MccA but might be substrates for cognate MccB adenylate transferases.
TABLE 1 .
Predicted MccA peptides encoded by mcc-like operons of various bacteriaa
| Organism/source | MccA peptide |
|---|---|
| Escherichia coli pMccC7 plasmid | MRTGNAN |
| Bartonella washoensis Sb944nv | MDHIGFN |
| Helicobacter pylori HPP12 and pHPM8 plasmids | MKLSYRN |
| Lactobacillus johnsonii NCC 533 | MHRIMKN |
| Streptococcus thermophilus LMD-9 | MKGTILN |
| Yersinia pseudotuberculosis IP32953 | MYQVGIILSIKCN |
| Yersinia pseudotuberculosis PB 1/+ | MHQSEIKLTKRLKIKRVDVNKVKEQQKKVLECGAATCGGGSN |
| Synechococcus sp. CC9605 | MTQPNDRQLSNEELSDVAAGLFRRTFFKPRTSRKTLLQPKRLDKVAKNQLWADMMN |
For each organism listed in the left column, the predicted MccB substrates are listed in the right (single-amino-acid code). Terminal asparagine residues are indicated in bold typeface.
In the genome of Synechococcus sp. CC9605, an mccB-like gene is clustered with an mccD-like gene and two genes coding for proteins homologous to E. coli mccE N- and C-terminal domains (Fig. 1B). While no mccC-like genes are present, three genes coding for ABC-like transporters are located upstream of the mccB homolog. Two open reading frames encoding identical 56-amino-acid-long peptides with terminal asparagines are located between the ABC-transporter gene cluster and the mccB-like gene (Fig. 1B and Table 1). These peptides have been proposed to be adenylation substrates for a cognate mccB gene product (16). A very similar operon is also encoded by Synechococcus sp. CC9616 (16).
In Yersinia pseudotuberculosis IP32953, an operon containing homologs of E. coli mccBCDE genes is present (Fig. 1B). The order of the mccB and mccC genes is inverted compared to that observed in E. coli mcc and in “minimal” mccABC operons, and two genes homologous to those coding for a predicted methylase and functionally uncharacterized yqcI- and ycgG-encoded family proteins are inserted between the mccB and mccD homologs. In addition, the homolog of Yersinia pseudotuberculosis IP32953 MccB is also fused to a methylase domain. An open reading frame encoding a 13-amino-acid-long peptide with terminal asparagine was identified upstream of the mccC homolog (Fig. 1B), and the corresponding peptide (Table 1) was proposed to be the substrate for cognate MccB (16).
E. coli McC can be prepared in vitro by adenylation of synthetic MRTGNAN peptide by recombinant E. coli MccB (8). Here, we used this approach to validate predicted mccA-mccB pairs from bacteria other than E. coli. We show that adenylated peptides can be prepared for most of predicted putative MccA-MccB pairs and that many of the resulting adenylated peptides inhibit the growth of E. coli in a YejABEF-dependent manner. By analogy with E. coli, we propose that many of these peptide adenylates inhibit the growth of cognate bacteria that lack mcc-like operons.
RESULTS
Enzymatic synthesis of McC-like peptide adenylates.
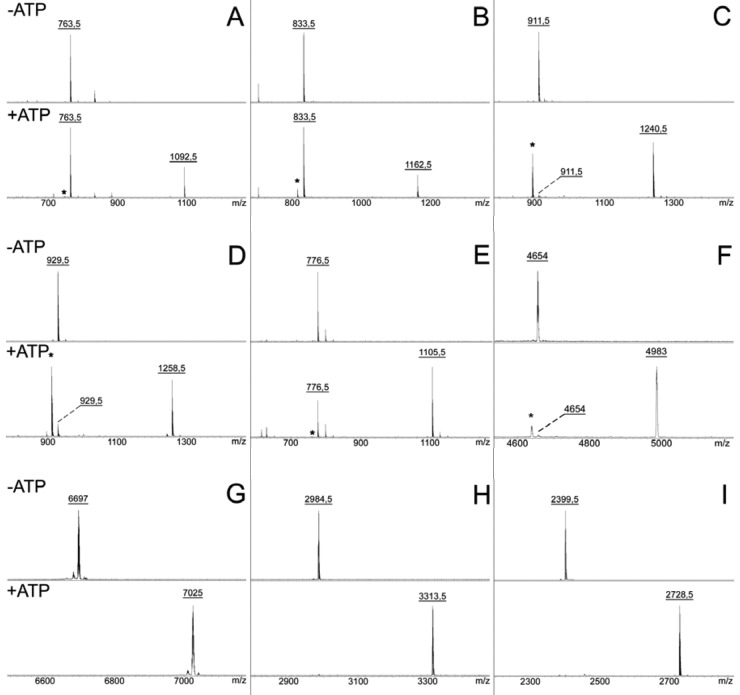
Walsh and colleagues previously reported the in vitro synthesis of an McC maturation intermediate, adenylated MccA peptide lacking the aminopropyl decoration and N-terminal formyl group, in reaction mixtures containing synthetic MRTGNAN peptide, recombinant E. coli MccB protein, and ATP (8). Using their conditions and mass-spectrometric analysis of reaction products, we too observed partial conversion of a mass peak corresponding to reaction substrate MRTGNAN (m/z = 763.5) into adenylated product (m/z = 1,092.5) (Fig. 2A). Accumulation of the reaction intermediate, a peptide containing a C-terminal succinimide (m/z = 745), was also observed (indicated by an asterisk in the mass spectrum presented in the bottom panel of Fig. 2A), in agreement with earlier observations (8).
FIG 2 .
In vitro
adenylation of predicted MccA peptides by cognate MccB homologs. Chemically synthesized peptides corresponding to 7-amino-acid-long MccA products of E. coli (A), B. washoensis Sb944nv (B), H. pylori plasmid HPP12 (C), L. johnsonii NCC 533 (D), S. thermophilus LMD-9 (E), Y. pseudotuberculosis PB1/+ (F), full-length Synechococcus sp. CC9605 peptide (G), and peptides corresponding to the last 25 (H) and 20 (I) amino acids of Synechococcus sp. CC9605 peptide (Table 1) were combined with the corresponding recombinant MccB homologs in the absence (top) and the presence (bottom) of ATP. Reaction products were analyzed by MALDI MS. Only relevant parts of the spectra are shown. Asterisks indicate peaks corresponding to adenylation reaction intermediates containing a terminal succinimide (Fig. 1A).
Next, recombinant versions of MccB homologues encoded by H. pylori plasmids pHPM8 and HPP12 and the genomes of S. thermophilus LMD-9, L. johnsonii NCC 533, B. washoensis Sb944nv, Y. pseudotuberculosis IP32953, and Synechococcus sp. CC9605 were purified using E. coli heterologous expression systems. The full-sized Y. pseudotuberculosis IP32953 MccB homolog was very poorly expressed. We considered that the unusual terminal fusion with a methylase-like protein (Fig. 1B) might have affected the folding of this protein. Accordingly, a recombinant version of Y. pseudotuberculosis MccB lacking the C-terminal methylase domain was prepared. This protein was soluble and expressed at high level and was used in subsequent experiments. Predicted peptide substrates were prepared by solid-phase chemical synthesis. The only exception was the putative substrate peptide encoded by Synechococcus sp. CC9605, which was prepared as a fusion with the MBP protein. Following tobacco etch virus (TEV) protease treatment, a 58-amino-acid peptide that lacked the N-terminal methionine residue of the predicted synechococcal MccA homolog and contained three extra N-terminal amino acids (SerGlySer) was obtained.
Recombinant MccB homologs were combined with predicted peptide substrates in the presence or in the absence of ATP under conditions optimized for E. coli MccB-catalyzed reactions. Equal amounts of synthetic peptides were used. The amount of recombinant Synechococcus sp. CC9605 peptide was at least 10 times lower, since we had difficulties obtaining sufficient amounts of this peptide. The results of mass-spectrometric analysis of reaction products are presented in Fig. 2.
The predicted seven-amino-acid MccA-like peptides encoded by B. washoensis Sb944nv (MDHIGFN), S. thermophilus LMD-9 (MKGTILN), and L. johnsonii NCC 533 (MHRIMKN) were readily adenylated by cognate enzymes (Fig. 2B, D, and E). The predicted seven-amino-acid MccA-like peptide MKLSYRN encoded by H. pylori plasmids pHPM8 and HPP12 was adenylated by MccB encoded by the HPP12 plasmid (Fig. 2C). The pHPM8-encoded protein MccB was inactive (data not shown). The pHPM8-encoded MccB lacks 46 N-terminal amino acids present in the HPP12-encoded homolog. The corresponding N-terminal amino acids of E. coli MccB form a peptide clamp domain that is involved in MccA binding (9). Analysis of mccB genes from H. pylori plasmids revealed that the pHPM8-borne copy of the gene had undergone an A-to-G substitution that destroyed the starting codon of the HPP12 mccB ORF as well as a 4-nucleotide insertion in an area between the annotated start codons of the HPP12 and pHPM8 homologs. The absence of intact N-terminal domain in pHPM8-encoded MccB explains its inactivity. The corresponding gene is thus defective and should be considered a pseudogene, while the mcc operon borne by the HPM8 plasmid must be inactive for production of McC-like compound.
The 58-amino-acid putative Synechococcus sp. CC9605 substrate peptide was fully adenylated by the cognate enzyme (Fig. 2G).
The 13-amino-acid-long Y. pseudotuberculosis IP32953 MccA peptide predicted earlier (16) was not modified by the cognate MccB protein (data not shown). A more current annotation of the Y. pseudotuberculosis PB1/+ genome (NC_010634.1) suggests a longer 42-codon peptide (YP_001872354.1) as a possible substrate of adenylation by MccB. The same polypeptide is also encoded by Y. pseudotuberculosis IP32953. When the longer peptide was prepared by chemical synthesis and tested in the adenylation reaction, highly efficient conversion into adenylated product was observed (Fig. 2F). The result thus validates the 42-amino-acid-long peptide as a substrate of Y. pseudotuberculosis IP32953 MccB.
In all cases where adenylation products were detected, corresponding succinimide-containing intermediates (-18 Da of input peptide) were also observed, indicating that the reaction mechanism of MccB-like enzymes is conserved. The only exception was Synechococcus sp. CC9605 substrate peptide, which was quantitatively converted to adenylated product with no reaction intermediate detected. Adenylation reactions were specific; i.e., peptides became adenylated when incubated with enzymes from the same species but remained unmodified when incubated with noncognate enzymes (data not shown). With the exception of Synechococcus sp. CC9605 peptide, no full modification of input peptides was achieved under our conditions (Fig. 2). Nevertheless, whenever modification was observed, a significant fraction of input peptide was adenylated that was comparable to what was observed in the control reaction with E. coli MccA-MccB.
Biological activity of peptide adenylates.
The results of enzymatic adenylation in vitro validated most of the earlier bioinformatics predictions. We envision that adenylated peptides with or without additional modifications, such as the propylamine decoration present in E. coli McC, are produced by bacteria containing validated mcc-like operons and inhibit the growth of bacteria that do not carry the mcc-like operon. While we lack the capacity to test the homologous biological activity of adenylation reaction products, we tested their ability to inhibit the growth of E. coli. In vitro adenylation reaction mixtures containing equal amounts of input substrate peptide and cognate adenylation enzymes were incubated with or without ATP, and aliquots of reaction mixtures were deposited on freshly seeded lawns of E. coli B cells (Table 2). Only reaction mixtures containing enzyme-substrate pairs that resulted in adenylated peptide production were tested. As a control, we used an McC-resistant isogenic strain lacking the yejB gene (4). As can be seen from Table 2, reaction mixtures containing adenylated peptides from E. coli, S. thermophilus LMD-9, L. johnsonii NCC 533, and H. pylori inhibited the growth of wild-type E. coli cells. While the efficiencies of adenylation of the various peptides differed (Fig. 2), there was no gross difference in the sizes of the growth inhibition zones (8 to 15 mm in diameter), suggesting that the ability of various adenylated peptides to inhibit E. coli is comparable to that of McC. No inhibition zones were observed on lawns of mutant cells, suggesting that adenylated peptides from S. thermophilus LMD-9, L. johnsonii NCC 533, and H. pylori enter E. coli through the YejABEF transporter.
TABLE 2 .
Antibiotic activity of enzymatically synthesized McC-like compoundsa
| Source | Peptide adenylate | E. coli B+ | E. coli B ΔyejB | E. coli B ΔsbmA |
|---|---|---|---|---|
| E. coli | MRTGNAD-AMP | + | − | + |
| B. washoensis Sb944nv | MDHIGFD-AMP | − | − | − |
| H. pylori P12 | MKLSYRD-AMP | + | − | + |
| L. johnsonii NCC 533 | MHRIMKD-AMP | + | − | + |
| S. thermophilus LMD-9 | MKGTILD-AMP | + | − | + |
| Yersinia pseudotuberculosis PB1/+ | MHQSEIKLTKRLKIKRVDVNKVKEQQKKVLECGAATCGGGSD-AMP | − | − | − |
| Synechococcus sp. CC9605* | SGSTQPNDRQLSNEELSDVAAGLFRRTFFKPRTSRKTLLQPKRLDKVAKNQLWADMMD-AMP | − | − | − |
| Synechococcus sp. CC9605* | SRKTLLQPKRLDKVAKNQLWADMMD-AMP | + | + | − |
| Synechococcus sp. CC9605* | LQPKRLDKVAKNQLWADMMD-AMP | − | − | − |
| E. coli mcb | Microcin B | + | + | − |
For each peptide adenylate listed, the results of biological activity testing of the indicated E. coli strain lawns are shown. For activity testing, 10 µl of the adenylation reaction mixture was deposited on freshly seeded lawns and, after an overnight incubation, growth inhibition zones around the positions where adenylation reaction mixture aliquots were deposited were recorded. +, clear growth inhibition zones were observed; −, no inhibition zones were observed. For Synechococcus sp. CC9605, an adenylation reaction was conducted with a recombinant peptide containing three nonnatural N-terminal amino acids (indicated in bold typeface; see Materials and Methods). The Synechococcus sp. CC9605 lines (marked with an asterisk) show results obtained with adenylated peptides corresponding to the last 25 and 20 amino acids of Synechococcus sp. CC9605 MccA.
Adenylated B. washoensis Sb944nv and Y. pseudotuberculosis IP32953 peptides produced no inhibition zones on either wild-type or mutant cell lawns, suggesting that these peptides either are not internalized or are not processed within the cell (see below). An adenylated MccA homolog from Synechococcus sp. CC9605 also produced no growth inhibition zones. However, as mentioned above, the amount of this peptide used in the adenylation reaction was much lower than the amount of synthetic peptides, which could have affected the result of biological activity testing. Synthetic peptides corresponding to 25, 20, 15, and 10 C-terminal amino acids of synechococcal MccA were prepared and tested in the adenylation reaction with Synechococcus sp. CC9605 MccB. Mass-spectrometric analysis indicated that the two longer peptides were efficiently adenylated (Fig. 2H and I), while the shorter peptides remained unmodified (data not shown). Reaction mixtures containing an adenylated 25-amino-acid peptide inhibited the growth of both the wild-type and yejB mutant E. coli strains with equal efficiencies (Table 2). In contrast, an adenylated 20-amino-acid peptide was inactive (Table 2). Since data determined with heptapeptide adenylates suggest that the YejABEF transporter appears to be sequence nonspecific, the result hints that there may be an additional route of cell entry for longer peptide adenylates. Indeed, E. coli cells with a disrupted sbmA gene coding for an inner-membrane transporter were sensitive to McC but were resistant to the action of an adenylated 25-amino-acid peptide. As expected, these cells were also resistant to microcin B, a much longer posttranslationally modified microcin that is known to be internalized through SbmA (17).
In vitro activity of adenylated peptides.
To determine if various adenylated peptides function similarly to the E. coli McC, aliquots of adenylation reaction mixtures were combined with S30 extracts of E. coli K-12 wild-type cells, incubated for 15 min, and tested for aminoacylation of tRNAAsp, a reaction catalyzed by the target of processed McC, AspRS. Elsewhere, we show that a 15-min incubation is sufficient for McC processing under our reaction conditions (5, 6). In Fig. 3, the ratios of AspRS activities in extracts to which cognate MccB-MccA pair reaction mixtures with or without ATP were added are shown. As can be seen, the addition of reaction mixtures containing E. coli MccA, MccB, and ATP inhibited AspRS activity at a level ca. 10 times higher than that seen with the control (no ATP). Comparable levels of inhibition were observed in reaction mixtures containing all other adenylated peptides with the exception of that from B. washoensis Sb944nv, which showed poor inhibition (55% AspRS activity compared to the no-ATP control), and that from Y. pseudotuberculosis (90% AspRS activity compared to the no-ATP control).
FIG 3 .
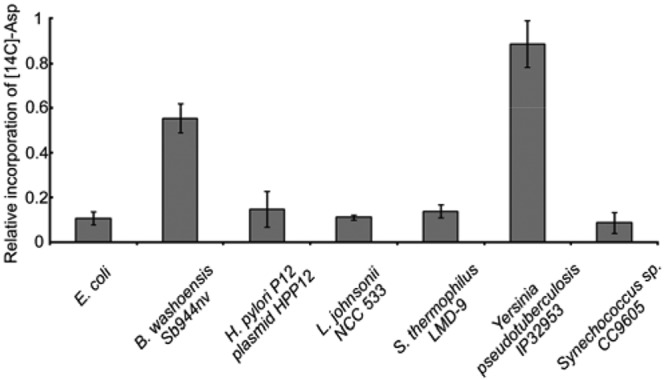
Adenylated peptides from various sources inhibit E. coli AspRS in vitro. AspRS-catalyzed aminoacylation of tRNAAsp in S30 extracts prepared from wild-type E. coli was performed in the presence of aliquots of the indicated adenylation reaction mixtures with or without ATP. Extracts were incubated for 15 min to allow processing. The ratios of acid-insoluble radioactivity in the corresponding reaction mixtures with and without ATP are presented (mean values and standard deviations of measurements taken from three independently performed experiments are shown).
We hypothesized that the poor inhibition of AspRS by adenylated B. washoensis Sb944nv and Y. pseudotuberculosis peptides was due to slow processing of the peptide in E. coli S30 extracts. This hypothesis was tested for the B. washoensis Sb944nv peptide by monitoring the fate of unmodified E. coli and B. washoensis Sb944nv MccA peptides in E. coli S30 extracts. As can be seen in Fig. 4, the N-terminal methionine is efficiently removed from both peptides (Fig. 4, second row). A mass peak corresponding to the resulting E. coli hexapeptide RTGNAN (m/z = 632.5) completely disappeared after a 15-minute incubation with the extract due to processing by aminopeptidases, as expected (5). In contrast, the B. washoensis Sb944nv DHIGFN hexapeptide (m/z = 702) remained essentially intact even after a 60-min incubation, indicating that processing (and, therefore, release of C-terminally located AspRS inhibitor in the case of adenylated peptide) was inefficient.
FIG 4 .
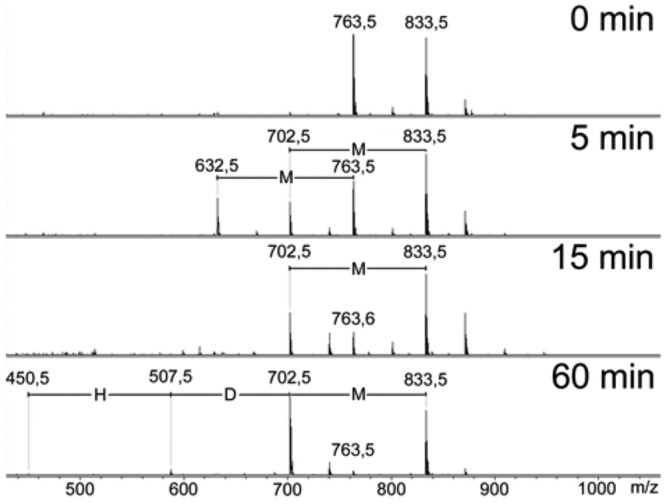
Differential processing rates of E. coli and B. washoensis Sb944nv MccA peptides in E. coli extracts. Equal amounts of E. coli and B. washoensis Sb944nv MccA peptides were combined with wild-type E. coli S30 extracts. At the times indicated, aliquots of reaction mixtures were removed and analyzed by mass spectrometry. H, D, and M stand for histidine, aspartate, and methionine residues, respectively.
Expanding the family of microcin maturation adenylate transferases.
Encouraged by successful verification of several predicted microcins, we decided to revisit the MccB-like family of enzymes in order to identify those that could be involved in production of McC-like compounds. MccB-like proteins belong to a ThiF/HesA/MoeB/E1 superfamily of phosphotransferases, which has been analyzed in detail previously (18). In particular, it has been shown that this superfamily has a complex evolutionary history and is so divergent that phylogenetic reconstructions based on sequence alignments alone are impossible. The relationships between the families must be therefore elucidated based on analysis of conserved sequence features, domain organization, and gene context (18). In particular, it has been indicated that the MccB family is related to the PaaA (pantocin A biosynthesis protein) family (18). Unlike MccB proteins, which adenylate terminal asparagines in peptides, the PaaA enzyme from Pantoea agglomerans appears to modify an internal asparagine residue of the substrate peptide (19, 20).
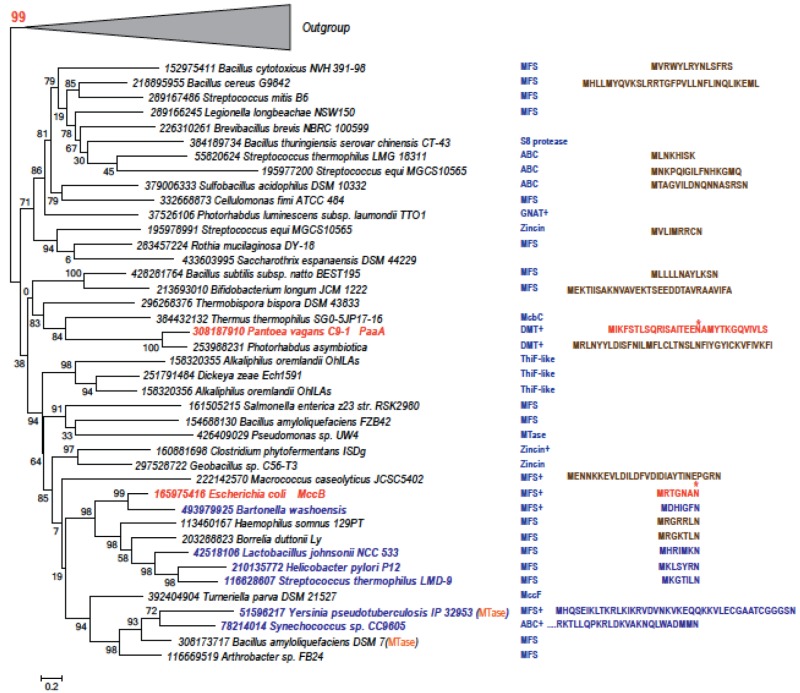
We systematically identified homologs of both MccB and PaaA proteins in fully sequenced bacterial genomes (see Materials and Methods). The resulting tree of the MccB and PaaA homologs is displayed in Fig. 5. All MccB and PaaA homologs included in the tree contain a predicted N-terminal peptide clamp domain that in E. coli MccB is required for MccA recognition (9). Further, the genes coding for MccB and PaaA homologs included in the tree have neighboring genes that encode either a potential efflux pump or proteins that could be involved in additional microcin modifications and/or self-immunity (Fig. 5). These features are not found in members from the outgroup, which includes sequences that are confidently separated in both FastTree and RAxML trees (the bootstrap values are 99 and 69, respectively) and that are likely involved in thiamine and molybdenum cofactor biosynthesis. We therefore predict that the proteins presented in Fig. 5 are involved in the synthesis of McC-like compounds. Indeed, while this was not implicit in our analysis, all operons encoding premicrocin peptides that were predicted before and verified in our work are present in the tree. The corresponding peptides are shown in the blue font in Fig. 5 (the prototypical E. coli MccA, and the pantocin precursor peptide are shown in the red font). Furthermore, in front of genes coding for some family members, we were able to identify short open reading frames coding for peptides of various lengths and containing terminal or internal asparagine residues (highlighted in the brown font). We predict that some of these peptides may be adenylation substrates for cognate MccB and PaaA homologs.
FIG 5 .
Phylogenetic tree and gene neighbors of predicted members of MccB/PaaA family. The maximum-likelihood phylogenetic unrooted tree was built using the FastTree program (33) and multiple alignment of conserved blocks (136 informative positions) of the ThiF/HesA/MoeB/E1 domain. The same program was also used to compute the bootstrap values indicated for all branches. The tree is rooted using outgroups (shown as triangles) of sequences with functions unrelated to microcin production; the bootstrap value confidently separating the outgroup is highlighted. Each terminal node of the tree is labeled by the GenBank Identifier number and the full systematic name of an organism. The founding MccB and PaaA genes are highlighted by red. Proteins that are discussed in this work are shown by blue highlights. Domain fusions are indicated in parentheses. One gene encoded in the same predicted operon suggesting relevance to microcin production is shown in the column on the right side of the tree. The symbol “+” indicates that there are other genes in the same operon that are related to microcin biosynthesis or immunity. Predicted and verified microcin precursor peptides are shown as follows: red, McC and pantocin precursors, with arrows indicating asparagine residues that are adenylated; blue, peptides validated in this work; brown, peptides predicted upstream of the corresponding MccB/PaaA family gene. Abbreviations: for the predicted efflux pumps, ABC, ABC-transported family, MFS, major facilitator transporter family, and DMT, divalent metal transporter family; for the predicted immunity genes, zincin, zincin superfamily Zn-dependent proteases, S8, S8 family of serine proteases, MccF, S66 family serine protease, and GNAT, GNAT family acetyltransferase; and for the predicted microcin modification enzymes, ThiF-like, ThiF/HesA/MoeB/E1 protein, MTase, S-adenosyl-l-methionine-dependent methyltransferase, and McbC, microcin B-processing oxidoreductase.
DISCUSSION
In this work, we validated bioinformatically predicted microcin C-like compounds encoded in the genomes and/or plasmids of several diverse bacteria. All four predicted heptapeptides tested, from H. pylori, S. thermophilus, L. johnsonii, and B. washoensis, were adenylated by cognate enzymes, and adenylated products inhibited the growth of McC-sensitive E. coli through the Trojan horse mechanism that was earlier described for E. coli McC (the mechanism facilitated transport through YejABEF transporter, intracellular processing by aminopeptidases, and inhibition of AspRS by a nonhydrolyzable aspartyl adenylate). In the case of E. coli McC, the enzymatic product of the MccB-catalyzed reaction mixture contains a direct N-P link that renders it a nonhydrolyzable analog of the normally hydrolyzable Asp-AMP. While none of the analogs obtained in our work have been directly shown to contain an N-P bond, the fact that robust inhibition of AspRS is observed upon processing strongly supports this assumption.
The B. washoensis McC-like compound was the least active of the heptapeptide adenylates. Apparently, a hexapeptide with an N-terminal aspartate that is produced after the N-terminal methionine of B. washoensis MccA is removed is poorly processed in E. coli. This conjecture agrees with the fact that PepN, which is considered the major aminopeptidase in E. coli (21), disfavors acidic residues and instead strongly prefers basic amino acids (21, 22). Indeed, amino acids in the second position of efficiently processed E. coli, H. pylori, S. thermophilus, and L. johnsonii MccA peptides are basic, and none of these peptides contain internal acidic residues that could hinder processing.
By analogy with E. coli McC, we hypothesize that the physiological function of adenylated heptapeptides validated in our work is to target bacteria closely related to the producing strain. The corresponding operons have the minimal mccABC arrangement, suggesting that heptapeptide adenylates function without the additional modification characteristic of E. coli McC. In the case of E. coli McC, the aminopropyl decoration increases the bioactivity ca. 10-fold by promoting the interaction of processed McC with its target through a favorable electrostatic interaction between the amine of aminoproryl and negatively charged residues of AspRS (10). The corresponding residues are present in AspRS enzymes from H. pylori, S. thermophilus, L. johnsonii, and B. washoensis, suggesting that the presence of aminopropyl would have increased the potency of McC-like molecules produced by these organisms. The absence of mccD and -NTD aminopropyl synthesis genes, as well as autoimmunity genes mccE-CTD and mccF, suggests that the minimal mccABC arrangement is ancestral and that the more complex arrangement of E. coli is derived. Stand-alone homologs of mccE and mccF are encoded in many bacterial genomes and could have been “adopted” by the ancestor of the E. coli mcc operon.
Perhaps the most surprising result of our work is the validation of adenylation of the much-longer MccA peptides from Synechococcus and Y. pseudotuberculosis. The mcc operon from Synechococcus is unique since the mccA gene is tandemly duplicated. In addition, the likely export pumps are encoded by transporters of a type different from those in other validated adenylated-peptide biosynthetic operons (Fig. 1B), which may have to do with the greater length of the peptide moiety. Structural analysis of E. coli MccB bound to its substrate shows that there is an opening in the enzyme that should allow the N-terminally extended 7-amino-acid MccA to bind to the enzyme (9). Our results show that in the case of synechococcal enzyme, the substrate must be at least 20 amino acids in length to be adenylated, indicating that enzyme-substrate interactions removed further from the catalytic center than in that of E. coli MccB are required. The presence of the mccD homolog and two genes corresponding to separate domains of MccE suggests that the McC-like compound produced by Synechococcus may be aminopropylated. A recent report suggests that Synechococcus sp. CC9605, when cocultured with other synechococci that lack the mcc operon, inhibits the growth of those organisms and that mcc genes are responsible for the inhibition (23). Mass-spectrometric analysis of Synechococcus sp. CC9605 cultured medium failed to reveal a mass peak corresponding to full-sized adenylated MccA (our unpublished observations). However, it is possible that the initial adenylated peptide produced in the reaction catalyzed by Synechococcus sp. CC9605 MccB is proteolytically modified and that the active compound is actually much smaller.
Phylogenetic analysis shows that Y. pseudotuberculosis MccB is most closely related to Synechococcus sp. CC9605 MccB. The Y. pseudotuberculosis mcc operon has several distinct features, including fusion of MccB with a predicted methylase domain and two additional genes (one of which is another predicted methylase gene) with an uncharacterized function(s). Since we were unable to show the in vitro function of full-sized Y. pseudotuberculosis MccB, it is formally possible that Y. pseudotuberculosis is inactivated by these unique insertions. An alternative and more interesting possibility is that extra methylase homologs participate in additional modifications of Y. pseudotuberculosis peptide adenylate. The adenylated 42-amino-acid Y. pseudotuberculosis MccA protein is inactive against E. coli. It is also unable to inhibit aminoacylation of tRNAAsp in E. coli extracts. The greater length of Y. pseudotuberculosis MccA is unlikely to be responsible, since the even longer Synechococcus sp. CC9605 peptide adenylate is readily processed. Thus, as in the case of B. washoensis, Y. pseudotuberculosis MccA sequence likely prevents initial processing. In this case, Y. pseudotuberculosis MccA processing is probably complicated by the formation of a disulfide bond between the two cysteines present in the peptide (see Table 1). Cocultivation experiments similar to those performed with Synechococcus (23) will be necessary to determine if the mcc operon mediates antagonistic interactions within the Yersinia genus, and comparative analysis of low-molecular-mass compounds produced by Y. pseudotuberculosis with and without the mcc operon should help to elucidate the structure of the active compound, which may differ from that of the adenylated peptide obtained here by the in vitro approach.
E. coli McC penetrates sensitive cells through the YejABEF inner-membrane transporter, and it was proposed by us earlier that the presence of such transporter is a good predictor of McC sensitivity (4). For organisms with validated MccA-MccB pairs, clear homologs of YejABEF are detected only in H. pylori. The transport systems responsible for uptake of McC-like compounds in B. washoensis, S. thermophilus, and L. johnsonii remain to be identified, but it is clear that they must be different from YejABEF. Cyanobacteria are highly sensitive to McC (23) (O. Bantysh, unpublished), despite the lack of clear YejABEF homologs, again emphasizing that transporters other than YejABEF can import peptide adenylates. Our analysis of shortened versions of Synechococcus sp. CC9605 peptide adenylate revealed, curiously, that when peptide length exceeds a certain critical value (20 amino acids in our case), adenylated peptides cease to be transported by the YejABEF transporter. On the other hand, when peptide length reaches a certain critical value (25 amino acids in our case), adenylated peptides can be transported by SbmA. When the peptide length becomes even longer (52 amino acids for full-length Synechococcus sp. CC9605 peptide adenylate), SbmA becomes unable to perform the transport function. While at the present stage these observations cannot exclude the effects of sequence and charge, they nevertheless suggest that, by alterations of the length of the transport moiety of the McC-type Trojan horse antimicrobials, one can prepare peptide adenylates that enter bacteria through different routes, which could help counter bacterial resistance to this class of compounds.
The results of our bioinformatics analysis suggest that enzymes of the MccB/PaaA family that are capable of adenylation of asparagine residues in short ribosomally synthetized peptides are very abundant in completely sequenced genomes and are present in many bacterial phyla, including numerous human parasites and symbionts. Our list is by no means exclusive, since it is entirely possible that we missed some of the most divergent members of the MccB/PaaA family in this analysis. We predict that several peptides with asparagine residues that are encoded directly upstream of some MccB/PaaA family genes are adenylation substrates. In vitro adenylation of synthetic peptides by cognate MccB enzymes provides a robust method for experimental validation of these predictions. Nevertheless, many MccB/PaaA family genes lacked identifiable neighboring premicrocin peptide genes. This suggests either that a different amino acid within an upstream peptide-encoding gene could be adenylated or that substrate peptides could be encoded elsewhere in respective genomes. The latter phenomenon was reported for thiazole-containing heterocyclic bacteriocins of bacilli (24).
MATERIALS AND METHODS
DNA, molecular cloning, and microbial techniques.
H. pylori plasmids pHMG8 and HPP12 were provided by Sarah A. McIntire (Texas Woman’s University) and Wolfgang Fischer (Max von Pettenkofer-Institut), respectively. L. johnsonii NCC 533 was provided by Stéphane Duboux and Raymond David Pridmore (Nestlé Research Center), Synechococcus sp. CC9605 was provided by Brian Palenik (Scripps Institution of Oceanography, University of California, San Diego [UCSD]), genomic DNA of Y. pseudotuberculosis IP32953 was provided by Elisabeth Carniel (Institut Pasteur), and genomic DNA of B. washoensis Sb944nv was provided by Michael Kosoy (Centers for Disease Control and Prevention, NIH).
ORFs of E. coli mccB orthologs were amplified using appropriate primers and cloned in pET28a(+) expression vector between polylinker restriction sites NdeI and BamHI (S. thermophilus LMD-9), NcoI and HindIII (H. pylori pHMG8), NcoI and XhoI (H. pylori pHPP12 and L. johnsonii NCC 533), NdeI and XhoI (B. washoensis Sb944nv), NdeI and BlpI (Synechococcus sp. CC9605), and NcoI and NotI (full-sized Y. pseudotuberculosis IP32953). E. coli mccB was cloned between NcoI and BamHI of pET32b vector. A fragment of Y. pseudotuberculosis mccB (codons 2 to 347) was expressed as a fusion with MalE. To obtain the fusion protein, a fragment of the E. coli malE gene coding for mature MBP was PCR amplified and inserted between the NdeI and BamHI restriction sites of the pET22a vector to generate pET22MBP. Next, the PCR-amplified DNA fragment of Y. pseudotuberculosis mccB was introduced between the BamHI and XhoI restriction sites of pET22MBP.
Plasmid p28MBP was a generous gift of Satish Nair. Cells harboring this plasmid produce an MBP C-terminally fused to Synechococcus sp. CC9605 mccA.
To disrupt the yejB and sbmA genes in a laboratory E. coli B strain hypersensitive to McC (6), we used a method described by Datsenko and Wanner (25) with pKD46, pKD3, pKD13, and pCP20 plasmids and the following primers: primer pair YejB_Del_For1 (5′ ATGGGCGCTTACCTGATTCGCCGTCTGTTGCTGGTGATCGTGTAGGCTGGAGCTGCTTC 3′) and YejB_Del_Rev1 (5′ TTAACGTCCCTCAAAATCAATACGCGGATCAACCAGCGTCATATGAATATCCTCCTTAG 3′) and primer pair SbmA_Del_For (5′ TGTTAAAACGATAAGAAGTTAGCAGGAGTGCATATGTTACACGTCTTGAGCGATTG 3′) and SbmA_Del_Rev (5′ GGTTACTTCCTGAATTTTGTCACCATCCAGCTCATGATTCCGGGGATCCGTCGACCTG 3′).
Peptides.
Synthetic peptides were purchased from LLC Syneuro, Russia, and were at least 98% pure. The 42-amino-acid-long MccA peptide of Y. pseudotuberculosis PB1/+ was purchased from GenScript USA Inc. and was more than 85% pure. Peptides were dissolved in deionized water to up to a 15 mM concentration and stored at −20°C until further use.
To prepare Synechococcus sp. CC9605 MccA peptide, the p28MBP plasmid was transformed in E. coli BL21(DE3) cells. Cells were grown at 37°C in LB broth with 1% glucose and 50 µg/ml kanamycin until the optical density at 600 nm (OD600) reached 0.6. Cells were harvested, washed thrice with fresh LB, and resuspended in 200 ml of fresh LB broth supplemented with kanamycin and 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After overnight growth at 18°C, cells were harvested and resuspended in 20 ml of column buffer (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA, 1 mM sodium azide, 0.01 M β-mercaptoethanol, and 0.1 mM phenylmethylsulfonyl fluoride [PMSF]). Cells were disrupted by sonication. After centrifugation (30,000 × g for 30 min at +4°C), the supernatant was combined with 500 µl of Amilose resin (NEB) equilibrated in the same buffer and proteins were allowed to bind for 40 min at 4°C with gentle agitation. The resin was settled by gravity and washed with column buffer, and bound protein was eluted in column buffer supplemented with 10 mM maltose. Eluted material was dialyzed against 50 mM Tris-HCl (pH 8.0)–0.1 mM EDTA at +4°C, supplemented with 5 mM dithiothreitol (DTT), and treated with AcTEV protease (Invitrogen) according to the manufacturer’s suggestions. The mixture was incubated at +4°C overnight and was then dialyzed against 75 mM Tris-HCl (pH 8.0)–50 mM MgCl2 at +4°C. The MccA peptide was separated from MBP by using an Ultracel-30K Amicon Ultra centrifugal filter (Millipore). The filter flowthrough containing the separated peptide was concentrated on a rotary vacuum evaporator. Dried peptide was dissolved in water and stored at −20°C until further use.
Protein expression and purification.
All MccB proteins were expressed in E. coli BL21(DE3) cells. To obtain soluble MccB proteins from H. pylori, L. johnsonii NCC 533, Synechococcus sp. CC9605, Y. pseudotuberculosis IP32953, and B. washoensis Sb944nv expressing cells were cotransformed with a compatible pG-KJE8 vector (from a Chaperone plasmid set; TaKaRa Bio inc.) expressing GroEL/GroES, DnaK, DnaJ, and GrpE chaperones. The chaperone-expressing plasmid was found to increase the yield of soluble MccB homologs. The BL21(DE3) cells were grown at 37°C on 200 ml LB medium supplemented with 1% glucose and necessary antibiotics until the OD600 reached 0.6. Cells were harvested by centrifugation, washed thrice with fresh LB medium, and resuspended in 200 ml of fresh LB with antibiotics–0.1 mM IPTG. Where needed, 5 ng/ml tetracycline and 2 mg/ml arabinose were added to induce chaperone expression. Cells were grown for 20 h at 18°C with vigorous agitation. Cells were harvested and resuspended in 8 ml of loading buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 10 mM MgCl2) and disrupted by sonication. The lysates were cleared by centrifugation at 30,000 × g for 30 min at +4°C. To the supernatants, 300 µl of His Bind resin (Novagen) equilibrated in the same buffer was added, and proteins were allowed to bind for 4 h at 4°C with gentle agitation. The resin was allowed to settle by gravity and washed with 15 ml of wash buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 10 mM MgCl2, 50 mM imidazole), and bound proteins were eluted with 0.5 ml of elution buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 10 mM MgCl2, 200 mM imidazole). Four consecutive elutions were performed with each resin sample. Fractions containing maximal adenylation activity (see below) were combined, supplemented with glycerol up to 50%, and stored at −20°C until further use. The final concentration of MccB proteins ranged between 1 and 5 mg/ml. Proteins were at least 90% pure as judged by visual inspection of overloaded Coomassie-stained SDS gels.
In vitro adenylation assays.
Reaction mixtures contained (in 100 µl) 7.5 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 4 mM ATP (where appropriate), 2.5 mM TCEP [Tris(2-carboxyethyl) phosphine hydrochloride], and a 225 µM concentration of the appropriate MccA peptide (a concentration of full-sized Synechococcus sp. CC9605 peptide that was 10 times lower was used). Reactions were initiated by the addition of an MccB protein (5 µM final concentration), and reaction mixtures were incubated at 23°C for 20 h. Reactions were terminated by transferring them to −20°C.
MALDI-MS and MS/MS analysis.
Mass spectra were recorded on an Ultraflextreme matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF-ToF) mass spectrometer (MS; Bruker Daltonik) equipped with an Nd laser (355 nm). The MH+ molecular ions were measured in reflector mode; the accuracy of monoisotopic mass peak measurement was 50 ppm. Aliquots (1 µl) of desalted adenylation reaction mixtures were mixed on a steel target with 0.5 µl of 2,5-dihydroxybenzoic acid (Aldrich) solution (20 mg/ml in 30% MeCN–0.5% trifluoroacetic acid [TFA]).
In vivo sensitivity test.
E. coli B wild-type, ΔyejB, or ΔsbmA cells were grown in 10 ml of M63 broth supplemented with yeast extract at 37°C to an OD600 of ~1. A 750-µl volume of cell culture was added to 15 ml of melted top agar (0.65 g/liter of agar in M63 broth) cooled to ~50°C. The mixture was poured on the surface of an LB agar plate. After the agar solidified, 10-µl drops of solutions of 50 µM McC1092 (used as a positive control) or adenylation reaction mixtures (see above) were placed on a plate surface and allowed to dry. Plates were incubated for 4 to 6 h at 37°C, and growth inhibition zones around the sites where samples were applied were visually detected.
Preparation of S30 cell extract.
McC-sensitive E. coli BW25113 and an McC-resistant ΔABN derivative lacking the pepA, pepB, and pepN genes (5) were grown in 200 ml of LB medium at 37°C until the OD600 reached 0.5. Cells were collected by centrifugation, washed with a buffer (20 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 100 mM KCl), resuspended in 4 ml of the same buffer with the addition of 1 mM DTT, and disrupted by sonication. The lysate was centrifuged at 30,000 × g for 30 min at +4°C. The supernatants (total protein concentrations of 4 and 6 mg/ml for BW25113 and ΔABN lysates, respectively) were divided into 200-µl aliquots and stored at −70°C until further use.
tRNA aminoacylation reactions.
S30 extracts (4 µl) were mixed with 2 µl of reaction mixtures that had completed adenylation (see above) or 0.1 mM McC1092 and incubated for 15 min to allow processing. At this point, 16 µl of reaction mix (30 mM Tris-HCl [pH 8.0], 30 mM KCl, 8 mM MgCl2, 1 mM DTT, 3 mM ATP), 5 mg/ml total tRNA (tRNA from E. coli MRE 600; Roche Diagnostics), and 40 µM 14C-Asp (6) were added and reactions proceeded at room temperature for 5 min. The reaction products were precipitated on filters with cold trichloroacetic acid (10% final concentration) and subjected to scintillation counting.
MccA peptide processing.
A 2.5-µl volume of 5 mM MRTGNAN peptide and a 2.5-µl volume of 5 mM MDHIGFN peptide were mixed and combined with 5 µl E. coli BW25113 30S extract. The reaction mixtures were incubated for 0, 5, 15, and 60 min at room temperature and terminated by combining 1-µl reaction aliquots with 99 µl of 0.5% trichloroacetic acid. The reactions were next directly analyzed by mass spectrometry.
Sequence analysis.
Sequences from 2,262 completely sequenced bacterial and archaeal genomes available in the Refseq database (26) (as of February 2013) were assigned to the ThiF/HesA/MoeB/E1 superfamily using the RPS-BLAST program (matching profiles for pfam00899 and COG0476 with an E value cutoff of 0.01 and with low-complexity filtering off) as implemented in a Conserved Domain Database search (27) The BLASTClust program (28) set up with a length coverage cutoff of 0.8 and a score coverage threshold (bit score divided by alignment length) of 0.8 was used for clustering. One sequence was chosen for each cluster for further analysis. Genes encoding MccB and PaaA as well as all those that were experimentally analyzed in this work were added to the sequence set. For all selected genes, five up- and downstream genes were collected for analysis of gene neighborhoods and assigned to profiles corresponding to those in the matching COG (29) and Pfam (30) databases using the RPS-BLAST program as indicated above. The Position-Specific Iterated (PSI)-BLAST program (31) with an inclusion E value cutoff of 0.01 was used to retrieve homologs of MccB and PaaA starting from several queries. For each query, the homologs with the best scores were collected until the first gene whose neighborhood suggested its involvement in biological processes unrelated to microcin production was identified. In addition, 10 more sequences most similar to those representative of all the queries taken together were added into the final set of 60 sequences. Multiple alignment was built for this set using the MUSCLE program (32) followed by minor manual corrections, and one incomplete sequence was discarded. The length of the region (a potentially corresponding peptide clamp domain) upstream of the ThiF/HesA/MoeB/E1 domain was also measured. The confidently aligned blocks corresponding to the ThiF/HesA/MoeB/E1 domain with 136 informative positions were used for maximum-likelihood tree reconstruction using the FastTree program (33) with default parameters: JTT evolutionary model, discrete gamma model with 20 rate categories. The RAxML program (34) with the WAG substitution matrix and gamma-distributed evolutionary rates was used for the same alignment to build the phylogenetic tree to verify the outgroup position. Both programs were employed to calculate the respective bootstrap values. Short peptides, the mccA candidates, were predicted if they were encoded immediately upstream of the respective MccB/PaaA-like genes and contained at least one asparagine residue.
ACKNOWLEDGMENTS
We thank all the colleagues who provided genomic DNA samples that were used in this study. O.B. thanks Evgeny Gordienko for help.
Work in K.S.’s laboratories is supported by Russian Academy of Sciences Presidium Nanotechnology and Molecular and Cellular Biology program grants and by the Ministry of Education and Science of the Russian Federation project 14.B25.31.0004 grant. The MALDI MS facility became available to us in the framework of the Moscow State University Development Program (PNG 5.13). K.M. is supported by intramural funds of the U.S. Department of Health and Human Services (allocated to the National Library of Medicine).
Footnotes
Citation Bantysh O, Serebryakova M, Makarova KS, Dubiley S, Datsenko KA, Severinov K. 2014. Enzymatic synthesis of bioinformatically predicted microcin C-like compounds encoded by diverse bacteria. mBio 5(3):e01059-14. doi:10.1128/mBio.01059-14.
REFERENCES
- 1. García-Bustos JF, Pezzi N, Asensio C. 1984. Microcin 7: purification and properties. Biochem. Biophys. Res. Commun. 119:779–785. 10.1016/S0006-291X(84)80318-6 [DOI] [PubMed] [Google Scholar]
- 2. Khmel IA, Bondarenko VM, Manokhina IM, Basyuk EI, Metlitskaya AZ, Lipasova VA, Romanova YM. 1993. Isolation and characterization of Escherichia coli strains producing microcins of B and C types. FEMS Microbiol. Lett. 111:269–274. 10.1111/j.1574-6968.1993.tb06397.x [DOI] [PubMed] [Google Scholar]
- 3. Guijarro JI, González-Pastor JE, Baleux F, San Millán JL, Castilla MA, Rico M, Moreno F, Delepierre M. 1995. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J. Biol. Chem. 270:23520–23532. 10.1074/jbc.270.40.23520 [DOI] [PubMed] [Google Scholar]
- 4. Novikova M, Metlitskaya A, Datsenko K, Kazakov T, Kazakov A, Wanner B, Severinov K. 2007. The Escherichia coli Yej transporter is required for the uptake of translation inhibitor microcin C. J. Bacteriol. 189:8361–8365. 10.1128/JB.01028-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kazakov T, Vondenhoff GH, Datsenko KA, Novikova M, Metlitskaya A, Wanner BL, Severinov K. 2008. Escherichia coli peptidase A, B, or N can process translation inhibitor microcin C. J. Bacteriol. 190:2607–2610. 10.1128/JB.01956-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Metlitskaya A, Kazakov T, Kommer A, Pavlova O, Praetorius-Ibba M, Ibba M, Krasheninnikov I, Kolb V, Khmel I, Severinov K. 2006. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic microcin C. J. Biol. Chem. 281:18033-18042. 10.1074/jbc.M513174200 [DOI] [PubMed] [Google Scholar]
- 7. González-Pastor JE, San Millán JL, Moreno F. 1994. The smallest known gene. Nature 369:281. 10.1038/369281a0 [DOI] [PubMed] [Google Scholar]
- 8. Roush RF, Nolan EM, Löhr F, Walsh CT. 2008. Maturation of an Escherichia coli ribosomal peptide antibiotic by ATP-consuming N-P bond formation in microcin C7. J. Am. Chem. Soc. 130:3603–3609. 10.1021/ja7101949 [DOI] [PubMed] [Google Scholar]
- 9. Regni CA, Roush RF, Miller DJ, Nourse A, Walsh CT, Schulman BA. 2009. How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic. EMBO J. 28:1953–1964. 10.1038/emboj.2009.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Metlitskaya A, Kazakov T, Vondenhoff GH, Novikova M, Shashkov A, Zatsepin T, Semenova E, Zaitseva N, Ramensky V, Van Aerschot A, Severinov K. 2009. Maturation of the translation inhibitor microcin C. J. Bacteriol. 191:2380–2387. 10.1128/JB.00999-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Novoa MA, Díaz-Guerra L, San Millán JL, Moreno F. 1986. Cloning and mapping of the genetic determinants for microcin C7 production and immunity. J. Bacteriol. 168:1384-1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. González-Pastor JE, San Millán JL, Castilla MA, Moreno F. 1995. Structure and organization of plasmid genes required to produce the translation inhibitor microcin C7. J. Bacteriol. 177:7131–7140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fomenko DE, Metlitskaya AZ, Péduzzi J, Goulard C, Katrukha GS, Gening LV, Rebuffat S, Khmel IA. 2003. Microcin C51 plasmid genes: possible source of horizontal gene transfer. Antimicrob. Agents Chemother. 47:2868–2874. 10.1128/AAC.47.9.2868-2874.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Novikova M, Kazakov T, Vondenhoff GH, Semenova E, Rozenski J, Metlytskaya A, Zukher I, Tikhonov A, Van Aerschot A, Severinov K. 2010. MccE provides resistance to protein synthesis inhibitor microcin C by acetylating the processed form of the antibiotic. J. Biol. Chem. 285:12662–12669. 10.1074/jbc.M109.080192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tikhonov A, Kazakov T, Semenova E, Serebryakova M, Vondenhoff G, Van Aerschot A, Reader JS, Govorun VM, Severinov K. 2010. The mechanism of microcin C resistance provided by the MccF peptidase. J. Biol. Chem. 285:37944–37952. 10.1074/jbc.M110.179135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Severinov K, Semenova E, Kazakov A, Kazakov T, Gelfand MS. 2007. Low-molecular-weight post-translationally modified microcins. Mol. Microbiol. 65:1380–1394. 10.1111/j.1365-2958.2007.05874.x [DOI] [PubMed] [Google Scholar]
- 17. Laviña M, Pugsley AP, Moreno F. 1986. Identification, mapping, cloning and characterization of a gene (sbmA) required formicrocin B17 action on Escherichia coli K12. J. Gen. Microbiol. 132:1685–1693 [DOI] [PubMed] [Google Scholar]
- 18. Burroughs AM, Iyer LM, Aravind L. 2009. Natural history of the E1-like superfamily: implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins 75:895-910. 10.1002/prot.22298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin M, Wright SA, Beer SV, Clardy J. 2003. The biosynthetic gene cluster of pantocin A provides insights into biosynthesis and a tool for screening. Angew. Chem. Int. Ed. Engl. 42:2902–2905. 10.1002/anie.200351054 [DOI] [PubMed] [Google Scholar]
- 20. Jin M, Liu L, Wright SA, Beer SV, Clardy J. 2003. Structural and functional analysis of pantocin A: an antibiotic from Pantoea agglomerans discovered by heterologous expression of cloned genes. Angew. Chem. Int. Ed. Engl. 42:2898–2901. 10.1002/anie.200351053 [DOI] [PubMed] [Google Scholar]
- 21. Chandu D, Nandi D. 2003. PepN is the major aminopeptidase in Escherichia coli: insights on substrate specificity and role during sodium-salicylate-induced stress. Microbiology 149:3437–3447. 10.1099/mic.0.26518-0 [DOI] [PubMed] [Google Scholar]
- 22. Addlagatta A, Gay L, Matthews BW. 2008. Structural basis for the unusual specificity of Escherichia coli aminopeptidase N. Biochemistry 47:5303–5311. 10.1021/bi7022333 [DOI] [PubMed] [Google Scholar]
- 23. Paz-Yepes J, Brahamsha B, Palenik B. 2013. Role of a microcin-C-like biosynthetic gene cluster in allelopathic interactions in marine Synechococcus. Proc. Natl. Acad. Sci. U. S. A. 110:12030–12035. 10.1073/pnas.1306260110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haft DH. 2009. A strain-variable bacteriocin in Bacillus anthracis and Bacillus cereus with repeated Cys-Xaa-Xaa motifs. Biol. Direct 4:15. 10.1186/1745-6150-4-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35:D61–D65. 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. 2009. CDD: specific functional annotation with the conserved domain database. Nucleic Acids Res. 37:D205–D210. 10.1093/nar/gkn845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wheeler D, Bhagwat M. 2007. BLAST QuickStart: example-driven web-based BLAST tutorial. Methods Mol. Biol. 395:149–176. 10.1007/978-1-59745-514-5_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138–D141. 10.1093/nar/gkh121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stamatakis A, Ludwig T, Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463. 10.1093/bioinformatics/bti191 [DOI] [PubMed] [Google Scholar]