ABSTRACT
Bacterial ParA and ParB proteins are best known for their contribution to plasmid and chromosome segregation, but they may also contribute to other cell functions. In segregation, ParA interacts with ParB, which binds to parS centromere-analogous sites. In transcription, plasmid Par proteins can serve as repressors by specifically binding to their own promoters and, additionally, in the case of ParB, by spreading from a parS site to nearby promoters. Here, we have asked whether chromosomal Par proteins can likewise control transcription. Analysis of genome-wide ParB1 binding in Vibrio cholerae revealed preferential binding to the three known parS1 sites and limited spreading of ParB1 beyond the parS1 sites. Comparison of wild-type transcriptomes with those of ΔparA1, ΔparB1, and ΔparAB1 mutants revealed that two out of 20 genes (VC0067 and VC0069) covered by ParB1 spreading are repressed by both ParB1 and ParA1. A third gene (VC0076) at the outskirts of the spreading area and a few genes further away were also repressed, particularly the gene for an outer membrane protein, ompU (VC0633). Since ParA1 or ParB1 binding was not evident near VC0076 and ompU genes, the repression may require participation of additional factors. Indeed, both ParA1 and ParB1 proteins were found to interact with several V. cholerae proteins in bacterial and yeast two-hybrid screens. These studies demonstrate that chromosomal Par proteins can repress genes unlinked to parS and can do so without direct binding to the cognate promoter DNA.
IMPORTANCE
Directed segregation of chromosomes is essential for their maintenance in dividing cells. Many bacteria have genes (par) that were thought to be dedicated to segregation based on analogy to their roles in plasmid maintenance. It is becoming clear that chromosomal par genes are pleiotropic and that they contribute to diverse processes such as DNA replication, cell division, cell growth, and motility. One way to explain the pleiotropy is to suggest that Par proteins serve as or control other transcription factors. We tested this model by determining how Par proteins affect genome-wide transcription activity. We found that genes implicated in drug resistance, stress response, and pathogenesis were repressed by Par. Unexpectedly, the repression did not involve direct Par binding to cognate promoter DNA, indicating that the repression may involve Par interactions with other regulators. This pleiotropy highlights the degree of integration of chromosomal Par proteins into cellular control circuitries.
INTRODUCTION
Many low-copy-number plasmids and bacterial chromosomes have parABS genes for segregating replicated sisters to opposite halves of dividing cells (1, 2). The products of parAB are two trans-acting proteins, ParA and ParB. parS is a cis-acting site which functions analogously to eukaryotic centromeres. ParA actively moves plasmids/chromosomes that have their parS bound by ParB.
In plasmids, the parAB genes comprise an operon, which is autorepressed either by ParA or ParB or by a ParA-ParB complex (1). When ParA serves as the repressor, it binds to operator sites unrelated to parS. This regulation can be further tightened by the participation of ParB and parS (3). In some plasmids, parS fulfills both the operator and centromere functions. The regulation of chromosomal par genes is known in Streptomyces coelicolor, where the parAB operon is autorepressed by ParB (4). This seems to be an isolated case, as parS sites are not usually found upstream of chromosomal par operons, and the domains of plasmid ParA proteins that specifically bind to operator sequences are usually missing from chromosomal ParA (1). The transcriptional regulation of chromosomal par genes remains largely unknown.
It is also not known whether Par proteins can regulate transcription of genes other than parAB. Such a possibility was suggested in Bacillus subtilis, where Soj and Spo0J (ParA and ParB homologues) appeared to control transcription of many sporulation genes (5). Subsequently, the results were reinterpreted to be due to Soj increasing the replication initiation activity of DnaA (6). The increase in DnaA activity turns on a cascade of events that lead to repression of sporulation genes (7). Thus, Soj control of transcription is considered to be indirect, occurring through DnaA.
ParB not only represses promoters that overlap parS but also represses promoters at a distance. The distal promoters are reached by spreading of ParB onto sequences that flank parS (8–11). The spreading can interfere with RNA polymerase interactions with promoter elements, a process termed (gene) silencing. The spreading has also been implicated in the control of DNA replication (8, 12, 13). The spreading can interfere with DNA-protein interactions involved in replication control, which can both promote and interfere with replication initiation, depending upon the situation.
Segregation, gene silencing, and DNA replication aside, Par proteins contribute to chromosome organization by loading condensin in the vicinity of the replication origin in B. subtilis and in Streptococcus pneumoniae (14–16) and contribute to cell cycle progression and cell division in Caulobacter crescentus (17, 18), cell growth in Mycobacterium smegmatis (19), cell growth and motility in Pseudomonas aeruginosa (20), cell morphology in Pseudomonas. putida (21), and cell division in Streptomyces coelicolor (22). It is clear that chromosomal par genes play pleiotropic roles.
Here, we have investigated whether Par proteins control transcription in Vibrio cholerae, a bacterium with two chromosomes (chrI and chrII). Both chromosomes have their own parABS genes (parABS1 for chrI and parABS2 for chrII) (23). Their role in chromosome segregation has been studied in detail (24, 25). The nature of regulation of chromosomal parAB operons, or whether the Par proteins can control transcription of genes other than their own, is largely unknown. We show that ParB1 binds specifically only to parS1 sites and can spread to flanking DNA, which results in transcriptional silencing in a minority of cases. Additionally, we found that both ParA1 and ParB1 proteins could silence genes unlinked to parS, apparently without direct binding to promoter DNA. The possibility of involvement of other factors is suggested by the finding that both Par proteins can interact with several V. cholerae proteins.
RESULTS
ParB1 binding to parS1 sites and flanking DNA.
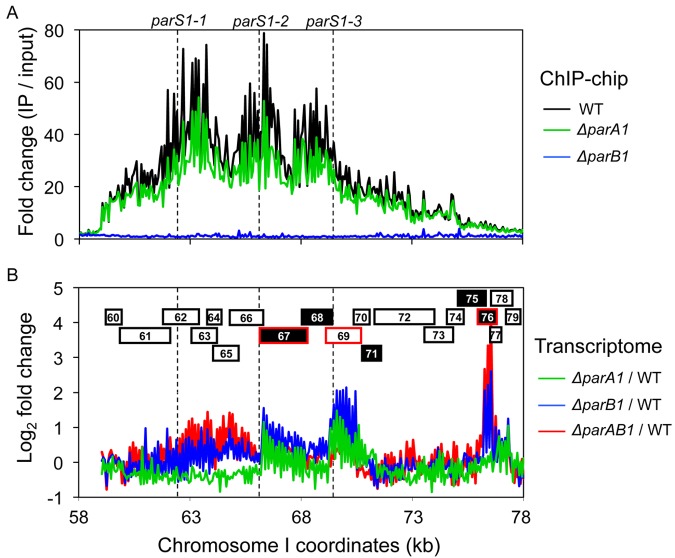
We determined genome-wide ParB1 binding in V. cholerae N16961 (CVC796) using chromatin immunoprecipitation with microarray technology (ChIP-chip). When the amount of DNA precipitated by ParB1 antibody was compared with total DNA from the whole-cell extract, DNA in the region containing the three known parS sites of chrI (parS1-1, parS1-2, and parS1-3) was selectively precipitated, indicating preferential ParB1 binding to those sites (Fig. 1A; see also Table S1 in the supplemental material). This result is consistent with an earlier finding that identified the sites by bioinformatics analysis and yellow fluorescent protein (YFP)-ParB1 focus formation (23). The modal position of the immunoprecipitated DNA peaks roughly corresponded to the three parS1 sites, although the two outer peaks (apparently representing binding to parS1-1 and parS1-3 sites) were shifted slightly toward the central peak, which apparently represents binding to the parS1-2 site. The peaks spread wider (2 to 3 kb at half-maximal height) than the average DNA fragment length (0.5 kb). The peaks were narrower when a similarly fragmented DNA preparation was precipitated with RctB antibody (see Fig. S1) (26). The RctB protein is a site-specific DNA binding protein and is not known to spread along DNA outside its specific binding sites (12). These results are consistent with ParB1 spreading on either side of parS1 sites. The total length covered by ParB1 over three parS1 sites was about 16 kb (chrI coordinates 59 to 75 kb), although the sites are located within a 7-kb region. This result is also consistent with spreading. The extent of Spo0J (ParB) spreading was about 18 kb in B. subtilis (9). Unlike the situation in B. subtilis, the parS1 sites in V. cholerae are located about 65 kb away from the origin, which is apparently too far to be reached by ParB1 spreading (14, 15).
FIG 1 .
(A) ChIP-chip binding profiles of ParB1 in region containing the three parS1 sites (dashed lines) in chrI of V. cholerae. The profiles are shown for wild-type (in black), ∆parA1 (in green), and ∆parB1 (in blue) cells growing exponentially in LB broth. The fold change represents the amount of immunoprecipitated (IP) DNA normalized with respect to input DNA. The profiles represent average signals from three independent experiments. (B) Changes in expression levels of genes around the three parS1 sites (dashed lines) in V. cholerae ∆parA1 (green profile), ∆parB1 (blue profile), and ∆parAB1 (red profile) cells compared with those in WT cells growing exponentially in LB broth. The fold change values are log2 ratios of expression levels in mutants over the WT cells. Data presented are the averages from three independent experiments. The numbers in the boxes indicate the locus tag from VC0060 to VC0079 (abbreviated as 60 to 79). The white and black boxes indicate genes on plus and minus strands, respectively. The boxes outlined in red indicate the genes whose expression was considered changed at least 2-fold in the absence of ParA1 and/or ParB1. The genes 60 to 66 constitute an operon whose promoter is upstream of gene 60. The promoter activity of the operon appears not to be affected by Par proteins, although transcription elongation appears to have been reduced, most likely by opposing ParB1 spreading.
We validated spreading of ParB1 in Escherichia coli by silencing of plasmid replication (Fig. 2). When the parS1-1 site was present in the pGB2 vector, transformants carrying such a plasmid could not be selected when ParB1 synthesis was induced. Under identical conditions, cells without induction of ParB1 could grow. In similar experiments, the growth failure was attributed to ParB spreading into the replication initiator gene and silencing its promoter, which is located only 200 bp away from parS (10, 27).
FIG 2 .
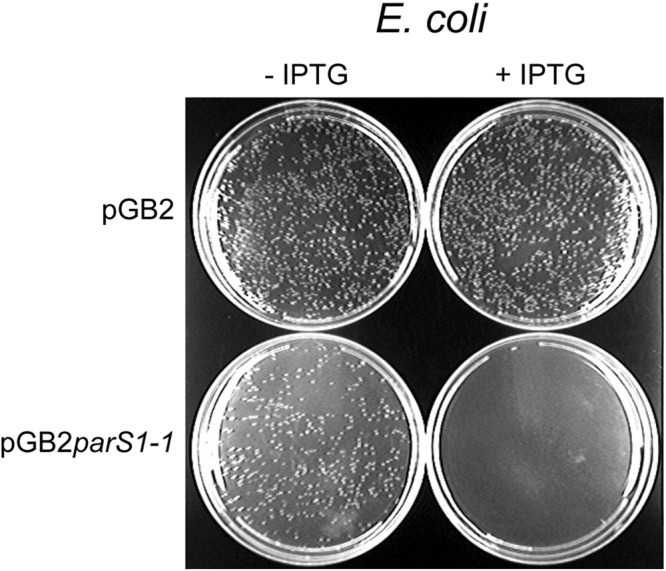
Effect of ParB1 on growth of E. coli carrying parS1 plasmids. The cells had either an empty vector (pGB2) or the same vector carrying the parS1-1 site (pGB2parS1-1). The cells also had another plasmid (pBJH15) carrying the parB1 gene under the control of an IPTG-inducible promoter. Cells were grown on LB agar plates with or without IPTG, under drug selection for both plasmids.
In order to determine the contribution of ParA1 to ParB1 binding and spreading, we performed ChIP-chip using a ∆parA1 strain (CVC797) (Fig. 1A). The ChIP-chip profile of the deletion strain was nearly identical to that of the wild-type (WT) strain, although the peak heights were reduced. This suggests that ParA1 might promote specific binding of ParB1. In contrast, no peaks could be seen in a ∆parB1 strain (CVC1123), as would be expected if the ChIP signals were due to ParB1 binding.
Gene silencing around parS1 sites.
We performed transcriptome analysis to determine expression levels of genes near the parS1 sites. We focused on 20 genes that were present within the region covered by ParB1 spreading. Only three genes were expressed at levels greater than 2-fold in the ∆parB1 strain compared to their expression in the WT. The genes encoded aminopeptidase P (VC0067), a putative multidrug resistance protein (VC0069), and a universal stress protein A (VC0076) (Fig. 1B). The upstream regions of all three genes (without including the parS1 sites) were tested for ParB1 binding by electrophoretic mobility shift assay (EMSA), but no specific binding was detected (see Fig. S2A in the supplemental material). We also tested the regulation of expression of the three genes by ParB1 by fusing their upstream regions (without parS1) to a promoterless lacZ gene and measuring β-galactosidase activity in WT and ∆parB1 cells (see Fig. S2B). The absence of parB1 did not influence the activity of the first two genes (VC0067 and VC0069) and only marginally increased the expression of the third gene (VC0076). The results of the first two genes are consistent with parS1-dependent spreading and silencing by ParB1. The fact that the third gene is located farthest from the parS1 sites in a region where ParB1 spreading was barely significant yet showed maximal ParB1-mediated silencing suggests that the third gene is silenced by ParB1 without requiring parS1. Why only two out of 20 genes were silenced in the region of significant spreading remains to be determined. The results do indicate that spreading and silencing need not be correlated, as was the case in B. subtilis (9).
We also performed transcriptome analysis in ∆parA1 and ∆parAB1 mutants (Fig. 1B). The expression level of VC0067 and VC0069, but not of VC0076, increased in ∆parA1 cells compared to WT. This again indicates that regulation by Par proteins differs between the first two genes and the third gene. Whereas the silencing of the first two genes appears to require both ParA1 and ParB1, the regulation by ParB1 appears more significant. ParA1 could act by increasing ParB1 binding, as the ChIP-chip data suggest (Fig. 1A). Since ParA1 does not influence ParB1 concentration (28), ParA1 could affect the first two genes by increasing specific DNA binding of ParB1. However, this inference is not supported by the results in ∆parAB1 cells. If ParA1 were acting through ParB1, then the derepression levels in ∆parB1 and ∆parAB1 cells should have been comparable, but they are not. Thus, the regulation may involve factors other than Par proteins, whose activity may depend on ParA1.
ParB1 appears to be the main regulator of the third gene, VC0076, since the regulation did not differ much between WT and ∆parA1 cells (Fig. 1B). We conclude that the requirements of spreading and ParA1 are considerably less for VC0076 than for the other two genes.
Gene silencing outside the ParB1 spreading region.
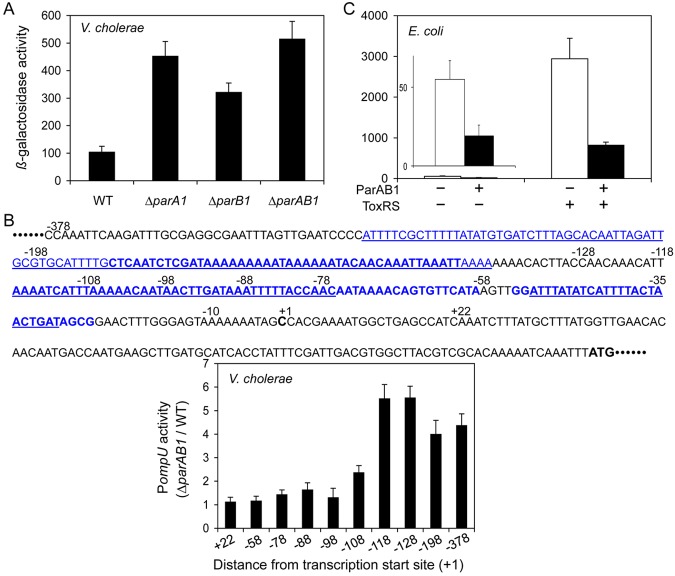
Transcriptome analysis also revealed that the expression levels of several genes outside the ParB1 spreading region can change in parA1, parB1, and parAB1 deletion strains (see Table S2 in the supplemental material). We selected six genes whose expression changed (up or down) most significantly in at least one of three mutants tested (Table 1). We validated expression level changes for these genes by fusing the gene regulatory regions to a promoterless lacZ gene. The change was most significant in the case of ompU (VC0633) in both transcriptome and lacZ fusion assays (Table 1). In the transcriptome analysis, the expression of the ompU gene increased in both ∆parA1 and ∆parB1 cells and synergistically in ∆parAB1 cells. These results indicate that both ParA1 and ParB1 can independently repress the expression of ompU (as in VC0067 and VC0069) and that the two proteins might cooperate in their repressive activities on ompU (unlike the situation in VC0067 and VC0069). Likewise, the lacZ fusion assays in ∆parA1, ∆parB1, and ∆parAB1 cells revealed that the ompU promoter activity increased in all three deletion strains, confirming that both ParA1 and ParB1 are involved in the repression of the ompU promoter (Fig. 3A). The synergistic effect seen in the transcriptome analysis was not evident in the gene fusion assay, the reason for which remains to be understood. Both assays, however, suggest that ParA1 and ParB1 can control ompU expression.
TABLE 1 .
Candidate genes showing different expression levels in ΔparA1, ΔparB1, and ΔparAB1 cells
| Gene | Function | Expression levela |
||||
|---|---|---|---|---|---|---|
| Transcriptome |
lacZ fusion |
|||||
| ∆parA1 | ∆parB1 | ∆parAB1 | ∆parA1 | ∆parAB1 | ||
| VC0321 (tufB) | Elongation factor Tu | 4.23 | 2.85 | 2.14 | 1.42 | |
| VC0470 (dns) | Extracellular DNase | 1.16 | 0.20 | 0.14 | 1.56 | |
| VC0633 (ompU) | Outer membrane protein | 4.86 | 3.89 | 120.26 | 4.72 | |
| VC0706 | Sigma-54 modulation protein, putative | 0.52 | 45.89 | 93.70 | 2.13 | |
| VCA0519 (fruR) | Fructose repressor | 0.83 | 0.92 | 0.21 | 1.95 | |
| VCA0676 (napF) | Iron-sulfur cluster-binding protein | 11.39 | 1.48 | 0.96 | 1.84 | |
Expression levels in mutant cells were in comparison to those in WT cells.
FIG 3 .
Regulation of ompU gene. (A) The activity of the ompU promoter fused to lacZ of the pMLB1109 vector, resulting in pBJH18, was tested in V. cholerae WT, ∆parA1, ∆parB1, and ∆parAB1 cells growing exponentially in LB broth. β-Galactosidase activities are averages from three experiments. (B) Ratios of activities of various lengths of the ompU promoter fused to lacZ of the pMLB1109 vector in V. cholerae WT and ∆parAB1 cells. The bold and underlined blue sequences represent published DNase I footprints of ToxR on the top and bottom strands, respectively (29). Also marked, in bold black, are the transcription (+1) and translation (ATG) start sequences. (C) Activity of a single copy of the ompU promoter fused to lacZ and integrated into the chromosome of E. coli. The resulting strain, CVC1882, contained two other plasmids: one was either an empty vector (pBAD24) (white bar) or the same vector supplying ParAB1 (pRN005) (black bar) and the other was a compatible vector (pKT25) or the same vector supplying ToxRS (pBJH172). The induction of Par proteins by arabinose and ToxRS proteins by IPTG was demonstrated previously (28, 40).
In order to determine the region required for repression by ParA1 and ParB1, we fused various lengths of the ompU promoter region to lacZ and compared promoter activities between WT and ∆parAB1 cells (Fig. 3B). We found that the repression is most efficient when the ompU promoter fragment contains 118 bp upstream of the transcription start site.
Notably, ompU is one of the genes whose promoter is significantly activated by ToxRS proteins (29). The ompU promoter contains three ToxR binding sites in the upstream regulatory region. To determine if the regulation of the promoter by Par proteins depends on ToxRS, we determined the promoter activity with and without ToxRS in E. coli (Fig. 3C). As reported, the activity of the ompU promoter increased dramatically in the presence of ToxRS, but the repression by ParAB1 could be seen in both the absence and the presence of ToxRS. These results indicate that ompU promoter repression by ParAB1 does not require ToxRS.
ParA1 and ParB1 interactions with other V. cholerae proteins.
Since direct binding of ParA1 and ParB1 without requiring parS1 in cis could not be demonstrated, we considered the alternate possibility that Par proteins function by interaction with other proteins. A candidate approach using the bacterial two-hybrid (B2H) assay confirmed that ParA1 and ParB1 do not interact with ToxR or OmpU (see Fig. S3 and Text S1 in the supplemental material). We showed above that Par regulation of ompU does not require ToxR and that the ompU gene is not autorepressed (data not shown). To identify interacting proteins, we took a global approach using both bacterial and yeast two-hybrid (Y2H) screening systems using ParA1 and ParB1 proteins as baits (see Table S3). We identified known interactions of ParA1 with ParB1, ParB1 with itself (28, 30), and ParA1 with HubP (VC0998) (30), indicating the effectiveness of our screening approach. The cyaA gene (VC0122) was also identified several times in our B2H screening system, which is based on reconstitution of CyaA activity. A number of other proteins were also found to interact with ParA1 and/or ParB1 (see Table S3). The relevance of these interactions in the regulation of ompU or other genes remains to be determined.
We note that although we used three different two-hybrid systems to achieve maximum coverage and reproduced several known interactions, overlapping interactions were not found among the three systems. Previous studies have shown that different versions of yeast two-hybrid (Y2H) vectors produce complementary interactions rather than overlapping interactions (31, 32), which may explain our findings. However, we cannot exclude the possibility of missing open reading frames (ORFs) among three different prey libraries.
parAB1 operon is not autorepressed.
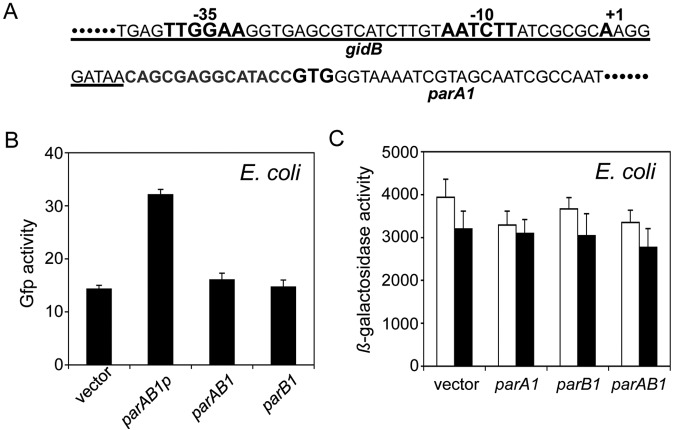
In plasmids such as P1 and F, the N-terminal region of ParA is important for transcriptional autorepression, but many chromosomal ParA proteins, including V. cholerae ParA1, lack this domain (1). How the chromosomal par genes are regulated remains unknown. To determine the regulation of parAB1 genes in V. cholerae, we located the promoter for parA1 within the upstream sequence of the gidB gene, using the 5′ RACE (rapid amplification of cDNA ends) method (Fig. 4A). To determine whether the promoter is the only one that directs transcription of both parAB genes, we created three separate fusions to a gfp reporter gene. The region fused consisted of either the parAB1 genes with the promoter identified by 5′ RACE (parAB1p) or parAB1 genes without that promoter or only the parB1 gene. Measurement of fluorescence intensities revealed that only the parAB1 fragment with the promoter showed increased fluorescence compared to that of the vector without any insert (Fig. 4B). When the inserts were parAB1 and parB1, the fluorescence intensities were comparable to that of the vector-alone control. These results suggest that the parB1 gene is transcribed from the parA1 promoter and that parAB1 genes constitute an operon.
FIG 4 .
Regulation of parAB1 genes. (A) Sequence of parAB1 promoter region. The −35 and −10 promoter elements (bold letters) reside within the C-terminal region of the gidB gene (underlined). Also marked in bold are the transcription (+1) and translation (GTG) start sequences of parA1. (B) Promoter activities from cloned fragments carrying parAB1 genes and the adjoining upstream region (parAB1p) or parAB1 genes without the upstream region (parAB1) or the parB1 gene only (parB1). The activities were measured after fusing the fragments to gfp and measuring fluorescence intensities in E. coli cells exponentially growing in LB broth. The intensities are averages from three experiments. The high fluorescence from the gfp vector alone (the bar marked “vector”) is attributed to autofluorescence because the cells without the gfp vector gave similar fluorescence values. (C) Activity of parAB1 promoter fused to lacZ integrated into the E. coli chromosome using a λ vector. The lysogens contained either an empty vector (pBAD24) or the same vector carrying parA1, parB1, or parAB1 genes (pRN006, pRKG212, or pRN005, respectively). The lysogens were grown in LB broth with 0 (white bar) or 0.02% (black bar) arabinose.
We then asked whether the parAB1 operon is autorepressed, as is usually the case in plasmids (1). We fused the parAB1 promoter to a promoterless lacZ gene present in pMLB1109 (resulting in pBJH17) and transferred it to the E. coli chromosome using phage λ (resulting in CVC1881). We determined the activity of the promoter both before and after induction of ParA1 only, ParB1 only, or both ParA1 and ParB1 (Fig. 4C). We previously showed that the induction increases the protein levels at least 16-fold (28). The promoter activity did not change upon induction of any of the proteins, indicating that ParA1 and ParB1 do not control their own promoter. ChIP-chip analysis indicated that ParA1 and ParB1 proteins also did not bind in the vicinity of the parAB1 promoter (see Table S1 in the supplemental material). The transcriptome analysis also revealed that the levels of parA1 transcript in ∆parB1 cells and parB1 transcript in ∆parA1 cells remain comparable to those in the WT, indicating that expression of parAB1 genes is not controlled by ParA1 and ParB1 proteins (see Table S2). The expression levels of the parA1 gene in ∆parA1 cells and the parB1 gene in ∆parB1 cells were very low, as would be expected. We previously showed that ParA1 and ParB1 protein levels do not change in ∆parB1 and ∆parA1 strains, respectively, compared to their levels in the WT (28). From these results, we conclude that the parAB1 operon is not autorepressed.
We note that the activity of the parAB1 promoter was high in both E. coli and V. cholerae. A single-copy promoter fusion showed about 4,000 Miller units of activity in E. coli (Fig. 4C), and the same fusion in multicopy showed about 2,500 Miller units of activity in V. cholerae (data not shown). The parAB1 genes could also be expressed from the upstream gidAB promoter as revealed by a reverse transcription-PCR (RT-PCR) assay (see Fig. S4 in the supplemental material). However, the activity of the gidAB promoter was about 10-fold lower than that of the parAB promoter, and the gidAB promoter was also not regulated by ParA1 and/or ParB1 (data not shown).
DISCUSSION
Par proteins as transcription factors.
Here, we show that in V. cholerae, parAB1 genes of chromosome I constitute an operon, which is usually the case in plasmids, but unlike the situation in plasmids, the operon is not autorepressed. Par proteins, however, can influence transcription of other genes by acting either separately or together. ParB1 silences VC0067 and VC0069 genes that are near parS1 sites, apparently by spreading. ParA1 could also influence the expression of these two genes either by reducing ParB1 binding to parS1 (Fig. 1A) or by unknown mechanisms. ParB1 can also repress genes not linked to parS1 (VC0076 and ompU). VC0076 was regulated mainly by ParB1, whereas ompU was regulated by both ParA1 and ParB1. To the extent studied, it is clear that the Par proteins can regulate transcription without necessarily requiring parS sites or each other and, remarkably, without direct binding to the promoter region.
Direct binding was considered unlikely for the following reasons. ParA1 ChIP-chip data did not reveal any significant binding above background (data not shown). ParA1 also does not contain the specific DNA binding domains that are often present in plasmid ParA proteins (1). ParB1 binding that could be detected unambiguously by ChIP-chip (Fig. 1A; see also Table S1 in the supplemental material) and EMSA (see Fig. S2A) was absent from the upstream regions of genes that it represses.
The upstream regions of VC0067, VC0069, VC0076, and ompU genes do not reveal any sequence similarity that would suggest the presence of a binding site for a common regulator. We considered whether the indirect regulation is through VC0067 and VC0069, the only two genes that ParA1 and ParB1 control in a parS-dependent manner, and that they in turn regulate other genes. VC0067 and VC0069 function as aminopeptidase P and multidrug resistance proteins, respectively, which are unlikely to be transcription regulators. Orthologous genes (ypdF and mdtL, respectively) which do not have a parABS system are present in E. coli, and yet the V. cholerae ompU gene was regulated by ParA1 and ParB1 when tested in E. coli. These reasons lead us to think that the regulation is indirect, the possible mechanisms of which are discussed below.
Par proteins are known to interact with two global regulators. In B. subtilis and S. pneumoniae, Spo0J/ParB loads SMC protein near the replication origin, which has been suggested to organize the origin region of the chromosome (14, 15). SMC proteins are known to condense chromosomes in general and therefore can have a global effect on DNA transactions by changing DNA topology. The effect can be more pronounced in local regions, as for example the origin and terminus regions, because of interactions with proteins such as ParB and TopoIV, respectively (14, 15, 33). ParA (Soj) of B. subtilis also interacts with DnaA, which can turn on a checkpoint response that ultimately controls the expression of many sporulation genes (7). Both ParA and ParB have significant nonspecific binding and in principle can have a role in transcriptional control, but this role remains to be demonstrated.
Pleiotropy of chromosomal Par proteins.
Plasmid and chromosomal Par proteins, although largely related, appear to be functionally distinct. The proteins are major players in plasmid segregation, whereas their contribution to chromosome segregation is often modest (2, 34, 35). The main distinguishing feature of chromosomal Par proteins is their ability to affect functions besides segregation, as discussed in the introduction. Here we show that ParA1 and ParB1 proteins control expression of genes that have been implicated in the control of drug resistance (VC0069), the stress response (VC0076), and pathogenesis (VC0633) of V. cholerae. In addition to interaction with SMC and DnaA, ParB binding to MipZ has been demonstrated to regulate cell division in C. crescentus (17, 18). Thus, DNA binding aside, protein-protein interactions contribute to the pleiotropy of Par proteins. Several proteins are found to interact with Par proteins in B2H and Y2H screening systems. Although these interactions need to be validated by independent methods, they suggest that there is a wider spectrum of cell functions regulated by Par proteins, involving both global and specific regulators.
Conservation of Par functions.
Here, we show that V. cholerae Par proteins share many of the properties of Par proteins seen in other bacteria. For example, ParB1 binds in a site-specific manner to its cognate centromeric sites and can spread to neighboring sequences, as was initially shown in B. subtilis (36). In both bacteria, ParA1 has little contribution to the spreading process. Spreading can silence some, but not all, genes encountered by spreading. As in B. subtilis, ParA1 and ParB1 do not regulate their own genes. The similarities, particularly with B. subtilis, suggest that the characteristics of chromosomal Par proteins have been conserved since the two bacteria diverged more than a billion years ago. Recently, Par proteins were shown to be global transcription regulators in P. aeruginosa (37). In this study, both ParA and ParB were shown to regulate many genes, apparently indirectly.
MATERIALS AND METHODS
Strains and plasmids.
V. cholerae and E. coli strains and plasmids used in this study are listed in Table S4 in the supplemental material.
ChIP-chip assay.
ChIP-chip assays were performed using a custom Agilent 8-by-60K V. cholerae oligonucleotide microarray, as described previously (12). Vibrio cholerae cells were cultivated in LB broth at 37°C to exponential phase and cross-linked with 1% formaldehyde for 30 min at room temperature. ParA1-DNA or ParB1-DNA complexes were immunoprecipitated using ParA1 or ParB1 antibody (Biosource International), respectively. Fold changes were calculated by dividing precipitated DNA (Cy5) signals by the total input (Cy3) signals from three independent experiments.
Transcriptome analysis.
Vibrio cholerae cells were cultivated in LB broth at 37°C to exponential phase. Total RNA from wild-type, ∆parA1, ∆parB1, or ∆parAB1 cells was purified using RNAprotect bacterial reagent and RNeasy minikits (Qiagen) according to the manufacturer’s protocol. Using the purified RNA, cDNA was synthesized, labeled, and hybridized to the Agilent V. cholerae microarray as described elsewhere (38). Fold changes were calculated by dividing the signals from deletion mutants (Cy5) by the signal from the WT (Cy3) from three independent experiments.
Plasmid replication assay.
Transformants containing an inducible source of parB1 (pBJH15) and either pGB2 vector or its parS1-1-carrying derivative (pBJH105) were grown on LB agar plates at 37°C for 1 day under drug selection for both plasmids. Isopropyl-β-d-thiogalactopyranoside (IPTG) (0.5 mM) was added to the plates when induction of ParB1 was desired (12).
Transcription assay by reporter gene fusion.
Promoter activities were measured using promoter-cloning vectors with a reporter gene, which was either lacZ (pMLB1109) or gfp (pBJH65), in exponentially growing cells in LB broth as described elsewhere (12). Fluorescence intensities were measured using a Victor2 (Wallac) microplate reader, and β-galactosidase activities were determined as described previously (39).
5′ RACE.
The 5′/3′ RACE kit was used according to the manufacturer’s protocol (Roche). cDNA was synthesized and amplified using primers BJH114 and BJH115 (see Table S5 in the supplemental material), respectively. RNAprotect bacterial reagent and RNeasy minikits (Qiagen) were used to purify RNA. The pGEM-T Easy vector system (Promega) was used for cloning amplified cDNA.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download
Comparison of ChIP-chip profiles of ParB1 (black, same as in Fig. 1A) and a nonspreading protein, RctB (orange). The RctB profile is shown for WT cells only in an origin-distal locus of chrII (coordinates 1024 to 1031 kb) (26) where RctB has specific binding sites spaced similarly to parS1 sites. Download
ParB1 interaction with promoter DNA in vitro and in vivo. (A) ParB1 binding to parS1 and promoter regions of VC0067, VC0069, VC0076, and ompU genes was determined by EMSA. DNA fragments were used at 10 nM each. (B) Promoter activities of genes VC0067, VC0069, and VC0076 in V. cholerae WT (white bars) and ∆parB1 (black bars) cells growing exponentially in LB broth. The activities were determined by lacZ fusion assay. Average β-galactosidase activities from three experiments are shown. Download
Interactions among OmpU, ParA1, ParB1, and ToxR determined by bacterial two-hybrid assay. E. coli BTH101 cells harboring pairs of bait and prey plasmids were cultured at 30°C for 2 days on LB agar plates containing IPTG and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Download
Transcription of gidAB-parAB1 region. The transcription of gidA, gidA-gidB, gidB-parA1, parA1-parB1, parB2, gidA-parA1, gidA-parB1, and gidB-parB1 was determined by RT-PCR. Arrows numbered 1 to 10 represent primers BJH183 to -192, respectively. Their sequences are in Table S5 in the supplemental material. The same analysis was performed without template (total RNA) or RT as negative controls. The 2-log DNA ladder was used as a size marker. The results show that parAB1 genes can be transcribed from promoters upstream of parAB1 as indicated by, e.g., the primer pairs 1 and 10. Download
Files of ChIP-chip data with P values from WT, ∆parA1, and ∆parB1 cells.
Files of transcriptome data with P values from ∆parA1, ∆parB1, and ∆parAB1 cells with respect to the WT.
Candidate proteins interacting with ParA1 or ParB1 as determined by bacterial and yeast two-hybrid analyses.
Bacterial strains and plasmids used in this study.
Primers used in this study.
ACKNOWLEDGMENTS
We are grateful to Lyn Sue Kahng and Michael Berman for strains, Jemima Barrowman for editing the manuscript, and Grazyna Jagura-Burdzy, Ole Skovgaard, and Michael Yarmolinsky for thoughtful comments on the manuscript.
This work was supported by the Intramural Research Program, Center for Cancer Research, the National Cancer Institute, and National Institutes of Health grant GM079710 to S.V.R.
Footnotes
Citation Baek JH, Rajagopala SV, Chattoraj DK. 2014. Chromosome segregation proteins of Vibrio cholerae as transcription regulators. mBio 5(3):e01061-14. doi:10.1128/mBio.01061-14.
REFERENCES
- 1. Gerdes K, Møller-Jensen J, Bugge Jensen R. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455–466 [DOI] [PubMed] [Google Scholar]
- 2. Wang X, Montero Llopis P, Rudner DZ. 2013. Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 14:191–203. 10.1038/nrg3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hao JJ, Yarmolinsky M. 2002. Effects of the P1 plasmid centromere on expression of P1 partition genes. J. Bacteriol. 184:4857–4867. 10.1128/JB.184.17.4857-4867.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim HJ, Calcutt MJ, Schmidt FJ, Chater KF. 2000. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J. Bacteriol. 182:1313–1320. 10.1128/JB.182.5.1313-1320.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quisel JD, Grossman AD. 2000. Control of sporulation gene expression in Bacillus subtilis by the chromosome partitioning proteins Soj (ParA) and Spo0J (ParB). J. Bacteriol. 182:3446–3451. 10.1128/JB.182.12.3446-3451.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murray H, Errington J. 2008. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135:74–84. 10.1016/j.cell.2008.07.044 [DOI] [PubMed] [Google Scholar]
- 7. Burkholder WF, Kurtser I, Grossman AD. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104:269–279. 10.1016/S0092-8674(01)00211-2 [DOI] [PubMed] [Google Scholar]
- 8. Rodionov O, Lobocka M, Yarmolinsky M. 1999. Silencing of genes flanking the P1 plasmid centromere. Science 283:546–549. 10.1126/science.283.5401.546 [DOI] [PubMed] [Google Scholar]
- 9. Breier AM, Grossman AD. 2007. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol. Microbiol. 64:703–718. 10.1111/j.1365-2958.2007.05690.x [DOI] [PubMed] [Google Scholar]
- 10. Bartosik AA, Lasocki K, Mierzejewska J, Thomas CM, Jagura-Burdzy G. 2004. ParB of Pseudomonas aeruginosa: interactions with its partner ParA and its target parS and specific effects on bacterial growth. J. Bacteriol. 186:6983–6998. 10.1128/JB.186.20.6983-6998.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kusiak M, Gapczynska A, Plochocka D, Thomas CM, Jagura-Burdzy G. 2011. Binding and spreading of ParB on DNA determine its biological function in Pseudomonas aeruginosa. J. Bacteriol. 193:3342–3355. 10.1128/JB.00328-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Venkova-Canova T, Baek JH, Fitzgerald PC, Blokesch M, Chattoraj DK. 2013. Evidence for two different regulatory mechanisms linking replication and segregation of Vibrio cholerae chromosome II. PLoS Genet. 9:e1003579. 10.1371/journal.pgen.1003579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamaichi Y, Gerding MA, Davis BM, Waldor MK. 2011. Regulatory cross-talk links Vibrio cholerae chromosome II replication and segregation. PLoS Genet. 7:e1002189. 10.1371/journal.pgen.1002189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sullivan NL, Marquis KA, Rudner DZ. 2009. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137:697–707. 10.1016/j.cell.2009.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gruber S, Errington J. 2009. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137:685–696. 10.1016/j.cell.2009.02.035 [DOI] [PubMed] [Google Scholar]
- 16. Minnen A, Attaiech L, Thon M, Gruber S, Veening JW. 2011. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol. Microbiol. 81:676–688. 10.1111/j.1365-2958.2011.07722.x [DOI] [PubMed] [Google Scholar]
- 17. Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134:945–955. 10.1016/j.cell.2008.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell 134:956–968. 10.1016/j.cell.2008.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jakimowicz D, Brzostek A, Rumijowska-Galewicz A, Zydek P, Dołzbłasz A, Smulczyk-Krawczyszyn A, Zimniak T, Wojtasz L, Zawilak-Pawlik A, Kois A, Dziadek J, Zakrzewska-Czerwinska J. 2007. Characterization of the mycobacterial chromosome segregation protein ParB and identification of its target in Mycobacterium smegmatis. Microbiology 153:4050–4060. 10.1099/mic.0.2007/011619-0 [DOI] [PubMed] [Google Scholar]
- 20. Lasocki K, Bartosik AA, Mierzejewska J, Thomas CM, Jagura-Burdzy G. 2007. Deletion of the parA (soj) homologue in Pseudomonas aeruginosa causes ParB instability and affects growth rate, chromosome segregation, and motility. J. Bacteriol. 189:5762–5772. 10.1128/JB.00371-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Godfrin-Estevenon AM, Pasta F, Lane D. 2002. The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Mol. Microbiol. 43:39–49. 10.1046/j.1365-2958.2002.02735.x [DOI] [PubMed] [Google Scholar]
- 22. Jakimowicz D, Zydek P, Kois A, Zakrzewska-Czerwinska J, Chater KF. 2007. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol. Microbiol. 65:625–641. 10.1111/j.1365-2958.2007.05815.x [DOI] [PubMed] [Google Scholar]
- 23. Yamaichi Y, Fogel MA, McLeod SM, Hui MP, Waldor MK. 2007. Distinct centromere-like parS sites on the two chromosomes of Vibrio spp. J. Bacteriol. 189:5314–5324. 10.1128/JB.00416-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fogel MA, Waldor MK. 2006. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 20:3269–3282. 10.1101/gad.1496506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamaichi Y, Fogel MA, Waldor MK. 2007. par genes and the pathology of chromosome loss in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 104:630–635. 10.1073/pnas.0608341104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baek JH, Chattoraj DK. 2014. Chromosome I controls chromosome II replication in Vibrio cholerae. PLoS Genet. 10:e1004184. 10.1371/journal.pgen.1004184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lobocka M, Yarmolinsky M. 1996. P1 plasmid partition: a mutational analysis of ParB. J. Mol. Biol. 259:366–382. 10.1006/jmbi.1996.0326 [DOI] [PubMed] [Google Scholar]
- 28. Kadoya R, Baek JH, Sarker A, Chattoraj DK. 2011. Participation of chromosome segregation protein ParAI of Vibrio cholerae in chromosome replication. J. Bacteriol. 193:1504–1514. 10.1128/JB.01067-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Crawford JA, Kaper JB, DiRita VJ. 1998. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol. Microbiol. 29:235–246. 10.1046/j.1365-2958.1998.00925.x [DOI] [PubMed] [Google Scholar]
- 30. Yamaichi Y, Bruckner R, Ringgaard S, Möll A, Cameron DE, Briegel A, Jensen GJ, Davis BM, Waldor MK. 2012. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev. 26:2348–2360. 10.1101/gad.199869.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen YC, Rajagopala SV, Stellberger T, Uetz P. 2010. Exhaustive benchmarking of the yeast two-hybrid system. Nat. Methods 7:667–668. 10.1038/nmeth0910-667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rajagopala SV, Hughes KT, Uetz P. 2009. Benchmarking yeast two-hybrid systems using the interactions of bacterial motility proteins. Proteomics 9:5296–5302. 10.1002/pmic.200900282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hayama R, Marians KJ. 2010. Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:18826–18831. 10.1073/pnas.1008140107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robertson GT, Reisenauer A, Wright R, Jensen RB, Jensen A, Shapiro L, Roop RM. 2000. The Brucella abortus CcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J. Bacteriol. 182:3482–3489. 10.1128/JB.182.12.3482-3489.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mierzejewska J, Jagura-Burdzy G. 2012. Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle. Plasmid 67:1–14. 10.1016/j.plasmid.2011.08.003 [DOI] [PubMed] [Google Scholar]
- 36. Berkmen MB, Grossman AD. 2007. Subcellular positioning of the origin region of the Bacillus subtilis chromosome is independent of sequences within oriC, the site of replication initiation, and the replication initiator DnaA. Mol. Microbiol. 63:150–165. 10.1111/j.1365-2958.2006.05505.x [DOI] [PubMed] [Google Scholar]
- 37. Bartosik AA, Glabski K, Jecz P, Mikulska S, Fogtman A, Koblowska M, Jagura-Burdzy G. 2014. Transcriptional profiling of ParA and ParB mutants in actively dividing cells of an opportunistic human pathogen Pseudomonas aeruginosa. PLoS One 9:e87276. 10.1371/journal.pone.0087276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baek JH, Lee SY. 2006. Novel gene members in the Pho regulon of Escherichia coli. FEMS Microbiol. Lett. 264:104–109. 10.1111/j.1574-6968.2006.00440.x [DOI] [PubMed] [Google Scholar]
- 39. Pal D, Venkova-Canova T, Srivastava P, Chattoraj DK. 2005. Multipartite regulation of rctB, the replication initiator gene of Vibrio cholerae chromosome II. J. Bacteriol. 187:7167-7175. 10.1128/JB.187.21.7167-7175.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morgan SJ, Felek S, Gadwal S, Koropatkin NM, Perry JW, Bryson AB, Krukonis ES. 2011. The two faces of ToxR: activator of ompU, co-regulator of toxT in Vibrio cholerae. Mol. Microbiol. 81:113–128. 10.1111/j.1365-2958.2011.07681.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download
Comparison of ChIP-chip profiles of ParB1 (black, same as in Fig. 1A) and a nonspreading protein, RctB (orange). The RctB profile is shown for WT cells only in an origin-distal locus of chrII (coordinates 1024 to 1031 kb) (26) where RctB has specific binding sites spaced similarly to parS1 sites. Download
ParB1 interaction with promoter DNA in vitro and in vivo. (A) ParB1 binding to parS1 and promoter regions of VC0067, VC0069, VC0076, and ompU genes was determined by EMSA. DNA fragments were used at 10 nM each. (B) Promoter activities of genes VC0067, VC0069, and VC0076 in V. cholerae WT (white bars) and ∆parB1 (black bars) cells growing exponentially in LB broth. The activities were determined by lacZ fusion assay. Average β-galactosidase activities from three experiments are shown. Download
Interactions among OmpU, ParA1, ParB1, and ToxR determined by bacterial two-hybrid assay. E. coli BTH101 cells harboring pairs of bait and prey plasmids were cultured at 30°C for 2 days on LB agar plates containing IPTG and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Download
Transcription of gidAB-parAB1 region. The transcription of gidA, gidA-gidB, gidB-parA1, parA1-parB1, parB2, gidA-parA1, gidA-parB1, and gidB-parB1 was determined by RT-PCR. Arrows numbered 1 to 10 represent primers BJH183 to -192, respectively. Their sequences are in Table S5 in the supplemental material. The same analysis was performed without template (total RNA) or RT as negative controls. The 2-log DNA ladder was used as a size marker. The results show that parAB1 genes can be transcribed from promoters upstream of parAB1 as indicated by, e.g., the primer pairs 1 and 10. Download
Files of ChIP-chip data with P values from WT, ∆parA1, and ∆parB1 cells.
Files of transcriptome data with P values from ∆parA1, ∆parB1, and ∆parAB1 cells with respect to the WT.
Candidate proteins interacting with ParA1 or ParB1 as determined by bacterial and yeast two-hybrid analyses.
Bacterial strains and plasmids used in this study.
Primers used in this study.