ABSTRACT
Distinct lineages of avian influenza viruses (AIVs) are harbored by spatially segregated birds, yet significant surveillance gaps exist around the globe. Virtually nothing is known from the Antarctic. Using virus culture, molecular analysis, full genome sequencing, and serology of samples from Adélie penguins in Antarctica, we confirmed infection by H11N2 subtype AIVs. Their genetic segments were distinct from all known contemporary influenza viruses, including South American AIVs, suggesting spatial separation from other lineages. Only in the matrix and polymerase acidic gene phylogenies did the Antarctic sequences form a sister relationship to South American AIVs, whereas distant phylogenetic relationships were evident in all other gene segments. Interestingly, their neuraminidase genes formed a distant relationship to all avian and human influenza lineages, and the polymerase basic 1 and polymerase acidic formed a sister relationship to the equine H3N8 influenza virus lineage that emerged during 1963 and whose avian origins were previously unknown. We also estimated that each gene segment had diverged for 49 to 80 years from its most closely related sequences, highlighting a significant gap in our AIV knowledge in the region. We also show that the receptor binding properties of the H11N2 viruses are predominantly avian and that they were unable to replicate efficiently in experimentally inoculated ferrets, suggesting their continuous evolution in avian hosts. These findings add substantially to our understanding of both the ecology and the intra- and intercontinental movement of Antarctic AIVs and highlight the potential risk of an incursion of highly pathogenic AIVs into this fragile environment.
IMPORTANCE
Avian influenza viruses (AIVs) are typically maintained and spread by migratory birds, resulting in the existence of distinctly different viruses around the world. However, AIVs have not previously been detected in Antarctica. In this study, we characterized H11N2 viruses sampled from Adélie penguins from two geographically different sites in Antarctica and show that the segmented AIV genome diverged between 49 and 80 years ago from other AIVs, with several genes showing similarity and shared ancestry with H3N8 equine influenza viruses. This study provides the first insight into the ecology of AIVs in Antarctica and highlights the potential risk of an introduction of highly pathogenic AIVs into the continent.
INTRODUCTION
Wild aquatic birds such as dabbling ducks, gulls, and other shorebirds are considered the natural reservoir for avian influenza viruses (AIVs). The ecology and migratory patterns of these birds therefore have a direct effect on the global distribution and diversity of AIVs (1). The ecology of AIVs in wild birds has been well studied in many regions of the Northern Hemisphere, but considerably less is known in the Southern Hemisphere. Of the publically available AIV sequences (n = 19,784), only 5.7%, 1%, and 0.1% are from Africa, Oceania, and South America, respectively, and none have previously been described from Antarctica (GISAID, http://www.gisaid.org). Wild migratory birds play a key role in the spread of AIVs on a local, regional, and intercontinental scale via broadly established flyways (1) (see Fig. S1 in the supplemental material). As a result, spatially segregated birds of the American and Eurasian landmasses harbor distinct lineages of AIVs (1). Traditionally, AIVs have been broadly separated into North American or Eurasian lineages, although this appears to be an oversimplification and less applicable to AIVs from the Southern Hemisphere (2). Recent genetic analysis of AIVs in Australia, located at the southern end of the East Asian-Australian flyway, showed some divergence from Eurasian strains, demonstrating that, although new strains have been introduced frequently from Europe and Asia, there is a high degree of endemic evolution that has produced a group of viruses that are genetically distinct from those circulating in Asia (2–4). Similar evolutionary divergence has also been observed in some South American AIVs, where internal gene segments form unique “South American-like” clades, although the hemagglutinin (HA) and/or neuraminidase (NA) gene segments are often “North American-like” (5–8). It is therefore possible that evolutionary divergence of AIVs may increase with further advancement south and that spatially segregated regions and continents such as Southern Patagonia and Antarctica may act as evolutionary “sinks” harboring highly diverged AIVs.
The major migratory flyways are not thought to extend to Antarctica, although each spring, over 100 million birds breed around the rocky Antarctic coastline and offshore islands (9). These include birds such as the Arctic tern (Sterna paradisaea) (10) and south polar skua (Stercorarius maccormicki) (11) that conduct transhemispheric migrations to Antarctica and thereby have the potential to facilitate movement of avian influenza viruses into and out of Antarctica. A small number of earlier AIV studies in Antarctica have detected influenza A antibodies in penguins and other birds (12–16), but none have detected or isolated any influenza viruses. The detection and subsequent analysis of AIVs from Antarctica would provide insight into the frequency and likely routes of AIV movement into and within the continent and help assess the potential risk of an incursion of highly pathogenic AIVs from neighboring regions.
RESULTS
Identification of influenza virus in penguins.
We took combined cloacal/tracheal swabs from 301 Adélie penguins (Pygoscelis adeliae) (adults and chicks), and blood from 270 of those, from two locations on the Antarctic Peninsula during January and February 2013 (Table 1 and Fig. 1). Eight samples (2.7%) (with cycle threshold [CT] values ranging from 25.4 to 36.9) were found to contain influenza virus RNA following a real-time reverse transcription (RT)-PCR assay for the influenza matrix gene. AIV was confirmed by sequence and BLAST analysis of at least one gene sequence (hemagglutinin [HA], nonstructural [NS], or matrix [M]) (Table 1). Six AIV-positive samples were from adult Adélie penguins and two from Adélie chicks, with all samples except for one collected from Rada Covadonga (Table 1). All eight influenza viruses, including the single virus from Admiralty Bay, had a high degree of homology (>99.7% nucleotide sequence identity) between respective genes. Following inoculation of these viruses into embryonated hens’ eggs, four viruses were successfully cultured (all from specimens with a CT value of <33 based on the matrix gene real-time RT-PCR assay). Full genome analysis of the four isolated viruses showed that they were all H11N2 influenza A viruses (GenBank sequence accession numbers KJ729348 to KJ729379). These four viruses were designated A/Adélie penguin/Antarctica/178/2013, A/Adélie penguin/Antarctica/184/2013, A/Adélie penguin/Antarctica/226/2013, A/Adélie penguin/Antarctica/270/2013 (abbreviated as A/peng/Ant/178/13, A/peng/Ant/184/13, A/peng/Ant/226/13, and A/peng/Ant/270/13, respectively).
TABLE 1 .
Prevalence of influenza virus and influenza antibodies in Adélie penguins
| Location | Age | Cloacal/tracheal swabs |
Serum samples |
||||
|---|---|---|---|---|---|---|---|
| No. of penguins sampled |
No. (%) of samples with AIV RNA detecteda |
No. of samples isolated in eggsb |
No. of penguins sampled |
No. (%) of samples with IAV antibodies detectedc |
No. (%) of samples with A(H11N2) antibodies detectedd |
||
| Admiralty Bay | Adult | 110 | 0 | 0 | 96 | 16 (16.7) | 0 |
| Chick | 40 | 1 (2.5) | 0 | 35 | 0 | 0 | |
| Rada Covadonga | Adult | 111 | 6 (5.4) | 3e | 99 | 27 (27.3) | 2 (1.8) |
| Chick | 40 | 1 (2.5) | 1f | 40 | 0 | 0 | |
| Total | 301 | 8 (2.7) | 4 | 270 | 43 (15.9) | 2 (0.7) | |
No. of samples where the HA, NS, or M gene sequence was obtained and confirmed to be influenza A virus by BLAST analysis.
Egg isolation was attempted only for those samples with a CT value of <38 (n = 14).
Determined using an anti-NP ELISA assay.
Determined using an HI assay with the H11N2 antigen A/peng/Ant/270/13.
Viruses were designated A/Adélie Penguin/Antarctica/226/2013, A/Adélie Penguin/Antarctica/184/2013, A/Adélie Penguin/Antarctica/178/2013.
Virus was designated A/Adélie Penguin/Antarctica/270/2013.
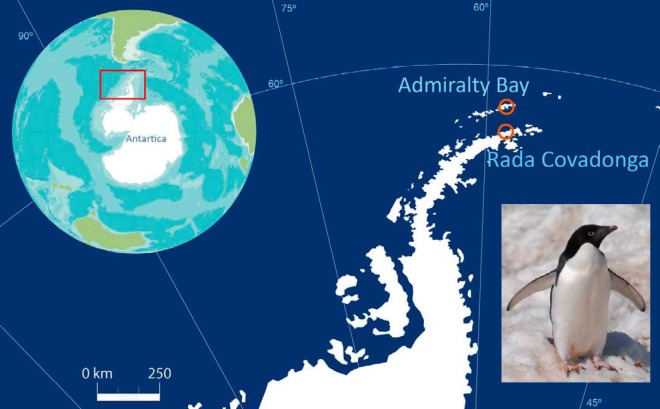
FIG 1 .
Map of sampling sites in Antarctica. Adélie penguins were sampled from two locations in Antarctica: Admiralty Bay, King George Island; and Kopaitik Island, Rada Covadonga, Antarctic Peninsula. Inset: An Adélie penguin. The map is courtesy of Marco Villarroel, reproduced with permission.
Evolutionary relationship and divergence time estimates.
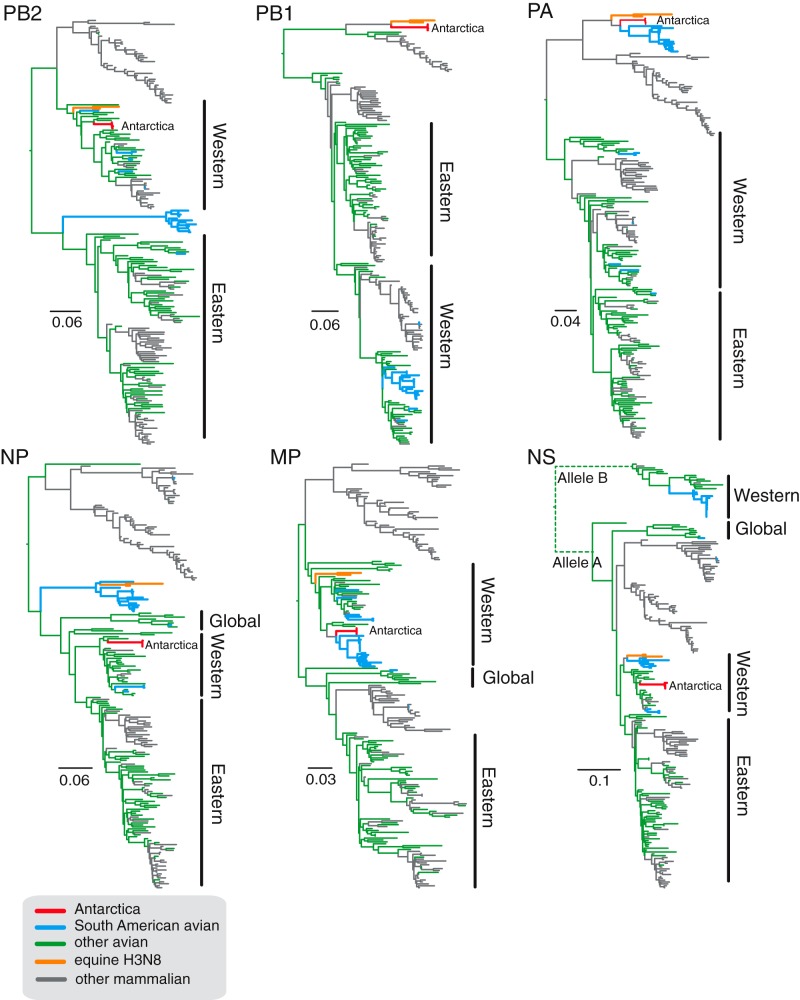
Full genome sequences of the four isolated viruses were compared with all available animal and human influenza virus sequences in public databases to determine the phylogenetic relationships and the evolutionary origin of each gene segment (Fig. 2 and 3). Detailed phylogenetic trees with strain designations are shown in Fig. S2 and S3 in the supplemental material. Unexpectedly, each AIV gene segment from the Adélie penguin viruses was highly different from that of contemporary AIVs circulating either in the Northern or Southern hemispheres (see Fig. S2 and S3 and Table S1 in the supplemental material). Four of the gene segments (HA, PB2, nucleoprotein [NP], and NS) were most closely related to North American avian lineage viruses from the 1960s to 1980s (90 to 96% identity) (Fig. 2 and 3; see also Table S1), while the N2 neuraminidase gene formed a basal position to the North American avian influenza lineage, indicating an early divergence from the known AIV gene pool (Fig. 2; see also Fig. S2). The MP gene of the penguin virus formed a close relationship with North American viruses from 1974 to 2001 but appears to be the closest ancestor to a large number of South American AIVs from Chile, Argentina, and Brazil (Fig. 3; see also Fig. S3E). The PA segments of the penguin AIVs also have a close ancestral relationship with South American viruses, which together appear on their own phylogenetic clade sharing ancestry with equine influenza H3N8 viruses from the 1960s (Fig. 3; see also Fig. S3C). Interestingly, the PB1 segments of the penguin AIVs are also most closely related to equine H3N8 viruses, with significant genetic distance from North American AIVs (Fig. 3; see also Fig. S3B).
FIG 2 .
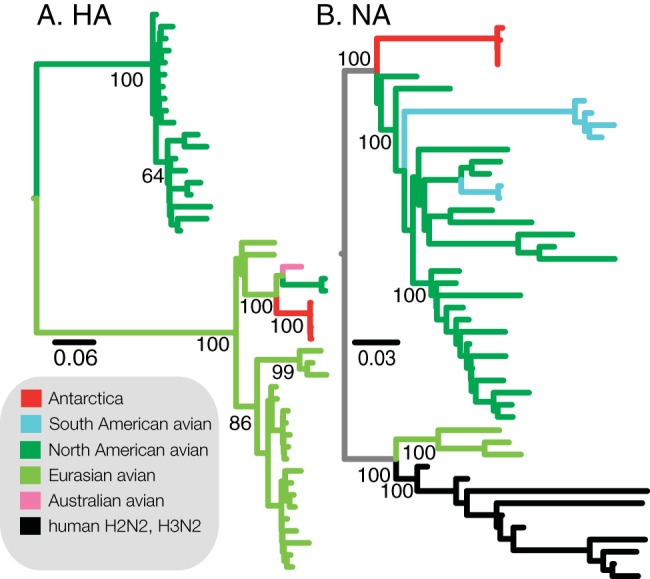
Phylogenetic tree of H11 (A) and N2 (B) sequences. Sequences from the viruses isolated from Adélie penguins in Antarctica are highly diverged from other circulating AIVs. The HA phylogenetic tree was constructed using maximum likelihood methodology and shows ancestry with Eurasian avian H11 viruses, while the NA sequence is highly diverged from all other clades but with shared ancestry with North American N2 viruses. Detailed phylogenetic trees with virus designations can be found in Fig. S2 in the supplemental material.
FIG 3 .
Phylogenetic trees of the internal gene sequences of avian, equine, and mammalian influenza A viruses. Major avian influenza clades are broadly classified into Western, Eastern, or global based on the geographic location of sample collection. Phylogenetic trees were constructed using maximum likelihood methodology. AIV sequences from the penguins are marked as “Antarctica,” with branches colored red. Blue branches indicate South American avian influenza sequences, green branches indicate avian influenza viruses from other regions, and orange branches indicate equine influenza viruses. Detailed phylogenetic trees with virus designations can be found in Fig. S3 in the supplemental material.
Using a molecular clock analysis that incorporates the evolutionary rate of each AIV gene segment, we estimated the time of the most recent common ancestor (TMRCA) of the penguin virus and its most closely related sequence. For the HA, NA, PB2, NP, PA, and NS genes, the TMRCA was between 1957 and 1964, whereas for the MP and PB1 genes, the estimated TMRCA was even earlier at 1933 and 1936, respectively (Table 2). Therefore, there has been an estimated 49 to 80 years of unsampled diversity and evolution leading to these previously unknown influenza viruses (Table 2).
TABLE 2 .
Summary of evolutionary relationships of each gene segment of influenza viruses isolated from Adélie penguins in Antarctica during 2013
| Gene | % identitya | Lineageb | Mean time of unsampled diversity in yrs (BCI)c |
|---|---|---|---|
| HA gene | 90 | North American avian | 50 (43-58) |
| NA gene | 90 | North American avian/ new lineage | 54 (50-60) |
| PB2 gene | 96 | North American avian | 50 (41-58) |
| PB1 gene | 89 | Equine H3N8 | 77 (61-100) |
| PA gene | 91 | Equine H3N8, South American avian | 56 (50-69) |
| NP gene | 96 | North American avian | 49 (41-58) |
| MP gene | 97 | North American avian, South American avian | 80 (56-112) |
| NS gene | 95 | North American avian | 56 (45-69) |
Percent identity was estimated using BLAST in NCBI influenza virus resources.
Virus lineages were determined using a maximum likelihood phylogeny generated for each gene segment of all influenza virus sequences that were >96% similar. Phylogenies with virus names and accession numbers are presented in Fig. S1A to H in the supplemental material.
Time to the most recent common ancestor (in years) of influenza viruses isolated from Adélie penguins in Antarctica during 2013 and the most closely related virus sequence; represents the mean time of unsampled diversity of this virus (orange bars in Fig. S2 and S3 in the supplemental material). Bayesian confidence intervals (BCI) for the time of unsampled diversity are shown in parentheses.
Molecular markers of host specificity and pathogenicity.
The HA of the Adélie penguin viruses did not contain the multibasic cleavage site observed in highly pathogenic H5 or H7 AIVs, nor did the genome contain other major mammalian adaptations, such as the PB2 E627K substitution, which is known to alter host range (17) and virulence (18). Infection of embryonated eggs was not pathogenic to the embryo. The receptor binding pocket residues have only one human-like amino acid at position A137 (H3 numbering), with the rest being avian-like with E190, G225, Q226, and G228, suggesting an α2,3-linked sialic acid receptor binding preference (see Fig. S4 in the supplemental material). Compared with the distantly related H11N9 virus isolated from a wild bird in Australia in 2004, the A/peng/Ant/178/13 HA sequence contained amino acid substitutions at I155T (at the base of the receptor binding site and within contact range to the sialic acid) and A186S and T187S (at the entry of the host receptor pocket), which may alter preferences for different variants of 2,3-linked glycans (Fig. 4). Furthermore, A/peng/Ant/178/13 had a deletion of the A134 residue compared to other wild bird H11 viruses and an insertion of G133 compared to H13 and H16 viruses with known crystal structures (see Fig. S4), which could alter receptor binding slightly due to relative shifts of the adjacent 160 loop. Even though their PB1 and PA genes were phylogenetically related to those of the equine viruses, the Adélie penguin viruses contained typical avian-like amino acid signatures (N375 [PB1], D55 and V100 [PA]), which differ from the equine-like signatures that presumably arose following adaptation to viral replication in horses (19). The viruses contained none of the mutations known to confer resistance to either the adamantane or neuraminidase inhibitor classes of influenza antiviral drugs.
FIG 4 .
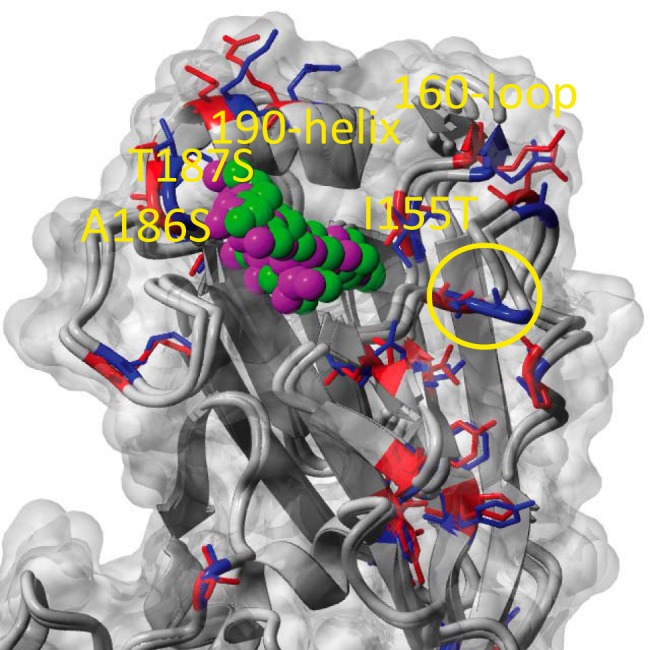
HA structural homology modeling. The structures of penguin and a related sandpiper H11 virus were modeled based on the HA crystal structures of H13 (PDB ID number 4kps), H16 (PDB ID number 4f23), and H2 (PDB ID number 2wr3) in complex with an α2,3Gal-linked sialic acid analogue (avian-like receptor). The receptor binding pocket residues for both H11s are typically avian, with E190, G225, Q226, and G228 (H3 numbering), suggesting α2,3-linked SA receptor (avian-like) binding (purple and green balls). Comparison of the receptor binding pocket in the A/peng/Ant/178/13 with that in A/sharp tailed sandpiper/Australia/10/2004 reveals differences at I155, A186, and T187 (red or blue sticks for penguin or sandpiper H11, respectively), which may influence preferences for different variants of 2,3-linked glycans. Several major antigenic sites also differ among the H11s; for example, epitope Sb (at the top of the 190 helix) and upper parts of Sa (the area around the 160 loop). Another unique structural feature distinguishing the penguin H11 from the nearest known H11 sequences and the H13 and H16 structures are different indel patterns around G133 (yellow circle).
Identification of influenza antibodies in penguins.
Sera from 270 of the 301 penguins, including the eight penguins that were AIV positive, were tested by enzyme-linked immunosorbent assay (ELISA) for cross-reactive NP antibodies. As the NP is highly conserved across all influenza A viruses, the assay is designed to detect any previous influenza A infection, regardless of subtype. In comparison, a hemagglutination inhibition (HI) assay, which detects antibodies to the highly variable HA protein, was used to detect previous infection with either the penguin H11N2 viruses or a distantly related H11N9 virus isolated from a wild bird in Australia in 2004. A total of 15.9% of penguins (43/270) were shown to have influenza A NP antibodies by ELISA. All NP antibody-positive penguins were adults, with a greater proportion from Rada Covadonga than from Admiralty Bay (27.3% compared to 16.7%) (Table 1). Only 2/270 (0.7%) penguins had positive HI titers against the penguin H11N2 virus (Table 1). One of the H11N2 antibody-positive serum samples (with a titer of 40) was from an H11N2 PCR- and isolation-positive penguin that yielded A/peng/Ant/226/13, while the other positive serum sample (with a titer of 80 to 160) was from a penguin that was RT-PCR negative for influenza virus. Neither these sera nor the remaining 268 sera inhibited the unrelated H11N9 wild bird virus from Australia. Overall, 5 of the 8 influenza virus-positive penguins had high NP antibody titers, but only one had specific H11 antibodies. This suggests that the detected NP antibodies may have been induced by prior infection(s), as the remaining three influenza virus-positive penguins (which included samples with the highest RNA copy number and two that were culture positive) were NP antibody negative.
Infectivity in ferrets and receptor binding.
To assess the potential risk of A/peng/Ant/270/13 infection in mammals, we administered the virus to ferrets via intranasal (i.n.) or intratracheal (i.t.) routes and compared infectivity with an Australian wild bird H11N9 virus and a contemporary human H3N2 influenza virus. Intranasal administration of A/peng/Ant/270/13 resulted in little or no viral replication, with detection of virus only on day 2 postinfection (at a low titer of 1 × 101 50% egg infectious doses [EID50]/ml) (see Fig. S5 in the supplemental material). The ferrets showed no clinical symptoms or significant weight loss and had no detectable H11 serum antibodies by HI assay on day 14 or at 21 days following an intramuscular boost on day 14. In comparison, i.n. delivery of an Australian wild bird H11 virus and the human H3N2 virus yielded productive infections (shedding virus from days 2 to 5, with peak viral titers of 1 × 104.5 and 1 × 105.5 EID50/ml, respectively) (see Fig. S5) and homologous HI antibody titers of 40 and 2,560, respectively, on day 14 postinfection. Following i.t. delivery of A/peng/Ant/270/13, a low virus titer (1 × 101 EID50/ml) was detected in one lung lobe but not in the other lobe at day 4, but again no H11 antibodies were detected on day 14. No viral titer was detected in either lung lobe following i.t. administration of ferrets with either the Australian wild bird H11 virus or a human H3N2 influenza virus (see Fig. S5). Using turkey erythrocytes enzymatically modified to contain either terminal SAα2,3Gal (avian-like receptors) or SAα2,6Gal (human-like receptors) linkages on cell surface oligosaccharides, we further demonstrated that the A/peng/Ant/270/13 primarily bound avian-like receptors, with only minor binding to α2,6-linked SA receptors (see Table S2). We also showed that, compared to the human H3N2 virus, the viral infectivity of A/peng/Ant/270/13 in vitro was considerably reduced at 33°C compared to that at 37°C, indicating a lack of adaptation to replication in the cooler mammalian upper airways (see Fig. S6).
DISCUSSION
Here, we describe the detection of AIVs in Antarctica, providing the first insights into their ecology in this isolated continent. Previous studies of birds and mammals in Antarctica have failed to detect either live AIV or influenza virus RNA. Although AIV antibodies have been detected in penguins at frequencies ranging from 11.8 to 12.5% in Gentoo penguins (Pygoscelis papua) (12, 15), from 3.7 to 58.4% in Adélie penguins (12, 14, 16), and 10.8% in chinstrap penguins (Pygoscelis antarcticus) (12), several other studies have failed to detect any influenza antibodies in these or other penguin species in Antarctica (14, 16, 20, 21), suggesting that influenza infections may be absent from penguin colonies in Antarctica for long periods of time. The detection of H11N2 virus in 2.7% of Adélie penguins sampled in this study, along with the substantially lower prevalence of penguins with detectable H11 antibody titers (0.7%), suggests either that we sampled during the acute phase of infection before antibodies to H11 had developed or that the H11N2 infections caused only a weak antibody response. The intensity and longevity of antibody responses in penguins infected with AIVs are currently unknown, highlighting the need for longitudinal repeat sampling studies of the same penguin colonies with tagged individuals to enable interpretation of serology data in these species. Although H11 AIVs have never been reported in wild birds from South America, they have been detected in migratory shorebirds in North America (where they were the second most frequently detected HA subtype) (22), as well as in Australia (23) and New Zealand (24). Additionally, H11 AIVs have been commonly detected in ducks in many other regions of the world (25, 26).
It is important to understand the frequency and magnitude of inter- and intracontinental movement of Antarctic AIVs. Long-distance migrants such as skuas and giant petrels that traverse large distances both within Antarctica and between continents have been shown to have a high frequency of AIV antibodies (skuas, 1.0 to 11.1% [12–14]; giant petrels, 51.5 to 58.8% [12]). These birds have very close interactions with penguins, often nesting alongside their colonies and attacking penguin chicks, providing a likely route for AIV transmission. The yellow-billed pintail (Anas georgica) is a species of duck that, while most commonly observed in South America, has occasionally been recorded on the Antarctic peninsula, making it the most southerly recorded waterfowl species (27). Given the high rates of AIV carriage in waterfowl compared to those in other orders of birds, it is possible that this species plays a role in AIV ecology in Patagonia, the South Shetland Islands, and the Antarctic Peninsula. The return of migratory birds to Antarctica each year between October and December (start of the austral summer period) coincides with penguins returning to colonies to nest and rear chicks. Because of the high population density of birds, summer is the most likely period for AIV transmission and infection in Antarctica. The large amount of penguin feces in colonies during summer, which in some cases is so significant it can be observed on satellite images (28), presumably facilitates AIV transmission by the fecal-oral route (29). Although the relative susceptibility of different penguin species to AIVs is unknown, we did take swabs from a small number of gentoo (n = 74) and chinstrap (n = 18) penguins in Rada Covadonga, but none were AIV positive. The different species typically form exclusive separate colonies, which may explain the lack of cross-infection between the three penguin species at the time of sampling in Rada Covadonga.
The detection of this highly divergent H11N2 AIV suggests that its existence in Antarctica is not transient. Although two gene segments share common ancestry with South American AIVs, the other remaining six genes shared ancestry with either North American avian or equine viruses. Antarctica therefore may be the most extreme example of an AIV “evolutionary sink,” where viruses are seeded into the continent on an infrequent basis and then become established, persisting as a local reservoir and evolving independently from AIVs in other areas of the globe due to a lack of interconnectedness. Future studies to understand which species or groups of animals are responsible for maintaining AIVs in Antarctica are needed. Based on all previous studies in Antarctica, the overall AIV antibody prevalence in penguins (3.3%; 82/2,500) (12, 14–16, 20, 21, 30, 31) is considerably lower than it is in skuas and giant petrels (13.3%; 47/353) (12–14), suggesting that the latter species may be more likely to maintain and transmit AIVs in Antarctica. An alternative hypothesis is that AIVs circulate between birds only in summer and then become cryopreserved in ice over winter (32), after which birds are again infected once the ice melts in the following summer. In addition, a number of Antarctic marine mammals, such as seals and whales, may also have the potential to become infected and spread influenza viruses given the previous reports of their susceptibility to influenza infection (33, 34). Although three reported studies of seals (n = 306) in Antarctica did not detect influenza viruses or antibodies (14, 35, 36), these animals appear to be susceptible to both avian and human influenza viruses, as their respiratory tracts contain both α2,3- and α2,6-linked sialic acid receptors (33). Seals have been infected with both AIV (33) and human influenza viruses (37), as well as infecting humans with an H7N7 virus following close experimental contact (38, 39). Whether these animals could act as a “mixing vessel” for avian or human influenza viruses in a natural environment in the same manner as pigs is unknown, although the bidirectional transmission of influenza between seals and humans may be a potential risk for both local wildlife and public health with increasing human activity in Antarctica. The H11N2 Antarctica AIV A/peng/Ant/270/13 appeared to be less infectious in ferrets than has been observed for other AIVs from wild birds. AIVs such as H6N1, H1N9, H9N2, H6N5, and H11N9 (used in this study) have readily infected ferrets and in many cases been able to successfully transmit between cohoused naive ferrets (40–42). Further work is needed to understand the key residues that may be responsible for these differences in replication.
While it has previously been postulated that AIV infection caused mortality of Adélie penguin chicks in Antarctica (16), we did not notice any clinical symptoms or below-average weight in the H11N2-infected penguins in this study. Given that highly pathogenic AIVs have been detected in both South America (43) and Australia (44), it is not inconceivable that such viruses could be transferred to the Antarctic continent by migratory birds, potentially resulting in catastrophic mass mortality, such as that seen with other pathogens such as avian cholera (Pasteurella multocida) (45, 46). This is the first insight into AIV ecology in Antarctica and provides further evidence, in addition to some South American studies (8, 47, 48), of an avian ancestry to H3N8 equine influenza viruses that emerged and spread globally among horses since 1963 (49). As such, this study and the recent detection of novel influenza A viruses in fruit bats from Central and South America (50, 51) fill important gaps in our understanding of the global movement of AIVs, the reservoir species that may maintain them, and their potential impact on animal and human health.
MATERIALS AND METHODS
Sampling locations and times.
Adélie penguins were sampled from two locations in Antarctica (Fig. 1): Admiralty Bay, King George Island (62°9′35″S, 58°28′17″W), 1 km east of Arctowski Polish Base, between 14 January and 31 January 2013; and Kopaitik Island, Rada Covadonga, 1 km west of Base General Bernardo O’Higgins Riquelme, Antarctic Peninsula (63°19′15″S, 57°53′55″W), between 1 February and 15 February 2013.
Sample collection from penguins.
Separate cloacal and tracheal swabs were taken from each Adélie penguin and then placed in the same tube containing viral transport medium (brain heart infusion [BHI] broth-based medium [Oxoid; catalog number CM1135] with 0.3 mg/ml penicillin, 5 mg/ml streptomycin, 0.1 mg/ml gentamicin, and 2.5 µg/ml amphotericin B). Swab specimens were kept on ice for up to 4 h before being frozen at −80°C. Blood was collected from penguins either from wing or foot veins and put into serum clot-activating tubes, centrifuged, and serum stored at either −80°C or −20°C. Approvals to conduct sampling from penguins in Antarctica were provided by the Universidad de Concepción, Facultad de Ciencias Veterinarias, Chillán, Chile (application number CE-3-2010), and Instituto Antártico Chileno, Chile (application number 03/2013).
RT-PCR and sequence analysis.
RNA was isolated from cloacal/tracheal swabs using the QIAGEN QIAextractor system. All samples were tested by a one-step RT‑PCR assay targeting the influenza matrix genome segment (52), using a SensiFast Probe Lo-ROX one-step kit (Bioline) on an Applied Biosystems 7500 Fast real-time PCR system (Life Technologies). Samples with a cycle threshold (CT) value of <42 cycles or cultured influenza viruses were analyzed further by conventional RT-PCR using universal primers targeting each of the eight influenza segments (53). Amplified RT-PCR products were sequenced using standard techniques on an Applied Biosystems 3500XL genetic analyzer.
Virus isolation.
Virus isolation was attempted from RT-PCR-positive samples by inoculating the swab specimen (diluted 1:1 with phosphate-buffered saline [PBS] containing 1% neomycin-polymyxin solution [bioCSL]) into the allantoic cavity of 11-day-old embryonated hens’ eggs. Allantoic fluid was harvested after 3 days, and influenza virus was detected by hemagglutination with turkey erythrocytes. The EID50/ml of each isolate was determined by performing log10 dilutions in PBS containing 1% neomycin-polymyxin solution and infecting the allantoic cavity of 11-day-old embryonated hens’ eggs. Calculation of infectious titers was performed according to the Reed and Muench method (54).
Serological analysis.
To test serum samples for broadly reactive influenza NP antibodies, a competitive ELISA with plates coated with the NP from A/turkey/Ontario/6213/66 (H5N1) was used as described by Selleck and Kirkland (55). Serum was considered antibody positive if the sample inhibited >60% of the monoclonal antibody (MAb) binding. To test serum samples for specific HA antibodies, a hemagglutination inhibition (HI) assay using turkey erythrocytes was used following previously described methods (56).
Dataset preparation and preliminary phylogenetic analysis.
To elucidate the evolutionary relationships of viruses isolated in this study, we first carried out a provisional phylogenetic analysis for the hemagglutinin and neuraminidase genes, including all available H11 and N2 subtype influenza gene sequences, and for the internal gene datasets, including viruses of all subtypes, using the maximum likelihood method with a general time reversible (GTR) nucleotide substitution model in FastTree 2 (57). The purpose of these large-scale phylogenetic analyses was to identify broad relationships between the new sequences and established influenza A viruses from multiple hosts. These analyses indicated that the Adélie penguin viruses were distantly related to previously detected avian influenza virus and their derived lineages; hence, for all subsequent analyses, the dataset sizes were reduced by identifying representative sequences at 96% identity using the software CD-HIT (58). Due to the geographical proximity of Antarctica to South America, sequences of all publicly available avian isolates from South America were included in our final datasets.
Maximum likelihood and Bayesian relaxed-molecular clock analyses.
For the final datasets, maximum likelihood phylogenies were generated using the GTR model with gamma rate heterogeneity, using RaXML (59). Statistical supports were estimated using 500 maximum likelihood bootstrap replicates. To estimate divergence times, we applied the log-normal relaxed-clock Bayesian Markov chain Monte Carlo method with an SRD06 codon substitution model (60), as implemented in BEAST version 1.7.5 (61). For each dataset, multiple analyses were conducted for 50 million generations, sampling every 5,000 generations, resulting in 10,000 trees. After convergence was ensured, the final 90% of trees were summarized using TreeAnnotator (61) and produced using Fig tree (http://tree.bio.ed.ac.uk/software/figtree/).
HA structural homology modeling.
Due to the lack of a published H11 crystal structure, numerous models were made for the H11 viruses based on the crystal structure of the most closely related HAs available in the RCSB protein databank (http://www.rcsb.org). The H11 structures were modeled based on the HA crystal structures of H13 (PDB ID number 4kps), H16 (PDB ID number 4f23), and H2 (PDB ID number 2wr3) in complex with an α2,3Gal-linked sialic acid analogue (avian-like receptor) using MODELLER (62) and YASARA (63).
Ferret infectivity.
Ferrets 6 to 18 months old were administered 1 × 107 EID50/ml egg-propagated virus via the i.n. or i.t. routes. Of two ferrets administered virus via the i.n. route, one was nasal washed on days 2, 3, 4, and 5 before being bled and euthanized on day 14, and the other was bled and given a subsequent intramuscular boost with inactivated virus on day 14 and then bled again and euthanized on day 21. Of two ferrets administered virus via the i.t. route, one was nasal washed on days 2, 3, 4, and 5 before being bled and euthanized on day 14, and the other ferret was culled on day 4 to analyze viral titers in lungs. Weights, temperatures, and clinical signs were monitored throughout the experiment.
Receptor binding assay.
The receptor specificity of viruses was assessed using methods described by Rogers and Paulson (64). Briefly, viruses were diluted to a hemagglutination titer of 64 hemagglutinating units (HAU)/25 µl in PBS. A 10% suspension of turkey erythrocytes was treated for 1 h at 37°C with 500 mU/ml Vibrio cholerae enzyme (Sigma). Erythrocytes were washed three times with PBS and resuspended to a 20% suspension. The erythrocytes were then mixed with an equal volume of recombinant beta-galactoside-alpha-2,3-sialtransferase (10 mU/µl) or recombinant beta-galactoside-alpha-2,6-sialtransferase (10 mU/µl) (Japan Tobacco, Inc.) and incubated for 3 to 4 h at 37°C. Erythrocytes were washed a further three times with PBS and resuspended to 1%. Hemagglutination assays were conducted using untreated erythrocytes, sialidase-treated erythrocytes (Asialo), and sialidase-treated erythrocytes resialylated to contain either terminal SAα2,3Gal or SAα2,6Gal linkages on cell surface oligosaccharides.
Cell culture infectivity assay at 33°C and 37°C.
Madin-Darby canine kidney (MDCK) cells (American Type Culture Collection; CCL-34) were grown to near confluence in 96-well plates (5% CO2, 37°C). Log10 virus dilutions were made in Dulbecco’s modified Eagle’s medium (SAFC Biosciences) containing sodium bicarbonate (3%) with the addition of 2 mM glutamine (SAFC Biosciences), 1% nonessential amino acids (Sigma), 0.05% NaHCO3 (Sigma), 0.02 M HEPES (Sigma), 4% penicillin and streptomycin (Sigma), and 4 µg/ml trypsin (Sigma). Cells were washed twice with PBS before infection with four replicates of 10-fold dilutions of each virus. After incubation for 5 days at 33°C or 37°C in 5% CO2, each well was scored for virus growth by cytopathic effect and hemagglutination of turkey erythrocytes, and the dose required to infect 50% of wells (50% tissue culture infectious dose [TCID50]) was determined by the Reed and Muench method (54).
Nucleotide sequence accession numbers.
Full genome sequences from the four H11N2 influenza viruses isolated in eggs have been deposited in GenBank under accession numbers KJ729348 to KJ729379.
SUPPLEMENTAL MATERIAL
The major wild bird global migratory pathways. Download
(A and B) Detailed phylogenetic trees of HA and NA sequences. Trees were drawn using maximum likelihood methodology. Red text indicates AIVs from Antarctica, blue text indicates AIVs from South America, and black text indicates AIVs from all other regions. Download
(A to F) Detailed phylogenetic trees of internal gene sequences. Trees were drawn using maximum likelihood methodology. Red text and branches indicate AIVs from Antarctica, blue text and branches indicate AIVs from South America, green text and branches indicate AIVs from all other regions, black text and branches indicate mammalian influenza viruses from all regions. Download
HA amino acid alignment of the penguin HA sequence compared to other H11 avian influenza sequences, more distant avian influenza sequences (H13, H16, and H5), and mammalian influenza sequences (H1, H2, and H3). Residues 155, 186, 187, 190, 225, 226, and 228, which are associated with sialic acid receptor binding preference, are indicated on the figure with their respective residue number. Download
Infectivity of A/peng/Ant/270/13 in ferrets compared with a different H11 wild bird virus (A/Sharptailed Sandpiper/Australia/6/2004) and a human A(H3N2) virus (A/Victoria/361/2011). Ferrets were infected intranasally and nasal washed on days 2 to 5 or infected intratracheally and lungs were taken on day 4 (UR, upper right lung lobe; LL, lower left lobe). Fifty-percent egg infectious dose (EID50) was determined for nasal washes and homogenized lung lobes. Download
In vitro
replication at 33°C and 37°C in MDCK cells of A/peng/Ant/270/13 compared with a different H11 wild bird virus (A/Sharptailed Sandpiper/Australia/6/2004) and a human A(H3N2) virus (A/Victoria/361/2011). Download
BLAST results showing the closest match and percent nucleotide identity to strains in GenBank.
Hemagglutination titers of viruses with resialylated erythrocytes containing terminal SAα2,3Gal or SAα2,6Gal linkages.
ACKNOWLEDGMENTS
The fieldwork was funded by the Instituto Antártico Chileno as part of the project INACH T-27-10: “The common seabird tick Ixodes uriae (White, 1852) as vector of pathogenic virus, bacteria and protozoa to penguins of the Antarctic environment.” The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health. D.V. is funded by the Agency of Science, Technology and Research, Singapore, and the Ministry of Health, Singapore, and Singapore Ministry of Education Academic Research Fund grant MOE2011-T2-2-049.
We thank the staff of the Chilean Antarctic bases Bernardo O’Higgins and Arctowski for their help and assistance during our field work and acknowledge Marco Villarroel for producing the map in Fig. 1 and Ross Lunt, CSIRO AAHL, Australia, for advice on the NP antibody ELISA.
Footnotes
Citation Hurt AC, Vijaykrishna D, Butler J, Baas C, Maurer-Stroh S, Silva-de-la-Fuente MC, Medina-Vogel G, Olsen B, Kelso A, Barr IG, González-Acuña D. 2014. Detection of evolutionarily distinct avian influenza A viruses in Antarctica. mBio 5(3):e01098-14. doi:10.1128/mBio.01098-14.
REFERENCES
- 1. Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus AD, Fouchier RA. 2006. Global patterns of influenza A virus in wild birds. Science 312:384–388. 10.1126/science.1122438 [DOI] [PubMed] [Google Scholar]
- 2. Vijaykrishna D, Deng YM, Su YC, Fourment M, Iannello P, Arzey GG, Hansbro PM, Arzey KE, Kirkland PD, Warner S, O’Riley K, Barr IG, Smith GJ, Hurt AC. 2013. The recent establishment of North American H10 lineage influenza viruses in Australian wild waterfowl and the evolution of Australian avian influenza viruses. J. Virol. 87:10182–10189. 10.1128/JVI.03437-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hansbro PM, Warner S, Tracey JP, Arzey KE, Selleck P, O’Riley K, Beckett EL, Bunn C, Kirkland PD, Vijaykrishna D, Olsen B, Hurt AC. 2010. Surveillance and analysis of avian influenza viruses, Australia. Emerg. Infect. Dis. 16:1896–1904. 10.3201/eid1612.100776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bulach D, Halpin R, Spiro D, Pomeroy L, Janies D, Boyle DB. 2010. Molecular analysis of H7 avian influenza viruses from Australia and New Zealand: genetic diversity and relationships from 1976 to 2007. J. Virol. 84:9957–9966. 10.1128/JVI.00930-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alvarez P, Mattiello R, Rivailler P, Pereda A, Davis CT, Boado L, D’Ambrosio E, Aguirre S, Espinosa C, La Torre J, Donis R, Mattion N. 2010. First isolation of an H1N1 avian influenza virus from wild terrestrial non-migratory birds in Argentina. Virology 396:76–84. 10.1016/j.virol.2009.10.009 [DOI] [PubMed] [Google Scholar]
- 6. Pereda AJ, Uhart M, Perez AA, Zaccagnini ME, La Sala L, Decarre J, Goijman A, Solari L, Suarez R, Craig MI, Vagnozzi A, Rimondi A, König G, Terrera MV, Kaloghlian A, Song H, Sorrell EM, Perez DR. 2008. Avian influenza virus isolated in wild waterfowl in Argentina: evidence of a potentially unique phylogenetic lineage in South America. Virology 378:363–370. 10.1016/j.virol.2008.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rimondi A, Xu K, Craig MI, Shao H, Ferreyra H, Rago MV, Romano M, Uhart M, Sutton T, Ferrero A, Perez DR, Pereda A. 2011. Phylogenetic analysis of H6 influenza viruses isolated from rosy-billed pochards (Netta peposaca) in Argentina reveals the presence of different HA gene clusters. J. Virol. 85:13354–13362. 10.1128/JVI.05946-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu K, Ferreri L, Rimondi A, Olivera V, Romano M, Ferreyra H, Rago V, Uhart M, Chen H, Sutton T, Pereda A, Perez DR. 2012. Isolation and characterization of an H9N2 influenza virus isolated in Argentina. Virus Res. 168:41–47. 10.1016/j.virusres.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shirihai H. 2008. The complete guide to the Antarctic wildlife. Princeton University Press, Princeton, NJ. [Google Scholar]
- 10. Egevang C, Stenhouse IJ, Phillips RA, Petersen A, Fox JW, Silk JR. 2010. Tracking of Arctic terns Sterna paradisaea reveals longest animal migration. Proc. Natl. Acad. Sci. U. S. A. 107:10182–10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kopp M, Peter HU, Mustafa O, Lisovski S, Ritz MS, Phillips RA, Hahn S. 2011. South polar skuas from a single breeding population overwinter in different oceans though show similar migration patterns. Mar. Ecol. Prog. Ser. 435:263–267. 10.3354/meps09229 [DOI] [Google Scholar]
- 12. Baumeister E, Leotta G, Pontoriero A, Campos A, Monalti D, Vig G, Pecoraro M, Savy V. 2004. Serological evidence of influenza A virus infection in Antarctica migratory birds. Int. Congr. Ser. 1263:737–740. 10.1016/j.ics.2004.02.099 [DOI] [Google Scholar]
- 13. Miller GD, Watts JM, Shellam GR. 2008. Viral antibodies in south polar skuas around Davis Station, Antarctica. Antarct. Sci. 20:455–461 [Google Scholar]
- 14. Austin FJ, Webster RG. 1993. Evidence of ortho- and paramyxoviruses in fauna from Antarctica. J. Wildl. Dis. 29:568–571. 10.7589/0090-3558-29.4.568 [DOI] [PubMed] [Google Scholar]
- 15. Wallensten A, Munster VJ, Osterhaus AD, Waldenstr J, Bonnedahl J, Broman T, Fouchier RA, Olsen B. 2006. Mounting evidence for the presence of influenza A virus in the avifauna of the Antarctic region. Antarct. Sci. 18:353–356. 10.1017/S095410200600040X [DOI] [Google Scholar]
- 16. Morgan IR, Westbury HA. 1981. Virological studies of Adelie penguins (Pygoscelis adeliae) in Antarctica. Avian Dis. 25:1019–1026. 10.2307/1590077 [DOI] [PubMed] [Google Scholar]
- 17. Subbarao EK, London W, Murphy BR. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842. 10.1126/science.1062882 [DOI] [PubMed] [Google Scholar]
- 19. Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437:889–893. 10.1038/nature04230 [DOI] [PubMed] [Google Scholar]
- 20. Morgan IR, Westbury HA. 1988. Studies of viruses in penguins in the Vestfold Hills. Hydrobiologia 165:263–269. 10.1007/BF00025595 [DOI] [Google Scholar]
- 21. Gauthier-Clerc M, Eterradossi N, Toquin D, Guittet Ml, Kuntz Gg, Le Maho Y. 2002. Serological survey of the king penguin, Aptenodytes patagonicus, in Crozet Archipelago for antibodies to infectious bursal disease, influenza A and Newcastle disease viruses. Polar Biol. 25:316–319. 10.1007/s00300-001-0346-7 [DOI] [Google Scholar]
- 22. Krauss S, Walker D, Pryor SP, Niles L, Chenghong L, Hinshaw VS, Webster RG. 2004. Influenza A viruses of migrating wild aquatic birds in North America. Vector Borne Zoonotic Dis. 4:177–189. 10.1089/vbz.2004.4.177 [DOI] [PubMed] [Google Scholar]
- 23. Hurt AC, Hansbro PM, Selleck P, Olsen B, Minton C, Hampson AW, Barr IG. 2006. Isolation of avian influenza viruses from two different transhemispheric migratory shorebird species in Australia. Arch. Virol. 151:2301–2309. 10.1007/s00705-006-0784-1 [DOI] [PubMed] [Google Scholar]
- 24. Langstaff IG, McKenzie JS, Stanislawek WL, Reed CE, Poland R, Cork SC. 2009. Surveillance for highly pathogenic avian influenza in migratory shorebirds at the terminus of the East Asian-Australasian flyway. N. Z. Vet. J. 57:160–165. 10.1080/00480169.2009.36896 [DOI] [PubMed] [Google Scholar]
- 25. Munster VJ, Baas C, Lexmond P, Waldenström J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WE, Schutten M, Olsen B, Osterhaus AD, Fouchier RA. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3:e61. 10.1371/journal.ppat.0030061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng Y, Xie ZX, Liu JB, Pang YS, Deng XW, Xie ZQ, Xie LJ, Fan Q, Luo SS. 2013. Epidemiological surveillance of low pathogenic avian influenza virus (LPAIV) from poultry in Guangxi Province, Southern China. PLoS One 8:e77132. 10.1371/journal.pone.0077132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ogilvie MA, Young S. 2002. Photographic handbook: wildfowl of the world. New Holland Publishers, London, United Kingdom [Google Scholar]
- 28. Fretwell PT, Trathan PN. 2009. Penguins from space: faecal stains reveal the location of emperor penguin colonies. Glob. Ecol. Biogeogr. 18:543–552. 10.1111/j.1466-8238.2009.00467.x [DOI] [Google Scholar]
- 29. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abad FX, Busquets N, Sanchez A, Ryan PG, Maj N, Gonzalez-Sols J. 2013. Serological and virological surveys of the influenza A viruses in Antarctic and sub-Antarctic penguins. Antarct. Sci. 25:339–344. 10.1017/S0954102012001228 [DOI] [Google Scholar]
- 31. Morgan IR, Westbury HA, Caple IW, Campbell J. 1981. A survey of virus infection in sub-Antarctic penguins on Macquarie Island, Southern Ocean. Aust. Vet. J. 57:333–335 [DOI] [PubMed] [Google Scholar]
- 32. Shoham D, Jahangir A, Ruenphet S, Takehara K. 2012. Persistence of avian influenza viruses in various artificially frozen environmental water types. Influenza Res. Treat. 2012:912326. 10.1155/2012/912326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anthony SJ, St, Leger JA, Pugliares K, Ip HS, Chan JM, Carpenter ZW, Navarrete-Macias I, Sanchez-Leon M, Saliki JT, Pedersen J, Karesh W, Daszak P, Rabadan R, Rowles T, Lipkin WI. 2012. Emergence of fatal avian influenza in New England harbor seals. mBio 3(4):e00166-12. 10.1128/mBio.00166-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hinshaw VS, Bean WJ, Geraci J, Fiorelli P, Early G, Webster RG. 1986. Characterization of two influenza A viruses from a pilot whale. J. Virol. 58:655–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McFarlane RA. 2009. Health assessment and diseases of the Weddell seal, Leptonochotes wedelli in Vestfold hills, east Antarctica, p 139–166 In Kerry KR, Riddle MJ. (ed), Health of Antarctic wildlife. Springer-Verlag, Berlin, Germany [Google Scholar]
- 36. Lynch MJ, Osterhaus ADME, Cousins DV, Selleck P, Williams P. 1999. Anaesthesia, hematology and disease investigation of free ranging crabeater seals (Lobodon carcinophagus), p 13–14 In Proceedings of the Wildlife Disease Association (Australasian Section) Annual Conference American Association of Zoo Veterinarians. University of Canberra Field Station, Jervis Bay, NSW, Australia [Google Scholar]
- 37. Goldstein T, Mena I, Anthony SJ, Medina R, Robinson PW, Greig DJ, Costa DP, Lipkin WI, Garcia-Sastre A, Boyce WM. 2013. Pandemic H1N1 influenza isolated from free-ranging Northern elephant seals in 2010 off the central California coast. PLoS One 8:e62259. 10.1371/journal.pone.0062259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Webster RG, Hinshaw VS, Bean WJ, Van Wyke KL, Geraci JR, St Aubin DJ, Petursson G. 1981. Characterization of an influenza A virus from seals. Virology 113:712–724. 10.1016/0042-6822(81)90200-2 [DOI] [PubMed] [Google Scholar]
- 39. Webster RG, Geraci J, Petursson G, Skirnisson K. 1981. Conjunctivitis in human beings caused by influenza A virus of seals. N. Engl. J. Med. 304:911. 10.1056/NEJM198104093041516 [DOI] [PubMed] [Google Scholar]
- 40. Driskell EA, Pickens JA, Humberd-Smith J, Gordy JT, Bradley KC, Steinhauer DA, Berghaus RD, Stallknecht DE, Howerth EW, Tompkins SM. 2012. Low pathogenic avian influenza isolates from wild birds replicate and transmit via contact in ferrets without prior adaptation. PLoS One 7:e38067. 10.1371/journal.pone.0038067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, Monne I, Stevens J, Cattoli G, Capua I, Chen LM, Donis RO, Busch J, Paulson JC, Brockwell C, Webby R, Blanco J, Al-Natour MQ, Perez DR. 2008. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS One 3:e2923. 10.1371/journal.pone.0002923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nam JH, Kim EH, Song D, Choi YK, Kim JK, Poo H. 2011. Emergence of mammalian species-infectious and -pathogenic avian influenza H6N5 virus with no evidence of adaptation. J. Virol. 85:13271–13277. 10.1128/JVI.05038-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Senne DA. 2007. Avian influenza in North and South America, 2002-2005. Avian Dis. 51:167–173. 10.1637/7621-042606R1.1 [DOI] [PubMed] [Google Scholar]
- 44. Selleck PW, Arzey G, Kirkland PD, Reece RL, Gould AR, Daniels PW, Westbury HA. 2003. An outbreak of highly pathogenic avian influenza in Australia in 1997 caused by an H7N4 virus. Avian Dis. 47:806–811. 10.1637/0005-2086-47.s3.806 [DOI] [PubMed] [Google Scholar]
- 45. Parmelee DF, Maxon SJ, Bernstein NP. 1979. Fowl cholera outbreak among brown skuas at Palmer Station. Antarct. J. U. S. 14:168–169 [Google Scholar]
- 46. Leotta GA, Chinen I, Vigo GB, Pecoraro M, Rivas M. 2006. Outbreaks of avian cholera in Hope Bay, Antarctica. J. Wildl. Dis. 42:259–270. 10.7589/0090-3558-42.2.259 [DOI] [PubMed] [Google Scholar]
- 47. Spackman E, McCracken KG, Winker K, Swayne DE. 2006. H7N3 avian influenza virus found in a South American wild duck is related to the Chilean 2002 poultry outbreak, contains genes from equine and North American wild bird lineages, and is adapted to domestic turkeys. J. Virol. 80:7760–7764. 10.1128/JVI.00445-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spackman E, McCracken KG, Winker K, Swayne DE. 2007. An avian influenza virus from waterfowl in South America contains genes from North American avian and equine lineages. Avian Dis. 51:273–274. 10.1637/7529-032106R.1 [DOI] [PubMed] [Google Scholar]
- 49. Waddell GH, Teigland MB, Sigel MM. 1963. A new influenza virus associated with equine respiratory disease. J. Am. Vet. Med. Assoc. 143:587–590 [PubMed] [Google Scholar]
- 50. Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 109:4269–4274. 10.1073/pnas.1116200109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New World bats harbor diverse influenza A viruses. PLoS Pathog. 9:e1003657. 10.1371/journal.ppat.1003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. CDC 2011. CDC protocol of real-time RT-PCR for influenza A(H1N1). World Health Organization, Geneva, Switzerland: http://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf [Google Scholar]
- 53. Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275–2289. 10.1007/s007050170002 [DOI] [PubMed] [Google Scholar]
- 54. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoint. Am. J. Hyg. 27:493–497 [Google Scholar]
- 55. Selleck PW, Kirkland PD. 2009. Avian influenza, p 1–32 In Sub-committee on animal health laboratory standards for animal health committee. Australian and New Zealand standard diagnostic procedures for animal diseases, Australia. Department of Agriculture, Fisheries and Forestry, ACT, Australia http://www.scahls.org.au/Procedures/Documents/ANZSDP/anzsdp-for-avian-influenza-05092012.pdf [Google Scholar]
- 56. WHO 2011. Global Influenza Surveillance Network, p 59–61 In Manual for the laboratory diagnosis and virological surveillance of influenza. World Health Organization, Geneva, Switzerland [Google Scholar]
- 57. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 59. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 60. Shapiro B, Rambaut A, Drummond AJ. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23:7–9. 10.1093/molbev/msj021 [DOI] [PubMed] [Google Scholar]
- 61. Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST. Mol. Biol. Evol. 1 7:29:1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Eswar N, Eramian D, Webb B, Shen MY, Sali A. 2008. Protein structure modeling with MODELLER. Methods Mol. Biol. 426:145–159. 10.1007/978-1-60327-058-8_8 [DOI] [PubMed] [Google Scholar]
- 63. Krieger E, Koraimann G, Vriend G. 2002. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins 47:393–402. 10.1002/prot.10104 [DOI] [PubMed] [Google Scholar]
- 64. Rogers GN, Paulson JC. 1983. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127:361–373. 10.1016/0042-6822(83)90150-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The major wild bird global migratory pathways. Download
(A and B) Detailed phylogenetic trees of HA and NA sequences. Trees were drawn using maximum likelihood methodology. Red text indicates AIVs from Antarctica, blue text indicates AIVs from South America, and black text indicates AIVs from all other regions. Download
(A to F) Detailed phylogenetic trees of internal gene sequences. Trees were drawn using maximum likelihood methodology. Red text and branches indicate AIVs from Antarctica, blue text and branches indicate AIVs from South America, green text and branches indicate AIVs from all other regions, black text and branches indicate mammalian influenza viruses from all regions. Download
HA amino acid alignment of the penguin HA sequence compared to other H11 avian influenza sequences, more distant avian influenza sequences (H13, H16, and H5), and mammalian influenza sequences (H1, H2, and H3). Residues 155, 186, 187, 190, 225, 226, and 228, which are associated with sialic acid receptor binding preference, are indicated on the figure with their respective residue number. Download
Infectivity of A/peng/Ant/270/13 in ferrets compared with a different H11 wild bird virus (A/Sharptailed Sandpiper/Australia/6/2004) and a human A(H3N2) virus (A/Victoria/361/2011). Ferrets were infected intranasally and nasal washed on days 2 to 5 or infected intratracheally and lungs were taken on day 4 (UR, upper right lung lobe; LL, lower left lobe). Fifty-percent egg infectious dose (EID50) was determined for nasal washes and homogenized lung lobes. Download
In vitro
replication at 33°C and 37°C in MDCK cells of A/peng/Ant/270/13 compared with a different H11 wild bird virus (A/Sharptailed Sandpiper/Australia/6/2004) and a human A(H3N2) virus (A/Victoria/361/2011). Download
BLAST results showing the closest match and percent nucleotide identity to strains in GenBank.
Hemagglutination titers of viruses with resialylated erythrocytes containing terminal SAα2,3Gal or SAα2,6Gal linkages.