Unlike classical class-switch recombination, class switching to IgD occurs independently of the IgH 3′ regulatory region.
Abstract
Classical class-switch recombination (cCSR) substitutes the Cμ gene with Cγ, Cε, or Cα, thereby generating IgG, IgE, or IgA classes, respectively. This activation-induced deaminase (AID)–driven process is controlled by the IgH 3′ regulatory region (3′RR). Regulation of rare IgD CSR events has been enigmatic. We show that μδCSR occurs in mouse mesenteric lymph node (MLN) B cells and is AID-dependent. AID attacks differ from those in cCSR because they are not accompanied by extensive somatic hypermutation (SHM) of targeted regions and because repaired junctions exhibit features of the alternative end-joining (A-EJ) pathway. In contrast to cCSR and SHM, μδCSR is 3′RR-independent, as its absence affects neither breakpoint locations in Sμ- and Sδ-like (σδ) nor mutation patterns at Sμ-σδ junctions. Although mutations occur in the immediate proximity of the μδ junctions, SHM is absent distal to the junctions within both Sμ and rearranged VDJ regions. In conclusion, μδCSR is active in MLNs, occurs independently of 3′RR-driven assembly, and is even dramatically increased in 3′RR-deficient mice, further showing that its regulation differs from cCSR.
After functional V(D)J recombination, bone marrow immature B cells express a BCR made up of surface IgM. In secondary lymphoid organs, mature B cells simultaneously express surface IgD of the same specificity as surface IgM through alternative splicing of a pre-mRNA composed of V(D)J and both Cμ and Cδ heavy chain constant exons (Moore et al., 1981; Preud’homme et al., 2000; Chen and Cerutti, 2010). After encountering antigen, in adequate lymphoid tissue structures providing accessory signals (direct interactions with T cells and follicular dendritic cells and binding of cytokines secreted by such cells), activated B cells undergo classical class-switch recombination (cCSR) and substitute the Cμ gene with Cγ, Cα, or Cε, thereby generating secondary IgG, IgA, and IgE BCR and antibody classes with the same antigenic specificity but new effector functions. This recombination targets specific stretches of repetitive DNA (S regions) preceding Cμ and all Cγ, Cα, and Cε genes and requires the DNA-editing enzyme activation-induced deaminase (AID; Pavri and Nussenzweig, 2011). Transcription and, to an even higher extent, accessibility of the IgH locus S regions to cCSR is under the control of the IgH 3′ regulatory region (3′RR), located downstream of Cα, and encompassing the four transcriptional enhancers hs3a, hs1,2, hs3b, and hs4 (Vincent-Fabert et al., 2010a; Pinaud et al., 2011). Genomic disruption or complete deletion of the 3′RR in the mouse abrogates cCSR to all IgG, IgA, and IgE classes (Vincent-Fabert et al., 2010a,b; Pinaud et al., 2011). Recently the 3′RR was also reported to be transcribed and itself undergo AID-mediated mutation and recombination around phylogenetically conserved switch-like DNA repeats, highlighting its close functional partnership with AID (Péron et al., 2012).
Despite the absence of a repetitive S region upstream of Cδ and similar to class switching from IgM to IgG, IgA, and IgE, some CD38+ human B cells from tonsils carry an IgH locus switched to IgD expression after an AID-mediated deletion of Cμ (Arpin et al., 1998; Brandtzaeg and Johansen, 2005; Johansen et al., 2005; Chen et al., 2009). Secreted IgD originating from such cells with CSR from Cμ to Cδ (μδCSR) enhances immune surveillance and can exert proinflammatory and antimicrobial effects (Chen et al., 2009). Although there is no canonical switch region 5′ to the Cδ gene in mammals (Preud’homme et al., 2000; Chen and Cerutti, 2010), rudimentary Sδ-like (σδ) sequences have been described at Cμ-Cδ junctions but CSR from μ to δ stands as a rare event whose regulation is obscure (Chen and Cerutti, 2010). To explore the role of the 3′RR on such Cμ-Cδ recombinations, in the context of the endogenous locus, we analyzed IgH 3′RR-deficient mice lacking the 30-kb extent of the 3′RR, a deletion which we previously characterized as inducing a severe cCSR defect toward IgG, IgA, and IgE (Vincent-Fabert et al., 2010b), a severe somatic hypermutation (SHM) defect (Rouaud et al., 2013) but with normal V(D)J recombination (Rouaud et al., 2012).
RESULTS
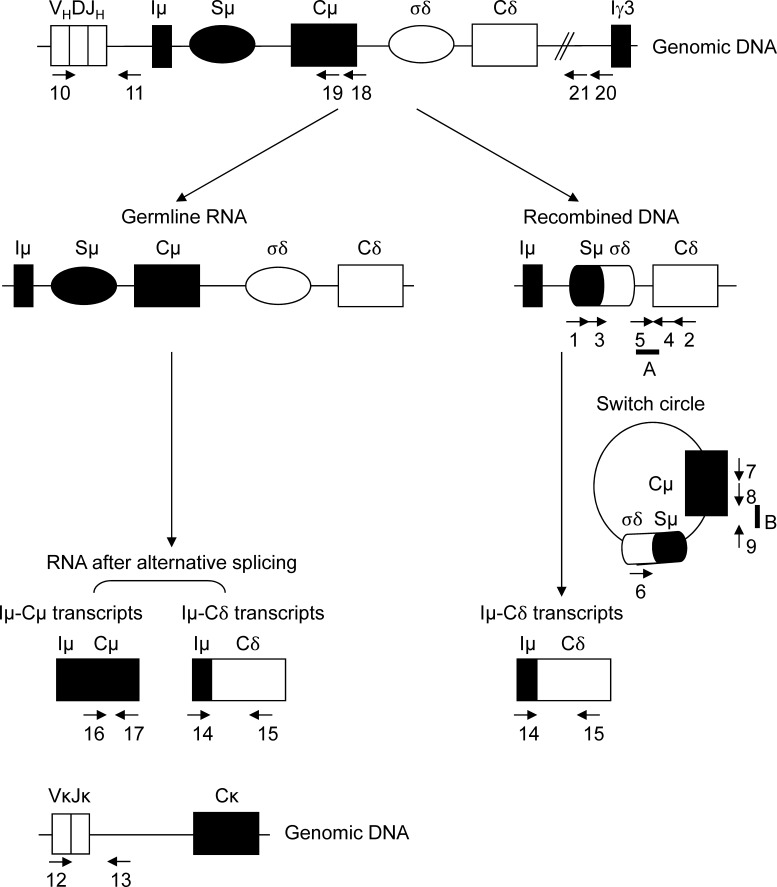
IgD secretion in mice occurs at low levels and is poorly understood. Germline transcription of Iμ-Cμ enrolls the Sμ regions as a substrate for DNA modification by the cCSR machinery including AID (Chen and Cerutti, 2010). Iμ-Cδ transcripts can originate either from alternate splicing of a pre-mRNA encompassing Cμ and Cδ exons or from primary Cδ transcripts after μδCSR. Fig. 1 schematizes these processes and locates PCR primers and probes used in this study.
Figure 1.
Two pathways for IgD synthesis. IgD can originate either from the alternate splicing of a pre-mRNA encompassing Cμ and Cδ exons (left) or from primary Cδ transcripts after μδCSR (right). CSR from Sμ to σδ includes germline Iμ-Cμ and Iμ-Cδ transcripts and σδ-Sμ switch circles. Arrows located primers used in the study. Numbers and letters refer to the indicated primers and probes, respectively, as reported in Materials and methods.
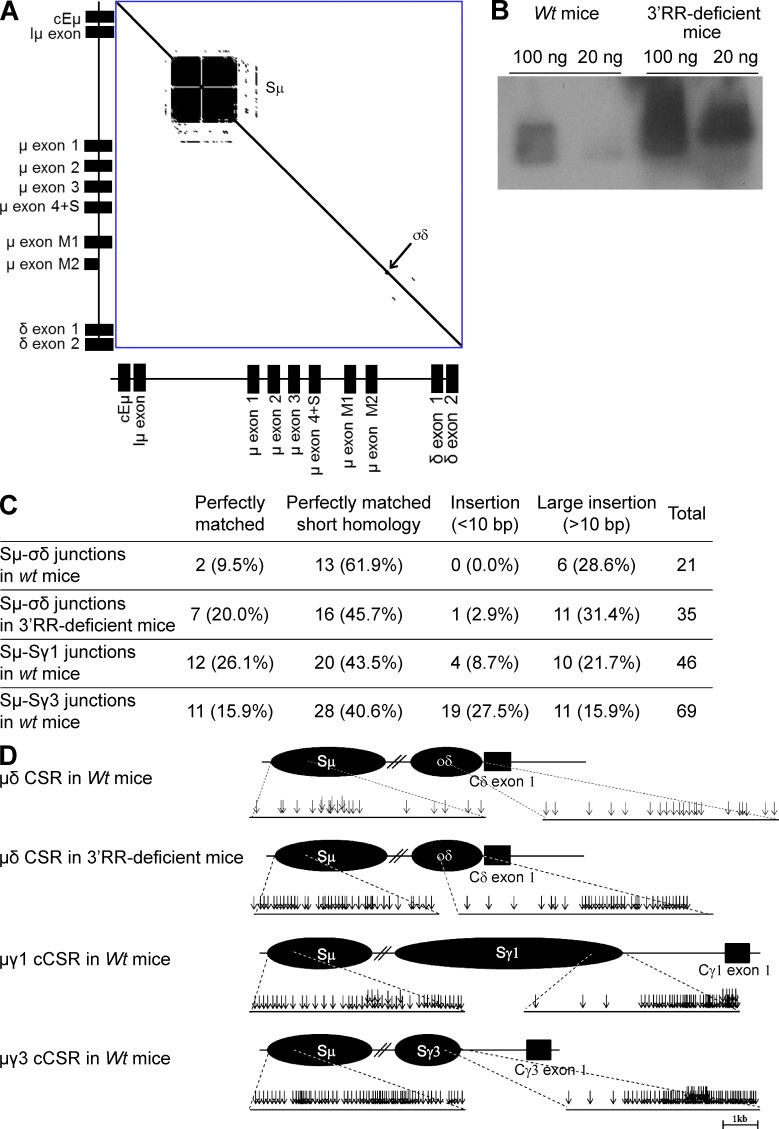
μδCSR occurs in normal mesenteric LNs (MLNs) of wt mice and 3′RR-deficient mice
μδCSR is currently not documented in mice (Chen and Cerutti, 2010). Although little is known about the role of soluble IgD in physiology, a well-known inflammatory disease in humans features high serum IgD levels and was thus called hyper-IgD syndrome, a disease also featuring enlargement of MLNs (Oretti et al., 2006). In a search for IgD+IgM− cells after μδCSR in wt mice, we thus focused our attention on MLN B cells. S regions consist of tandem repeats of short G-rich sequences. While readily showing Sμ repeats, dot plot analysis of the mouse DNA fragment encompassing the Eμ enhancer to Cδ only revealed few DNA repeats upstream of Cδ in a short 0.5-kb-long region that was thereby named σδ (Preud’homme et al., 2000; Fig. 2 A). Using a nested PCR-based strategy (location of primers in Fig. 1) followed by Southern blotting (location of the probe in Fig. 1) of PCR products, we were able to detect Sμ-σδ junctions as a result of Sμ to σδ CSR (Fig. 2 B). We definitely authenticated Sμ-σδ junctions by cloning them and analyzing their sequences (Fig. 2 C and Table S1). Because 3′RR-deficient mice are known to be almost completely defective for cCSR (Vincent-Fabert et al., 2010b), we also checked them for μδCSR. To our surprise, Southern blotting of PCR product revealed the presence of Sμ-σδ junctions as a result of Sμ to σδ CSR (Fig. 2 B). These junctions were again authenticated by cloning and sequencing (Fig. 2 C and Table S1).
Figure 2.
MLN B cells undergo μδCSR. (A) DNA dot plot analysis of the genomic sequence encompassing a fragment from the Eμ enhancer to the δ exon 2 from a 129 wt mouse (GenBank accession no. AJ851868.3). The sequence was compared with itself and dots off the diagonal represent repetitive patterns within the sequence. Sµ and σδ regions are indicated. (B) Southern blot analysis of Sμ-Cδ junctions amplified by PCR and hybridized with a 5′Cδ probe from MLN B cells of 3′RR-deficient mice and wt mice. 100 and 20 ng DNA were used for PCR experiments. Data are representative of 6 independent experiments with 1 mouse per group. (C) μδ junctions obtained from wt mice (data are pooled from 13 independent experiments with 1 mouse per experiment) and 3′RR-deficient mice (data are pooled from 10 independent experiments with 1 mouse per experiment) were cloned and sequenced. Percentages of junctions with insertion and junctional microhomology are reported. μγ1 and μγ3 junctions from wt mice (data are pooled from 6 independent experiments with 1 mouse per experiment) obtained after in vitro LPS ± IL4 stimulation of splenocytes are reported. The sequenced junctions depicted from all the mice combined. (D) Location of Sμ-σδ breaks in 13 wt and 10 3′RR-deficient mice and Sμ-Sγ1 and Sμ-Sγ3 breaks during in vitro cCSR in 6 wt mice (same junctions and mice as in Fig. 2C).
Molecular features of μδCSR junction sequences
The locations of Sμ-σδ breaks in Sμ and σδ for wt mice and 3′RR-deficient animals are reported in Fig. 2 D. Among Sμ-σδ junctions in wt mice (Fig. 2 C and Table S1), 13/21 (61.9%) showed 1–8-bp-long junctional microhomology. Complex junctions were also frequent (6/21, 28.6%). Similarly to wt mice, junctions with short nucleotide insertions and complex junctions were found in 3′RR-deficient mice. Junctions with microhomologies 1–8 bp long represented 45.7% (16/35) of junctions (Fig. 2 C and Table S1). Complex junctions represented 31.4% (11/35) of junctions. Direct Sμ-σδ joining was slightly increased in 3′RR-deficient mice (7/35, 20%) as compared with wt mice (2/21, 9.2%). To compare μδCSR junctions cloned from MLN cells from wt mice with cCSR junctions, splenocytes from wt mice were stimulated with LPS or LPS + IL4 to generate Cμ-Cγ3 and Cμ-Cγ1 cCSR, respectively (Vincent-Fabert et al., 2009). Sequences of Sμ-Cγ3 and Sμ-Cγ1 junctions resulting from Sμ to Sγ3 and Sγ1 cCSR were reported in Fig. 2 C and Table S1. Features of Sμ-Sγ CSR junctions (i.e., relative frequencies of direct joints, microhomologies, insertions, and complex junctions) were similar to those classically reported (Stavnezer et al., 2010). Sequences from Sμ-σδ junctions of wt mice showed strikingly different features in that they more rarely exhibited a direct joining (2/21, 9.5%) than either Sμ-Sγ1 (12/46, 26.1%) or Sμ-Sγ3 (11/70, 15.9%) junctional sequences. Apart from that, the location of Sμ breaks was similar in Cμ-Cδ, Cμ-Cγ1, and Cμ-Cγ3 junctions (Fig. 2 D). Finally, the cloned σδ breaks were mostly located in the 3′ part of the σδ region similar to breaks in the downstream part of Sγ1 and Sγ3 for cCSR junctions (Fig. 2 D). The increased representation of DNA breaks in the 5′ part of Sμ and the 3′ part of σδ or Sγ might involve a methodological bias whereby PCR cloning of the shortest recombined DNA segment would be favored.
Identical phenotype of IgD+IgM− B cells in 3′RR-deficient and wt mice
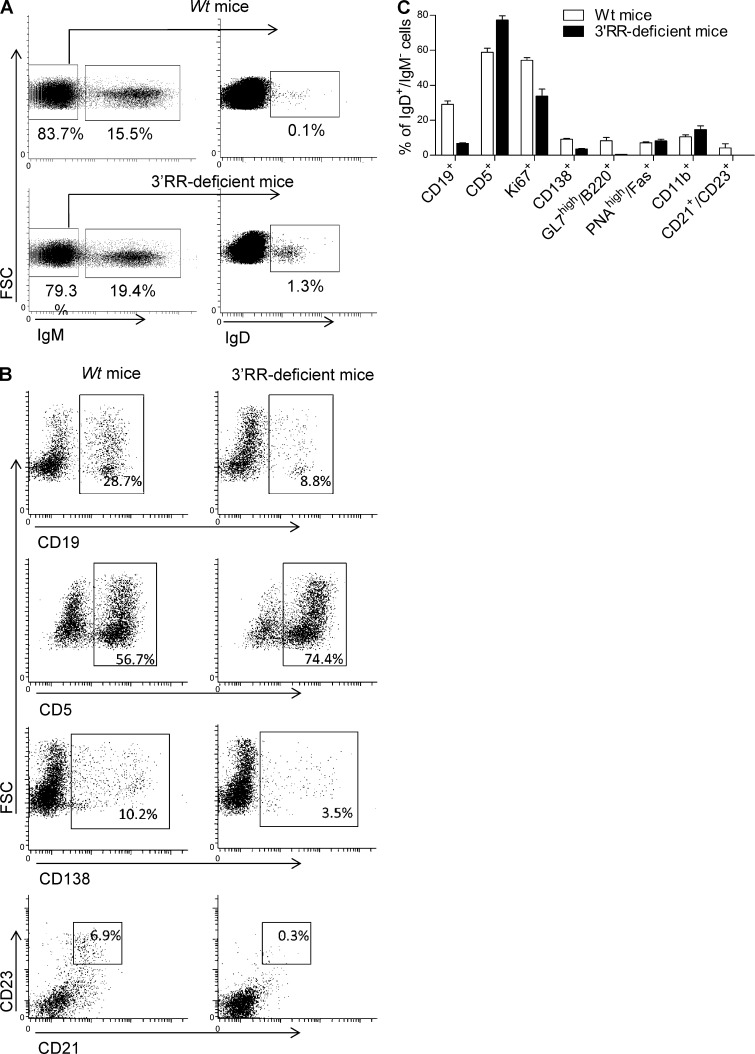
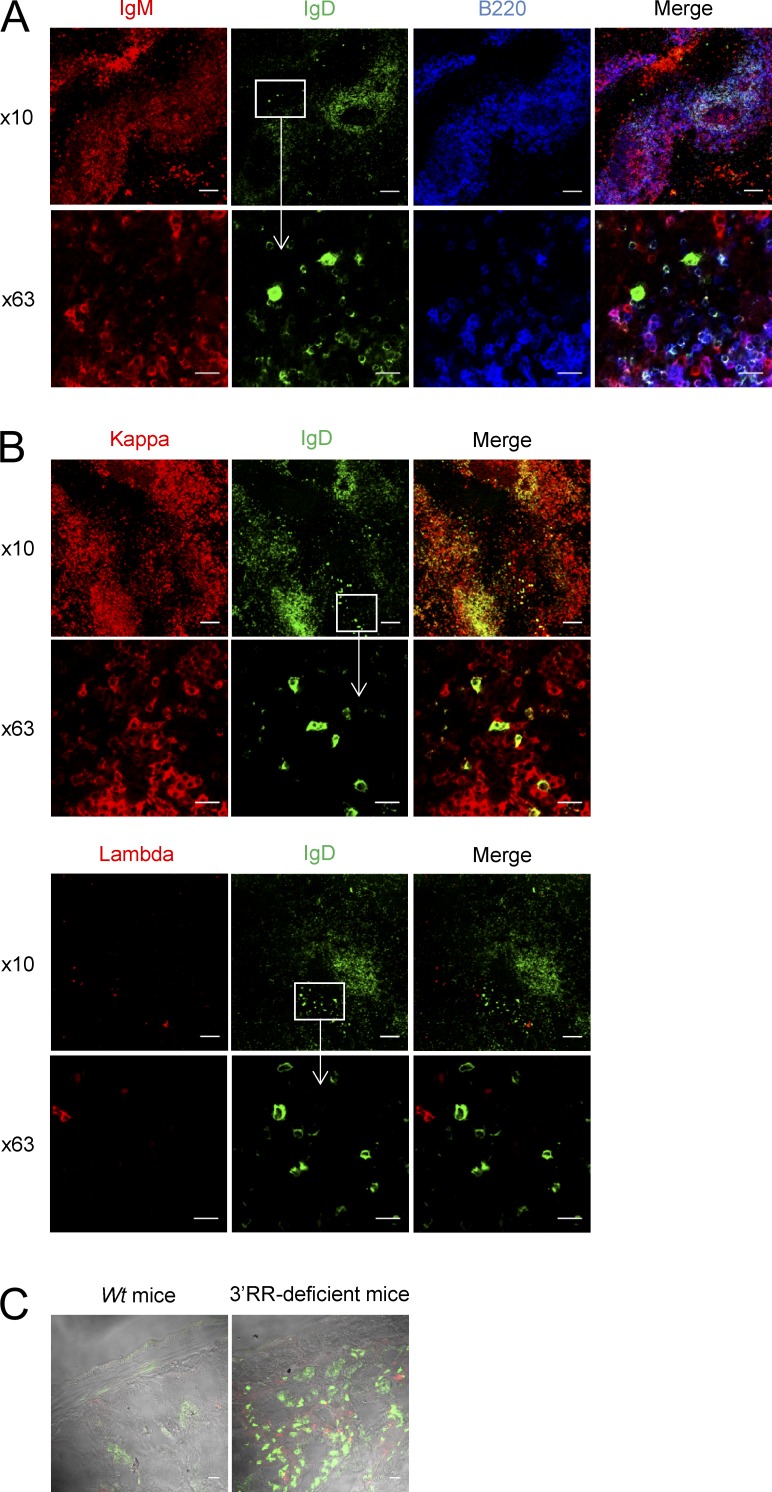
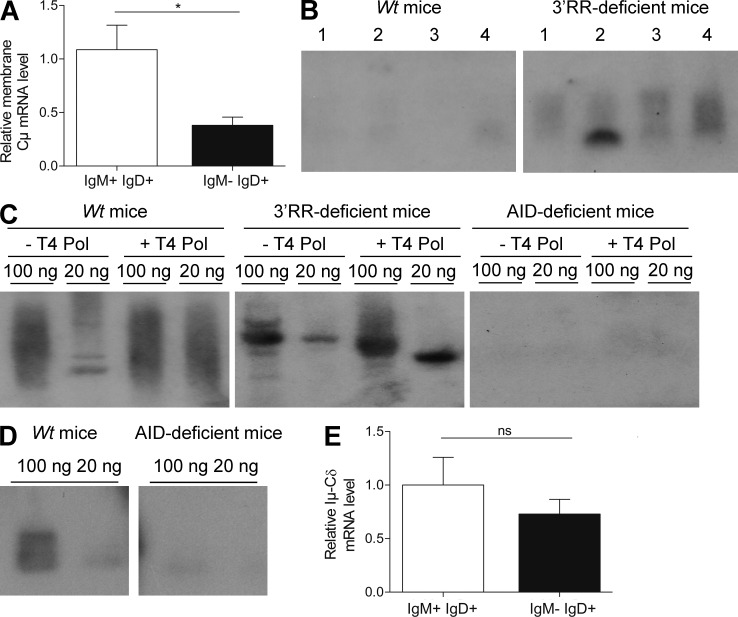
Flow cytometry analysis indicated that IgD+IgM− B cells accounted for only ∼0.2% of total MLN cells in wt mice (Fig. 3 A). Consistent with published human data (Chen et al., 2009), a low percentage of IgD+IgM− B cells expressed CD138 (syndecan-1), a hallmark of mature plasma cells (Fig. 3, B and C). Furthermore, the vast majority of IgD+IgM− B cells did not express surface B220, CD11b, CD19, CD21, and CD23 markers which are usually down-regulated by differentiated plasma cells (Fig. 3, B and C). IgD+IgM− B cells also lacked PNA and GL7 staining and thus differed from germinal center (GC) B cells (Fig. 3 C). In contrast to human IgD+IgM− B cells (Arpin et al., 1998; Chen et al., 2009), no bias for surface λ-light chain was found (not depicted). Similarly to human IgD+IgM− B cells (Chen et al., 2009), mouse IgD+IgM− B cells expressed CD5, an activation-induced B cell molecule (Fig. 3, B and C), and had a low proliferation rate (Ki67 index of ∼50%; Fig. 3 C). IgD+IgM− B cells were not detected in splenocytes, bone marrow B cells, and circulating B cells of wt mice (unpublished data). Flow cytometry analysis indicated the presence of IgD+IgM− B cells in MLNs of 3′RR-deficient mice and that they accounted for ∼2% of total MLN cells (Fig. 3 A), i.e., a 10-fold increase as compared with wt mice. Increased IgD switching was also confirmed after Southern blotting of Sμ-σδ junctions (Fig. 2 B). Besides their increased abundance, IgD+IgM− B cells from 3′RR-deficient animals qualitatively differed from those in wt mice by showing a lower expression of the B cell surface markers B220, CD19, CD21, and CD23 (Fig. 3, B and C), although they overwhelmingly (80%) expressed CD5 and poorly proliferated (<40% were Ki67+) as for wt IgD+IgM− cells. Due to their low levels in MLNs, IgD+IgM− B cells did not show up in wt mice using immunohistochemistry but were detected in 3′RR-deficient mice. In agreement with their PNA− and GL7− status, IgD+IgM− cells were located in the marginal zone of GC (Fig. 4 A) and had a typical ovaloid-elongated plasma cell shape, medium to large size, and an abundant cytoplasm containing Cδ IgH chains (Fig. 4 A). In contrast to humans (Chen et al., 2009), and confirming results with wt mice, we did not find any bias in the κ+/λ+ ratio when specifically studying those cells that express IgD in the absence of IgM expression (Fig. 4 B). Similarly to wt mice, such cells were not detected in blood, bone marrow, spleen, and Peyer’s patches of 3′RR-deficent mice (unpublished data). A large fraction of B cells from the anti-mesenteric wall of the mouse small intestine displays an IgD+IgM− phenotype (Hamada et al., 2002). We confirmed the presence of IgD+ B cells by immunostaining all along the mouse small intestine but, again, more noticeably in 3′RR-deficient mice than in wt mice (Fig. 4 C).
Figure 3.
IgD+IgM− B cell phenotype. (A) Flow cytometry analysis of IgM−IgD+ cells in MLN of wt mice and 3′RR-deficient mice. Cells were fixed and permeabilized before IgM and IgD staining. Cells were first gated on IgM− cells and second on IgD+ cells highlighting the presence of IgM−IgD+ cells. Plots are representative of 6 independent experiments with 1 mouse per group. (B) Cytometry analysis of CD19, CD5, CD138, and CD23/CD21 cell surface markers in IgD+IgM− cells from wt mice and 3′RR-deficient mice. Plots are representative of 6 independent experiments with 1 mouse per group. (C) Percentage of mouse IgD+IgM− cells expressing CD19, CD5, Ki67, CD138, GL7/B220, PNA/Fas, CD11b, and CD21/CD23 in wt and 3′RR-deficient mice. Data are the mean ± SEM from 6 independent experiments with 1 mouse per group.
Figure 4.
Immunohistochemistry analysis of IgD+IgM− cells. (A) MLN tissue of 3′RR-deficient mice stained for IgM (red), IgD (green), and B220 (blue). Images are representative of 4 independent experiments with 1 mouse per experiment. Bars: (10×) 100 µm; (63×) 10 µm. (B) MLN tissue stained for IgD (green), κ light chain (red), and λ light chain (red) in 3′RR-deficient mice. Images are representative of 4 independent experiments with 1 mouse per experiment. Bars: (10×) 100 µm; (63×) 10 µm. (C) Intestine tissue stained for IgD (green) and IgM (red) in 3′RR-deficient mice and wt animals. Images are representative of 4 independent experiments with 1 mouse per group. Bar, 100 µm.
μδCSR has numerous characteristics of cCSR
To ensure whether the IgD+ phenotype is linked to μδCSR, PCR analysis of μδCSR in sorted IgD+IgM− cells (from three mice) was performed in 3′RR-deficient mice (where these cells are more abundant and accessible to cell sorting). After cloning and sequencing we detected 11 different μδ sequences among 22 analyzed sequences (i.e., 50%). No junctions were found with sorted IgD−IgM+ cells, reinforcing the hypothesis that the IgD+IgM− phenotype is linked to μδCSR. Real-time PCR analysis indicated a dramatic decrease (62%, P = 0.005, Mann-Whitney U test) of Cμ transcripts in sorted IgD+IgM− cells as compared with sorted IgD+IgM+ cells, reinforcing the hypothesis that the IgD+IgM− phenotype is linked to μδCSR (Fig. 5 A). During CSR, there are abundant double-strand breaks (DSBs) in Sμ. A fraction of these DSBs may be joined to rare DSBs in other parts of the genome, especially within flanking regions on the same chromosome. To demonstrate a specific association of σδ with μδCSR, PCR analysis was performed with other reverse PCR primers (location in Fig. 1) corresponding to Cμ and a region between Cδ and Iγ3. Using these primers, we were unable to detect any μδ junctions (out of 200 independent junk sequences analyzed) in MLN DNA of four 3′RR-deficient mice with detectable μδCSR, demonstrating that the joining between Sμ and σδ represents a specific recombination event. Using a PCR amplification (location of primers in Fig. 1), followed by Southern blotting (location of the probe in Fig. 1), we identified extrachromosomal σδ-Sμ circles in MLN B cells of 3′RR-deficient mice (Fig. 5 B), indicating ongoing μδCSR and a looping-out and deletion mechanism analogous to cCSR (in conditions where ongoing μδCSR was not detectable in MLN DNA from wt mice). The identity of the PCR product was confirmed by sequencing (location of primers in Fig. 1; not depicted). The hallmark of cCSR is its dependence on AID for DSBs formation in S regions. Ligation-mediated PCR (LM-PCR) highlighted the presence of DSBs in σδ in wt mice and 3′RR-deficient mice but not in AID-deficient mice (Fig. 5 C). Using the same experimental strategy as for wt and 3′RR-deficient mice, we were unable to detect any Sμ-σδ junction in genomic DNA from MLNs of AID-deficient mice by Southern blotting of PCR products designed to amplify such junctions (Fig. 5 D). Despite the lack of significant hybridization, we further tried to clone potential junctions from AID-deficient mice (by the same protocol as for wt mice) and failed to identify any (out of 288 independent junk sequences analyzed) indicating that μδCSR in mice requires AID as previously suggested by analyses in human AID-deficient patients (Arpin et al., 1998; Chen et al., 2009). Finally, an Iμ-Cδ transcript was previously identified in human (Chen et al., 2009). Real-time PCR analysis confirmed its presence not only in IgD+IgM+ (where it can originate from alternate splicing of a pre-mRNA encompassing Cμ and Cδ exons) but also in IgD+IgM− where it is the reflect of the post-IgD CSR transcription (Fig. 5 E).
Figure 5.
μδCSR in 3′RR-deficient mice. (A) Real-time PCR analysis of membrane Cμ transcripts in sorted IgD+IgM+ and IgD+IgM− cells from 3′RR-deficient mice. Data are the mean ± SEM of 7 independent experiments with 1 mouse and 8 independent experiments with 1 mouse for IgD+IgM+ and IgD+IgM− cells, respectively. *, P < 0.01 (Mann-Whitney U test). (B) Southern blot analysis of σδ-Sμ switch circles amplified by PCR and hybridized with a Cμ-labeled probe from MLN B cells of 3′RR-deficient and wt mice. Results from four mice (lines 1–4) in each group are reported. Data are representative of 6 independent experiments with 1 mouse per group. (C) LM-PCR. Genomic DNA from wt, 3′RR-deficient, and AID-deficient mice were left untreated or treated with T4 DNA polymerase (T4 Pol), ligated with T4 DNA ligase and probed for double-stranded breaks in σδ after by semi-nested PCR. Reactions with 100 and 20 ng DNA are shown. Data are representative of 6 independent experiments each with 1 mouse per genotype. (D) Southern blot analysis of Sμ-Cδ junctions amplified by PCR and hybridized with a 5′Cδ probe from MLN B cells of wt mice and AID-deficient mice. 100 and 20 ng DNA were used for PCR experiments. Data are representative of 6 independent experiments with 1 mouse per group. (E) Real-time PCR analysis of Iμ-Cδ transcript in sorted IgD+IgM+ and IgD+IgM− cells from 3′RR-deficient mice. Data are of the mean ± SEM of 6 independent experiments with 1 mouse and 8 independent experiments with 1 mouse for IgD+IgM+ and IgD+IgM− cells, respectively. ns = not significant.
Mutations in IgD+IgM− cells
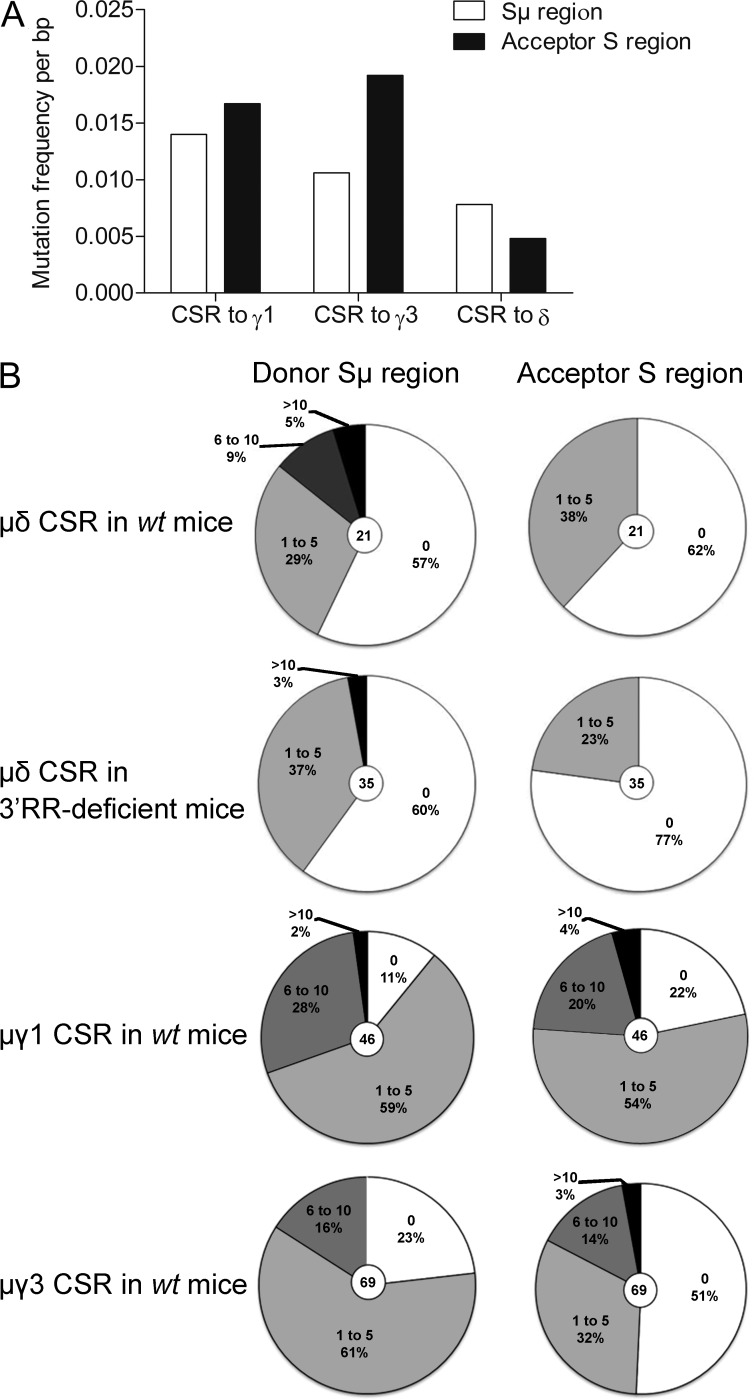
Interactions with cognate antigens recruit activated B cells into GC and can induce AID-mediated modifications. These include SHM in V(D)J exons for the generation of high-affinity antibodies and cCSR (Henderson and Calame, 1998; Durandy, 2003; Hackney et al., 2009). cCSR and SHM are linked to transcription, dependent from some common actors, and both blocked by AID deficiency. SHM can also be considered as the usual mark for antigen-experienced B cells having participated to a GC reaction (Chan and Brink, 2012). We purified IgD+IgM− cells from 3′RR-deficient mice (where these cells are more abundant and accessible to cell sorting) to evaluate VκJκ SHM and identify them as pre- or post-GC B cells. To stimulate SHM, mice were immunized both orally with sheep red blood cells and intraperitoneally with LPS (i.e., in conditions known to yield in vivo activated spleen and Peyer’s patches B cells with VκJκ SHM both in wt and 3′RR-deficient mice; Péron et al., 2012; Rouaud et al., 2013). Rearranged IgH VDJ and VκJκ regions from MLN IgD+IgM− cells of 3′RR-deficient mice were cloned and sequenced. No SHM could be documented out of 90 VDJ sequences analyzed (3 mutations among 68,582 bp analyzed, 0.0044%) and 58 κ sequences analyzed (7 mutations among 36,134 bp analyzed, 0.0194%). Although the absence of SHM in VDJ rearranged region was expected in 3′RR-deficient mice recently demonstrated with and IgH locus-specific SHM defect (Rouaud et al., 2013), the lack of SHM in the κ light chain locus clearly indicate that those B cells with μδCSR have not undergone conventional antigen-driven maturation as usually occurring into GC. By comparison, 3′RR-deficient animals recently analyzed after the same immunization protocol showed a >70-fold higher (1.39%) SHM rate in VκJκ segment from Peyer’s patches GC B cells (Rouaud et al., 2013). The lack of mutation in VDJ segments fitted well with their PNA−GL7− status and their localization in the marginal zone of GC. Contrasting with mice, human IgD+IgM− had extensive SHM in their IgV genes that indicated their clonal relatedness (Arpin et al., 1998). Mouse IgD+IgM− had no SHM in their IgV genes and the use of a JH4 probe on Southern blots from IgD+IgM− cell DNA revealed no rearranged bands in addition to the germline band indicating no potential clonal relatedness (unpublished data). We finally analyzed mutations during μδCSR in wt mice as compared with cCSR to find if intensity of AID attack is similar between them. The mutation frequency at Sμ breakpoints and 5′ to Sμ breakpoints were lower in μδCSR than in IgG1 and IgG3 cCSR (although the latter control was chosen using in vitro conditions poorly inducing SHM; Fig. 6, A and B). A decrease of 25 and 52% in the frequency of mutations in the Sμ donor region was found during μδCSR as compared with IgG3 and IgG1 cCSR, respectively. The numbers of mutations at σδ breakpoints and 3′ to σδ breakpoints were also much lower than in target Sγ region of cCSR (a decrease of 89 and 86% of those found in Sγ1 and Sγ3, respectively; Fig. 6, A and B). Thus, not only the Sμ donor region but also the σδ acceptor region implicated in μδCSR exhibited lower AID-induced mutations than Sμ, Sγ1, and Sγ3 during IgG1 and IgG3 cCSR. Interestingly, the pattern of mutations at Sμ-σδ junctions, 5′ to Sμ, and 3′ to σδ was similar in wt mice and 3′RR-deficient animals (Fig. 6 B), showing that the absence of the 3′RR did not affect the AID-targeting at the Sμ donor and σδ acceptor regions during μδCSR.
Figure 6.
Mutation in Sμ and σδ during μδCSR. Mutation frequency in the donor region (Sμ) and the acceptor region (σδ, Sγ1, or Sγ3) during μδCSR and cCSR. (A) Mutation frequency per mutated bp. 46 Sμ-Cγ1, 69 Sμ-Cγ3, and 21 Sμ-Cδ junctions from wt mice were investigated. Sμ-Cγ1 and Sμ-Cγ3 junctions were obtained after in vitro stimulation of splenocytes from wt mice (data are pooled from 6 independent experiments with 1 mouse). Sμ-Cδ junctions were obtained after genomic extraction of MLN from wt mice (data are pooled from 13 independent experiments with 1 mouse). (B) Number of mutation in each S acceptor and S donor regions. 35 Sμ-Cδ junctions were obtained after genomic extraction of MLN from 3′RR-deficient mice (data are pooled from 10 independent experiments with 1 mouse).
DISCUSSION
cCSR is a very complex process during which accessibility to recombination is regulated by both cis-acting factors (such as germline I promoters and 3′RR enhancers) and trans-acting factors (most of which are induced by cytokine-dependent signals and/or B cell activation; Stavnezer et al., 2010; Boboila et al., 2012). Analysis of DNA sequence at or around the recombination sites has helped us to understand the mechanisms first generating DSBs after targeting of S regions by AID and then allowing synapsis and repair of chromosomal breaks mostly through a classical nonhomologous end joining (C-NHEJ) pathway or to a lower extend through the alternative end-joining (A-EJ) pathway (Yan et al., 2007; Boboila et al., 2012). Although various disruptions of the 3′RR partially compromised cCSR (Cogné et al., 1994; Manis et al., 1998; Pinaud et al., 2001; Vincent-Fabert et al., 2009; Bébin et al., 2010), its complete deletion has clarified its mandatory role for cCSR toward IgG, IgA, and IgE isotypes (Vincent-Fabert et al., 2010b) as well as for SHM in rearranged VDJ exons (Rouaud et al., 2013).
Phylogenetic studies indicate that IgD is as ancient as IgM but, in contrast to IgM, IgD is evolutionarily labile showing, for instance, many duplications/deletions of domains and splice forms (Ohta and Flajnik, 2006; Rauta et al., 2012). Due to the lack of an authentic Sδ region, B cells exclusively expressing IgD are extremely rare in humans and almost absent in mice (Owens et al., 1991). Our study clearly shows that despite the lack of canonical Sδ region, CSR from μ to δ occurs in MLNs from wt mice. Sequences of μδCSR junctions reveal a lower percentage of direct end junctions in μδCSR than in cCSR together with a higher percentage of junctions with either microhomologies between recombined ends or showing insertions of various lengths at the recombination site. Notably, this pattern is strongly reminiscent of the junctions affected by the A-EJ pathway and previously reported during cCSR in B cells lacking key components of the C-NHEJ pathway, its main characteristic being a dramatic reduction of CSR direct joints and frequent junctions with microhomologies and insertions (Yan et al., 2007). Although the A-EJ pathway has only recently been shown to contribute to cCSR, it has been shown that in the absence of C-NHEJ, cCSR can be performed by A-EJ at up to 50% of wt levels (Yan et al., 2007). Notably, the same A-EJ–based pattern of junctions is also observed when referring to a few mouse IgD-secreting hybridomas documented in the past (Owens et al., 1991).
Similarly to cCSR, the absence of μδCSR in AID-deficient mice confirms it also requires AID. The hallmark of CSR is its dependence on AID for DSBs formation in S regions. LM-PCR experiments found the presence of DSBs in σδ in wt mice and 3′RR-deficient mice but not in AID-deficient mice. μδCSR is also AID-dependent in human (Chen et al., 2009). However, this does not imply that all IgD-producing plasma cells have undergone μδCSR. Indeed, serum IgD persists and is even augmented in some AID-deficient patients, probably as a result of the activation of an alternative pathway involving the emergence of plasma cells that produce both IgM and IgD via alternative splicing, particularly at mucosal sites (Chen et al., 2009). Our data indicated that during μδCSR the Sμ and (still more clearly) the σδ acceptor region are less actively targeted by AID than Sμ, Sγ1, and Sγ3 during IgG1 and IgG3 cCSR. This suggests, aside from differences in junction diversity, that μδCSR is regulated differently from cCSR. Individual S regions range in size from 10 kb for Sγ1 (IgG1 being the most abundant Ig class) to 2.5 kb for Sγ3 and to only 1 kb for Sε. Studies pointed the S region size as an important factor in determining endogenous cCSR efficiency because cCSR frequencies correlated with S region length (Zarrin et al., 2005). The very low level of μδCSR in wt mice may be linked to the short length (0.5 kb) of the σδ region and to a less active targeting by AID than Sγ1 and Sγ3 during cCSR.
An intronic (I) exon is positioned upstream of each CH gene except Cδ both in mice and humans. IH-CH germinal transcripts render SH regions substrate for DNA modifications by the CSR machinery, including AID. As previously identified in humans (Chen et al., 2009), an Iμ-Cδ transcript is identified in mice. This Iμ-Cδ transcript is of importance to render both Sμ and σδ regions substrate for DNA modifications, especially because no Iδ exon is documented in the mouse and the human IgH locus. In contrast to mice and humans, an Iδ exon has been recently reported in the bovine IgH locus (Xu et al., 2012). This Iδ exon derives from the recent duplication of an Iμ-Cμ1 genomic fragment (Zhao et al., 2002). Of interest, the bovine δ gene can be expressed by CSR; analysis of Sμ-Sδ junctions demonstrated, as in mice, a preference for using the microhomology-based end-joining pathway (Xu et al., 2012). After μδCSR, Iμ-Cδ transcripts are still detected and are the reflection of the post-IgD CSR transcription and not simply a germinal transcript of importance to render both Sμ and σδ regions substrate to CSR-induced DNA modifications.
The 3′RR is mandatory for cCSR toward IgG, IgA, and IgE isotypes (Vincent-Fabert et al., 2010b). This is obviously not the case for IgD. Patients with hyper-IgM syndromes have higher μδCSR that may stand as a compensatory response due to the absence of other switched isotypes (Chen and Cerutti, 2010). It appears that deletion of the 3′RR, most probably through a compensatory response due to the absence of other switched isotypes, enhances IgD CSR without widely impacting the patterns of Sμ-σδ junctions. Thus, AID-induced μδCSR occurs independently of the IgH 3′RR, a result that markedly contrasts with Sμ-Sγ, Sμ-Sε, and Sμ-Sα cCSR. The lower AID-induced mutations both in Sμ and σδ region during μδCSR (as compared with those in Sμ, Sγ1, and Sγ3 during Sμ-Sγ1 and Sμ-Sγ3 cCSR) suggest that AID might be recruited to the IgH locus by different means than cCSR. This result partially fits with recent data showing, in addition to cCSR, that the 3′RR is also a key element for AID recruitment during AID-induced SHM both in VDJ-expressed sequences and Sμ (Rouaud et al., 2013). The analysis of SHM in VDJ expressed sequences of 3′RR-deficient B cells that have undergone μδCSR confirms that the 3′RR is essential to the SHM process on V(D)J exons and that a physiological dissociation exists between SHM and CSR in primary B cells of 3′RR-deficient mice as in T cell–independent responses (Phan et al., 2005).
CSR to IgD is obviously a rare event and we now show that it occurs independently of the IgH 3′RR. It might be of interest to notice that in this special case, and in contrast to SHM of VDJ and of Sμ regions during cCSR, the rate of mutations affecting the target regions of μδCSR is not affected by the 3′RR deletion. This observation, together with the unique location of IgD switching only in the mucosa-associated lymphoid tissue, argues for a specific regulation of this event, allowing AID recruitment to the IgH locus in a 3′RR-independent manner and in response to stimuli that remains to be determined. Transcription being required for AID targeting, the simultaneous accessibility of Sμ and σδ to AID likely needs constitutive Iμ-Cδ transcription (encompassing both Sμ to σδ). However, because Iμ-Cδ transcription is common in all naive B cells and during their initial entry into GC, it is obviously not sufficient by itself for inducing accessibility to μδCSR and another stimulus uniquely occurring into mouse MLNs and into human tonsils likely allows the 3′RR-independent recruitment of AID on the IgH locus in some B cells. Until now, our knowledge concerning μδCSR in mice only derived from studies of myeloma and hybridoma cell lines (Owens et al., 1991; Preud’homme et al., 2000). 3′RR-deficient mice give us the first opportunity to investigate it in primary B cells.
Location of IgD+IgM− cells in the marginal zone, but not in the follicular zone, together with their PNA−GL7− status and their lack of SHM in the VDJ segment might be suggestive of T cell–independent non-GC origin for cells with μδCSR, as previously documented in physiology for some mucosa-associated lymphoid tissue B cells undergoing T cell–independent IgA CSR (Bergqvist et al., 2010). In addition, cCSR in the absence of SHM was reported in some cases of chronic lymphocytic leukemia B cells (Oppezzo et al., 2003) upon N-terminal mutation of AID (Shinkura et al., 2004) and, recently, in Ung−/− mice during an acute antigenic challenge (Zahn et al., 2013). Our data with μδCSR further indicates that AID can be recruited to the IgH locus under various different forms, eventually associated with some different partners to mediate either cCSR and VDJ SHM (both processes requiring the 3′RR), or in the present case μδCSR (where the 3′RR is at least superfluous; Ronai et al., 2007). According to the concept that such IgD-only cells may locally participate in immune surveillance against pathogens present at mucosal sites, their increase in 3′RR-deficient mice with an IgA production defect might be a compensatory mechanism as previously suggested in IgA-deficient patients (Brandtzaeg and Johansen, 2005).
IgD+IgM− B cells in mice exhibited numerous similarities with human ones. Consistent with published human data (Chen et al., 2009), they were CD138−, expressed the activation-induced (or B1 compartment-related) molecule CD5, and down-regulated several B cell–specific markers usually down-regulated by differentiated plasma cells. Whether human IgD+IgM− cells show a strong bias toward the preferential expression of the λ-light chains (Arpin et al., 1998; Chen et al., 2009), it is not the case in mice, a result which might be linked to the κ+/λ+ ratio, globally reported in mouse B cells around 95/5 (Arakawa et al., 1996). Whether human IgD+IgM− cells are reported to have undergone extensive SHM and display clonal relatedness (Arpin et al., 1998), mice IgD+IgM− cells are unmutated in their VDJ regions with no evident sign of clonality. These latter results are in accordance with their localization in the marginal zone of GC, a region with cells not submitted to an intensive SHM process or exhibiting a high rate of proliferation (mice IgD+IgM− cells had a low Ki67 index).
In conclusion, IgD CSR occurs in B cells of MLNs of wt mice. There is extensive somatic mutation of the IgH V region in human IgD+ cells (Arpin et al., 1998) but not in mouse. The effects of physiological location of switching cells might explain this difference as well as the structural differences in mouse and human 3′ RR. The molecular features of Sμ-σδ junctions suggest a predominant involvement of the A-EJ pathway in this process. In contrast to IgG, IgA, and IgE classes, IgD CSR occurs independently of the IgH 3′RR and now stands as an amazing exception contrasting with all the other AID-dependent modifications of the IgH locus previously shown to formally involve the 3′RR in mouse B cells.
MATERIALS AND METHODS
Mice.
Generation of 3′RR-deficient mice has been previously reported (Vincent-Fabert et al., 2010b). wt mice and 3′RR-deficient mice were 8–12 wk old except when specified. Mice were bred and maintained under specific pathogen-free conditions. Mice immunizations were done orally with sheep red blood cells for 2 wk and intraperitoneally with 10 µg LPS for 3 d.
Cell cytometry analysis.
Single-cell suspensions from MLNs were fixed and permeabilized with the Intraprep permeabilization reagent (Beckman Coulter) before incubation with FITC-labeled IgD (SouthernBiotech) or irrelevant antibody and analyzed on a Fortessa LSR2 (BD; Truffinet et al., 2007; Vincent-Fabert et al., 2009). Phenotyping of IgD+IgM− cells were made with the following antibodies: v450-IgD, APC-IgM, FITC-B220, FITC-CD5, PE-CD11b, PE-CD19, PE-CD21, PC7-CD23, PE-CD138, PE-GL7 and FITC-PNA, and PE-Fas and FITC-Ki67.
Dot plot analysis.
Dot plot analysis of the mouse DNA fragment encompassing the 5′ Eμ intronic enhancer and the Cδ exon 2 was done with the Nucleic Acid Dot Plot Program. Window size and mismatch limit were set to 23 and 3, respectively.
Amplification, cloning, and sequencing of Sμ-σδ junctions.
Sμ-σδ junctions were studied using a touchdown PCR, followed by a nested PCR on fresh MLN cells using the following parameters. Touchdown PCR: primer 1, 5′-CAGTTGAGGCCAGCAGGT-3′ and primer 2, 5-CCAATTACTAAACAGCCCAGGT-3′ (1 cycle, 98°C for 3 min; 3 cycles, 98°C for 30 s, 64°C for 40 s, and 72°C for 90 s; 3 cycles, 98°C for 30 s, 62°C for 40 s, and 72°C for 90 s; 25 cycles, 98°C for 30 s, 60°C for 40 s, and 72°C for 90 s; and 1 cycle, 72°C for 7 min). Nested PCR: primer 3, 5′-CAGGTCGGCTGGACTAACTC-3′ and primer 4, 5′-CAGCCCAGGTTTATCTTTTCA-3′ (1 cycle, 98°C for 3 min; 35 cycles, 98°C for 30 s, 65°C for 40 s, and 72°C for 90 s; and 1 cycle, 72°C for 7 min). The PCR products were cloned into the Zero Blunt Topo PCR cloning (Invitrogen). Plasmids were isolated using the NucleoSpin kit (Macherey-Nagel Eurl) and sequenced using an automated laser fluorescent ANA ABI-PRISM sequencer (Perkin-Elmer; Fiancette et al., 2011). To demonstrate a specific association of σδ with μδCSR, nested PCR analysis was carry out with sense 1 and sense 3 primers and other reverse PCR primers corresponding to Cμ (primer 18, 5′-GTGGGACGAACACATTTACATTGG-3′ and primer 19, 5′-GTTCATCTCTGCGACAGCTG-3′) and a region between Cδ and Iγ3 (primer 20, 5′-GCTCATAGCCTCCTTAGGTTCC-3′ and primer 21, 5′-GAGACTGTTCACTCTATCTTTACCAC-3′).
Amplification, Southern blotting, and sequencing of μδCSR circles.
μδCSR circles were amplified by PCR using the following parameters: primer 6, 5′-GGACTCGTCACCAAATTCCA-3′ (located at the beginning of σδ) and primer 7, 5′-GAAGACATTTGGGAAGGACTGACT-3′ (located in Cμ exon 1; 1 cycle, 95°C for 2 min; 40 cycles, 95°C for 15 s, 55°C for 20 s, and 72°C for 180 s; and 1 cycle, 72°C for 7 min). Hybridization of μδCSR circles was performed with a 240 bp probe (probe B) located in the Cμ exon1 (Péron et al., 2012). The Cμ exon1 probe was cloned as a PCR fragment according to the following parameters: primer 8, 5′-CGTTCGAAGAAGGCTTCCAAAGTC-3′ and primer 9, 5′-TTATCGATGAGGACCAGAGAGGG-3′ (1 cycle, 94°C for 3 min; 30 cycles, 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and 1 cycle, 72°C for 7 min). The circle PCR products were cloned into the Zero Blunt Topo PCR cloning (Invitrogen) and sequenced with and ANA ABI-PRISM sequencer (Perkin-Elmer; Fiancette et al., 2011).
Southern blotting of Cμ-Cδ junctions.
Cμ-Cδ junctions were studied after hybridization with a 32P-labeled probe (460 bp length, probe A). This Cδ probe was cloned as a PCR fragment located in-between primer 5 (5′-CCCAGAACCTGAGAAGGAAG-3′), located in the intron 5′ of Cδ and primer 4 (5′-CAGCCCAGGTTTATCTTTTCA-3′), located in Cδ exon 1 (1 cycle, 94°C for 3 min; 30 cycles, 93°C for 45 s, 52°C for 45 s, and 72°C for 30 s; and 1 cycle, 72°C for 7 min).
DNA extraction and amplification for SHM experiments.
Genomic DNA was extracted from sorted B220+IgD+IgM− cells (85% of purity). IgH VDJ-rearranged fragments were amplified by PCR using the following primers and multistep programs as previously reported (Rouaud et al., 2013): primer 10, 5′-GCGAAGCTTARGCCTGGGRCTTCAGTGAAG-3′ complementary to the VHJ558 segment and primer 11, 5′-AGGCTCTGAGATCCCTAGACAG-3′ corresponding to a sequence 517 bp downstream of the JH4 segment using 1 cycle (98°C for 30 s), 33 cycles (98°C for 10 s, 67°C for 30 s, and 72°C for 90 s), and 1 cycle (72°C for 10 min). Igκ light chain VJ-rearranged fragments were amplified by PCR as previously reported (Rouaud et al., 2013) using the following primers and multistep programs: primer 12, 5′-GGCTGCAGSTTCAGTGGCAGTGGRTCWGGRAC-3′ (consensus for Vκ) and primer 13, 5′-AGCGAATTCAACTTAGGAGACAAAAGAGAGAAC-3′ (found 557 bp downstream of the Jκ5 segment) using 1 cycle at 98°C for 30s, 33 cycles (98°C for 10 s, 66°C for 30 s and 72°C for 90 s), and 1 cycle at 72°C for 10 min. PCR products were cloned and sequenced as described (Fiancette et al., 2011).
Immunohistochemistry.
Frozen 8-µm sections were thawed at room temperature and dried before being hydrated in PBS. Slides were fixed with cold acetone for 20 min at −20°C, dried for 10 min, and incubated with PBS BSA 3% for 45 min. Rat anti–mouse APC-CD45R (BD; clone RA3-6B2, 560472), rat anti–mouse Alexa Fluor 488–IgD (BioLegend; clone 11-26c-2a, 405718), and rat anti–mouse PE-IgM (eBioscience; clone II/41, 17–5790-82) were used at 1:50 for 1 h at 37°C in PBS BSA 0.3%. Goat anti–mouse κ (Beckman Coulter; 731871) or anti–mouse λ (SouthernBiotech; 1060–01) was used at 1:100 and revealed with a DyLight 594 rabbit anti–goat IgG (Thermo Fisher Scientific; 072–09-13-06) used at 1:250. Slides were mounted with moviol medium.
Real-time PCR analysis of membrane Cμ and Iμ-Cδ transcripts.
Total RNA was prepared from freshly isolated IgD+IgM− and IgD+IgM+ MLN B cell. Reverse transcription was performed with 500 ng of total RNA. RT-PCR experiments were done with SuperScript III reverse transcription (Invitrogen). Real-time PCR analysis (on an ABI Prism 7000 system; Applied Biosystems) was made with the Syber Green method using 10 ng cDNA using the following primer: Iμ primer 14, 5′-CTCTGGCCCTGCTTATTGTTG-3′ and Cδ primer 15, 5′-GCTCCCAGCTGATTTTCAGT-3′; μ membrane primer 16 (in exon μ 4), 5′-TGGAACTCCGGAGAGACCTA-3′ and μ membrane primer 17 (in exon μ membrane 1), 5′-TTCCTCCTCAGCATTCACCT-3′. GAPDH was used for normalization of gene expression levels (TaqMan reference Mm99999915-g1).
Spleen cell cultures for CSR.
Single-cell suspensions of spleen cells were cultured 3 d at 106 cells/ml in RPMI 1640 with 10% FCS and 20 µg/ml LPS, with or without addition of 20 ng/ml IL-4 (PeproTech). 3-d in vitro–stimulated splenocytes were harvested and DNA was extracted for investigation of Sμ-Sγ1 and Sμ-Sγ3 junctions (Vincent-Fabert et al., 2009).
LM-PCR.
Genomic DNA from wt mice, 3′RR-deficient mice, and AID-deficient mice was extracted and purified using Phase Lock gel. 10 µg was treated with T4 DNA polymerase (New England Biolabs, Inc.), which trims back 3′ overhangs while filling in 3′-recessed ends, thereby yielding blunt 5′-phosphorylated DNA ends. 1 µg DNA was then ligated with T4 DNA ligase (New England Biolabs, Inc.) in a 20-µl reaction volume with the double-strand anchor linker BW (Zan and Casali, 2008). DSBs were detected by semi-nested LM-PCR using BW1 primer targeting the BW linker sequence (5′-GCGGTGACCCGGGAGATCTGAATTC-3′) and primer 2, specific to σδ (5′-CCAATTACTAAACAGCCCAGGT-3′), using the following parameters: (1 cycle, 94°C for 30 s; 25 cycles, 94°C for 10 s, 55°C for 20 s, and 65°C for 2 min; and 1 cycle, 65°C for 7 min). DSBs were studied after hybridization with the same 32P-labeled probe as for Southern blotting of Cµ-Cδ junctions.
Clonality assay.
Genomic DNA prepared from IgD+IgM− cells of 3′RR-deficient mice was digested with EcoRI and analyzed by Southern blot with a 32P-labeled JH probe (Truffinet et al., 2007).
Online supplemental material.
Table S1 shows Sμ-σδ, Sμ-Sγ1, and Sμ-Sγ3 junctions.
Supplementary Material
Acknowledgments
This paper is dedicated to Michael S. Neuberger in memoriam.
We thank S. Desforges and B. Remerand for help with animal care.
P. Rouaud was supported by a grant from ARC. This work was supported by grants from Conseil Régional du Limousin, Association pour la Recherche sur le Cancer (ARC SL 220100601332) and ANR (Projets Blanc 2011).
The authors declare no competing financial interests.
Contributions: P. Rouaud, A. Saintamand, S. Lecardeur, F. Saad, C. Carrion, M. Cogné, and Y. Denizot actively participated to the experimental design of the study. M. Cogné and Y. Denizot participated in the scientific discussion for manuscript writing, and obtained financial grants and agreement of the ethics committee of our institution to perform the study.
Footnotes
Abbreviations used:
- 3′RR
- 3′ regulatory region
- A-EJ
- alternative end-joining
- AID
- activation-induced deaminase
- cCSR
- classical class-switch recombination
- C-NHEJ
- classical nonhomologous end joining
- DSB
- double-strand break
- GC
- germinal center
- LM-PCR
- ligation-mediated PCR
- MLN
- mesenteric LN
- SHM
- somatic hypermutation
References
- Arakawa H., Shimizu T., Takeda S. 1996. Re-evaluation of the probabilities for productive arrangements on the κ and λ loci. Int. Immunol. 8:91–99 10.1093/intimm/8.1.91 [DOI] [PubMed] [Google Scholar]
- Arpin C., de Bouteiller O., Razanajaona D., Fugier-Vivier I., Brière F., Banchereau J., Lebecque S., Liu Y.J. 1998. The normal counterpart of IgD myeloma cells in germinal center displays extensively mutated IgVH gene, Cμ-Cδ switch, and λ light chain expression. J. Exp. Med. 187:1169–1178 10.1084/jem.187.8.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bébin A.G., Carrion C., Marquet M., Cogné N., Lecardeur S., Cogné M., Pinaud E. 2010. In vivo redundant function of the 3′ IgH regulatory element HS3b in the mouse. J. Immunol. 184:3710–3717 10.4049/jimmunol.0901978 [DOI] [PubMed] [Google Scholar]
- Bergqvist P., Stensson A., Lycke N.Y., Bemark M. 2010. T cell-independent IgA class switch recombination is restricted to the GALT and occurs prior to manifest germinal center formation. J. Immunol. 184:3545–3553 10.4049/jimmunol.0901895 [DOI] [PubMed] [Google Scholar]
- Boboila C., Alt F.W., Schwer B. 2012. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv. Immunol. 116:1–49 10.1016/B978-0-12-394300-2.00001-6 [DOI] [PubMed] [Google Scholar]
- Brandtzaeg P., Johansen F.E. 2005. Mucosal B cells: phenotypic characteristics, transcriptional regulation, and homing properties. Immunol. Rev. 206:32–63 10.1111/j.0105-2896.2005.00283.x [DOI] [PubMed] [Google Scholar]
- Chan T.D., Brink R. 2012. Affinity-based selection and the germinal center response. Immunol. Rev. 247:11–23 10.1111/j.1600-065X.2012.01118.x [DOI] [PubMed] [Google Scholar]
- Chen K., Cerutti A. 2010. New insights into the enigma of immunoglobulin D. Immunol. Rev. 237:160–179 10.1111/j.1600-065X.2010.00929.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Xu W., Wilson M., He B., Miller N.W., Bengtén E., Edholm E.S., Santini P.A., Rath P., Chiu A., et al. 2009. Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nat. Immunol. 10:889–898 10.1038/ni.1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogné M., Lansford R., Bottaro A., Zhang J., Gorman J., Young F., Cheng H.L., Alt F.W. 1994. A class switch control region at the 3′ end of the immunoglobulin heavy chain locus. Cell. 77:737–747 10.1016/0092-8674(94)90057-4 [DOI] [PubMed] [Google Scholar]
- Durandy A. 2003. Activation-induced cytidine deaminase: a dual role in class-switch recombination and somatic hypermutation. Eur. J. Immunol. 33:2069–2073 10.1002/eji.200324133 [DOI] [PubMed] [Google Scholar]
- Fiancette R., Rouaud P., Vincent-Fabert C., Laffleur B., Magnone V., Cogné M., Denizot Y. 2011. A p53 defect sensitizes various stages of B cell development to lymphomagenesis in mice carrying an IgH 3′ regulatory region-driven c-myc transgene. J. Immunol. 187:5772–5782 10.4049/jimmunol.1102059 [DOI] [PubMed] [Google Scholar]
- Hackney J.A., Misaghi S., Senger K., Garris C., Sun Y., Lorenzo M.N., Zarrin A.A. 2009. DNA targets of AID: evolutionary link between antibody somatic hypermutation and class switch recombination. Adv. Immunol. 101:163–189 10.1016/S0065-2776(08)01005-5 [DOI] [PubMed] [Google Scholar]
- Hamada H., Hiroi T., Nishiyama Y., Takahashi H., Masunaga Y., Hachimura S., Kaminogawa S., Takahashi-Iwanaga H., Iwanaga T., Kiyono H., et al. 2002. Identification of multiple isolated lymphoid follicles on the antimesenteric wall of the mouse small intestine. J. Immunol. 168:57–64 [DOI] [PubMed] [Google Scholar]
- Henderson A., Calame K. 1998. Transcriptional regulation during B cell development. Annu. Rev. Immunol. 16:163–200 10.1146/annurev.immunol.16.1.163 [DOI] [PubMed] [Google Scholar]
- Johansen F.E., Baekkevold E.S., Carlsen H.S., Farstad I.N., Soler D., Brandtzaeg P. 2005. Regional induction of adhesion molecules and chemokine receptors explains disparate homing of human B cells to systemic and mucosal effector sites: dispersion from tonsils. Blood. 106:593–600 10.1182/blood-2004-12-4630 [DOI] [PubMed] [Google Scholar]
- Manis J.P., van der Stoep N., Tian M., Ferrini R., Davidson L., Bottaro A., Alt F.W. 1998. Class switching in B cells lacking 3′ immunoglobulin heavy chain enhancers. J. Exp. Med. 188:1421–1431 10.1084/jem.188.8.1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K.W., Rogers J., Hunkapiller T., Early P., Nottenburg C., Weissman I., Bazin H., Wall R., Hood L.E. 1981. Expression of IgD may use both DNA rearrangement and RNA splicing mechanisms. Proc. Natl. Acad. Sci. USA. 78:1800–1804 10.1073/pnas.78.3.1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y., Flajnik M. 2006. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc. Natl. Acad. Sci. USA. 103:10723–10728 10.1073/pnas.0601407103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppezzo P., Vuillier F., Vasconcelos Y., Dumas G., Magnac C., Payelle-Brogard B., Pritsch O., Dighiero G. 2003. Chronic lymphocytic leukemia B cells expressing AID display dissociation between class switch recombination and somatic hypermutation. Blood. 101:4029–4032 10.1182/blood-2002-10-3175 [DOI] [PubMed] [Google Scholar]
- Oretti C., Barbi E., Marchetti F., Lepore L., Ventura A., D’Osualdo A., Gattorno M., Martelossi S., Tommasini A. 2006. Diagnostic challenge of hyper-IgD syndrome in four children with inflammatory gastrointestinal complaints. Scand. J. Gastroenterol. 41:430–436 10.1080/00365520500327743 [DOI] [PubMed] [Google Scholar]
- Owens J.D., Jr, Finkelman F.D., Mountz J.D., Mushinski J.F. 1991. Nonhomologous recombination at sites within the mouse JH-C δ locus accompanies C μ deletion and switch to immunoglobulin D secretion. Mol. Cell. Biol. 11:5660–5670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R., Nussenzweig M.C. 2011. AID targeting in antibody diversity. Adv. Immunol. 110:1–26 10.1016/B978-0-12-387663-8.00005-3 [DOI] [PubMed] [Google Scholar]
- Péron S., Laffleur B., Denis-Lagache N., Cook-Moreau J., Tinguely A., Delpy L., Denizot Y., Pinaud E., Cogné M. 2012. AID-driven deletion causes immunoglobulin heavy chain locus suicide recombination in B cells. Science. 336:931–934 10.1126/science.1218692 [DOI] [PubMed] [Google Scholar]
- Phan T.G., Gardam S., Basten A., Brink R. 2005. Altered migration, recruitment, and somatic hypermutation in the early response of marginal zone B cells to T cell-dependent antigen. J. Immunol. 174:4567–4578 [DOI] [PubMed] [Google Scholar]
- Pinaud E., Khamlichi A.A., Le Morvan C., Drouet M., Nalesso V., Le Bert M., Cogné M. 2001. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity. 15:187–199 10.1016/S1074-7613(01)00181-9 [DOI] [PubMed] [Google Scholar]
- Pinaud E., Marquet M., Fiancette R., Péron S., Vincent-Fabert C., Denizot Y., Cogné M. 2011. The IgH locus 3′ regulatory region: pulling the strings from behind. Adv. Immunol. 110:27–70 10.1016/B978-0-12-387663-8.00002-8 [DOI] [PubMed] [Google Scholar]
- Preud’homme J.L., Petit I., Barra A., Morel F., Lecron J.C., Lelièvre E. 2000. Structural and functional properties of membrane and secreted IgD. Mol. Immunol. 37:871–887 10.1016/S0161-5890(01)00006-2 [DOI] [PubMed] [Google Scholar]
- Rauta P.R., Nayak B., Das S. 2012. Immune system and immune responses in fish and their role in comparative immunity study: a model for higher organisms. Immunol. Lett. 148:23–33 10.1016/j.imlet.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Ronai D., Iglesias-Ussel M.D., Fan M., Li Z., Martin A., Scharff M.D. 2007. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J. Exp. Med. 204:181–190 10.1084/jem.20062032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaud P., Vincent-Fabert C., Fiancette R., Cogné M., Pinaud E., Denizot Y. 2012. Enhancers located in heavy chain regulatory region (hs3a, hs1,2, hs3b, and hs4) are dispensable for diversity of VDJ recombination. J. Biol. Chem. 287:8356–8360 10.1074/jbc.M112.341024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaud P., Vincent-Fabert C., Saintamand A., Fiancette R., Marquet M., Robert I., Reina-San-Martin B., Pinaud E., Cogné M., Denizot Y. 2013. The IgH 3′ regulatory region controls somatic hypermutation in germinal center B cells. J. Exp. Med. 210:1501–1507 10.1084/jem.20130072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkura R., Ito S., Begum N.A., Nagaoka H., Muramatsu M., Kinoshita K., Sakakibara Y., Hijikata H., Honjo T. 2004. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol. 5:707–712 10.1038/ni1086 [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Björkman A., Du L., Cagigi A., Pan-Hammarström Q. 2010. Mapping of switch recombination junctions, a tool for studying DNA repair pathways during immunoglobulin class switching. Adv. Immunol. 108:45–109 10.1016/B978-0-12-380995-7.00003-3 [DOI] [PubMed] [Google Scholar]
- Truffinet V., Pinaud E., Cogné N., Petit B., Guglielmi L., Cogné M., Denizot Y. 2007. The 3′ IgH locus control region is sufficient to deregulate a c-myc transgene and promote mature B cell malignancies with a predominant Burkitt-like phenotype. J. Immunol. 179:6033–6042 [DOI] [PubMed] [Google Scholar]
- Vincent-Fabert C., Truffinet V., Fiancette R., Cogné N., Cogné M., Denizot Y. 2009. Ig synthesis and class switching do not require the presence of the hs4 enhancer in the 3′ IgH regulatory region. J. Immunol. 182:6926–6932 10.4049/jimmunol.0900214 [DOI] [PubMed] [Google Scholar]
- Vincent-Fabert C., Fiancette R., Cogné M., Pinaud E., Denizot Y. 2010a. The IgH 3′ regulatory region and its implication in lymphomagenesis. Eur. J. Immunol. 40:3306–3311 10.1002/eji.201040778 [DOI] [PubMed] [Google Scholar]
- Vincent-Fabert C., Fiancette R., Pinaud E., Truffinet V., Cogné N., Cogné M., Denizot Y. 2010b. Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. 116:1895–1898 10.1182/blood-2010-01-264689 [DOI] [PubMed] [Google Scholar]
- Xu B., Wang J., Zhang M., Wang P., Wei Z., Sun Y., Tao Q., Ren L., Hu X., Guo Y., et al. 2012. Expressional analysis of immunoglobulin D in cattle (Bos taurus), a large domesticated ungulate. PLoS ONE. 7:e44719 10.1371/journal.pone.0044719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.T., Boboila C., Souza E.K., Franco S., Hickernell T.R., Murphy M., Gumaste S., Geyer M., Zarrin A.A., Manis J.P., et al. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 449:478–482 10.1038/nature06020 [DOI] [PubMed] [Google Scholar]
- Zahn A., Daugan M., Safavi S., Godin D., Cheong C., Lamarre A., Di Noia J.M. 2013. Separation of function between isotype switching and affinity maturation in vivo during acute immune responses and circulating autoantibodies in UNG-deficient mice. J. Immunol. 190:5949–5960 10.4049/jimmunol.1202711 [DOI] [PubMed] [Google Scholar]
- Zan H., Casali P. 2008. AID- and Ung-dependent generation of staggered double-strand DNA breaks in immunoglobulin class switch DNA recombination: a post-cleavage role for AID. Mol. Immunol. 46:45–61 10.1016/j.molimm.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrin A.A., Tian M., Wang J., Borjeson T., Alt F.W. 2005. Influence of switch region length on immunoglobulin class switch recombination. Proc. Natl. Acad. Sci. USA. 102:2466–2470 10.1073/pnas.0409847102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Kacskovics I., Pan Q., Liberles D.A., Geli J., Davis S.K., Rabbani H., Hammarstrom L. 2002. Artiodactyl IgD: the missing link. J. Immunol. 169:4408–4416 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.