Interaction between the inhibitory molecule PD-L2 on dendritic cells and repulsive guidance molecule b (RGMb) on lung macrophages is required to establish respiratory tolerance.
Abstract
We report that programmed death ligand 2 (PD-L2), a known ligand of PD-1, also binds to repulsive guidance molecule b (RGMb), which was originally identified in the nervous system as a co-receptor for bone morphogenetic proteins (BMPs). PD-L2 and BMP-2/4 bind to distinct sites on RGMb. Normal resting lung interstitial macrophages and alveolar epithelial cells express high levels of RGMb mRNA, whereas lung dendritic cells express PD-L2. Blockade of the RGMb–PD-L2 interaction markedly impaired the development of respiratory tolerance by interfering with the initial T cell expansion required for respiratory tolerance. Experiments with PD-L2–deficient mice showed that PD-L2 expression on non–T cells was critical for respiratory tolerance, but expression on T cells was not required. Because PD-L2 binds to both PD-1, which inhibits antitumor immunity, and to RGMb, which regulates respiratory immunity, targeting the PD-L2 pathway has therapeutic potential for asthma, cancer, and other immune-mediated disorders. Understanding this pathway may provide insights into how to optimally modulate the PD-1 pathway in cancer immunotherapy while minimizing adverse events.
Programmed death 1 (PD-l, CD279) and its ligands PD-L1 (B7-H1, CD274) and PD-L2 (B7-DC, CD273) are key inhibitory molecules in immune regulation (Keir et al., 2008; Pardoll, 2012). This pathway provides particularly promising targets for cancer immunotherapy (Topalian et al., 2012). There is considerable evidence that PD-L2 inhibits immunity by binding to the PD-1 co-inhibitory receptor (Latchman et al., 2001; Zhang et al., 2006). However, several studies have shown that PD-L2 can function to stimulate T cell proliferation and cytokine production, even in PD-1–deficient T cells or with PD-L2 mutants that did not bind to PD-1 (Liu et al., 2003; Shin et al., 2003; Wang et al., 2003). These findings suggest that PD-L2 may function through a receptor other than PD-1. Most studies using blocking mAbs show a dominant role for PD-L1 in inhibiting immune responses; however, PD-L2 plays a dominant role in responses such as airway hypersensitivity, experimental allergic conjunctivitis and nematode infection (Ritprajak et al., 2012). In some situations, PD-L2 dominance may be explained by preferential PD-L2 up-regulation by IL-4, but other instances may be explained by the binding of PD-L2 to a receptor other than PD-1.
Here, we demonstrate that PD-L2 binds to a second receptor, repulsive guidance molecule b (RGMb). RGMb, also known as DRAGON, is a member of the RGM family which consists of RGMa, RGMb, and RGMc/hemojuvelin (Severyn et al., 2009). RGMs are glycosylphosphatidylinositol-anchored membrane proteins that bind bone morphogenetic proteins (BMPs) and neogenin (Conrad et al., 2010). RGMs do not directly signal but can act as co-receptors that modulate BMP signaling (Samad et al., 2005). RGMb is expressed and functions in the nervous system (Severyn et al., 2009). In addition, RGMb expression is observed in macrophages and other cells of the immune system (Xia et al., 2011). However, the function of RGMb in the immune system is only beginning to emerge (Galligan et al., 2007; Xia et al., 2011). RGMb-deficient mice have an early lethal phenotype (Xia et al., 2011).
Here, we characterize RGMb binding to PD-L2 and identify RGMb protein expression in mouse hematopoietic cells and human cancer cell lines. Based on the critical role of PD-L2 in lung immune regulation (Akbari et al., 2010; Singh et al., 2011) and RGMb expression in the lung, we investigated the function of RGMb and PD-L2 in respiratory tolerance. Blockade of PD-L2 and RGMb interaction prevented the development of respiratory tolerance.
RESULTS
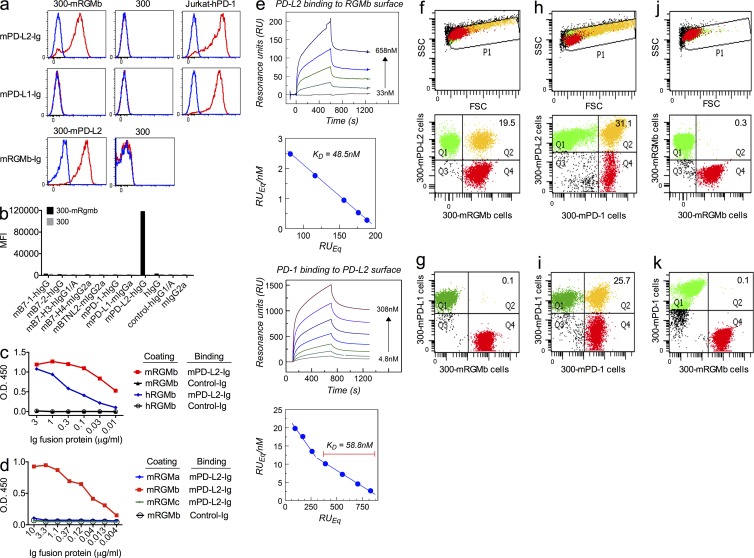
RGMb binds to PD-L2, but not to PD-L1 or other related molecules
We identified RGMb as a novel binding partner for PD-L2 using COS cell expression cloning with PD-L2-Ig fusion protein. Using flow cytometry with stably transfected 300 cells and Ig fusion proteins, we found that mRGMb binds to mPD-L2 but not mPD-L1 or other proteins of the B7 family (Fig. 1, a and b). ELISA with purified proteins showed that mRGMb binds to mPD-L2 and hPD-L2, and that hRGMb binds to hPD-L2 and mPD-L2 (Fig. 1 c and not depicted). Thus, the RGMb–PD-L2 interaction occurs in both mice and humans. Further studies showed that PD-L2 does not bind to RGMa or RGMc (Fig. 1 d). Biacore data showed that PD-L2 bound to RGMb with a similar affinity as to PD-1, Kd = 48.5 and 58.8 nM, respectively (Fig. 1 e).
Figure 1.
RGMb binds to PD-L2, but not PD-L1 or other related molecules. (a and b) mRGMb- or mPD-L2–transfected 300 cells, untransfected 300 cells, and PD-1–transfected Jurkat T cells were stained with the indicated Ig fusion proteins (red) or control-Ig (blue) and analyzed by flow cytometry. MFI, mean fluorescence intensity. (c and d) Binding of mPD-L2-Ig or control-Ig to plates coated with mRGMb-HIS, hRGMb-HIS, mRGMa-HIS or mRGMc-HIS, was analyzed by ELISA. (e) Biacore analysis of the interaction of RGMb with PD-L2 and of PD-1 with PD-L2. (f–k) Cell–cell binding of the indicated transfected cells was analyzed by cell conjugation assay. In f, h, and j, the top panels show the FSC-SSC plots and the bottom panels show the corresponding dot plots. g, i, and k show only the dot plots. All data are representative of 2–11 independent experiments.
To test the RGMb–PD-L2 interaction under more physiological conditions, we used a cell conjugation assay where one transfected cell was labeled with a red dye and the other transfected cell with a green dye. The binding of the two cells was assessed by flow cytometry and indicated by double positive events (yellow dots). As expected, mRGMb cells bound to mPD-L2 cells (Fig. 1 f), but not mPD-L1 cells (Fig. 1 g). As positive controls, mPD-1 cells bound to both mPD-L2 and mPD-L1 cells (Fig. 1, h and i). Negative control binding assays had few conjugates (<0.3%; Fig. 1, j and k; and not depicted). These results show that mRGMb binds specifically to mPD-L2, but not to mPD-L1, and the structural orientation is compatible with RGMb and PD-L2 cell surface-to-cell surface binding.
Characterization of RGMb and PD-L2 mAbs
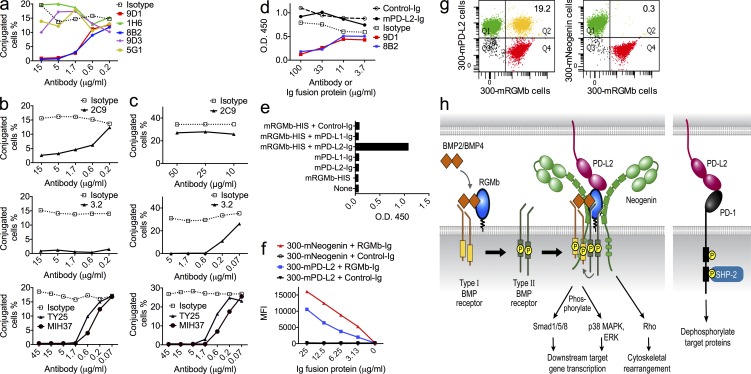
We characterized a panel of mRGMb mAbs and identified 6 RGMb mAbs showing specificity by flow cytometry on mRGMb- and/or hRGMb-transfected 300 cells, ELISA, and Western blotting (unpublished data). We identified two mRGMb mAbs, 9D1 and 8B2, that blocked interaction of RGMb with PD-L2 (Fig. 2 a). These RGMb antibodies do not bind to mRGMa or mRGMc (unpublished data).
Figure 2.
Blocking capacities of RGMb and PD-L2 mAbs and a model for RGMb and PD-L2 interactions. (a–c) The blocking capacity of RGMb mAbs and PD-L2 mAbs was determined by cell conjugation assay. After cells were stained with dyes, (a) 300-mRGMb cells were incubated with the indicated concentrations of RGMb mAbs before incubation with 300-mPD-L2 cells; (b) 300-mPD-L2 cells were incubated with the indicated concentrations of PD-L2 mAbs before incubation with 300-mRGMb cells; or (c) before incubation with 300-mPD-1 cells. (d) mRGMb-HIS was preincubated with mAbs or Ig fusion proteins and then added to BMP-4–coated plates. Binding of mRGMb-HIS was detected by anti–penta-HIS-HRP in an ELISA. Similar results were seen with BMP-2 (not depicted). (e) mPD-L2-hIgG or control-hIgG were preincubated alone or with monomeric mRGMb-HIS, and then added to BMP-4–coated plates. Binding of Ig fusion proteins was detected with anti–hIgG-HRP in an ELISA. Similar results were also seen for BMP-2 (not depicted). (f) mNeogenin- or mPD-L2–transfected 300 cells were stained with indicated Ig fusion proteins or control-Ig and analyzed by flow cytometry. (g) Binding of 300-mRGMb cells to 300-mPD-L2 or 300-mNeogenin cells was analyzed by cell conjugation assay. Cells stained with a red dye were incubated with cells stained with a green dye and binding was measured by flow cytometry. (h) Molecular model depicting PDL2–BMP–BMPR–RGMb–neogenin and PD-L2–PD-1 pathways. All data are representative of two or more independent experiments.
We also determined whether mPD-L2 mAbs would block PD-L2 interactions with RGMb. We identified one mPD-L2 mAb, 2C9, that blocked PD-L2–RGMb but not PD-L2–PD-1 interactions, and three mPD-L2 mAbs, 3.2, TY25, and MIH37, that blocked both PD-L2–RGMb and PD-L2–PD-1 interactions (Fig. 2, b and c). TY25 and 3.2 share the same epitope, whereas 2C9 recognizes a different epitope (unpublished data). The existence of both single and double blocker PD-L2 mAbs suggests that the PD-1– and RGMb-binding sites on PD-L2 are close but distinct. Properties of RGMb and PD-L2 mAbs are summarized in Table I.
Table 1.
Blocking capacities of mRGMb and mPD-L2 mAbs
| Blocking | mRGMb to mPD-L2 | mPD-L2 to mPD-1 | mRGMb to mBMP-2/4 | mRGMb to mNeogenin |
| mRGMb mAbs | ||||
| 307.9D1, 307.8B2 | Yes | N.A. | Yes | Weak |
| 307.1H6, 307.9D3, 307.5G1 | No | N.A. | No | Weak |
| mPD-L2 mAbs | ||||
| GF17.2C9 | Yes | No | N.A. | N.A. |
| 3.2, TY25, MIH37 | Yes | Yes | N.A. | N.A. |
N.A., not applicable.
Interactions of RGMb with PD-L2, BMPs, and neogenin
RGMb is known to bind directly to BMP-2/4 (Samad et al., 2005; Wu et al., 2012). BMPs consist of a family of >20 members related to the TGF-β family (Bragdon et al., 2011; Yoshioka et al., 2012). We analyzed the capacities of RGMb antibodies to block RGMb binding to BMP-2/4. 9D1 and 8B2 blocked RGMb binding to BMP-2/4 in an ELISA (Fig. 2 d and not depicted) and thus are dual blockers of RGMb interactions with PD-L2 and BMP-2/4. However, PD-L2-Ig fusion protein did not block RGMb binding to BMP-2/4 in an ELISA (Fig. 2 d and not depicted). These data suggest that the binding sites on RGMb for PD-L2 and BMP are close but distinct.
To test if RGMb can bind both PD-L2 and BMP at the same time, we performed an ELISA to analyze the binding of PD-L2-Ig fusion protein to immobilized BMP-2/4 in the presence or absence of RGMb. PD-L2 could not directly bind to BMP-2/4, but in the presence of RGMb could form a complex with BMP when RGMb and PD-L2-Ig were added simultaneously or sequentially to BMP-2/4 (Fig. 2 e and not depicted). These data are consistent with RGMb having distinct sites for PD-L2 and BMP binding, and show that RGMb has the capacity to form a trimeric complex with BMP and PD-L2.
RGMb has also been reported to bind to neogenin (Conrad et al., 2010; Bell et al., 2013). Furthermore, neogenin has been shown to directly bind BMP-2/4/6/7, and to modulate BMP signaling (Hagihara et al., 2011; Tian and Liu, 2013). We found that soluble RGMb-mIgG2a bound to neogenin-transfected 300 cells (Fig. 2 f). RGMb mAbs weakly blocked RGMb-mIgG2a binding to neogenin (unpublished data). Surprisingly, cell conjugation assays showed that 300-mRGMb cells did not bind to 300-neogenin cells (Fig. 2 g), suggesting that the structural orientation of RGMb and neogenin binding is not compatible with cell to cell binding but can support binding in cis on the same cell surface or of soluble RGMb to cell surface neogenin.
Model of RGMb interactions
Based on previous studies and our findings, we propose a model for RGMb–PD-L2 signaling (Fig. 2 h, left). RGMb directly binds to BMP-2 or BMP-4, which bind to type I BMP receptors (BMPR1a, BMPR1b, ACVR1, and ACVRL1) and recruit type II BMP receptors (BMPR2, ACVR2a, and ACVR2b; Corradini et al., 2009; Yoshioka et al., 2012). Then type II BMP receptors phosphorylate type I BMP receptors, which phosphorylate Smad1/5/8 or p38 mitogen-activated protein kinase (MAPK) and extracellular signal-regulated protein kinase (ERK), leading to downstream target gene transcription (Corradini et al., 2009; Xia et al., 2011). RGMs facilitate the utilization of ACVR2a by BMP-2/4. In the absence of an RGM, BMP-2/4 preferentially use BMPR2 (Corradini et al., 2009). RGMb may also signal through neogenin and downstream effector Rho, triggering cytoskeletal rearrangement (Conrad et al., 2007; Bell et al., 2013). We propose that RGMb can form a signaling supercomplex of BMP–BMP receptors–RGMb–neogenin (BBRN supercomplex). We would caution that this model pictures all the possible players and not all may be complexed simultaneously. PD-L2 may interact with this BBRN supercomplex by binding to RGMb, and modulate these signaling pathways. Fig. 2 h (right) shows PD-L2 binding to PD-1, which results in tyrosine phosphorylation of the PD-1 cytoplasmic domain, recruitment of tyrosine phosphatases, particularly SHP-2, and attenuation of antigen receptor signals. Thus, PD-L2 may participate in three important signaling circuits, the PD-1, BMP, and neogenin signaling pathways, by binding to either PD-1 or RGMb.
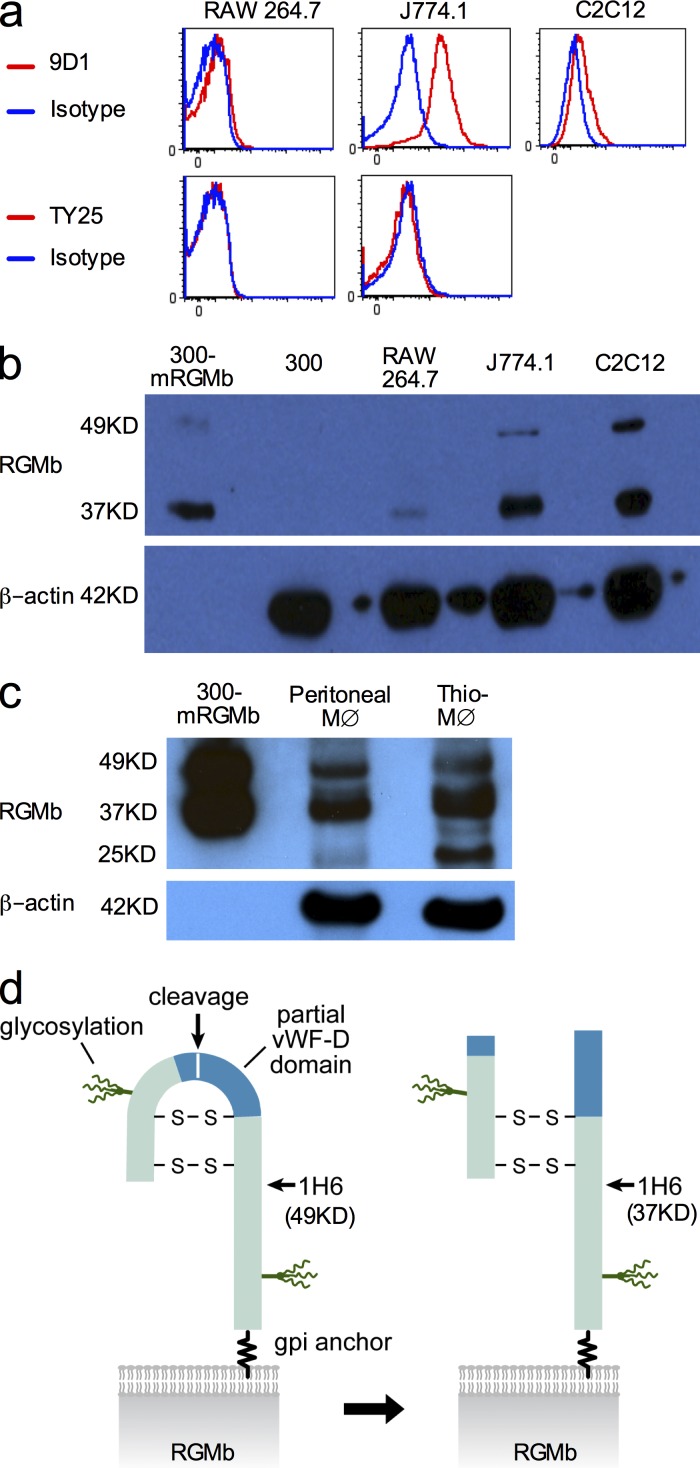
RGMb expression in mouse macrophages and RGMb protein structure
RGMb mRNA has been reported in mouse lung macrophages, macrophage cell line RAW264.7 and myoblast cell line C2C12 (Xia et al., 2011), but protein expression was not determined. We confirmed RGMb mRNA expression (not depicted) and showed cell surface expression of RGMb protein on these cell lines by flow cytometry using RGMb mAb 9D1 (Fig. 3 a).
Figure 3.
RGMb expression in mouse macrophages and RGMb protein structure. (a) Flow cytometric analysis of cell surface expression of mRGMb and mPD-L2 on indicated cells using PE-conjugated mAbs. (b and c) RGMb expression in the indicated cell type by Western blotting using mAb 1H6. The loading amount was 60–80 µg/lane except for 0.5 or 1 µg/lane for 300-mRGMb. MØ indicates macrophages. Thio-MØ indicates thioglycollate-induced peritoneal macrophages. (d) Model of RGMb protein structure. Data in a–c are representative of two or more experiments.
We also analyzed RGMb protein expression using RGMb mAb 1H6, which Western blots both mouse and human RGMb. One major band (∼37 kD) and one minor band (∼49 kD) were detected in RAW264.7, J774.1, and C2C12 cells as well as mRGMb-transfected 300 cells (Fig. 3 b). No such bands were observed in 300 cells (Fig. 3 b), consistent with the absence of RGMb mRNA (unpublished data). Western blotting demonstrated RGMb expression in mouse peritoneal macrophages and thioglycollate-induced peritoneal macrophages (Fig. 3 c).
Fig. 3 d shows how proteolytic cleavage of RGMb accounts for these multiple protein bands. RGMb contains a portion of a von Willebrand Factor type D domain with a proteolytic cleavage site between Asp171 and Pro172. After cleavage, the two fragments of RGMb remain connected by disulfide bonds (Severyn et al., 2009). The molecular weight of uncleaved RGMb is predicted to be 40 kD, and cleaved RGMb will have N- and C-terminal fragments of 13 and 27 kD, respectively. Each fragment has one predicted N-linked glycosylation site, which should increase the molecular weight of each fragment by 5–10 kD. The 1H6 mAb reacts with an epitope in the C-terminal fragment and recognizes the 37-kD cleaved form as well as the 49-kD uncleaved form. Our Western blotting analysis shows that most native RGMb is the cleaved form.
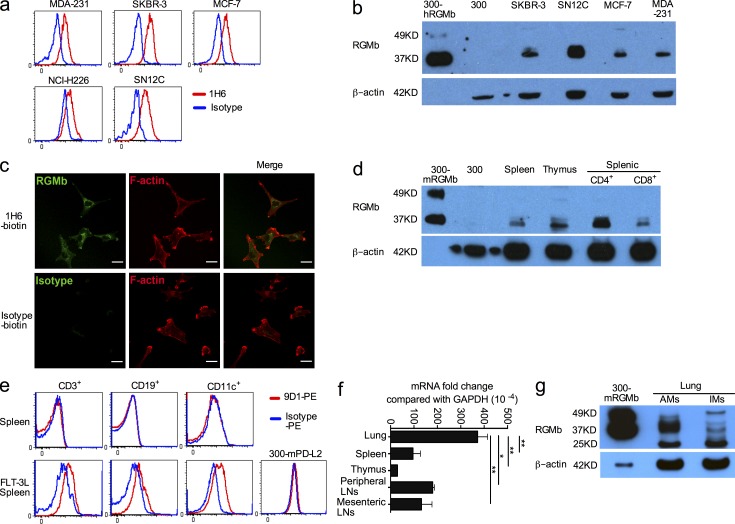
RGMb can be expressed on the cell surface, but is primarily localized intracellularly
Previous studies have reported RGMb in breast and prostate tumor cells (Li et al., 2012a,b). We detected RGMb protein on the cell surface of human breast cancer cell lines, MDA-231, SKBR-3, and MCF-7, nonsmall cell lung cancer cell line NCI-H226, and renal cancer cell line SN12C (Fig. 4 a), and this expression was confirmed by Western blotting (Fig. 4 b). Because a previous immunohistochemical study showed RGMb primarily in the cytoplasm of prostate tumor cells (Li et al., 2012a), we also evaluated the cellular localization of RGMb in SKBR-3 cells. Confocal microscopy showed substantial amounts of RGMb inside SKBR-3 cells (Fig. 4 c), consistent with the published study, but also revealed detectable RGMb on the cell surface (Fig. 4 a).
Figure 4.
RGMb can be expressed on the cell surface but is primarily localized intracellularly. (a) RGMb cell surface expression on human breast cancer cell lines (MDA-231, SKBR-3, and MCF-7), the nonsmall cell lung cancer cell line NCI-H226, and the renal cancer cell line SN12C was analyzed by flow cytometry with mAb 1H6 (red) or isotype control (blue). (b) RGMb expression by Western blotting using mAb 1H6. The loading amount was 60–80 µg/lane except for 0.5 µg/lane for 300-hRGMb. (c) SKBR-3 cells were stained for RGMb (1H6-biotin) or isotype control plus streptavidin-Alexa Fluor 488 (green), and then with Phalloidin-TRIC to label F-actin, and analyzed by confocal microscopy. Bars, 25 µm. (d) RGMb expression by Western blotting using mAb 1H6. The loading amount was 60–80 µg/lane, except for 0.5 µg/lane for 300-mRGMb cells. (e) Splenocytes from naive mice and mice treated with FLT-3L as well as 300-mPD-L2 cells (negative control) were stained for surface markers CD3, CD19, and CD11c. Then intracellular flow cytometry staining with PE-conjugated RGMb mAb 9D1 (red) or rat IgG2a (blue) was used to analyze RGMb expression in these cells. (f) RGMb expression in unstimulated cells from the indicated mouse organs by qRT-PCR. Data are mean ± SEM; n = 2; *, P < 0.05; **, P < 0.01. Ordinary one-way ANOVA followed by Dunnett’s multiple comparison test. (g) RGMb expression in lung AM and IM by Western blotting as in d. All data are representative of two or more experiments.
Western blotting showed positive RGMb expression in cells from spleen, thymus, purified splenic CD4+, and CD8+ T cells from naive mice (Fig. 4 d). Cell surface RGMb expression was not detectable in primary hematopoietic cells by flow cytometry with PE-conjugated RGMb mAb 9D1 (unpublished data). RGMb mRNA and protein levels were not up-regulated in T cells by CD3 and/or CD28 activation (unpublished data), suggesting that RGMb is not a T cell activation molecule. Intracellular flow cytometry staining using PE-conjugated RGMb mAb 9D1 did not detect RGMb expression in splenic T cells (CD3+), B cells (CD19+), or DCs (CD11c+) from naive mice (Fig. 4 e, top), but detected low levels of RGMb expression in these cells from mice treated with FMS-like tyrosine kinase 3 ligand (FLT-3L) to expand DC populations (Fig. 4 e, bottom). Our findings of intracellular RGMb expression are in agreement with the intracellular expression seen in confocal microscopy of SKBR-3 cells (Fig. 4 c) and immunohistochemical staining of cancer cells (Li et al., 2012a). The intracellular localization of RGMb along with the variability in RGMb cell surface expression suggests additional levels of regulation of RGMb cell surface expression.
qRT-PCR showed positive RGMb expression in cells from lung, spleen, thymus, peripheral, and mesenteric LNs with much higher levels of RGMb mRNA in lung cells (Fig. 4 f). Western blotting demonstrated RGMb expression in mouse lung alveolar macrophages (AMs) and interstitial macrophages (IMs; Fig. 4 g).
Lung IMs, alveolar epithelial cells (AECs), and DCs may be involved in RGMb–PD-L2 interaction in the lung
PD-L2 blockade has particularly strong effects on immune responses in the lung (Akbari et al., 2010; Singh et al., 2011), and RGMb is highly expressed in the lung (Fig. 4 f). Notably, lung inflammation (pneumonitis) is the most severe adverse event reported in human clinical trials of PD-1 mAb (Topalian et al., 2012). Therefore, we investigated the roles of PD-L2 and RGMb, and of RGMb–PD-L2 interactions in a mouse model of OVA-induced respiratory tolerance. Lung IMs, but not AMs, have been reported to produce high levels of IL-10 and inhibit LPS-induced maturation and migration of DCs, thereby preventing airway allergy in mice (Bedoret et al., 2009). In addition, epithelial cells have been shown to interact with DCs to maintain respiratory tolerance through production of a diverse array of mediators that modulate the activity of DCs (Strickland et al., 2010).
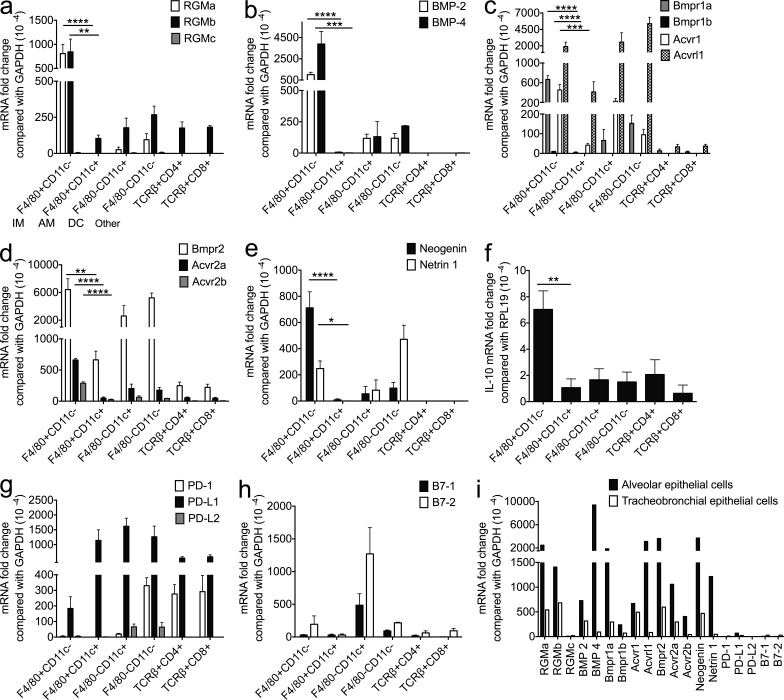
To identify the lung cell subsets involved in RGMb–PD-L2 interaction that regulate respiratory tolerance, we examined mRNA expression of RGMb, PD-L2, and related proteins in unstimulated lung IMs (F4/80+CD11c−), AMs (F4/80+CD11c+), DCs (F4/80−CD11c+), other cells (F4/80−CD11c−), as well as CD4+ T cells (TCRβ+CD4+), CD8+ T cells (TCRβ+CD8+), AECs, and tracheobronchial epithelial cells (TECs; Fig. 5, a–i). IMs and AECs expressed the highest levels of RGMb, BMP-2/4, BMP type I receptors (Bmpr1a, Bmpr1b, and Acvr1), BMP type II receptors (Bmpr2, Acvr2a, and Acvr2b), neogenin and its ligand netrin 1 (Fig. 5, a–e and i). IMs also expressed the highest levels of IL-10 (Fig. 5 f). PD-L2 expression on IMs and AECs was barely detectable (Fig. 5 g). DCs expressed readily detectable levels of PD-L2, B7-1, and B7-2, but low levels of most other molecules (Fig. 5, a–h). IMs had very different expression profiles than AMs (Fig. 5, a–h). Lung CD4+ and CD8+ T cells expressed very low levels of all the molecules examined except for PD-1 and PD-L1 (Fig. 5, a–h). These expression data support our proposed BBRN model with RGMb associating in a supercomplex with BMP receptors and/or neogenin on IMs and/or AECs, then interacting with PD-L2 on DCs (Fig. 2 h).
Figure 5.
Expression of RGMb, PD-L2, BMPs, BMPRs, and related molecules in resting lung cell populations and airway epithelial cells. Expression of indicated mRNAs in resting lung cell populations and airway epithelial cells by qRT-PCR. (a–h) IMs (F4/80+CD11c−), AMs (F4/80+CD11c+), DCs (F4/80−CD11c+), and other cells (F4/80−CD11c−) were sorted from lung cells pooled from five to eight mice; CD4+ T cells (TCRβ+CD4+) and CD8+ T cells (TCRβ+CD8+) were sorted from lung cells pooled from two mice. (i) AECs and TECs were pooled from three and eight mice, respectively. Data are mean ± SEM; n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001. Ordinary one-way ANOVA followed by Dunnett’s multiple comparison test. All data are representative of two or more experiments.
Blockade of RGMb–PD-L2 interaction inhibits the induction of respiratory tolerance
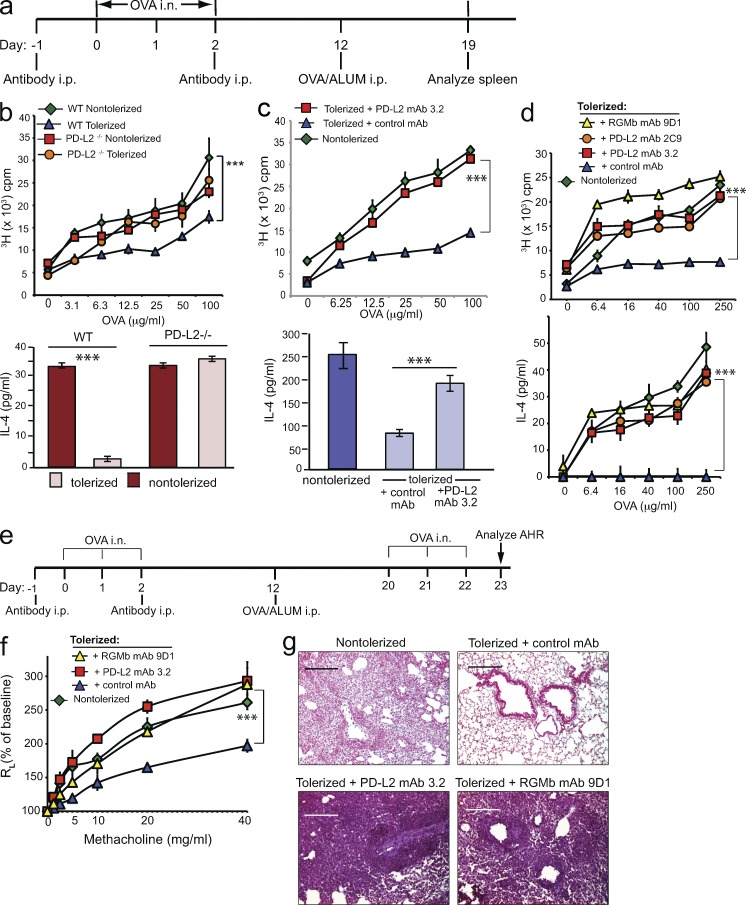
Respiratory tolerance is a state of antigen-specific immunological nonresponsiveness induced by exposure to innocuous antigens inhaled in the respiratory tract. To induce tolerance, mice were exposed intranasally (i.n.) to OVA or PBS control on days 0, 1, and 2. Mice were challenged by immunization with OVA in ALUM i.p. on day 12, and splenic T cell responses were examined 1 wk later by in vitro restimulation of T cells with antigen (Fig. 6 a). We first compared the development of respiratory tolerance in WT and PD-L2–deficient mice. T cells from WT mice exposed to intranasal OVA were tolerized, as indicated by significantly reduced T cell proliferation and IL-4 responses compared with control mice that did not receive i.n. OVA (Fig. 6 b). Strikingly, PD-L2–deficient mice showed resistance to the development of respiratory tolerance. T cells from PD-L2–deficient mice that received OVA i.n. displayed similar levels of proliferation and IL-4 production as T cells from control PD-L2–deficient mice that received PBS i.n. Similarly, treatment of WT mice with a PD-L2 mAb (3.2) that blocks both PD-L2–RGMb and PD-L2–PD-1 interaction prevented the development of respiratory tolerance, resulting in increased proliferation and IL-4 production compared with tolerized mice treated with control mAb (Fig. 6 c). These results suggest that PD-L2 is critical for the development of respiratory tolerance.
Figure 6.
PD-L2 or RGMb blockade inhibits induction of respiratory tolerance. (a) Experimental protocol for induction of respiratory tolerance. (b–d) PD-L2–deficient or WT mice in b, and WT mice injected with the indicated mAb in c and d, were exposed to OVA i.n. (tolerized) or PBS (nontolerized) and subsequently received 50 µg OVA/ALUM i.p. T cell proliferation (top) and IL-4 production (bottom) in response to restimulation with OVA in vitro are shown. n = 2–3. Data are representative of two to five experiments. Data are mean ± SEM, *, P < 0.05; **, P < 0.01; ***, P < 0.005. (b, top) two-way ANOVA followed by Tukey’s multiple comparisons test. (c, top, and d) Two-way ANOVA followed by Dunnett’s multiple comparison test. (b and c, bottom) Two-tailed Student’s t test. (e) Experimental protocol used in f and g. (f) WT mice injected with the indicated mAb were exposed to OVA i.n. or PBS, immunized with 50 µg OVA/ALUM i.p. Mice were subsequently challenged with OVA i.n. on three consecutive days and assessed for AHR by measuring lung resistance (RL) and dynamic compliance (Cdyn). Data are the mean ± SEM of 4 mice/group. (g) Lung histopathology of mice from panel f. Lung tissue was stained with H&E and analyzed for cell infiltration. Data in f and g are representative of two experiments.
Because PD-L2 can interact with both PD-1 and RGMb, we used specific mAbs to evaluate the contribution of RGMb in development of respiratory tolerance. We treated mice that received OVA i.n. with RGMb mAb 9D1 (blocks RGMb–PD-L2 and RGMb–BMP) or with PD-L2 mAb 2C9 (blocks RGMb–PD-L2 but not PD-1–PD-L2) or with isotype control mAb. Administration of either 2C9 or 9D1 mAb inhibited the development of respiratory tolerance and led to higher levels of T cell proliferation and IL-4 production compared with control mAb-treated mice (Fig. 6 d). These results support a role for the RGMb–PD-L2 interaction in promoting the development of respiratory tolerance.
To further evaluate the effects of the RGMb–PD-L2 interaction in tolerance, we rechallenged mice that were exposed to antigen i.n during RGMb or PD-L2 blockade, and evaluated airway hyperreactivity (AHR), a cardinal feature of asthma. Previous work has shown that tolerization via i.n. administration of OVA can protect from OVA-induced AHR. We exposed mice to OVA i.n. or PBS control, and then immunized with OVA in ALUM i.p., challenged with OVA i.n. on days 20–22, and assessed for the development of AHR on day 23 (Fig. 6 e). As expected, mice that received PBS i.n. (nontolerized) developed high levels of AHR measured as airway resistance (RL) and dynamic compliance (Cdyn) in anesthetized, tracheostomized, and mechanically ventilated mice. Administration of OVA i.n. induced respiratory tolerance, such that the tolerized mice were protected and developed only mild levels of AHR. However, treatment of the mice with RGMb mAb 9D1 or with PD-L2 mAb 3.2 abolished the induction of tolerance, and upon rechallenge with OVA, these mice developed severe AHR which was comparable to that of nontolerized mice (Fig. 6 f). The lungs of tolerized mice showed low levels of cellular infiltration surrounding the airways in contrast to nontolerized mice, which showed substantial airway inflammation. Mice treated with PD-L2 mAb or RGMb mAb demonstrated even more extensive cellular infiltration than nontolerized mice (Fig. 6 g). Thus, respiratory tolerance promoted by RGMb–PD-L2 interaction protects against the development of airway hyperreactivity and lung inflammation after antigenic challenge.
To determine if RGMb blockade would impact systemic immunization, mice treated with RGMb mAb were immunized in the footpad with OVA in IFA, and T cell responses in the draining lymph nodes were evaluated 9 d later. There were no differences in proliferation, IL-4 or IFN-γ production between mice treated with RGMb mAb 9D1 or control mAb (unpublished data). These results demonstrate that treatment with RGMb mAb does not have a global impact on in vivo immune responses.
Tolerized mice have fewer OVA-responsive T cells than control mice or RGMb mAb-treated mice
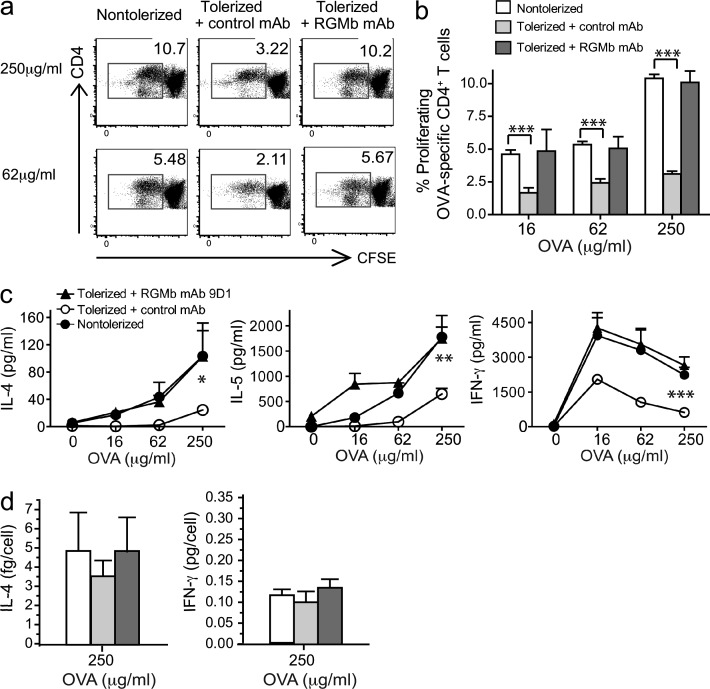
To further understand the mechanisms underlying this model of respiratory tolerance induction and how blockade of RGMb–PD-L2 interaction abrogates tolerance in this model, we compared the number and phenotype of T cells responding to challenge with OVA after tolerance induction in the presence or absence of RGMb mAb. Splenocytes from RGMb mAb-treated and control mice were labeled with CFSE before culture with OVA. Tolerized mice had a lower percentage of proliferating OVA-specific CD4+ T cells (CFSE low cells) than nontolerized mice (Fig. 7, a and b). Treatment with RGMb mAb prevented tolerance induction, as shown by a restoration of the percentage of CD4+ T cells proliferating to OVA to levels observed in nontolerized mice. Cells from tolerized mice produced less IL-4, IL-5, and IFN-γ than nontolerized mice, whereas cells from tolerized, RGMb mAb-treated mice had the same high levels of cytokine as nontolerized mice (Fig. 7 c). Cytokine production per cell was calculated by dividing total IL-4 or IFN-γ production by the number of responding (CFSE low) T cells. No significant differences were found in cytokine per cell between nontolerized, tolerized, and tolerized plus RGMb mAb-treated groups at 250 μg/ml OVA in culture (Fig. 7 d). Thus, tolerization primarily affects the quantity of responsive T cells. These data suggest that the primary mechanism of tolerance in this model is deletion or anergy of OVA-specific cells and that treatment with RGMb mAb during tolerance induction impairs this process.
Figure 7.
Tolerized mice have fewer OVA-responsive T cells than control mice or RGMb mAb–treated mice. B cell–depleted splenocytes prepared on day 19 from mice treated as in Fig. 6 a were labeled with CFSE and cultured with OVA for 4 d. Cells were stained for CD3 and CD4 and subgated on CD3+CD4+ cells for analysis of CFSE dilution. (a) Flow cytometry plots show data from one representative mouse and (b) shows mean ± SEM of three mice/group. (c) On day 4, cultures were harvested as in panel a and supernatants were collected and assayed for cytokine production. Total IL-4, IL-5, and IFN-γ in culture supernatants is shown. (d) Cytokine per cell was calculated as total cytokine amount in supernatant divided by the number of CFSE low cells. Data are mean ± SEM; **, P < 0.01; ***, P < 0.005; n = 3, Student’s t test. Data are representative of two experiments.
Lung macrophages and DCs are not depleted by treatment with PD-L2 or RGMb mAbs
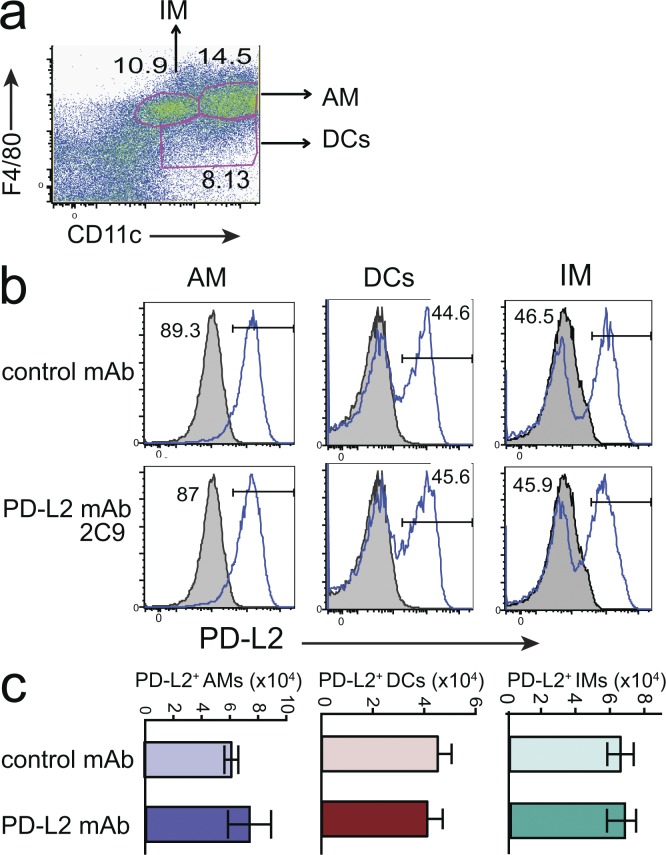
To determine if PD-L2 mAb 2C9 inhibited tolerance induction by depleting PD-L2–expressing cells, mice primed with OVA in ALUM were treated with mAb 2C9 on the day of the second administration of OVA i.n. Previous work had shown that OVA-primed mice express high levels of PD-L2 on lung DCs and macrophages. The number and intensity of PD-L2–positive cells in the lung were examined 48 h later using a PD-L2 mAb (TY25) that recognizes a different epitope than mAb 2C9. Under these inflammatory conditions, PD-L2 was expressed by alveolar macrophages (F4/80+CD11c+) and about half of dendritic cells (F4/80−CD11c+) and interstitial macrophages (F4/80+CD11clo). The PD-L2 intensity and number of cells was not different in mice treated with mAb 2C9 (Fig. 8, b and c), indicating that treatment with mAb 2C9 did not deplete PD-L2–expressing cells. Similarly, treatment of mice with RGMb mAb 9D1 did not deplete these populations (not depicted).
Figure 8.
Impact of RGMb–PD-L2 blockade on tolerance induction is not caused by depletion of APC. (a–c) Mice immunized with OVA in ALUM on day 0 received OVA i.n. on days 7–9 and were treated with PD-L2 mAb 2C9 or isotype control (500 µg i.p.) on day 8. Lung cells were dispersed on day 10 and the expression of PD-L2 analyzed on AMs (F4/80+CD11c+), DCs (F4/80−CD11c+), and IMs (F4/80+CD11clo). (a) Gating strategy used in b–d. (b) Solid line indicates staining with PD-L2 mAb TY25; shaded histogram indicates isotype control. One mouse/group representative of three is shown. (c) The number of PD-L2+ macrophages and DCs was quantified. Data are representative of two experiments.
Blockade of RGMb–PD-L2 interaction impairs T cell expansion to antigen
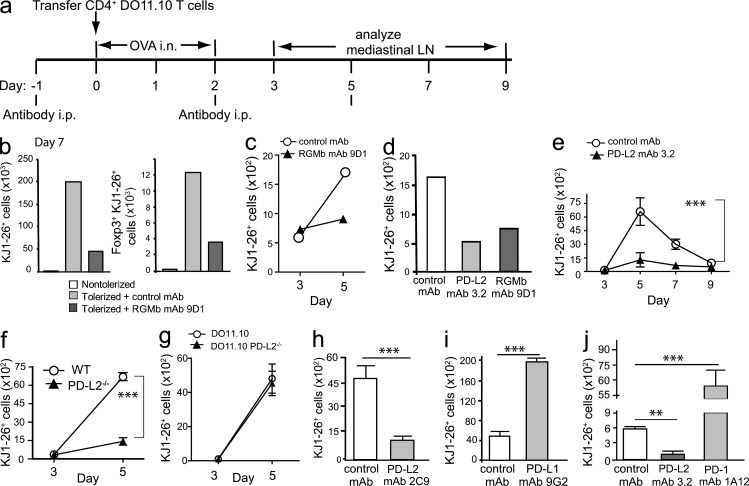
Previous studies of respiratory tolerance showed that intranasal administration of antigens such as house dust mite (Hoyne et al., 1996) or OVA (Tsitoura et al., 1999; Albacker et al., 2012) resulted in a strong transient CD4+ T cell response, followed by deletion of most of the antigen-specific T cells, with a small population of unresponsive T cells remaining. Inhaled antigen is sampled by immature DCs in the lung, which migrate to the draining mediastinal LNs, where they encounter antigen-specific T cells. In the absence of inflammatory stimuli, DCs induce transient antigen-specific T cell activation followed by T cell deletion and unresponsiveness (Hawiger et al., 2001). Respiratory tolerance involves multiple mechanisms including deletion of antigen-specific T cells, the development of anergy and regulatory T (T reg) cells and these processes may occur concurrently (Holt and Upham, 2004; GeurtsvanKessel and Lambrecht, 2008; Bedoret et al., 2009; Albacker et al., 2012). To explore the mechanism whereby RGMb and PD-L2 interaction enhances respiratory tolerance, we examined the effect of blocking RGMb–PD-L2 interaction on the activation and expansion of transferred OVA-specific DO11.10 CD4+ T cells. Recipient mice were treated with RGMb mAb 9D1 or control mAb, subsequently given OVA or PBS i.n. on days 0, 1, and 2, and the fate of DO11.10 T cells was monitored using the clonotypic mAb KJ1-26 (Fig. 9 a). As expected, exposure of the mice to i.n. OVA resulted in marked expansion of the DO11.10 cells in the mediastinal LN on day 7 compared with mice that received i.n. PBS. Notably, this expansion of KJ1-26+ T cells was greatly diminished in tolerized mice treated with anti-RGMb compared with the control mAb treated mice (Fig. 9 b, left). The number of OVA-specific T reg cells (KJ1-26+ Foxp3+) was similarly reduced in RGMb mAb-treated mice, indicating that the reduced expansion of KJ1-26+ T cells was not due to increased numbers of OVA-specific T reg cells (Fig. 9 b, right).
Figure 9.
Blockade of PD-L2–RGMb interaction reduces the initial expansion of antigen-specific T cells after intranasal OVA administration. (a) Experimental protocol for Fig. 9. Mice received DO11.10 T cells on day 0 and OVA i.n. The number of KJ1-26+ cells in mediastinal LNs was determined on the indicated days. (b) Mice treated with RGMb mAb 9D1 or control mAb on day −1 received 5 × 106 DO11.10 T cells. Left, TCRβ+ KJ1-26+ CD4+; Right, FoxP3+ TCRβ+ KJ1-26+ CD4+ cells. For each group, mediastinal LNs of 3–5 mice were combined; data are representative of two experiments. (c–e) Mice received 2 × 105 DO11.10 T cells after (c) RGMb mAb 9D1 or control mAb; (d) RGMb mAb 9D1, PD-L2 mAb 3.2, or control mAb; (e) PD-L2 mAb 3.2 or control mAb. (f) WT DO11.10 T cells were transferred into WT or PD-L2−/− mice; (g) WT or PD-L2−/− DO11.10 T cells were transferred into WT mice. (h–j) Mice received 2 × 105 DO11.10 T cells after (h) PD-L2 mAb 2C9 or control mAb; (i) PD-L1 mAb 9G2 or control mAb; (j) PD-L2 mAb 3.2, PD-1 mAb 1A12, or control mAb. The number of KJ1-26+ T cells in mediastinal lymph nodes on day 5 (h–j) was determined by flow cytometry. (b–d) Each point represents a pool of LNs from three mice. Data are representative of two to four experiments. (e–j) Each point represents the mean and SEM of KJ1-26+ T cells in LNs of two or three groups of two mice each. Data are representative of two experiments. **, P < 0.01; ***, P < 0.005, two-tailed Student’s t test.
We next examined the effect of RGMb mAb on the expansion of KJ1-26+ T cells at earlier time points. Similar numbers of KJ1-26+ cells were detected in LNs of RGMb mAb and control treated mice on day 3. However, by day 5 the number of KJ1-26+ cells increased in the control mice, but showed little expansion in the RGMb mAb-treated group (Fig. 9 c). These findings suggest that blockade of RGMb inhibits the induction of tolerance by impairing the expansion of T cells that normally occurs after respiratory administration of OVA.
To determine if treatment with PD-L2 mAb would similarly inhibit the response of OVA-specific CD4+ T cells after exposure to OVA i.n., mice were treated with PD-L2 mAb, RGMb mAb, or control mAb and the number of KJ1-26+ cells examined on day 5. Reduced numbers of KJ1-26+ cells were detected in both PD-L2 mAb and RGMb mAb-treated groups compared with control mAb-treated mice, indicating that expansion of KJ1-26+ cells was inhibited (Fig. 9 d). To determine if anti–PD-L2 mAb was altering the time course of the T cell response, we examined the in vivo response of DO11.10 T cells to i.n. OVA over a 9-d period in mice treated with PD-L2 mAb or control mAb (Fig. 9 e). In control mAb-treated mice, the number of KJ1-26+ cells in mediastinal LNs increased substantially after day 3, peaking at day 5 and declining thereafter with a small but detectable population remaining on day 9, as previously described (Tsitoura et al., 1999). In mice treated with PD-L2 mAb, a limited increase in the number of KJ1-26+ cells was observed between days 3 and 5 that was significantly lower than the increase observed in control mAb treated mice. Numbers subsequently declined, indicating that blocking PD-L2 inhibited the expansion of OVA-specific T cells throughout the response.
We then determined if expression of PD-L2 on T cells, non-T cells, or both was required for T cell expansion. We compared expansion of WT KJ1-26+ T cells after transfer to PD-L2–deficient or WT mice and exposure to OVA i.n. Between days 3 and 5, the WT KJ1-26+ cells underwent a 72-fold expansion in WT recipients but only a 9.7-fold expansion in PD-L2−/− recipients (Fig. 9 f), indicating a requirement for PD-L2 expression on non–T cells. Consistent with this, DO11.10 T cells from PD-L2−/− and WT mice expanded similarly when transferred to WT mice (Fig. 9 g). These experiments demonstrate the importance of PD-L2 expression on non–T cells for the initial expansion of T cells to OVA i.n.
To determine the involvement of the PD-1–PD-L2 versus RGMB–PD-L2 pathways in this process, mice were treated with PD-L2 mAb 2C9, which blocks only the RGMb–PD-L2 interaction or with control mAb. The expansion of KJ1-26+ cells on day 5 was significantly reduced in mice treated with mAb 2C9 compared with control-treated mice (Fig. 9 h), suggesting that PD-1 is not required for this effect because 2C9 does not block the interaction of PD-L2 with PD-1. Together, these data indicate that blocking RGMb–PD-L2 interaction prevents the induction of respiratory tolerance, and does so by reducing the initial expansion of CD4+ T cells in response to OVA by a pathway that does not involve PD-1.
We then examined the effects of blocking PD-L1 or PD-1 compared with PD-L2 on the initial activation and expansion of transferred DO11.10 OVA-specific CD4+ T cells. On day 5, mice treated with the blocking PD-L1 mAb 9G2 or PD-1 mAb 1A2 had substantially higher numbers of KJ1-26+ cells in the mediastinal LNs than control mAb-treated mice (Fig. 9, i and j), which indicated that PD-1 or PD-L1 blockade significantly increased the expansion of OVA-specific T cells in response to OVA i.n. In contrast, PD-L2 mAb treatment led to significantly lower numbers of KJ1-26+ cells than in control mAb-treated mice (Fig. 9 j). These data further suggest that RGMb–PD-L2 interaction reduces the initial expansion of CD4+ T cells in response to OVA by a pathway that does not involve PD-1.
DISCUSSION
Here, we demonstrate that RGMb is a novel binding partner of PD-L2, and that this interaction regulates the development of respiratory tolerance in the lung. PD-L2 has been reported to have co-stimulatory or co-inhibitory function (Latchman et al., 2001; Tseng et al., 2001; Liu et al., 2003; Shin et al., 2003; Wang et al., 2003; Pfistershammer et al., 2006; Zhang et al., 2006). There is now substantial agreement that the PD-1–PD-L2 interaction inhibits immune activation but the mechanism and outcome of other PD-L2 interactions is unclear. Our data show that the RGMb–PD-L2 interaction promotes the development of respiratory tolerance, as PD-L2 or RGMb blockade abrogated the induction of respiratory tolerance. The RGMb–PD-L2 interaction facilitates the initial T cell expansion in draining LNs during respiratory tolerance induction (co-stimulatory function) because PD-L2 or RGMb blockade leads to greatly reduced T cell numbers between days 3–7 after tolerance induction. This expansion is required for tolerance induction and is followed by deletion of Ag-specific T cells. The outcome of PD-L2 or RGMb blockade is that the T cells are not tolerized, thus the final effect of the RGMb–PD-L2 interaction could be interpreted as co-inhibitory. Therefore, PD-L2 could have both co-stimulatory and co-inhibitory functions depending on the receptor and context, as illustrated by the network of multiple interactions shown in Fig. 2 h. In this network model, PD-L2 bridges PD-1, BMP, and neogenin signaling pathways, accounting for the multifaceted functions of its components.
Our data on RGMb and PD-L2 pathway expression in lung cell populations, together with previous findings (Bedoret et al., 2009; Strickland et al., 2010) suggest that IM, AECs, and DCs are involved in the induction of respiratory tolerance. We propose that RGMb on IMs and/or AECs forms a BBRN supercomplex with BMP-2/4, BMP receptors, and neogenin and interacts with PD-L2 on DCs. This may signal directly or induce the production of mediators such as IL-10 in IMs or AECs to inhibit the maturation and migration of DCs, thereby promoting respiratory tolerance. In support of this, our data show that PD-L2 expression on non-T cells provides the required signal for the induction of respiratory tolerance because WT T cells transferred into PD-L2–deficient mice do not undergo the initial T cell expansion involved in tolerance induction. BMP signaling could be involved in the induction of respiratory tolerance, as the RGMb mAb 9D1 that blocks both RGMb–PD-L2 and RGMb–BMP-2/4 interactions impaired respiratory tolerance.
Components of the BBRN complex have been shown to have immune function. Previous studies have shown that BMP signaling has immunoregulatory effects on T cells (Graf et al., 2002; Hager-Theodorides et al., 2002; Lu et al., 2010; Yoshioka et al., 2012). BMP-7 was recently shown to be an instructive factor for Langerhans cell differentiation (Yasmin et al., 2013). Elevated BMP/Smad signaling has been found during airway inflammation (Rosendahl et al., 2002). RGMb-mediated BMP signaling was reported to inhibit IL-6 production by lung macrophages (Xia et al., 2011). Neogenin has been shown to be involved in pulmonary inflammation and acute inflammatory peritonitis (König et al., 2012; Mirakaj et al., 2012), so neogenin signaling could also be involved in the induction of respiratory tolerance. RGMb interaction with neogenin has been shown to control aggregation and migration of neogenin-positive neuroepithelial cells (Conrad et al., 2010). Another member of the RGM family, RGMa, was also highly expressed in IMs and AECs (Fig. 5, a and i), and was shown to be involved in immune responses (Muramatsu et al., 2011). Because our mRGMb mAb does not bind to mRGMa, the effects of our RGMb mAb on respiratory tolerance are independent of RGMa.
Our model suggests that PD-L2 binding to RGMb could signal through BMP and/or neogenin signaling pathways. This hypothesis may offer insight into the signaling conundrum of RGMb and PD-L2, i.e., RGMb and PD-L2 do not appear to be able to signal directly because RGMb is a glycosylphosphatidylinositol-anchored protein and PD-L2 has only a short cytoplasmic tail without obvious signaling motifs. To signal, RGMb–PD-L2 may need to form a complex with BMPs, BMP receptors, and/or neogenin. BMP and neogenin signaling may act together because neogenin can directly bind BMP-2/4/6/7 and modulate BMP signaling (Hagihara et al., 2011; Tian and Liu, 2013). Nevertheless, these two signaling pathways may also function separately because a recent study showed that RGMb induces apoptosis in mouse renal tubular epithelial cells through neogenin and not the BMP pathway (Liu et al., 2013). RGMb is the pivot joining all these proteins, but the molecular details of the cross-talk between the components of the BBRN signaling supercomplex remain to be determined.
Though early work emphasized the restricted expression of PD-L2 on DCs and macrophages, recent work has identified PD-L2 expression on human primary nasal epithelial cells (Kim et al., 2005), Th2 cells (Lesterhuis et al., 2011), activated T cells (Messal et al., 2011), and some B cell subsets (Zhong and Rothstein, 2011). In the BBRN model, PD-L2 on any of these cells might interact with RGMb on various cells and signal through BMP and/or neogenin pathways.
Antibodies blocking the PD-1 pathway have shown promising results in cancer immunotherapy clinical trials. The most severe adverse event with PD-1 mAb treatment is pneumonitis (3%) leading to 3 deaths (1%; Topalian et al., 2012). The role of the different PD-1 ligands in pneumonitis is not yet understood. PD-1 blockade may shift the balance in PD-L2 interaction with its binding partners, increasing PD-L2 availability for binding to RGMb, leading to pneumonitis. Additional work will need to be done to distinguish which therapeutic modality has the optimal balance of efficacy and minimal adverse events. Treatment with PD-L2-Ig decreased murine TC-1 lung tumor burden and increased survival, possibly by depletion of T cells expressing high levels of PD-1 (Mkrtichyan et al., 2012); however, these PD-L2-Ig effects could also be through RGMb and the BBRN complex. Further studies are needed to determine if RGMb–PD-L2 interaction serves as a target for immunotherapy.
In summary, we discovered that RGMb is a binding partner for PD-L2. RGMb may coordinate with multiple receptors in the BMP and neogenin pathways to form a BBRN signaling supercomplex. Engagement of PD-L2 with RGMb in this supercomplex may impact BMP and neogenin signaling. We detected high mRNA levels of RGMb and components of the BBRN complex in resting lung IMs and AECs. Our data suggest that the novel engagement of RGMb and PD-L2 promotes the development of respiratory tolerance by facilitating the initial T cell expansion in draining LNs. Targeting the PD-L2 pathway has therapeutic potential for asthma, cancer, and other immune disorders, and may help better use the PD-1 pathway in cancer immunotherapy while minimizing adverse events.
MATERIALS AND METHODS
Mice.
WT BALB/cJ mice and BALB/cByJ mice were purchased from The Jackson Laboratory. PD-L2−/− mice on BALB/c background have been previously described (Keir et al., 2008). OVA TCR-transgenic DO11.10 mice and PD-L2−/− mice crossed to OVA TCR-transgenic DO11.10 mice were used as donors of OVA-specific CD4+ T cells (Tsitoura et al., 1999). Age-matched female mice were used at 6–12 wk. Animal protocols were approved by The Animal Care and Use Committees at Boston Children’s Hospital, the Dana-Farber Cancer Institute, and Harvard Medical School.
Cells and culture media.
The mouse 300.19 pre–B cell line was transfected by electroporation with mRGMb, hRGMb, or mNeogenin cDNA in the pEF-Puro expression vector. Cells were selected in media containing puromycin, sorted, and subcloned. Cell surface expression of mRGMb, hRGMb, or mNeogenin was verified by flow cytometry using an mRGMb polyclonal antibody (R&D Systems), an hRGMb mAb (R&D Systems), or mRGMb-Ig, respectively. Other transfected cells, such as 300-mPD-L2, 300-mPD-L1, 300-mPD-1, and Jurkat-hPD-1, have been made previously using similar methods. Cells were cultured in RPMI-1640 (Mediatech) supplemented with 10% heat-inactivated FBS (Invitrogen), 1% streptomycin/penicillin, 15 µg/ml gentamicin (Invitrogen), 1% glutaMAX (Invitrogen), and 50 µM β-mercaptoethanol (Sigma-Aldrich) at 37°C with 5% CO2.
The cell lines used in this study were purchased from American Type Culture Collection, except for SN12C which was obtained from the National Cancer Institute. Mouse spleen or LN cells for tolerance experiments were cultured in high-glucose DMEM (Invitrogen) supplemented with NEAA, sodium pyruvate, MEM vitamins, 10% FBS, 1% glutaMAX, 50 µM β-mercaptoethanol at 37°C in a 10% CO2 incubator.
COS cell expression cloning of RGMb.
A panel of murine-activated lymphocyte cDNA expression libraries were transiently transfected into COS cells using DEAE-Dextran or Lipofectamine as facilitator (Freeman et al., 1989). After 44 h, COS cells were harvested and panned on mPD-L2-mIgG2a/IgA–coated Petri plates. Episomal DNA was recovered and electroporated into Escherichia coli DH10B/P3. Spheroplast fusion was used to reintroduce the plasmids into COS cells for subsequent rounds of expression and panning. Before the second and third rounds, COS cells expressing PD-1 were removed by incubating with PD-1 mAb, followed by depletion with goat anti–rat IgG magnetic beads. After the third round of panning, individual plasmids were transfected into COS cells and binding of PD-L2-Ig to RGMb was verified by flow cytometry on mRGMb transiently transfected COS cells. At least four independent mRGMb cDNA clones were isolated.
Generation of antibodies.
Rats were immunized three times via intramuscular and intravenous injection of mRGMb plasmid cDNA (Latchman et al., 2001), and boosted three times with recombinant mRGMb-HIS (R&D Systems) via i.p. and s.c. injection. Hybridoma supernatants were screened by flow cytometry on mRGMb transfected 300 cells or ELISA on plates coated with recombinant mRGMb (R&D Systems). Hybridomas were subcloned to stability, and antibodies were purified from culture supernatants by protein G affinity chromatography, and verified to have endotoxin levels less than 2EU/mg protein. Clones 307.9D1 (rat IgG2a) and 307.1H6 (rat IgG2a) were selected for use in this study.
Generation of Ig fusion proteins.
mRGMb-Ig fusion proteins were generated by joining the extracellular domain of mRGMb to the Fc portion of mouse IgG2a protein, mutated to reduce FcR binding (Latchman et al., 2001). mPD-L2-hIgG1/IgA fusion proteins were generated by joining the extracellular domain of PD-L2 with the Fc portion of hIgG1 and the tail piece of hIgA (Hirano et al., 2006). Fc fusion proteins were purified from CHO cell culture supernatants by protein A or protein G affinity chromatography and verified to have endotoxin levels less than 2 EU/mg protein. Other Ig fusion proteins used have been described previously (Latchman et al., 2001).
Cell conjugation assay.
An assay for cell surface receptor-ligand binding was developed based on cell–cell binding as determined by flow cytometry. Cells transfected with cell surface gene 1 were labeled with the red dye PKH26 (Sigma-Aldrich), and cells transfected with cell surface gene 2 were labeled with the green dye PKH67 (Sigma-Aldrich). Red dye-labeled and green dye-labeled cells (105 each cell type) were incubated in 200 µl of HBSS buffer without calcium and magnesium (Invitrogen) supplemented with 10% heat-inactivated FBS (Invitrogen), 15 µg/ml gentamicin (Invitrogen), and 1% Hepes (Invitrogen) in a round-bottom 96-well plate for 45 min at 37°C. Conjugate formation was analyzed immediately by flow cytometry using the PE channel for the red dye and the FITC channel for the green dye. The automated plate harvester of the FACSCanto (BD) was used for uniformity of cell processing. The instrument settings were as follows: throughput mode, standard; loader settings, sample flow rate at 0.5 µl/second; sample volume and mixing volume at 100 µl; mixing speed at 50 µl/second; number of mixes at 2; and wash volume at 400 µl. When the cell conjugation assay was used to test the blocking capacity of antibodies, the relevant transfected cell was preincubated 30 min at room temperature with antibodies before adding candidate binding cells.
Flow cytometry.
Cells were stained with target antibodies and isotype controls using standard flow cytometry procedures, and analyzed on a FACSCanto (BD) using FlowJo 9.2 software (Tree Star).
To initially verify RGMb expression on mRGMb- or hRGMb-transfected cells, sheep anti-mRGMb (R&D Systems) or sheep IgG (SouthernBiotech) plus donkey anti–sheep IgG-PE (Jackson ImmunoResearch Laboratories) and mouse anti-hRGMb (mAb; R&D Systems) plus goat anti–mouse IgG-PE (SouthernBiotech) were used, respectively, all at 10 µg/ml.
To test the binding specificities of mRGMb antibodies, mRGMb- or hRGMb-transfected 300 cells were incubated with serial dilutions of sera, culture supernatants, or purified antibodies, and binding was detected with 5 µg/ml of goat anti–rat IgG-PE (SouthernBiotech). For biotin-conjugated mRGMb mAb 9D1, 1.4 µg/ml of streptavidin-PE was used.
For receptor-ligand binding assay, mRGMb-, mPD-L2–, or mNeogenin-transfected 300 cells, 300 cells, and hPD-1–transfected Jurkat T cells were stained with serial dilutions of specific protein-Ig or control-Ig plus 5 µg/ml of Fab2 goat anti–hIgG-PE (mouse-absorbed; SouthernBiotech), or 10 µg/ml of goat anti–mIgG2a-PE (SouthernBiotech), or 5 µg/ml of Fab2 goat anti-mIgG2a-PE (SouthernBiotech).
To test the capacities of RGMb mAbs to block mRGMb binding to mNeogenin, RGMb-mIgG2a at 5 µg/ml was incubated with serial dilutions of RGMb mAbs, and then added to neogenin-transfected 300 cells. Binding was detected using 10 µg/ml of goat anti–mIgG2a-PE (SouthernBiotech). To determine the background binding, 300-mNeogenin cells were stained with serial dilutions of mIgG2a isotype control.
To assess cell surface expression of RGMb and PD-L2 on mouse cell lines RAW264.7, J774.1, and C2C12, cells were preincubated with mouse Fc receptor mAb (2.4G2), and then stained with PE-conjugated RGMb mAb 9D1, mPD-L2 mAb TY25, or rat IgG2a at 5 µg/ml.
To analyze cell surface expression of RGMb on human cancer cell lines, RGMb mAb 1H6 or rat IgG2a at 10 µg/ml plus goat anti–rat IgG-PE (SouthernBiotech) at 5 µg/ml were used.
For intracellular flow cytometry analyses of RGMb expression in primary cells from spleen, cells were first stained with LIVE⁄DEAD Fixable Near-IR (Invitrogen) at 1:1,000. After preincubation with mouse Fc receptor mAb (2.4G2), cells were stained with CD3-Pacific blue, CD19-PE-cy7 and CD11c-APC (BioLegend). Then cells were permeabilized using BD Cytofix/Cytoperm Fixation/Permeabilization kit, preincubated with mouse Fc receptor mAb (2.4G2), and stained with PE-conjugated RGMb mAb 9D1 or rat IgG2a at 5 µg/ml. PBS containing 3% BSA and 0.1% Triton X-100 was used for wash and antibody incubation. 300-mPD-L2 cells were used as negative controls.
DO11.10 T cells were identified by staining with TCRβ-APC-eFluor 780, CD4-PerCP, and KJ1-26-APC (eBioscience). DO11.10 T reg cells were identified using CD25-FITC (BioLegend), followed by intracellular staining with Foxp3-PE (BioLegend).
ELISA.
To examine specificity of mRGMb mAbs, 96-well plates were coated with 2 µg/ml of recombinant mRGMa-HIS, mRGMb-HIS, or mRGMc-HIS (R&D Systems). Then serial dilutions of mRGMb mAbs and isotype controls were added and incubated for 1 h at 37°C. Mouse anti–rat IgG (γ-specific) HRP (Jackson ImmunoResearch Laboratories) at 1:2,500 was used for detection.
To examine RGMa/RGMb/RGMc and PD-L2 interaction, 96-well ELISA plates were coated with 2 or 5 µg/ml of recombinant mRGMa-HIS, mRGMb-HIS, mRGMc-HIS, or hRGMb-HIS (R&D Systems). Then serial dilutions of mPD-L2-hIgG1/IgA, mPD-L2-mIgG2a/IgA, hPD-L2-mIgG2a, or control-Ig fusion proteins were added and incubated for 1 h at 37°C. Fab2 goat anti–hIgG-HRP (Jackson ImmunoResearch Laboratories) or rat anti–mIgG2a-HRP (BD) at 1:1,000 or 1:10,000 were used for detection.
To test the capacities of mRGMb antibodies and mPD-L2 fusion proteins to block RGMb binding to BMP-2/4, 96-well ELISA plates were coated with 1 µg/ml of recombinant mouse BMP-2 (Invitrogen) or BMP-4 (R&D Systems). mRGMb antibodies, isotype controls, mPD-L2-hIgG1/IgA, mPD-L2-mIgG2a/IgA, or control Ig fusion proteins at the indicated concentrations were preincubated with 20 µg/ml mRGMb-HIS (R&D Systems) for 45 min at 4°C, then added to the plates and incubated for 1 h at 37°C. Anti–penta-HIS-HRP (QIAGEN) at 1:1,000 was used for detection.
To determine if RGMb can bind PD-L2 and BMP-2/4 simultaneously, 96-well ELISA plates were coated with BMP-2/4 as above. mPD-L2-hIgG1/IgA, mPD-L2-hIgG (R&D Systems), mPD-L1-hIgG (R&D Systems), or control-Ig fusion proteins at 10 µg/ml were preincubated with 10 µg/ml mRGMb-HIS (R&D Systems) or buffer alone for 15 min at room temperature, and then added to the plates and incubated for 1 h at 37°C. Alternatively, 10 µg/ml mRGMb-HIS (R&D Systems) or buffer alone was added first to the plates and incubated for 1 h at 37°C. After wash, mPD-L2-Ig or control-Ig fusion proteins were added and incubated for 1 h at 37°C. Fab2 goat anti–hIgG-HRP (Jackson ImmunoResearch Laboratories) at 1:10,000 was used for detection.
The substrate for HRP was TMB using the microwell peroxidase substrate system (KPL).
Biacore.
Interactions were determined by analysis of surface plasmon resonance on a BIAcore 3000 instrument. Flow cells within a CM5 biosensor chip (GE Healthcare) were activated with N-hydroxysuccinimide (NHS) in the presence of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride, generating an NHS ester that bound to free amines on RGMb or PD-L2-hIgG passed over the surface (R&D Systems; 25 µg/ml in 100 mM sodium acetate, pH 4.5), followed by blocking of free NHS ester with 1 M ethanolamine. A further cell was exposed to immobilizing reagents and blocking reagents in the absence of protein as a control surface. After extensive washing of the surfaces with binding buffer (10 mM Hepes, 150 mM NaCl, pH 7.0, and 0.05% Tween 20), PD-L2-hIgG fusion protein binding was assessed by injecting varying concentrations (0–658 nM) simultaneously over the control and RGMb flow cell surfaces. PD-1-hIgG (R&D Systems; 4.8–308 nM) was similarly passed over immobilized PD-L2 and control surfaces. Between each cycle, the surface was regenerated using 20 mM glycine-HCl, pH 2.8. Values shown in RU have been corrected for nonspecific binding by subtracting the SPR of the control flow chamber exposed to the same injected material, followed by subtraction of buffer alone passing over the RGMb or PD-L2 surface, respectively. Data were analyzed using BIAevaluation 3.2 software and fitted to a 1:1 Langmuir binding model with separate kd and ka determinations. The dissociation constant (Kd) was determined as kd/ka, and confirmed by manual linear transformation of the binding isotherms. For PD-1 binding to PD-L2 surface, the kinetic parameters were derived for monovalent binding of the Fc dimer at higher concentrations (red bar in Fig. 1 e), but data contributing to high-affinity bivalent interactions at lower concentrations were not included in the analysis.
Cell isolation and stimulation.
Spleen and thymus cells were isolated by mechanically disrupting the tissues. For FACS analysis, splenocytes were treated with red blood cell lysing buffer (Sigma-Aldrich).
To obtain lung cells or splenocytes to analyze splenic DCs, the lung (perfused with PBS) or spleen was cut into small pieces, digested in RPMI 1640 with 5% FBS, 1 mg/ml collagenase IV (Sigma-Aldrich), and 200 U/ml DNase I (Roche), and then treated with red blood cell lysing buffer (Sigma-Aldrich).
Lung cell populations from naive mice were sorted by flow cytometry. Whole lung cells were isolated as above, and stained with surface markers. Cell populations were defined as follows: IMs (F4/80+CD11c−), AMs (F4/80+CD11c+), DCs (F4/80−CD11c+; Bedoret et al., 2009), other cells (F4/80−CD11c−), CD4+ T cells (TCRβ+CD4+), CD8+ T cells (TCRβ+CD8+). IMs from OVA primed and challenged mice were identified as F4/80+CD11clow.
AECs and TECs were isolated from naive mice as described previously (Lam et al., 2011; Chuquimia et al., 2012). Thioglycollate-induced peritoneal macrophages were obtained from mice on day 4 after i.p. injection with 3% thioglycollate (DIFCO). CD4+ and CD8+ cells from mouse splenocytes were purified using CD4+ T cell isolation kit and CD8+ T cell isolation kit (Miltenyi Biotec). FLT-3L–stimulated splenocytes were obtained from mice 2 wk after s.c. injection of FLT-3L–transfected RENCA tumor line.
qRT-PCR.
Total RNA samples were isolated using the RNeasy mini kit (QIAGEN). Reverse transcription was performed using the QuantiTect reverse transcription kit (QIAGEN). qPCR using TaqMan gene expression assays (Applied Biosystems) was performed in a 7300 Real-Time PCR system (Applied Biosystems). Fold change compared with GAPDH was calculated using the ΔCt method.
Western blotting.
Cell lysates were prepared using RIPA buffer with complete ULTRA protein inhibitors tablets (Roche). Lysates (60–80 µg/lane for cell lines and primary cells, and 0.5 or 1 µg/lane for RGMb-transfected 300 cells) were run on SDS-PAGE under reducing conditions. Western blotting was performed using RGMb mAb 1H6 (10 µg/ml) plus goat anti–rat IgG-HRP (Santa Cruz Biotechnology Inc.; 1:5,000). To blot for loading control, the membranes were treated with Restore Plus Western Blot Stripping Buffer (Thermo Fisher Scientific) and blotted with mouse anti–mouse β-actin (Abcam; 1:5,000) plus goat anti–mouse IgG-HRP (Santa Cruz Biotechnology Inc.; 1:4000). Protein bands on the membranes were visualized using standard chemiluminescent techniques.
Immunohistochemistry staining and confocal microscopy.
Cells were seeded on coverslips in culture medium the day before staining. Cells were fixed with 3.7% formaldehyde for 15 min at room temperature. After three washes with PBS containing 0.5% BSA, cells were permeabilized in PBS containing 0.5% BSA and 0.5% Triton X-100 for 30 min. Endogenous biotin was blocked with Endogenous Biotin-Blocking kit (Molecular Probes), followed by blocking in PBS containing 3% BSA and 0.1% Triton X-100 for 30 min at room temperature and three washes in PBS containing 0.5% BSA and 0.1% Triton X-100. Cells were then incubated with biotin-conjugated RGMb mAb 1H6 or biotin-conjugated rat IgG2a at 0.5 µg/ml in PBS containing 1% BSA and 0.1% Triton X-100 for 2 h at room temperature, followed by three washes as above. Subsequently, cells were incubated with Alexa Fluor 488–conjugated streptavidin (Jackson ImmunoResearch Laboratories) at 0.5 µg/ml in PBS containing 1% BSA and 0.1% Triton X-100 for 1 h at room temperature, followed by three washes as above. Finally, cells were stained with Phalloidin-TRIC (Sigma-Aldrich) at 50 µg/ml to label F-actin, and coverslips were mounted on slides with mounting media. Images were taken using a Nikon d-Eclipse C1 confocal microscope equipped with a Melles Griot 488 Ion Laser (with a 515/30 emission filter), a Melles Griot 543 Laser (with a 590/50 emission filter), a Melles Griot 640 Laser, and the Confocal Acquisition Software Nikon EZ-C1 version 3.90.
Respiratory tolerance and mAb treatment.
To induce tolerance, lightly anesthetized WT BALB/cByJ or PD-L2−/− mice received 100 µg of LPS-free OVA (Worthington Biochemical) in PBS or PBS alone (control) by intranasal instillation on days 0, 1, and 2. In some experiments, mice were treated with RGMb, PD-L2, PD-L1, or PD-1 blocking mAb or control antibody i.p. on day −1 (450 µg/mouse) and day 2 (200 µg/mouse). On day 12, mice were immunized with 50 µg OVA (ICN Biomedical) adsorbed in 2 mg ALUM. Spleens were harvested on day 19 and splenocytes or B cell–depleted splenocytes were restimulated in vitro with OVA. Cultures were pulsed with 1 µCi of 3[H]-thymidine for the final 17 h and harvested at 96 h. Culture supernatants were harvested at 96 h. IL-4, IFN-γ, and IL-5 were quantitated by ELISA. In some experiments, cells were labeled with CFSE before culture, stained on day 4 with CD3-PerCPCy5.5, and CD4-Alexa Fluor 700, and analyzed for CFSE dilution.
Measurement of airway hyperreactivity.
Mice tolerized and immunized with OVA/ALUM i.p. as above were given OVA i.n. on day 20, 21, and 22. 1 d after the final dose, mice were sedated, tracheostomized, and ventilated. Direct measurement of airway resistance and dynamic compliance was performed by invasive plethysmography (BUXCO systems). Two pressure sensors measured changes in lung volume and tracheal pressure in response to increasing concentrations of aerosolized methacholine. From these measurements, average lung resistance was calculated for 3 min per dose of methacholine. For histopathology, lungs were flushed with PBS to remove blood, infused with 10% (vol/vol) formalin, and embedded in paraffin. 5-µm lung sections were cut and stained with hematoxylin and eosin.
Transfer of DO11.10 T cells and respiratory tolerance.
T cells from DO11.10 or DO11.10 PD-L2−/− mice were positively selected from spleens by incubating with CD4+ magnetic beads and sorting using MACS columns (Miltenyi Biotec). Each recipient received 2 × 105 or 5 × 106 DO11.10 T cells i.v., followed by 3 i.n. doses of 100 µg LPS-free OVA on days 0, 1, 2 and mAbs as indicated. Cells from mediastinal LNs were analyzed by flow cytometry on days 3, 5, and/or 7.
Evaluation of antibodies for in vivo cell depletion.
To determine if PD-L2 or RGMb mAbs inhibited tolerance induction by depleting cells, mice immunized on day 0 with OVA in ALUM received OVA i.n. on days 7–9, and were treated with PD-L2 mAb 2C9 or isotype control (500 µg i.p.) on day 8 and dispersed lung cells were examined by FACS on day 10. Expression of PD-L2 was analyzed on AMs (F4/80+CD11c+), IMs (F4/80+CD11clo) and DCs (F4/80−CD11c+), and cell numbers of PD-L2+ and total cells were enumerated. In a separate experiment, mice were treated with RGMb mAb 9D1 or isotype control and the same analysis was performed.
Statistical analysis.
Two-tailed Student’s t test, two-way ANOVA followed by Tukey’s or Dunnett’s multiple comparisons test and ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test were performed using Prism version 6.00 for MacOS X (GraphPad Software). P < 0.05 was considered as significant. Data are mean ± SEM.
Acknowledgments
This study was supported by National Institutes of Health grants P01 AI056299, R01 A1051559, P01 AI080192, P01 AI0178897, U19AI082630, and HHSN272201100018C (G.J. Freeman and A.H. Sharpe) and R01 AI089955 and P01 AI054456 (G.J. Freeman, R.H. DeKruyff, and D.T. Umetsu).
G.J. Freeman and A.H. Sharpe have patents and receive patent royalties on the PD-1 pathway from Bristol-Myers-Squibb, Merck, Roche, EMD-Serrono, Boehringer-Ingelheim, Amplimmune, and CoStim. Gordon Freeman and Arlene Sharpe are scientific founders and scientific advisory board members of CoStim Pharmaceuticals. A patent application on uses of the RGMb–PD-L2 interaction including Y. Xiao, L.M. Francisco, D.T. Umetsu, A.H. Sharpe, R.H. DeKruyff, and G.J. Freeman has been filed. The authors declare no other conflicting financial interests.
Footnotes
Abbreviations used:
- AEC
- alveolar epithelial cell
- AM
- alveolar macrophage
- APC
- allophycocyanin
- BBRN
- BMP-BMP receptor–RGMb–neogenin supercomplex
- BMP
- bone morphogenetic protein
- FLT-3L
- Fms-like tyrosine kinase 3 ligand
- IM
- interstitial macrophage
- PD-l
- programmed death 1
- PD-L
- programmed death ligand
- RGMb
- repulsive guidance molecule b
- TEC
- tracheobronchial epithelial cell
- T reg cell
- regulatory T cell
References
- Akbari O., Stock P., Singh A.K., Lombardi V., Lee W.L., Freeman G.J., Sharpe A.H., Umetsu D.T., Dekruyff R.H. 2010. PD-L1 and PD-L2 modulate airway inflammation and iNKT-cell-dependent airway hyperreactivity in opposing directions. Mucosal Immunol. 3:81–91 10.1038/mi.2009.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albacker L.A., Yu S., Bedoret D., Lee W.L., Umetsu S.E., Monahan S., Freeman G.J., Umetsu D.T., Dekruyff R.H. 2013. TIM-4, expressed by medullary macrophages, regulates respiratory tolerance by mediating phagocytosis of antigen-specific T cells. Mucosal Immunol. 6:580–590 10.1038/mi.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoret D., Wallemacq H., Marichal T., Desmet C., Quesada Calvo F., Henry E., Closset R., Dewals B., Thielen C., Gustin P., et al. 2009. Lung interstitial macrophages alter dendritic cell functions to prevent airway allergy in mice. J. Clin. Invest. 119:3723–3738 10.1172/JCI39717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C.H., Healey E., van Erp S., Bishop B., Tang C., Gilbert R.J., Aricescu A.R., Pasterkamp R.J., Siebold C. 2013. Structure of the repulsive guidance molecule (RGM)-neogenin signaling hub. Science. 341:77–80 10.1126/science.1232322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragdon B., Moseychuk O., Saldanha S., King D., Julian J., Nohe A. 2011. Bone morphogenetic proteins: a critical review. Cell. Signal. 23:609–620 10.1016/j.cellsig.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Chuquimia O.D., Petursdottir D.H., Rahman M.J., Hartl K., Singh M., Fernández C. 2012. The role of alveolar epithelial cells in initiating and shaping pulmonary immune responses: communication between innate and adaptive immune systems. PLoS ONE. 7:e32125 10.1371/journal.pone.0032125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad S., Genth H., Hofmann F., Just I., Skutella T. 2007. Neogenin-RGMa signaling at the growth cone is bone morphogenetic protein-independent and involves RhoA, ROCK, and PKC. J. Biol. Chem. 282:16423–16433 10.1074/jbc.M610901200 [DOI] [PubMed] [Google Scholar]
- Conrad S., Stimpfle F., Montazeri S., Oldekamp J., Seid K., Alvarez-Bolado G., Skutella T. 2010. RGMb controls aggregation and migration of Neogenin-positive cells in vitro and in vivo. Mol. Cell. Neurosci. 43:222–231 10.1016/j.mcn.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Corradini E., Babitt J.L., Lin H.Y. 2009. The RGM/DRAGON family of BMP co-receptors. Cytokine Growth Factor Rev. 20:389–398 10.1016/j.cytogfr.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman G.J., Freedman A.S., Segil J.M., Lee G., Whitman J.F., Nadler L.M. 1989. B7, a new member of the Ig superfamily with unique expression on activated and neoplastic B cells. J. Immunol. 143:2714–2722 [PubMed] [Google Scholar]
- Galligan C.L., Baig E., Bykerk V., Keystone E.C., Fish E.N. 2007. Distinctive gene expression signatures in rheumatoid arthritis synovial tissue fibroblast cells: correlates with disease activity. Genes Immun. 8:480–491 10.1038/sj.gene.6364400 [DOI] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Lambrecht B.N. 2008. Division of labor between dendritic cell subsets of the lung. Mucosal Immunol. 1:442–450 10.1038/mi.2008.39 [DOI] [PubMed] [Google Scholar]
- Graf D., Nethisinghe S., Palmer D.B., Fisher A.G., Merkenschlager M. 2002. The developmentally regulated expression of Twisted gastrulation reveals a role for bone morphogenetic proteins in the control of T cell development. J. Exp. Med. 196:163–171 10.1084/jem.20020276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager-Theodorides A.L., Outram S.V., Shah D.K., Sacedon R., Shrimpton R.E., Vicente A., Varas A., Crompton T. 2002. Bone morphogenetic protein 2/4 signaling regulates early thymocyte differentiation. J. Immunol. 169:5496–5504 [DOI] [PubMed] [Google Scholar]
- Hagihara M., Endo M., Hata K., Higuchi C., Takaoka K., Yoshikawa H., Yamashita T. 2011. Neogenin, a receptor for bone morphogenetic proteins. J. Biol. Chem. 286:5157–5165 10.1074/jbc.M110.180919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawiger D., Inaba K., Dorsett Y., Guo M., Mahnke K., Rivera M., Ravetch J.V., Steinman R.M., Nussenzweig M.C. 2001. Dendritic cells induce peripheral T cell unresponsiveness under steady state conditions in vivo. J. Exp. Med. 194:769–779 10.1084/jem.194.6.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano N., Butler M.O., Xia Z., Ansén S., von Bergwelt-Baildon M.S., Neuberg D., Freeman G.J., Nadler L.M. 2006. Engagement of CD83 ligand induces prolonged expansion of CD8+ T cells and preferential enrichment for antigen specificity. Blood. 107:1528–1536 10.1182/blood-2005-05-2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt P.G., Upham J.W. 2004. The role of dendritic cells in asthma. Curr. Opin. Allergy Clin. Immunol. 4:39–44 10.1097/00130832-200402000-00009 [DOI] [PubMed] [Google Scholar]
- Hoyne G.F., Askonas B.A., Hetzel C., Thomas W.R., Lamb J.R. 1996. Regulation of house dust mite responses by intranasally administered peptide: transient activation of CD4+ T cells precedes the development of tolerance in vivo. Int. Immunol. 8:335–342 10.1093/intimm/8.3.335 [DOI] [PubMed] [Google Scholar]
- Keir M.E., Butte M.J., Freeman G.J., Sharpe A.H. 2008. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 26:677–704 10.1146/annurev.immunol.26.021607.090331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Myers A.C., Chen L., Pardoll D.M., Truong-Tran Q.A., Lane A.P., McDyer J.F., Fortuno L., Schleimer R.P. 2005. Constitutive and inducible expression of b7 family of ligands by human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 33:280–289 10.1165/rcmb.2004-0129OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- König K., Gatidou D., Granja T., Meier J., Rosenberger P., Mirakaj V. 2012. The axonal guidance receptor neogenin promotes acute inflammation. PLoS ONE. 7:e32145 10.1371/journal.pone.0032145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H.C., Choi A.M., Ryter S.W. 2011. Isolation of mouse respiratory epithelial cells and exposure to experimental cigarette smoke at air liquid interface. J. Vis. Exp. 48:2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman Y., Wood C.R., Chernova T., Chaudhary D., Borde M., Chernova I., Iwai Y., Long A.J., Brown J.A., Nunes R., et al. 2001. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2:261–268 10.1038/85330 [DOI] [PubMed] [Google Scholar]
- Lesterhuis W.J., Steer H., Lake R.A. 2011. PD-L2 is predominantly expressed by Th2 cells. Mol. Immunol. 49:1–3 10.1016/j.molimm.2011.09.014 [DOI] [PubMed] [Google Scholar]
- Li J., Ye L., Kynaston H.G., Jiang W.G. 2012a. Repulsive guidance molecules, novel bone morphogenetic protein co-receptors, are key regulators of the growth and aggressiveness of prostate cancer cells. Int. J. Oncol. 40:544–550 [DOI] [PubMed] [Google Scholar]
- Li J., Ye L., Sanders A.J., Jiang W.G. 2012b. Repulsive guidance molecule B (RGMB) plays negative roles in breast cancer by coordinating BMP signaling. J. Cell. Biochem. 113:2523–2531 10.1002/jcb.24128 [DOI] [PubMed] [Google Scholar]
- Liu X., Gao J.X., Wen J., Yin L., Li O., Zuo T., Gajewski T.F., Fu Y.X., Zheng P., Liu Y. 2003. B7DC/PDL2 promotes tumor immunity by a PD-1-independent mechanism. J. Exp. Med. 197:1721–1730 10.1084/jem.20022089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Li X., Zhao Y., Meng X.M., Wan C., Yang B., Lan H.Y., Lin H.Y., Xia Y. 2013. Dragon (RGMb) inhibits E-Cadherin expression and induces apoptosis in renal tubular epithelial cells. J. Biol. Chem. 288:31528–31539 10.1074/jbc.M113.517573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Ma J., Wang X., Wang J., Zhang F., Yu J., He G., Xu B., Brand D.D., Horwitz D.A., et al. 2010. Synergistic effect of TGF-beta superfamily members on the induction of Foxp3+ Treg. Eur. J. Immunol. 40:142–152 10.1002/eji.200939618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messal N., Serriari N.E., Pastor S., Nunès J.A., Olive D. 2011. PD-L2 is expressed on activated human T cells and regulates their function. Mol. Immunol. 48:2214–2219 10.1016/j.molimm.2011.06.436 [DOI] [PubMed] [Google Scholar]
- Mirakaj V., Jennewein C., König K., Granja T., Rosenberger P. 2012. The guidance receptor neogenin promotes pulmonary inflammation during lung injury. FASEB J. 26:1549–1558 10.1096/fj.11-200063 [DOI] [PubMed] [Google Scholar]
- Mkrtichyan M., Najjar Y.G., Raulfs E.C., Liu L., Langerman S., Guittard G., Ozbun L., Khleif S.N. 2012. B7-DC-Ig enhances vaccine effect by a novel mechanism dependent on PD-1 expression level on T cell subsets. J. Immunol. 189:2338–2347 10.4049/jimmunol.1103085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu R., Kubo T., Mori M., Nakamura Y., Fujita Y., Akutsu T., Okuno T., Taniguchi J., Kumanogoh A., Yoshida M., et al. 2011. RGMa modulates T cell responses and is involved in autoimmune encephalomyelitis. Nat. Med. 17:488–494 10.1038/nm.2321 [DOI] [PubMed] [Google Scholar]
- Pardoll D.M. 2012. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 12:252–264 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfistershammer K., Klauser C., Pickl W.F., Stöckl J., Leitner J., Zlabinger G., Majdic O., Steinberger P. 2006. No evidence for dualism in function and receptors: PD-L2/B7-DC is an inhibitory regulator of human T cell activation. Eur. J. Immunol. 36:1104–1113 10.1002/eji.200535344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritprajak P., Hashiguchi M., Akiba H., Yagita H., Okumura K., Azuma M. 2012. Antibodies against B7-DC with differential binding properties exert opposite effects. Hybridoma (Larchmt). 31:40–47 10.1089/hyb.2011.0087 [DOI] [PubMed] [Google Scholar]
- Rosendahl A., Pardali E., Speletas M., Ten Dijke P., Heldin C.H., Sideras P. 2002. Activation of bone morphogenetic protein/Smad signaling in bronchial epithelial cells during airway inflammation. Am. J. Respir. Cell Mol. Biol. 27:160–169 10.1165/ajrcmb.27.2.4779 [DOI] [PubMed] [Google Scholar]
- Samad T.A., Rebbapragada A., Bell E., Zhang Y., Sidis Y., Jeong S.J., Campagna J.A., Perusini S., Fabrizio D.A., Schneyer A.L., et al. 2005. DRAGON, a bone morphogenetic protein co-receptor. J. Biol. Chem. 280:14122–14129 10.1074/jbc.M410034200 [DOI] [PubMed] [Google Scholar]
- Severyn C.J., Shinde U., Rotwein P. 2009. Molecular biology, genetics and biochemistry of the repulsive guidance molecule family. Biochem. J. 422:393–403 10.1042/BJ20090978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin T., Kennedy G., Gorski K., Tsuchiya H., Koseki H., Azuma M., Yagita H., Chen L., Powell J., Pardoll D., Housseau F. 2003. Cooperative B7-1/2 (CD80/CD86) and B7-DC costimulation of CD4+ T cells independent of the PD-1 receptor. J. Exp. Med. 198:31–38 10.1084/jem.20030242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Stock P., Akbari O. 2011. Role of PD-L1 and PD-L2 in allergic diseases and asthma. Allergy. 66:155–162 10.1111/j.1398-9995.2010.02458.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland D.H., Upham J.W., Holt P.G. 2010. Epithelial-dendritic cell interactions in allergic disorders. Curr. Opin. Immunol. 22:789–794 10.1016/j.coi.2010.10.019 [DOI] [PubMed] [Google Scholar]
- Tian C., Liu J. 2013. Repulsive guidance molecules (RGMs) and neogenin in bone morphogenetic protein (BMP) signaling. Mol. Reprod. Dev. 80:700–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian S.L., Hodi F.S., Brahmer J.R., Gettinger S.N., Smith D.C., McDermott D.F., Powderly J.D., Carvajal R.D., Sosman J.A., Atkins M.B., et al. 2012. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366:2443–2454 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng S.Y., Otsuji M., Gorski K., Huang X., Slansky J.E., Pai S.I., Shalabi A., Shin T., Pardoll D.M., Tsuchiya H. 2001. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J. Exp. Med. 193:839–846 10.1084/jem.193.7.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitoura D.C., DeKruyff R.H., Lamb J.R., Umetsu D.T. 1999. Intranasal exposure to protein antigen induces immunological tolerance mediated by functionally disabled CD4+ T cells. J. Immunol. 163:2592–2600 [PubMed] [Google Scholar]
- Wang S., Bajorath J., Flies D.B., Dong H., Honjo T., Chen L. 2003. Molecular modeling and functional mapping of B7-H1 and B7-DC uncouple costimulatory function from PD-1 interaction. J. Exp. Med. 197:1083–1091 10.1084/jem.20021752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Sun C.C., Lin H.Y., Babitt J.L. 2012. Repulsive guidance molecule (RGM) family proteins exhibit differential binding kinetics for bone morphogenetic proteins (BMPs). PLoS ONE. 7:e46307 10.1371/journal.pone.0046307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Cortez-Retamozo V., Niederkofler V., Salie R., Chen S., Samad T.A., Hong C.C., Arber S., Vyas J.M., Weissleder R., et al. 2011. Dragon (repulsive guidance molecule b) inhibits IL-6 expression in macrophages. J. Immunol. 186:1369–1376 10.4049/jimmunol.1002047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmin N., Bauer T., Modak M., Wagner K., Schuster C., Köffel R., Seyerl M., Stöckl J., Elbe-Bürger A., Graf D., Strobl H. 2013. Identification of bone morphogenetic protein 7 (BMP7) as an instructive factor for human epidermal Langerhans cell differentiation. J. Exp. Med. 210:2597–2610 10.1084/jem.20130275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka Y., Ono M., Osaki M., Konishi I., Sakaguchi S. 2012. Differential effects of inhibition of bone morphogenic protein (BMP) signalling on T-cell activation and differentiation. Eur. J. Immunol. 42:749–759 10.1002/eji.201141702 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chung Y., Bishop C., Daugherty B., Chute H., Holst P., Kurahara C., Lott F., Sun N., Welcher A.A., Dong C. 2006. Regulation of T cell activation and tolerance by PDL2. Proc. Natl. Acad. Sci. USA. 103:11695–11700 10.1073/pnas.0601347103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Rothstein T.L. 2011. L2pB1: a new player in autoimmunity. Mol. Immunol. 48:1292–1300 10.1016/j.molimm.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]