Granzyme B is expressed by hematopoietic stem cells (HSCs) and stromal cells in response to bacterial products or chemotherapy agents and limits HSC reconstitution potential.
Abstract
The serine protease granzyme B (GzmB) is stored in the granules of cytotoxic T and NK cells and facilitates immune-mediated destruction of virus-infected cells. In this study, we use genetic tools to report novel roles for GzmB as an important regulator of hematopoietic stem cell (HSC) function in response to stress. HSCs lacking the GzmB gene show improved bone marrow (BM) reconstitution associated with increased HSC proliferation and mitochondrial activity. In addition, recipients deficient in GzmB support superior engraftment of wild-type HSCs compared with hosts with normal BM niches. Stimulation of mice with lipopolysaccharide strongly induced GzmB protein expression in HSCs, which was mediated by the TLR4–TRIF–p65 NF-κB pathway. This is associated with increased cell death and GzmB secretion into the BM environment, suggesting an extracellular role of GzmB in modulating HSC niches. Moreover, treatment with the chemotherapeutic agent 5-fluorouracil (5-FU) also induces GzmB production in HSCs. In this situation GzmB is not secreted, but instead causes cell-autonomous apoptosis. Accordingly, GzmB-deficient mice are more resistant to serial 5-FU treatments. Collectively, these results identify GzmB as a negative regulator of HSC function that is induced by stress and chemotherapy in both HSCs and their niches. Blockade of GzmB production may help to improve hematopoiesis in various situations of BM stress.
Hematopoietic stem cells (HSCs) are on top of the hierarchically organized hematopoietic system as they have the ability to long-term self-renew while giving rise to progeny that can generate all mature blood cell types throughout adult life (Chao et al., 2008; Trumpp et al., 2010; Doulatov et al., 2012). Given that several hematopoietic diseases are only curable by allogenic HSC transplantation, extensive effort is currently focused on understanding the mechanisms by which HSCs maintain their self-renewal and multipotent properties after transplantation into conditioned recipients to be able to robustly reconstitute the hematopoietic system.
During homeostasis the most primitive HSCs reside in a dormant state while the majority of HSCs are slowly cycling (Sudo et al., 2000; Wilson et al., 2008; Foudi et al., 2009; Trumpp et al., 2010). The different HSC populations reside in specialized BM niches comprised of different hematopoietic and stromal cell types controlling HSC cycling, self-renewal, and differentiation (Morrison and Spradling, 2008; Wilson et al., 2008; Méndez-Ferrer et al., 2009; Ehninger and Trumpp, 2011). Several stimuli have been shown to activate HSCs in vivo, including the chemotherapeutic agent 5-fluorouracil (5-FU; Randall and Weissman, 1997; Venezia et al., 2004) and the cytokines G-CSF (Wilson et al., 2008) and IFN-α and IFN-γ (Essers et al., 2009; Baldridge et al., 2010). Recently, sublethal doses of LPS have also been shown to induce HSC and progenitor proliferation in vivo (Chen et al., 2010; Scumpia et al., 2010; Esplin et al., 2011; Takizawa et al., 2011), suggesting that bacterial and viral infections can lead to HSC activation.
Our laboratory has previously shown that Myc oncoproteins are crucial regulators of hematopoiesis (Wilson et al., 2004; Laurenti et al., 2008, 2009). Deletion of both c-myc and N-myc alleles cause rapid severe BM failure associated with apoptosis of all hematopoietic cell types except dormant HSCs (Laurenti et al., 2008). Mechanistically, c-Myc– and N-Myc–deficient HSCs showed a global reduction in ribosome biogenesis and a striking 150-fold increase in granzyme B (GzmB) transcripts preceding progenitor apoptosis (Laurenti et al., 2008, 2009). These data raise the possibility that high expression of cytoplasmic GzmB protein is the apoptotic mechanism leading to HSC death in response to loss of Myc activity (Laurenti et al., 2008).
Members of the granzyme family of cytotoxic serine proteases are released by cytoplasmic granules within NK and cytotoxic T cells to induce apoptotic cell death of viral infected cells or tumor cells. There are 11 known granzymes in mice and 5 in humans divided into three clusters (Grossman et al., 2003; Boivin et al., 2009). GzmA and GzmB are the most abundant ones causing apoptosis by various pathways, although the role of GzmA in apoptosis has become controversial. Both GzmA and GzmB KO mice are healthy and fertile but present susceptibility to ectromelia infection (Müllbacher et al., 1999). The serine protease GzmB is known to cleave and activate several proapoptotic proteins in response to infection by the perforin-dependent cytotoxic response pathway (Chowdhury and Lieberman, 2008). Its function has been well established in the adaptive immune system in mechanisms involved in graft-versus-host disease (Graubert et al., 1996, 1997), control of viral infections (Müllbacher et al., 1999), and tumor clearance (Cao et al., 2007). In addition, extracellular roles including cytokine modulation have also been suggested for granzymes (Boivin et al., 2009; Froelich et al., 2009). Although the function of GzmB in NK and cytotoxic T cells is well described, it has so far not been reported to play a role in regulating HSC biology. Following up on our unexpected finding that HSCs can express high levels of GzmB in vivo (Laurenti et al., 2008), we used genetic tools to investigate the role of GzmB in HSC function during homeostasis and in response to BM stress.
RESULTS AND DISCUSSION
GzmB-deficient mice have normal hematopoiesis
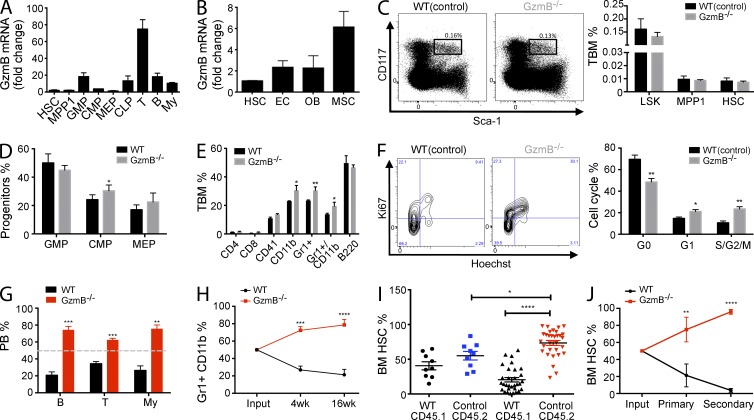
To investigate the role of GzmB in HSCs and progenitors, we analyzed its expression in hematopoietic and stromal cells of the BM. During homeostatic conditions, GzmB is robustly expressed in lymphoid cells but is expressed only at low levels in either HSCs or multipotent progenitor population 1 (MPP1) cells (Fig. 1 A and Table S1). Within the stromal compartment, the mesenchymal stromal cells express the highest level of GzmB mRNA and protein (Fig. 1 B and not depicted). GzmB−/− mice showed normal marrow cellularity and morphology (not depicted) and normal frequencies of LSK, MPP1, and HSC populations when compared with WT controls (Fig. 1 C). Analysis of the progenitor populations revealed increased common myeloid progenitor and decreased common lymphoid progenitor frequency in mutant mice (Fig. 1 D and not depicted). Accordingly, a significant increase in myeloid cells in the BM of GzmB−/− mice was notable (Fig. 1 E). To rule out the possibility that other granzymes could compensate for the lack of GzmB, the phenotype of GzmA−/−B−/− double KO (DKO) was examined (Simon et al., 1997). GzmA expression in the hematopoietic system is largely restricted to lymphoid cells and not detected in HSCs and MPP1 (not depicted), and overall, no difference between DKO and GzmB−/− mice in the hematopoietic system has been observed (not depicted). Given the putative extracellular roles of GzmB, we analyzed mobilization of GzmB-deficient HSCs; however, G-CSF–induced mobilization was unaffected in both GzmB−/− and DKO mice in comparison with controls, as assessed by the number of LSKs in the peripheral blood and CFU in BM, blood, and spleens (not depicted).
Figure 1.
GzmB−/− HSCs are more proliferative and show a superior reconstitution capacity because of an increase in functional HSCs. Cell surface marker definition of all cell populations is indicated in Table S1. (A and B) GzmB mRNA expression levels were assessed by RT-PCR in hematopoietic cells (A; n = 6) and in HSCs, endothelial cells (EC), mesenchymal stromal cells (MSC), and osteoblasts (OB) during homeostatic conditions in WT cells (B; n = 3). Fold change was calculated in relation to HSC GzmB mRNA levels. (C) BM cells from WT or GzmB−/− mice were analyzed for the indicated cell populations by flow cytometry. (left) Representative FACS plot showing Sca-1 and CD117 (c-Kit) staining of lin− cells; box shows gating on LSK (Lin−CD117+Sca1+) cells; (right) calculated frequencies of LSK, MPP1, and HSC cells (n = 6). (D and E) Relative frequencies of the indicated progenitor and mature populations in the BM WT and GzmB−/− mice, as assessed by flow cytometry (n = 6 each). (F) Cell cycle status of WT(control) and GzmB−/− HSCs was assessed by Ki67/Hoechst staining. The left panel shows a representative flow cytometry plot, and the right panel shows the quantification of data (n = 6). (G) CD45.1+ WT BM cells were mixed in a 1:1 ratio either with CD45.2+ WT or GzmB−/− BM cells. The relative contribution of donor-derived GzmB−/− CD45.2 and WT CD45.1 BM cells to T, B, and myeloid (My) cells in the circulation was assessed 16 wk after transplantation (n = 6). Gray dashed line indicates 50% reconstitution. (H) Frequency of WT and GzmB−/− myeloid cells in the circulation was assessed at 4 and 16 wk after transplantation (n = 6). (I) CD45.2+ GzmB−/− (red) or WT (blue) BM cells were mixed with CD45.1+ WT HSCs and injected into WT recipient mice, and the frequency of CD45.2 donor cells in the BM was assessed by flow cytometry (n > 9). Graphs in A–I are representative of three independent experiments. (J) CD45.2+ GzmB−/− and CD45.1+ WT HSCs from primary recipients were injected into secondary hosts, and their frequency after primary and secondary BM transplantation was assessed by flow cytometry compared with the input (n = 6). Results in all panels represent means ± SEM of at least two experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
The function of HSCs is closely related to their proliferation status. Analysis of GzmB-deficient HSCs revealed that they are less quiescent and are more actively cycling, as shown by a significant decrease of HSCs in G0 (Ki67−; Fig. 1 F). In summary, we observe that the hematopoietic compartment of adult GzmB−/− mice show only minor alterations during homeostasis, although the balance between HSC quiescence and cycling is significantly disturbed.
GzmB−/− HSCs show increased reconstitution ability in competitive BM transplantations
To determine the reconstitution capability of GzmB−/− HSCs, competitive repopulating assays were performed comparing engraftment of GzmB−/− with control BM cells. As early as 4 wk after transplantation, 75.47% (±4.63) of the total myeloid and 62.27% (±2.14) of T cell populations in the peripheral blood were of GzmB−/− origin (Fig. 1, G and H), and similarly, GzmB−/− HSCs had largely outcompeted WT HSCs in the recipient BM at 16 wk (Fig. 1 I). The competitive advantage of mutant HSCs was even more prominent in secondary BM transplantations (Fig. 1 J). The enhanced engraftment of GzmB−/− HSCs was not caused by improved homing ability (not depicted). In contrast, limiting dilution assays (LDAs) revealed a more than fourfold increase of functional HSCs in GzmB−/− mice compared with controls (Table 1). Collectively, these results suggest that GzmB-deficient mice harbor an increased number of repopulating functional HSCs, providing an explanation for their superior performance in the competitive setting.
Table 1.
LDA
| Group | Number of cells transplanted | Number of reconstituted mice/number of primary recipients | HSC frequency (95% CI) |
| Control | 1,000 | 3/10 | 1 in 26,559 (54,453–12,954) |
| Control | 10,000 | 4/10 | |
| Control | 100,000 | 9/10 | |
| GzmB−/− | 1,000 | 2/10 | 1 in 5,354 (10,164–2,820) |
| GzmB−/− | 10,000 | 10/12 |
Serially diluted control or GzmB−/− total BM cells were transplanted with supportive WT total BM cells, and HSC engraftment was quantified 14 wk after transplantation. Frequency of HSCs was calculated, according to Poisson statistics using ELDA software, from combined results of two independent experiments with n = 10/group (two-tailed Student’s t test; P = 0.026). Positive: >1% engraftment BM (LSK SLAM). CI, confidence interval.
To date, only a small number of genetic mutants have been identified that show improved HSC function in vivo (Rossi et al., 2012). Examples are mutants for Lnk (Ema et al., 2005), DNMT3a (Challen et al., 2012), or TNF receptors (Pronk et al., 2011). Remarkably, our experiments demonstrate that GzmB is one of the few known proteins suppressing HSC function under stress conditions, such as reconstitution after lethal irradiation. One reason for the positive effect observed in GzmB-deficient cells may be an increased resistance to stress-induced cell death. It has recently been shown that activation of the intrinsic apoptosis pathway occurs during transplantation, leading to a relevant loss of HSCs (Labi et al., 2013). Moreover, our results show that GzmB-deficient HSCs are more proliferative compared with normal HSCs. This is remarkable as increased HSC cycling typically goes along with loss of self-renewal and subsequent stem cell exhaustion (Wilson et al., 2009; Rossi et al., 2012). Possible mechanisms to explain the phenotype of GzmB−/− HSCs could be protection from ageing and senescence, increased metabolism, and lack of a fail-safe mechanism to halt proliferation uncoupled from self-renewal capacity.
A GzmB-deficient microenvironment promotes proliferation and improves HSC function
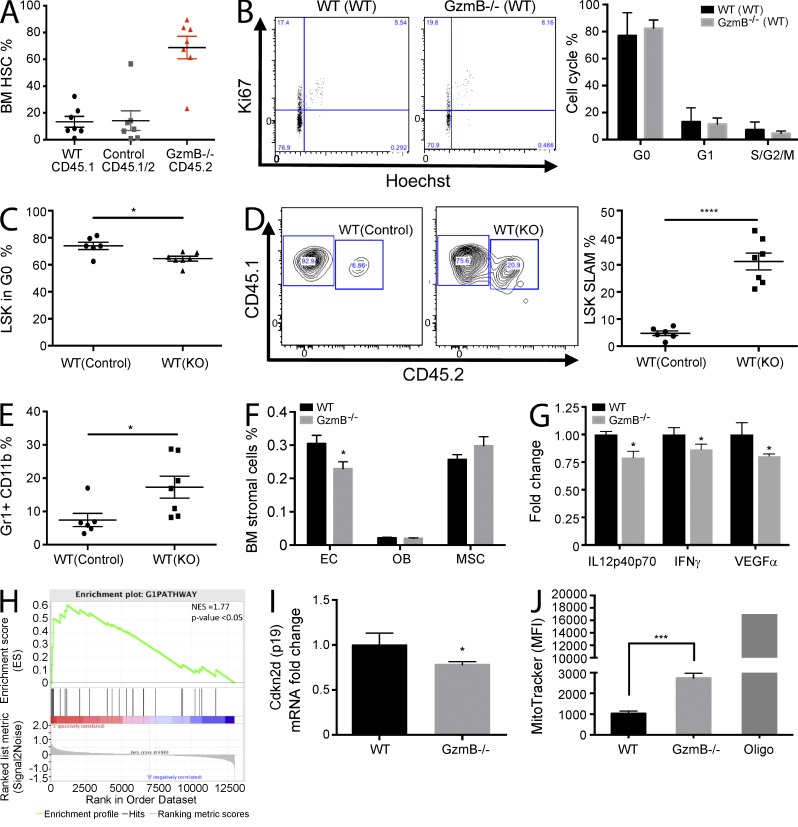
The cycling differences of GzmB−/− HSCs could be exclusively cell autonomous or also be mediated by cell-extrinsic cues derived from the microenvironment. In agreement with our data in Fig. 1 (G–I), GzmB-deficient HSCs out-competed WT HSCs if transplanted at a 1:1 ratio into a WT microenvironment (Fig. 2 A). However, the cell cycle behavior of mutant HSCs in this setting is indistinguishable from control HSCs (Fig. 2 B). Although here GzmB-deficient HSCs are growing in a WT microenvironment, both compartments lack this gene in GzmB mutant animals (Fig. 1 F). These results indicate that the augmented HSC proliferation observed in GzmB mutant mice is likely caused by the conditions present in GzmB-deficient BM niches.
Figure 2.
GzmB-deficient BM microenvironment promotes stem and progenitor cell proliferation and improves HSC function. (A and B) CD45.2 GzmB−/− and WT CD45.1/2 (control) BM was transplanted into WT CD45.1 recipient mice. (A) Engraftment of HSCs was assessed by flow cytometry (black; n = 7). (B) The cell cycle status of transplanted WT CD45.1/2 and GzmB−/− CD45.2 HSCs was assessed by Ki67/Hoechst staining. The left panel shows representative flow cytometry plots, and the right panel shows quantification from n = 7 mice. (C) Reverse chimeras were generated by transplanting 106 CD45.1/2 WT BM cells into WT (WT(control)) or GzmB-deficient mice WT(KO) (n = 7). Percentage of LSK cells in G0 phase of the cell cycle was assessed by Ki67/Hoechst staining 16 wk after transplantation (n = 7). (D) Total BM was harvested from the primary recipients described in C, mixed at a 1:1 ratio with fresh CD45.1 WT total BM, and injected into lethally irradiated secondary CD45.1 WT recipients. LSK SLAM cells (CD45.1/2) were quantified in secondary recipients (CD45.1) by flow cytometry 16 wk after transplantation (n = 7). Representative flow cytometry contour plots (left) and quantification from n = 7 mice/group (right) are shown. (E) The frequency of Gr1+CD11b+ myeloid progeny was assessed by flow cytometry in the reverse chimeras generated in D (n = 7). (F) Frequency of the indicated BM stromal cell types in control and GzmB−/− mice was assessed by flow cytometry (n = 3). (G) Protein levels of IL12, IFN-γ, and VEGF were measured in the BM of WT and GzmB−/− mice by cytokine array (n = 3). (H) HSCs were isolated from GzmB-deficient mice, and RNA was subject to microarray analysis. A GSEA plot of cell cycle G1-phase genes is shown. P < 0.05 and normalized enrichment score (NES) = 1.77. Positively (red) and negatively (blue) correlated genes are shown. (I) HSCs were purified from WT and GzmB−/− mice, and Cdkn2d (p19) mRNA levels were measured by RT-PCR (n = 6). (J) Mitochondrial mass analysis of homeostatic WT and GzmB−/− HSCs as assessed by MitoTracker staining (n = 6). MFI, mean fluorescence intensity. Data in A–G, I, and J represent means ± SEM of two independent experiments. *, P < 0.05; ***, P < 0.001; and ****, P < 0.0001.
Given the known role of GzmB in apoptosis, we reasoned that in a WT microenvironment HSCs are more susceptible to proapoptotic signals. To address this possibility, we have measured cleaved caspase-3 in long-term transplanted HSCs (not depicted) and mRNA levels of key pro- and antiapoptotic proteins in HSCs 3 wk after transplantation; however, no changes could be detected (not depicted). Nevertheless, because of technical constraints (e.g., fast in vivo dead cell clearance), we cannot formally rule out the possibility that GzmB-mediated apoptosis may play a role in the maintenance of engrafted HSCs.
To further investigate the role of GzmB in the BM niche, primary and secondary BM reconstitution assays were performed. First, WT total BM cells (WT) were transplanted into either WT (control) or GzmB−/− (KO) recipient mice. We observed that WT LSK cells in the GzmB-deficient BM setting (WT(KO)) were less quiescent and more proliferative compared with WT cells transplanted into a WT control niche (WT(control); Fig. 2 C). Interestingly, in competitive secondary transplantations, MPP1 and HSCs derived from the WT(KO) setting represented 31.26% (±3.11) of the secondary recipient BM, whereas HSCs derived from the WT(control) setting only contributed to 4.79% (±0.86) of total BM cells (Fig. 2 D). Similar results were obtained analyzing the myeloid progeny in the peripheral blood of the recipient mice (Fig. 2 E). These results suggest that the GzmB-deficient microenvironment enhances the expansion and engraftment capacity of normal HSCs. Thus, GzmB deficiency not only displayed a positive effect on the ability of HSCs to engraft, but a GzmB-deficient microenvironment was also beneficial to normal HSC engraftment and reconstitution of the hematopoietic system in a transplant setting. Possibly, GzmB might exert a negative effect on HSCs by modulating cytokine function by proteolysis (Boivin et al., 2012), cleaving extracellular matrix components (Froelich et al., 1993; Birdsall et al., 2004; Buzza et al., 2005) or receptors important for cell migration and proliferation such as FGFR1 and Notch1 (van Tetering et al., 2011; Chioni and Grose, 2012).
Because a GzmB-deficient microenvironment enhances HSC function in vivo, we analyzed the BM niche with respect to stromal cell types or presence of circulating cytokines. This revealed a mild decrease in endothelial cells (Fig. 2 F) and lower levels of the immune modulatory cytokines IL12, IFN-γ, and VEGF-α (Fig. 2 G). In summary, the results show that alterations within the GzmB−/− BM milieu facilitate HSC proliferation and improve the functional capacity of HSCs, suggesting that GzmB influences HSCs not only in an intrinsic but also in an extrinsic manner.
GzmB−/− HSCs show increased cell cycle and mitochondrial activity in vivo
To further elucidate the molecular mechanisms involved in the improved function of GzmB-deficient HSCs, their gene expression profile was determined. Gene set enrichment analysis (GSEA) indicated that cell cycle G1-phase genes are enriched in GzmB−/− HSCs (Fig. 2 H), in agreement with the increased proliferation of BM HSCs during homeostatic conditions. GzmB−/− HSCs express slightly lower mRNA levels of p19INK4d (Fig. 2 I), an inhibitor of CDK4/CDK6 and early G1-S transition which has also been implicated in senescence and protection from DNA damage and apoptosis (Stepanova and Sorrentino, 2005; Oguro et al., 2006). Additionally, functional annotation (DAVID) showed that GzmB−/− HSCs are enriched for mitochondrial genes (not depicted). Indeed, mitochondrial mass/activity was increased about threefold in GzmB−/− HSCs (Fig. 2 J), suggesting that GzmB is a negative regulator of mitochondrial activity in homeostatic HSCs. The increased mitochondrial mass in mutant HSCs is consistent with their increased cycling activity. Interestingly, in cytotoxic T cells, GzmB contributes to mitochondrial damage during the apoptotic response and is involved in Bcl2 inhibition, cytochrome c release, mitochondria swelling, transmembrane potential loss, and reactive oxygen species production (Chowdhury and Lieberman, 2008). Therefore, the lack of GzmB could potentially protect HSCs from mitochondrial injury in response to stress, allowing them to perform better in BM reconstitution assays after lethal irradiation.
LPS induces GzmB in HSCs and progenitors in vivo
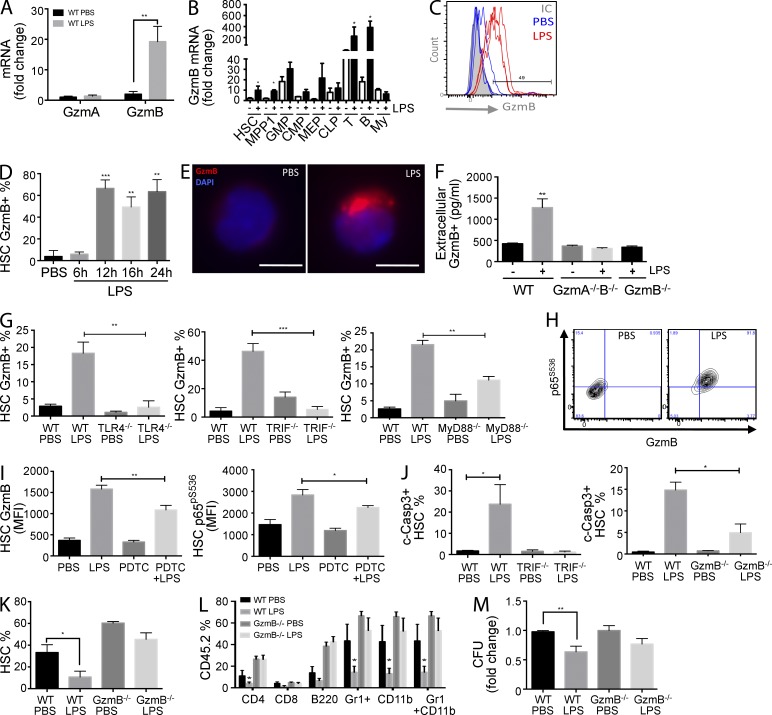
Next we asked whether other stress stimuli such as bacterial or viral infections would also trigger a GzmB response in vivo. LPS is the major component of the outer membrane of Gram-negative bacteria and binds to the TLR4–MD2 receptor complex, which leads to the production of inflammatory cytokines (Hennessy et al., 2010). High-dose LPS treatment of mice is often used as a model to study endotoxic shock (Akira and Takeda, 2004). Here we treated mice with low doses of LPS to mimic common bacterial infections, which can also lead to HSC activation (Takizawa et al., 2011). LPS treatment induced a robust 10-fold increase in GzmB but not GzmA transcripts in HSCs (Fig. 3 A). GzmB expression was also induced in progenitors and, as expected, also in T and B cells (Fig. 3 B). Not only mRNA, but also GzmB protein was increased in HSCs (Fig. 3 C), and the levels remained high for at least 24 h (Fig. 3 D). Our results show that in response to LPS, and thus bacterial infections, GzmB is not only induced in lymphocytes, but also in HSCs and progenitors in vivo.
Figure 3.
LPS-induced GzmB expression in HSCs is dependent on TLR4–TRIF–NF-κB signaling, is secreted, and impairs HSC function in vivo. WT mice were injected with 5 µg LPS i.p. or PBS, and BM was isolated 16 h later. (A and B) GzmA and GzmB mRNA levels in HSCs (A; n = 6) and GzmB mRNA in BM hematopoietic cells (B; n = 6) were measured by RT-PCR. (C) Representative flow cytometry histogram showing GzmB protein expression in HSCs from mice injected with LPS or PBS (n = 3). (D) GzmB expression in HSCs was measured by flow cytometry at the indicated times after LPS treatment (n = 6). (E) LSK SLAM cells were purified from in vivo LPS-stimulated WT mice for 16 h, and GzmB cellular localization was assessed by immunofluorescence using anti-GzmB antibodies and DAPI nuclear staining (n = 4). Bars, 5 µm. (F–H) Mice of the indicated genotype were injected with LPS or PBS as in A (n = 6). (F) Extracellular GzmB (pg/ml) in the BM niche was measured by ELISA. (G) Percentage of GzmB-expressing HSCs (GzmB+) in TLR4−/−, TRIF−/−, and MyD88−/− mice was assessed by flow cytometry. (H) Representative flow cytometry contour plot showing p65pS536 and GzmB expression in HSCs from WT mice 16 h after LPS stimulation. (I) WT mice were pretreated or not with 10 mg/kg PDTC i.p., followed by LPS 10 h later. GzmB and p65pS536 expression in HSCs was measured by flow cytometry 16 h after LPS injection (n = 6). (J) WT, TRIF−/−, or GzmB−/− mice were injected with LPS or PBS as in A, and levels of active caspase-3 (c-Casp3) were measured in HSCs by flow cytometry (n = 6). (K) CD45.2 GzmB−/− or WT mice were injected with LPS or PBS, and HSCs were purified 16 h later, mixed with 106 WT CD45.1/2 BM cells, and injected i.v. into lethally irradiated WT recipients. HSC engraftment was measured by flow cytometry 16 wk after transplant (n = 6). (L) Frequency of the indicated hematopoietic cell types in the mice described in K. (M) GzmB−/− and WT mice were treated with LPS or PBS as in A, and CFUs of total BM cells were assessed. Three plates per point were scored per experiment, and cells were pooled from three different mice. Data represent means ± SEM of at least three (A–F) or two (G–M) independent experiments. *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
In activated T and NK cells, GzmB is compartmentalized within cytoplasmic granules to protect the host cells from immediate apoptosis (Chowdhury and Lieberman, 2008). In analogy, GzmB was also localized in cytoplasmic granules in LPS-stimulated HSCs (Fig. 3 E), consistent with the hypothesis that this molecule is secreted into the BM microenvironment upon LPS stimulation by HSCs and its progeny. Indeed, the levels of extracellular GzmB in the BM were threefold increased after LPS treatment in vivo (Fig. 3 F). These data indicate that GzmB could act as a niche remodeling protease secreted by HSCs and progeny to alter inflammatory cues by modulating cytokine levels in response to LPS. In support of this, we found lower levels of IL-12 in the BM milieu of GzmB-deficient mice (not depicted), a cytokine know to promote immune defense by inducing the T helper 1 phenotype, enhancing NK cell cytotoxicity and IFN-γ production (Trinchieri, 2003). Further studies are required to evaluate the kinetics, release, and processing of these cytokines related to extracellular GzmB function upon stress.
GzmB expression in HSCs is dependent on TLR4–TRIF–NF-κB signaling
To further dissect the mechanisms involved in LPS-induced GzmB expression in HSCs, we used KO models, including the ones for the LPS receptor TLR4 and the adaptor proteins MyD88 and TRIF (Hennessy et al., 2010). LPS-dependent GzmB induction was completely abrogated in the TLR4−/− and TRIF−/− mice and partially reduced in MyD88−/− mice (Fig. 3 G). Interestingly, neither lack of IL6 production nor the abrogation of IFN-α response to LPS (Baldridge et al., 2011) affected the increased GzmB expression in LPS-stressed BM HSCs (not depicted). Together these results suggest that GzmB expression in HSCs is dependent on LPS-TLR4–mediated signaling in HSCs and/or myeloid cells but independent of the cytokines IL6 and type-1 IFNs.
Expression of human GzmB in NK cells can be regulated by NF-κB signaling (Huang et al., 2006), which is also known as an important component downstream of LPS signaling in myeloid cells in vivo (Kawai and Akira, 2006). We observed a strong correlation between GzmB expression and NF-κB effector p65 (Rel A) induction in HSCs. At 12 h after LPS treatment, 80.97% (±3.84) of HSCs were positive for both p65 and GzmB, which was sustained at later time points (not depicted). Moreover, coexpression of GzmB and the nuclear active form of p65 (p65pS536) was found in 92.87 ± 3.02% of HSCs after LPS treatment, whereas virtually no such cells were detectable in control animals (Fig. 3 H). Pretreatment of mice with pyrrolidine dithiocarbamate (PDTC), an antioxidant known to inhibit p65 accumulation in the nucleus (Ziegler-Heitbrock, 1993), significantly attenuated the expression of GzmB and p65 activation in HSCs in vivo (Fig. 3 I, left and right). These data provide evidence that GzmB expression in HSCs is at least partially dependent on LPS-mediated NF-κB activation. Although the data are consistent with the view that LPS-TRIF signaling within HSCs leads to NF-κB activation, it remains possible that myeloid cells may serve as an intermediate that trigger NF-κB activity in HSCs.
Our data suggest that LPS-induced GzmB expression could mediate cell-autonomous apoptotic mechanisms in HSCs. GzmB deficiency improved the resistance of mice to septic shock induced by LPS (Metkar et al., 2008), and patients undergoing sepsis showed an increase in circulating levels of GzmA and GzmB in the blood (Spaeny-Dekking et al., 1998; Lauw et al., 2000), demonstrating a role for GzmB in the inflammatory response to septic shock–induced cytotoxicity. Indeed, 23.73% (±5.34) of the LPS-stimulated HSCs showed an increase in active cleaved Caspase-3, whereas this induction was totally suppressed in TRIF−/− HSCs and partially suppressed in the GzmB-deficient HSCs (Fig. 3 J). To functionally explore whether GzmB mediates the deleterious effects of LPS on HSCs and progenitors, we isolated LPS-stimulated CD45.2 WT or GzmB−/− BM HSCs and performed 1:1 competitive transplantations with untreated CD45.1/2 WT total BM into CD45.1 WT recipients. As shown in Fig. 3 K, pretreatment with LPS significantly reduces the competitiveness of HSCs in this experiment from 33.20 ± 7.2% to 10.71 ± 5.3%. Strikingly, the negative effect of LPS was not observed in the GzmB-deficient setting, as neither HSCs (Fig. 3 K) nor differentiated hematopoietic cells showed a statistically significant difference in the chimerism compared with the PBS-treated controls (Fig. 3 L). In agreement, the colony-forming ability after in vivo LPS treatment was significantly reduced in a WT but not GzmB mutant setting (Fig. 3 M). In summary, LPS treatment negatively affects HSC and progenitor function in vivo and in vitro, which is rescued in the absence of induced GzmB.
GzmB deficiency confers resistance to serial 5-FU challenge
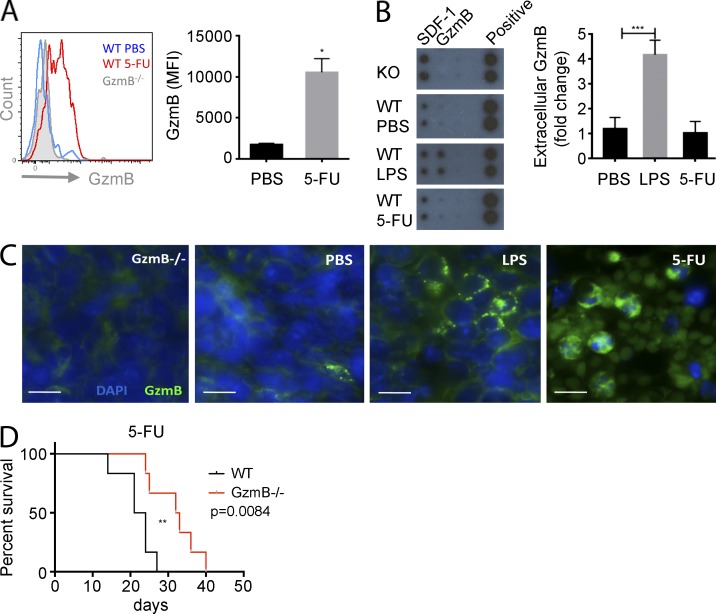
Elimination of proliferative myeloid cells by the chemotherapeutic drug 5-FU leads to activation of HSCs, which is triggered by an as yet undefined feedback loop (Randall and Weissman, 1997; Venezia et al., 2004; Wilson et al., 2008). A single dose of 5-FU triggered a strong induction of GzmB protein expression in HSCs (Fig. 4 A), suggesting that chemotherapeutic BM stress also triggers GzmB expression in HSCs. Interestingly, and contrary to the LPS response, no significant secretion of GzmB into the BM was observed upon 5-FU treatment (Fig. 4 B). This finding correlates with the subcellular localization of GzmB proteins in 5-FU– and LPS-treated mice. Although GzmB is stored in distinct granules upon LPS treatment, it is diffusely distributed in the cytoplasm of BM cells in response to 5-FU treatment (Fig. 4 C). To further address the contribution of extracellular GzmB in the regulation of the hematopoietic BM niche in response to stress, we administered 5-FU once every week (Cheng et al., 2000; Essers et al., 2009) to cohorts of control and GzmB−/− mice and monitored them for survival. Interestingly, GzmB-deficient mice showed a better survival under these harsh conditions despite the increased proliferative status of their HSCs. This is consistent with an improved chemoresistance of mutants compared with control mice, which is likely related to a decreased apoptotic response (Fig. 4 D). Strikingly, the cellular localization of GzmB upon 5-FU treatment strongly resembles the cytotoxic distribution pattern observed in the c-Myc/N-Myc DKO HSCs (Laurenti et al., 2008). To address whether there is a contribution of the niche cells to the observed HSC loss in response to 5-FU challenge, we examined the expression of GzmB in stromal cells. However, GzmB mRNA is down-regulated and barely detectable in such conditions (not depicted). Collectively, these data suggest that cytoplasmic GzmB expression contributes to the cytotoxic effects of 5-FU and its inhibition significantly attenuates its hematotoxic effects. This supports the hypothesis that in both stress scenarios, LPS and 5-FU, GzmB executes an intrinsic cytotoxic function and may function as a fail-safe mechanism to eliminate damaged HSCs in vivo. Future studies are needed to elucidate its possible function and its therapeutic value as a target in either augmenting myeloablative therapy in leukemias or attenuating 5-FU–mediated BM side effects in the treatment of nonhematological diseases.
Figure 4.
GzmB deficiency increases 5-FU chemoresistance in vivo. (A) WT and GzmB−/− mice were treated with a single dose of 5-FU, and LSK SLAM cells were quantified after 4 d. (left) Representative flow cytometry histogram; (right) bar graph showing quantitative data from n = 3 mice/group. (B and C) WT mice (n = 3) were injected with a single dose of 5-FU or LPS, and BM was analyzed at 4 d and 16 h, respectively. (B) GzmB secreted in the BM was assessed by dot blot (left) and quantified in n = 3 mice/group (right). (C) GzmB expression in the BM was assessed by immunofluorescence staining with anti-GzmB antibodies and DAPI nuclear stain (n = 3). Bars, 10 µm. (D) GzmB−/− and WT mice (n = 7/group) were injected weekly with 5-FU, and survival was monitored. Data in graphs represent means ± SEM of three independent experiments. *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
In summary, our finding that GzmB deficiency increased HSC function and likely self-renewal associated with increased mitochondrial function is of significant relevance for further studies, which should address its role in initiation and progression of leukemias. Moreover, our results suggest that under stress conditions, there might be two different mechanisms by which GzmB regulates HSC function (Fig. S1). The first mechanism, mediated by LPS, involves GzmB expression in granules and GzmB secretion into the BM niche by HSCs and its progeny, including myeloid and lymphoid cells. In this scenario, GzmB may act by remodeling extracellular matrix proteins or processing cytokines, similar to its proposed roles in chronic inflammatory diseases (Hendel et al., 2010) and in promoting aggressive T-ALL (Winrow et al., 2005). The second mode of action, as observed in response to 5-FU treatment, uses GzmB as a cell-autonomous cytotoxic protein leading to intrinsic hematopoietic cell death, similarly to the intrinsic cytotoxic mechanism observed in HSCs lacking c-Myc and N-Myc (Laurenti et al., 2008). Finally, our data suggest that both mechanisms of GzmB function may be used to either improve HSC engraftment in patients or for treatment of chronic inflammatory situations such as those observed in hematological malignancies.
MATERIALS AND METHODS
Mice.
8–12-wk-old female C57BL/6 mice purchased from Harlan Laboratories were used throughout the study. The following mouse lines were used: GzmB−/− (provided by T. Ley, Washington University in St. Louis, St. Louis, MO; Graubert et al., 1996); IFNRA−/− (Müller et al., 1994), TLR4−/− (Poltorak et al., 1998), TRIF−/− (provided by M. Pasparakis; Ermolaeva et al., 2008), Myd88−/− (Adachi et al., 1998), and p65-GFP (De Lorenzi et al., 2009). All animal protocols were approved by the Regierungspräsidium Karlsruhe Landwirtschaft, Ländlicher Raum, Veterinär- und Lebensmittelwesen of Baden-Württemberg (G-150-12) and according to the German laws for animal protection.
In vivo LPS, PDTC, and 5-FU treatment.
Mice were injected i.p. with PBS or 5 µg LPS from Escherichia coli (Sigma-Aldrich) once and were analyzed 16 h after the injection (unless otherwise indicated). Mice were injected i.p. with 10 mg/kg PDTC (Sigma-Aldrich). For 5-FU challenge, mice were injected i.p. with 150 mg/kg 5-FU (Sigma-Aldrich) once a week, and animal survival was recorded.
Isolation of BM cells.
For the isolation of total BM, mice were sacrificed and the BM was extracted under sterile conditions by crushing the bones in RPMI + 2% FCS using a mortar and pestle. The BM suspension was filtered through a 40-µm cell strainer and centrifuged for 5 min at 1,500 rpm. Subsequently, the cell pellet was resuspended in 1 ml ACK lysing buffer (Lonza) and incubated for 5 min at room temperature to allow lysis of red blood cells. Cells were washed in 5 ml RMPI + 2% FCS, centrifuged, and resuspended in up to 5 ml RMPI + 2% FCS. A 1:50 dilution of the cells was counted using the Vi-CELL cell viability analyzer (Beckman Coulter).
Lineage depletion.
Lineage depletion was performed using the Dynal Mouse CD4 Negative Isolation kit (Invitrogen) according to the manual instructions. CD4-Bio (eBioscience) was added to lineage antibody mix.
Flow cytometry and cell sorting.
For analysis of HSCs by flow cytometry, Lin− enriched BM cells were stained with antibodies against the following surface markers: CD4–PE-Cy7, CD8–PE-Cy7, CD11b–PE-Cy7, CD45R (B220)–PE-Cy7, Ter119–PE-Cy7, Gr-1–PE-Cy7, Sca1–APC-Cy7, Sca1–Alexa Fluor 700, Sca1–Pacific blue, CD117 (c-Kit)-APC, CD117 (c-Kit)–APC-Cy7, CD117 (c-Kit)–PE, CD150–PE-Cy5, CD48–Pacific blue, CD34–Alexa Fluor 700, IL7Ra-PE, and FcyRII/III-APC. All antibody conjugates were purchased from eBioscience. Antibodies were diluted in a 1:1 mix of 2.4G2 blocking buffer and RMPI + 2% FCS. For intracellular staining, surface-stained cells were fixed with BD Cytofix/Cytoperm for 15 min on ice and subsequently permeabilized with BD Perm/Wash. Cells were then incubated overnight with anti–human GzmB-PE or -APC (Invitrogen) or anti–mouse GzmB-FITC (eBioscience) in combination with either anti–active Caspase-3–FITC or –PE (BD) or p65pS536 Alexa Fluor 488 (Cell Signaling Technology). Before analysis, the cells were washed and resuspended in BD Perm/Wash. Flow cytometry analysis was performed on a BD LSRII flow cytometer or a BD LSRFortessa cell analyzer, and cell sorting was executed on a BD FACSAria II. FACS data were analyzed with FlowJo software (Tree Star).
BM transplantations.
For generation of competitive chimeras, mice were lethally irradiated with 11–12 Gy split-dose 4 h apart, and BM transplants were performed within 24 h by tail vein injection. Engraftment was measured monthly through facial vein bleed and FACS analysis with CD45.1-FITC, CD45.2-PB, CD4-PECy7, B220-PE, CD11b-PECy5, Gr1-APC, and Ter119-APCCy7. All antibodies were ordered from eBioscience unless otherwise specified.
LDA.
Serial dilutions of WT and GzmB−/− total BM (CD45.2) were transplanted by tail vein injection together with 2.105 supportive total BM cells (CD45.1) and transplanted into lethally irradiated WT CD45.1 recipient mice. Peripheral blood was analyzed for engraftment at 8 and 12 wk for myeloid and lymphoid reconstitution, and HSC engraftment in BM was analyzed at 14 wk. Mice for which engraftment was <1% were considered nonengrafted and were not taken into account for calculation of stem cell frequencies. Calculation of stem cell frequencies and statistics was performed using ELDA software (Hu and Smyth, 2009).
Cell culture.
Lineage-depleted BM cells were seeded in at a density of 1–2 × 106 cells per well in 500 µl HSC medium. The medium was composed of StemPro-34 SFM (Gibco) supplemented with StemPro-34 Nutrient (Gibco), 100 U/ml Pen/Strep (Sigma-Aldrich), 2 mM l-glutamine (Gibco), 50 ng/ml mSCF, 25 ng/ml mTPO, 30 ng/ml Flt3, and 105 U/ml IL-11 (R&D Systems). The cells were cultured at 37°C and 5% CO2.
Hematopoietic colony-forming cell assay.
Fresh total BM or cultured Lin− BM cells were counted and diluted to a concentration of 105 cells per milliliter in sterile PBS. For duplicates, 260 µl of the cell suspension was added to 2.6 ml MethoCult GF M3434, containing recombinant mouse SCF, recombinant mouse IL-3, recombinant human IL-6, and recombinant human EPO (STEMCELL Technologies). For triplicates, 350 µl of the cell suspension was added to 3.5 ml of MethoCult GF M3434. After 7–10 d, the dishes were scored for different hematopoietic colonies.
Mobilization assays.
Mice were injected with PBS or 300 µg huG-CSF (Amgen) for a consecutive 6 d subcutaneously. On day 6, mice were sacrificed, and BM, spleens, and peripheral blood were harvested. Single cell suspensions were analyzed by flow cytometry and stained with Lineage marker, Sca-1, and c-Kit.
Homing assay of hematopoietic progenitor cells.
C57BL/6 Ly5.1 mice were lethally irradiated (6 Gy) 24 h before transplantation. 6 or 16 h after injection, BM cells were harvested from femurs stem cells, and progenitors were quantified and analyzed by flow cytometry. Homing of hematopoietic progenitor cells is calculated relative to the total number of LSK SLAM cells.
GzmB intracellular staining.
For FACS analyses, total BM or Lineage-negative cells were stained for surface markers, and intracellular GzmB protein was detected with a directly conjugated GzmB-PE or -APC antibody (Invitrogen). Mouse IgG1-PE was used as the appropriate IgG control. For immunofluorescence assays, Lineage-negative or sorted LSK SLAM cells were fixed in 3.7% PFA and then cytospun onto slides. After permeabilization in PBS + 0.2% Triton X-100 and blocking with PBS + 15% goat serum, the slides were stained overnight with anti–mouse GrB biotinylated antibody (R&D Systems) and then for 1 h with SAV–Alexa Fluor 488 or 546. Nuclei were counterstained with DAPI.
Cell proliferation assays.
Surface staining and intracellular staining of Lin− BM cells were performed as described in Flow cytometry and cell sorting. For Ki67 staining, cells were fixed and permeabilized and further incubated with anti–Ki67-FITC or -APC antibody (BD) at 4°C overnight. Hoechst 33342 was added right before FACS analysis.
Mitochondrial mass.
Mitochondrial content was investigated using MitoTracker green FM (Invitrogen). In brief, total BM cells were incubated at 37°C for 30 min with 50 nM MitoTracker green, washed and resuspended in PBS, and analyzed by flow cytometry. As a positive control, cells were treated with 1 µM oligomycin for 1 h ex vivo before MitoTracker labeling.
BM histology.
Fresh femurs were collected for paraffin blocks or cryosections. Frozen sections were prepared by the tape transfer method (Section-Lab Co Ltd). Sections were subjected to hematoxylin and eosin (H&E) staining. For immunofluorescence experiments, sections were treated with 100% ethanol and blocked in 2% horse serum.
Immunohistochemistry.
Cytospins from fixed Lin− or Lin− Sca-1+ c-Kit+ CD48− CD150+ (LSK SLAM) sorted cells were prepared by diluting 1–2 × 104 cells in 100 µl PBS and transferring them into a preconditioned Shandon EZ Cytofunnel (Thermo Fisher Scientific) containing a glass microscope slide. The cells were spun onto the slide by centrifugation in the Cytospin 4 (Thermo Fisher Scientific) for 5 min at 1,000 rpm. Subsequently, the cytospins were subjected to immunohistochemical staining. Cells were blocked with goat serum + Triton X-100 in PBS for 60 min and then incubated overnight at 4°C with GzmB-Biotin (R&D Systems). After three times of washing, the respective secondary antibodies were added: streptavidin–Alexa Flour 546. After two washing steps, cells were incubated with a 1:500 dilution of DAPI for 5 min. After one last washing step, the slides were mounted with coverslips using Faramount Aqueous Mounting Medium (Dako). Fluorescence microscopy was performed on a Cell Observer or LSM700 using the AxioVision software (all Carl Zeiss).
ELISA and cytokine array.
Extracellular mouse GzmB was measured in an ELISA. In short, supernatant from fresh BM was collected and subjected to analysis with the mouse GzmB ELISA Ready-SET-Go! kit (eBioscience) or Cytokine array membranes (RayBiotech).
Microarray analysis.
Homeostatic and transplanted HSCs were sorted and RNA isolated using Arcturus PicoPure (Applied Biosystems), and 2.5 ng of total RNA was used for hybridization on Illumina Mouse Sentrix-6 BeadChips. For statistical quantification, permutation testing in GSEA was performed with 1,000 permutations by gene set. Microarray data are deposited in the Gene Expression Omnibus under accession no. GSE56574.
Real-time quantitative RT-PCR.
Total RNA was extracted from sorted HSCs using the Arcturus PicoPure (Applied Biosystems). Complementary DNA was synthesized from total RNA with the Vilo SuperScript III First-Strand Synthesis System for Q-PCR (Invitrogen). The real-time PCR measurement of individual cDNAs was performed using SYBR green dye to measure duplex DNA formation with the ViiA 7 (Applied Biosystems) and normalized to the expression of Oaz or 18S ribosomal RNA. The primers used in the real-time RT-PCR were the following: Oaz1 FW, 5′-TTTCAGCTAGCATCCTGTACTCC-3′; Oaz1 RV, 5′-GACCCTGGTCTTGTCGTTAGA-3′; r18S FW, 5′-GTAACCCGTTGAACCCCATT-3′; r18S RV, 5′-CCATCCAATCGGTAGTAGCG-3′; GzmB FW, 5′-TGTGAAGCCAGGAGATGTGTGCTA-3′; GzmB RV, 5′-TCAGCTCAACCTCTTGTAGCGTGT-3′; GzmA FW, 5′-ACACGGTTGTTCCTCACTCAAGAC-3′; GzmA RV, 5′-TCAATCAAAGCGCCAGCACAGATG-3′; Cdkn2d FW, 5′-GCCTTGCAGGTCATGATGTTTGGA-3′; Cdkn2d RV, 5′-AGTACCGGAGGCATCTTGGACATT-3′; IL12 FW, 5′-GCGTTCCAACAGCCTCAC-3′; IL12 RV, 5′-TGGCCAAAAAGAGGAGGTAG-3′; VEGF FW, 5′-CAGGCTGCTGTAACGATGAA-3′; and VEGF RV, 5′-CTCCTATGTGCTGGCTTTGG-3′.
Statistical analysis.
Data were processed using Prism 6 (GraphPad Software). All analyses were performed using two-tailed Student’s t tests (unless otherwise specified). For determination of stem cell frequency, Poisson test was performed. Statistical significance is indicated by *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material.
Fig. S1 shows a model depicting GzmB function in HSCs in response to stress. Table S1 lists cell surface marker phenotypes. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20131072/DC1.
Supplementary Material
Acknowledgments
We want to thank Andrea Takacs, Petra Zeisberger, Katja Müder, and Melanie Neubauer for technical assistance. We thank Dr. Timothy Ley for kindly providing the GzmB−/− mice. We would also like to thank Dr. Michael Milsom for comments on the manuscript. We would like to thank the German Cancer Research Center (DKFZ) Flow Cytometry Core facility and the DKFZ Light Microscopy facility for their assistance. We thank the microarray unit of the DKFZ Genomics and Proteomics Core Facility for providing the Illumina Whole-Genome Expression BeadChips and related services.
This work was supported by the BioRN Spitzencluster “Molecular and Cell-Based Medicine” supported by the German Bundesministerium für Bildung und Forschung, the EU-FP7 Program “EuroSyStem,” the SFB 873 funded by the Deutsche Forschungsgemeinschaft, and the Dietmar Hopp Foundation (all grants to A. Trumpp).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- 5-FU
- 5-fluorouracil
- DKO
- double KO
- GSEA
- gene set enrichment analysis
- GzmB
- granzyme B
- HSC
- hematopoietic stem cell
- LDA
- limiting dilution assay
- PDTC
- pyrrolidine dithiocarbamate
References
- Adachi O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., Nakanishi K., Akira S. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 9:143–150 10.1016/S1074-7613(00)80596-8 [DOI] [PubMed] [Google Scholar]
- Akira S., Takeda K. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499–511 10.1038/nri1391 [DOI] [PubMed] [Google Scholar]
- Baldridge M.T., King K.Y., Boles N.C., Weksberg D.C., Goodell M.A. 2010. Quiescent haematopoietic stem cells are activated by IFN-γ in response to chronic infection. Nature. 465:793–797 10.1038/nature09135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge M.T., King K.Y., Goodell M.A. 2011. Inflammatory signals regulate hematopoietic stem cells. Trends Immunol. 32:57–65 10.1016/j.it.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsall H.H., Porter W.J., Green D.M., Rubio J., Trial J., Rossen R.D. 2004. Impact of fibronectin fragments on the transendothelial migration of HIV-infected leukocytes and the development of subendothelial foci of infectious leukocytes. J. Immunol. 173:2746–2754 [DOI] [PubMed] [Google Scholar]
- Boivin W.A., Cooper D.M., Hiebert P.R., Granville D.J. 2009. Intracellular versus extracellular granzyme B in immunity and disease: challenging the dogma. Lab. Invest. 89:1195–1220 10.1038/labinvest.2009.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin W.A., Shackleford M., Vanden Hoek A., Zhao H., Hackett T.L., Knight D.A., Granville D.J. 2012. Granzyme B cleaves decorin, biglycan and soluble betaglycan, releasing active transforming growth factor-β1. PLoS ONE. 7:e33163 10.1371/journal.pone.0033163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzza M.S., Zamurs L., Sun J., Bird C.H., Smith A.I., Trapani J.A., Froelich C.J., Nice E.C., Bird P.I. 2005. Extracellular matrix remodeling by human granzyme B via cleavage of vitronectin, fibronectin, and laminin. J. Biol. Chem. 280:23549–23558 10.1074/jbc.M412001200 [DOI] [PubMed] [Google Scholar]
- Cao X., Cai S.F., Fehniger T.A., Song J., Collins L.I., Piwnica-Worms D.R., Ley T.J. 2007. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity. 27:635–646 10.1016/j.immuni.2007.08.014 [DOI] [PubMed] [Google Scholar]
- Challen G.A., Sun D., Jeong M., Luo M., Jelinek J., Berg J.S., Bock C., Vasanthakumar A., Gu H., Xi Y., et al. 2012. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 44:23–31 10.1038/ng.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M.P., Seita J., Weissman I.L. 2008. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb. Symp. Quant. Biol. 73:439–449 10.1101/sqb.2008.73.031 [DOI] [PubMed] [Google Scholar]
- Chen C., Liu Y., Liu Y., Zheng P. 2010. Mammalian target of rapamycin activation underlies HSC defects in autoimmune disease and inflammation in mice. J. Clin. Invest. 120:4091–4101 10.1172/JCI43873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T., Rodrigues N., Shen H., Yang Y., Dombkowski D., Sykes M., Scadden D.T. 2000. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 287:1804–1808 10.1126/science.287.5459.1804 [DOI] [PubMed] [Google Scholar]
- Chioni A.M., Grose R. 2012. FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J. Cell Biol. 197:801–817 10.1083/jcb.201108077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D., Lieberman J. 2008. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu. Rev. Immunol. 26:389–420 10.1146/annurev.immunol.26.021607.090404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzi R., Gareus R., Fengler S., Pasparakis M. 2009. GFP-p65 knock-in mice as a tool to study NF-κB dynamics in vivo. Genesis. 47:323–329 10.1002/dvg.20468 [DOI] [PubMed] [Google Scholar]
- Doulatov S., Notta F., Laurenti E., Dick J.E. 2012. Hematopoiesis: a human perspective. Cell Stem Cell. 10:120–136 10.1016/j.stem.2012.01.006 [DOI] [PubMed] [Google Scholar]
- Ehninger A., Trumpp A. 2011. The bone marrow stem cell niche grows up: mesenchymal stem cells and macrophages move in. J. Exp. Med. 208:421–428 10.1084/jem.20110132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema H., Sudo K., Seita J., Matsubara A., Morita Y., Osawa M., Takatsu K., Takaki S., Nakauchi H. 2005. Quantification of self-renewal capacity in single hematopoietic stem cells from normal and Lnk-deficient mice. Dev. Cell. 8:907–914 10.1016/j.devcel.2005.03.019 [DOI] [PubMed] [Google Scholar]
- Ermolaeva M.A., Michallet M.C., Papadopoulou N., Utermöhlen O., Kranidioti K., Kollias G., Tschopp J., Pasparakis M. 2008. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat. Immunol. 9:1037–1046 10.1038/ni.1638 [DOI] [PubMed] [Google Scholar]
- Esplin B.L., Shimazu T., Welner R.S., Garrett K.P., Nie L., Zhang Q., Humphrey M.B., Yang Q., Borghesi L.A., Kincade P.W. 2011. Chronic exposure to a TLR ligand injures hematopoietic stem cells. J. Immunol. 186:5367–5375 10.4049/jimmunol.1003438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., Trumpp A. 2009. IFNα activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Foudi A., Hochedlinger K., Van Buren D., Schindler J.W., Jaenisch R., Carey V., Hock H. 2009. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27:84–90 10.1038/nbt.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich C.J., Zhang X., Turbov J., Hudig D., Winkler U., Hanna W.L. 1993. Human granzyme B degrades aggrecan proteoglycan in matrix synthesized by chondrocytes. J. Immunol. 151:7161–7171 [PubMed] [Google Scholar]
- Froelich C.J., Pardo J., Simon M.M. 2009. Granule-associated serine proteases: granzymes might not just be killer proteases. Trends Immunol. 30:117–123 10.1016/j.it.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Graubert T.A., Russell J.H., Ley T.J. 1996. The role of granzyme B in murine models of acute graft-versus-host disease and graft rejection. Blood. 87:1232–1237 [PubMed] [Google Scholar]
- Graubert T.A., DiPersio J.F., Russell J.H., Ley T.J. 1997. Perforin/granzyme-dependent and independent mechanisms are both important for the development of graft-versus-host disease after murine bone marrow transplantation. J. Clin. Invest. 100:904–911 10.1172/JCI119606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman W.J., Revell P.A., Lu Z.H., Johnson H., Bredemeyer A.J., Ley T.J. 2003. The orphan granzymes of humans and mice. Curr. Opin. Immunol. 15:544–552 10.1016/S0952-7915(03)00099-2 [DOI] [PubMed] [Google Scholar]
- Hendel A., Hiebert P.R., Boivin W.A., Williams S.J., Granville D.J. 2010. Granzymes in age-related cardiovascular and pulmonary diseases. Cell Death Differ. 17:596–606 10.1038/cdd.2010.5 [DOI] [PubMed] [Google Scholar]
- Hennessy E.J., Parker A.E., O’Neill L.A. 2010. Targeting Toll-like receptors: emerging therapeutics? Nat. Rev. Drug Discov. 9:293–307 10.1038/nrd3203 [DOI] [PubMed] [Google Scholar]
- Hu Y., Smyth G.K. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 347:70–78 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Huang C., Bi E., Hu Y., Deng W., Tian Z., Dong C., Hu Y., Sun B. 2006. A novel NF-κB binding site controls human granzyme B gene transcription. J. Immunol. 176:4173–4181 [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. 2006. TLR signaling. Cell Death Differ. 13:816–825 10.1038/sj.cdd.4401850 [DOI] [PubMed] [Google Scholar]
- Labi V., Bertele D., Woess C., Tischner D., Bock F.J., Schwemmers S., Pahl H.L., Geley S., Kunze M., Niemeyer C.M., et al. 2013. Haematopoietic stem cell survival and transplantation efficacy is limited by the BH3-only proteins Bim and Bmf. EMBO Mol. Med. 5:122–136 10.1002/emmm.201201235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E., Varnum-Finney B., Wilson A., Ferrero I., Blanco-Bose W.E., Ehninger A., Knoepfler P.S., Cheng P.F., MacDonald H.R., Eisenman R.N., et al. 2008. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 3:611–624 10.1016/j.stem.2008.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E., Wilson A., Trumpp A. 2009. Myc’s other life: stem cells and beyond. Curr. Opin. Cell Biol. 21:844–854 10.1016/j.ceb.2009.09.006 [DOI] [PubMed] [Google Scholar]
- Lauw F.N., Simpson A.J., Hack C.E., Prins J.M., Wolbink A.M., van Deventer S.J., Chaowagul W., White N.J., van Der Poll T. 2000. Soluble granzymes are released during human endotoxemia and in patients with severe infection due to gram-negative bacteria. J. Infect. Dis. 182:206–213 10.1086/315642 [DOI] [PubMed] [Google Scholar]
- Méndez-Ferrer S., Chow A., Merad M., Frenette P.S. 2009. Circadian rhythms influence hematopoietic stem cells. Curr. Opin. Hematol. 16:235–242 10.1097/MOH.0b013e32832bd0f5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metkar S.S., Menaa C., Pardo J., Wang B., Wallich R., Freudenberg M., Kim S., Raja S.M., Shi L., Simon M.M., Froelich C.J. 2008. Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity. 29:720–733 10.1016/j.immuni.2008.08.014 [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Spradling A.C. 2008. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 132:598–611 10.1016/j.cell.2008.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müllbacher A., Waring P., Tha Hla R., Tran T., Chin S., Stehle T., Museteanu C., Simon M.M. 1999. Granzymes are the essential downstream effector molecules for the control of primary virus infections by cytolytic leukocytes. Proc. Natl. Acad. Sci. USA. 96:13950–13955 10.1073/pnas.96.24.13950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U., Steinhoff U., Reis L.F., Hemmi S., Pavlovic J., Zinkernagel R.M., Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science. 264:1918–1921 10.1126/science.8009221 [DOI] [PubMed] [Google Scholar]
- Oguro H., Iwama A., Morita Y., Kamijo T., van Lohuizen M., Nakauchi H. 2006. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J. Exp. Med. 203:2247–2253 10.1084/jem.20052477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak A., He X., Smirnova I., Liu M.Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., et al. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 282:2085–2088 10.1126/science.282.5396.2085 [DOI] [PubMed] [Google Scholar]
- Pronk C.J., Veiby O.P., Bryder D., Jacobsen S.E. 2011. Tumor necrosis factor restricts hematopoietic stem cell activity in mice: involvement of two distinct receptors. J. Exp. Med. 208:1563–1570 10.1084/jem.20110752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall T.D., Weissman I.L. 1997. Phenotypic and functional changes induced at the clonal level in hematopoietic stem cells after 5-fluorouracil treatment. Blood. 89:3596–3606 [PubMed] [Google Scholar]
- Rossi L., Lin K.K., Boles N.C., Yang L., King K.Y., Jeong M., Mayle A., Goodell M.A. 2012. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 11:302–317 10.1016/j.stem.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scumpia P.O., Kelly-Scumpia K.M., Delano M.J., Weinstein J.S., Cuenca A.G., Al-Quran S., Bovio I., Akira S., Kumagai Y., Moldawer L.L. 2010. Cutting edge: bacterial infection induces hematopoietic stem and progenitor cell expansion in the absence of TLR signaling. J. Immunol. 184:2247–2251 10.4049/jimmunol.0903652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M.M., Hausmann M., Tran T., Ebnet K., Tschopp J., ThaHla R., Müllbacher A. 1997. In vitro– and ex vivo–derived cytolytic leukocytes from granzyme A × B double knockout mice are defective in granule-mediated apoptosis but not lysis of target cells. J. Exp. Med. 186:1781–1786 10.1084/jem.186.10.1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaeny-Dekking E.H., Hanna W.L., Wolbink A.M., Wever P.C., Kummer J.A., Swaak A.J., Middeldorp J.M., Huisman H.G., Froelich C.J., Hack C.E. 1998. Extracellular granzymes A and B in humans: detection of native species during CTL responses in vitro and in vivo. J. Immunol. 160:3610–3616 (published erratum appears in J. Immunol. 2009. 182:5152) [PubMed] [Google Scholar]
- Stepanova L., Sorrentino B.P. 2005. A limited role for p16Ink4a and p19Arf in the loss of hematopoietic stem cells during proliferative stress. Blood. 106:827–832 10.1182/blood-2004-06-2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo K., Ema H., Morita Y., Nakauchi H. 2000. Age-associated characteristics of murine hematopoietic stem cells. J. Exp. Med. 192:1273–1280 10.1084/jem.192.9.1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa H., Regoes R.R., Boddupalli C.S., Bonhoeffer S., Manz M.G. 2011. Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J. Exp. Med. 208:273–284 10.1084/jem.20101643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3:133–146 10.1038/nri1001 [DOI] [PubMed] [Google Scholar]
- Trumpp A., Essers M., Wilson A. 2010. Awakening dormant haematopoietic stem cells. Nat. Rev. Immunol. 10:201–209 10.1038/nri2726 [DOI] [PubMed] [Google Scholar]
- van Tetering G., Bovenschen N., Meeldijk J., van Diest P.J., Vooijs M. 2011. Cleavage of Notch1 by granzyme B disables its transcriptional activity. Biochem. J. 437:313–322 10.1042/BJ20110226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venezia T.A., Merchant A.A., Ramos C.A., Whitehouse N.L., Young A.S., Shaw C.A., Goodell M.A. 2004. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2:e301 10.1371/journal.pbio.0020301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Murphy M.J., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., Trumpp A. 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 18:2747–2763 10.1101/gad.313104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E., et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 135:1118–1129 10.1016/j.cell.2008.10.048 [DOI] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Trumpp A. 2009. Balancing dormant and self-renewing hematopoietic stem cells. Curr. Opin. Genet. Dev. 19:461–468 10.1016/j.gde.2009.08.005 [DOI] [PubMed] [Google Scholar]
- Winrow C.J., Pankratz D.G., Vibat C.R., Bowen T.J., Callahan M.A., Warren A.J., Hilbush B.S., Wynshaw-Boris A., Hasel K.W., Weaver Z., et al. 2005. Aberrant recombination involving the granzyme locus occurs in Atm−/− T-cell lymphomas. Hum. Mol. Genet. 14:2671–2684 10.1093/hmg/ddi301 [DOI] [PubMed] [Google Scholar]
- Ziegler-Heitbrock H.W., Sternsdorf T., Liese J., Belohradsky B., Weber C., Wedel A., Schreck R., Bäuerle P., Ströbel M. 1993. Pyrrolidine dithiocarbamate inhibits NF-κB mobilization and TNF production in human monocytes. J. Immunol. 151:6986–6993 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.