Strict, lineage-intrinsic requirement for continuous adult Gfi-1b expression at two distinct critical stages of erythropoiesis and megakaryopoiesis.
Abstract
The zinc finger transcriptional repressor Gfi-1b is essential for erythroid and megakaryocytic development in the embryo. Its roles in the maintenance of bone marrow erythropoiesis and thrombopoiesis have not been defined. We investigated Gfi-1b’s adult functions using a loxP-flanked Gfi-1b allele in combination with a novel doxycycline-inducible Cre transgene that efficiently mediates recombination in the bone marrow. We reveal strict, lineage-intrinsic requirements for continuous adult Gfi-1b expression at two distinct critical stages of erythropoiesis and megakaryopoiesis. Induced disruption of Gfi-1b was lethal within 3 wk with severely reduced hemoglobin levels and platelet counts. The erythroid lineage was arrested early in bipotential progenitors, which did not give rise to mature erythroid cells in vitro or in vivo. Yet Gfi-1b−/− progenitors had initiated the erythroid program as they expressed many lineage-restricted genes, including Klf1/Eklf and Erythropoietin receptor. In contrast, the megakaryocytic lineage developed beyond the progenitor stage in Gfi-1b’s absence and was arrested at the promegakaryocyte stage, after nuclear polyploidization, but before cytoplasmic maturation. Genome-wide analyses revealed that Gfi-1b directly regulates a wide spectrum of megakaryocytic and erythroid genes, predominantly repressing their expression. Together our study establishes Gfi-1b as a master transcriptional repressor of adult erythropoiesis and thrombopoiesis.
Continuous, high-rate production of red blood cells and platelets is required to sustain vertebrate life. The erythroid and megakaryocytic lineages are thought to share initial differentiation steps from hematopoietic stem cells (HSCs; Akashi et al., 2000; Pronk et al., 2007). After loss of other fate potentials and passage through a bipotent progenitor stage, the lineages segregate into distinct terminal maturation pathways, culminating in the production of erythrocytes and platelets.
During maturation, cells of both lineages execute complex lineage-specific programs. In erythroid cells, these include coordinated heme biosynthesis and globin production, as well as nuclear condensation and the terminal expulsion of the nucleus (Hattangadi et al., 2011). In megakaryocytic differentiation, polyploid, multilobulated nuclei are generated as a result of endomitosis, and a large cytoplasm is formed, which provides a reservoir for platelet-specific granules, a system of demarcation membranes, and microtubules (Schulze and Shivdasani, 2005; Chang et al., 2007; Tijssen and Ghevaert, 2013). These cytoplasmic elements are ultimately consumed in the formation of proplatelets; thin megakaryocyte extensions that protrude into the intravascular space, where they segment and separate, releasing platelets into the blood stream (Kaushansky, 2008; Machlus and Italiano, 2013).
The erythroid and megakaryocytic lineages share a cadre of common transcriptional regulators, including Gata1, Nf-e2, Fog1/Zfpm1, Scl/Tal1, and Gfi-1b, all of which are preferentially expressed in both lineages and exert important roles in erythroid and/or megakaryocytic development (Kerenyi and Orkin, 2010). In addition, some factors are expressed and function in just one of the lineages, specifically Klf1 (formerly Eklf), an essential driver of erythropoiesis (Yien and Bieker, 2013), and Fli-1, which promotes megakaryopoiesis and antagonizes Klf1 (Starck et al., 2003, 2010). Gene-targeting studies in mice have shown that bilineage expression does not always predict prominent functional roles in both lineages. Thus, severe blocks in erythroid development at the progenitor and erythroblast stages were observed after Gata1 loss (Pevny et al., 1991; Gutiérrez et al., 2008; Mancini et al., 2012). However, absence of Gata1 did not abrogate megakaryopoiesis, even if it was associated with significantly reduced blood platelet counts and abnormal megakaryocytes (Vyas et al., 1999; Gutiérrez et al., 2008). Conversely, Nf-e2 was largely dispensable for erythroid development, whereas its disruption caused severe thrombocytopenia with abnormal, mature megakaryocytes (Shivdasani et al., 1995; Lecine et al., 1998). Gata1’s cofactor Fog1 is essential for the maintenance of both lineages. In the erythroid lineage, Fog1 disruption resulted in phenotypes similar to those found after Gata1 loss (Tsang et al., 1998; Mancini et al., 2012). However, unlike Gata1, Fog1 is required for megakaryocytic development at a very early stage, preceding the formation of committed progenitors (Tsang et al., 1998; Mancini et al., 2012). In distinction from the above factors, Scl/Tal1, essential for embryonic specification of all hematopoietic lineages (Porcher et al., 1996), is not strictly required for adult bone marrow erythropoiesis or thrombopoiesis. Its loss was associated with reduced blood counts and abnormal colony formation ex vivo (Mikkola et al., 2003), but production of mature cells was sufficient to prevent severe cytopenias and morbidity (Hall et al., 2005; McCormack et al., 2006; Chagraoui et al., 2011). Likely, Scl’s important adult role is partially obscured by redundancy with the closely related Lyl-1, which also supports erythropoiesis (Souroullas et al., 2009; Capron et al., 2011). Finally, Lmo2 and Ldb1, constituents of pentameric complexes with Scl and Gata1 (Wadman et al., 1997; El Omari et al., 2013), are indispensable for erythropoiesis and thrombopoiesis (Warren et al., 1994; Li et al., 2010, 2013).
In this study, we address the role of Gfi-1b in adult erythropoiesis and thrombopoiesis. An essential role for Gfi-1b in megakaryocytic and erythroid development has been defined in the embryo (Saleque et al., 2002). Studies in murine and human erythroid and megakaryocytic cell culture systems suggested that Gfi-1b’s role persisted in adult cells, but a requirement for Gfi-1b expression has not been demonstrated in the bone marrow (Osawa et al., 2002; Garçon et al., 2005; Saleque et al., 2007; Randrianarison-Huetz et al., 2010). In addition, the precise stage of Gfi-1b’s role within the two lineages and its relation to other transcription factor requirements have not been defined. Finally, it was recently reported that a conditional KO of Gfi-1b in adult HSCs was not associated with severe, early erythroid, or megakaryocytic phenotypes (Khandanpour et al., 2010). As Gfi-1b disruption in embryos resulted in lethal anemia and defective erythroid and megakaryocytic colony formation (Saleque et al., 2002), these findings raised the question of whether the requirement for Gfi-1b was less stringent in the adult than in the embryo. Here, we clarify these issues, identify strict, lineage-intrinsic requirements for Gfi-1b at distinct stages of adult erythropoiesis and thrombopoiesis, and illuminate Gfi-1b’s position in the network of lineage-specific regulators.
RESULTS
Gfi-1b loss interrupts adult bone marrow red blood cell and platelet production at different stages
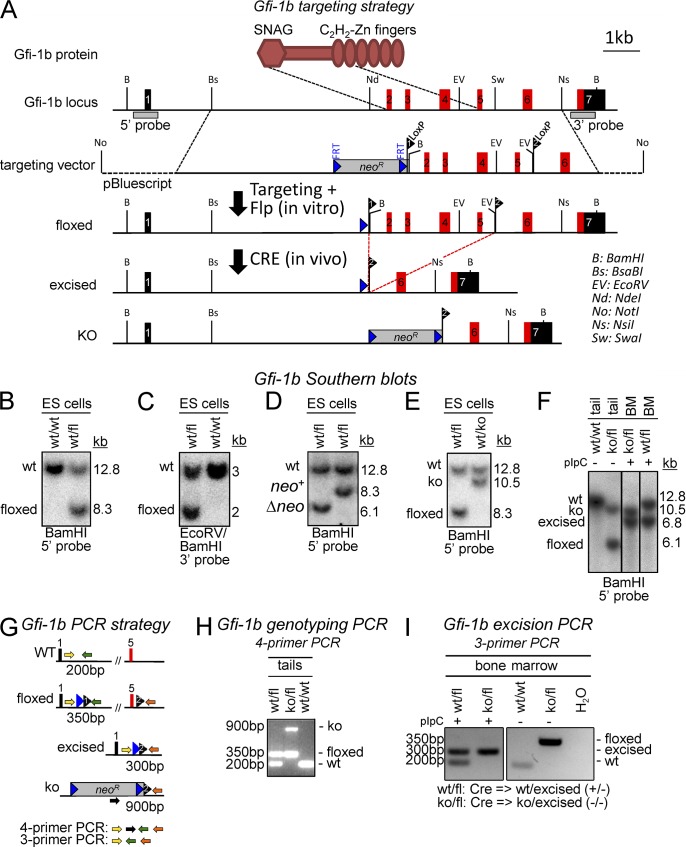
We generated a conditional mouse strain, in which exons 2–5 of Gfi-1b (encoding the N-terminal portion of the protein; start codon to zinc finger 2) are flanked by loxP sites (floxed; Fig. 1 A, top). For conditional disruption, a floxed Gfi-1b allele was combined with a germline-excised Gfi-1b KO allele (Fig. 1 A, bottom), so that single excision events achieved biallelic absence of intact Gfi-1b (KO/floxed → Cre → −/−). In control mice, a floxed allele was combined with a WT allele (WT/floxed → Cre → +/−). Excision was mediated either by the Mx-Cre transgene, which efficiently excises floxed alleles in the bone marrow upon injection of polyinosinic:polycytidylic acid (pIpC; Kühn et al., 1995; Hock et al., 2004b), or a novel doxycycline-inducible Cre transgene (TetO-Cre; Fig. 2). With either system, conditional disruption of Gfi-1b resulted in similar hematopoietic abnormalities and was uniformly lethal in less than 3 wk (Fig. 3, A and B). Analysis of complete blood counts from moribund animals revealed profound anemia with hemoglobin levels as low as 1–2 g/dl, severe thrombocytopenia, and in some cases no detectable blood platelets (Fig. 3 B). In contrast, lymphocyte counts and granulocyte counts were not significantly changed compared with controls (Fig. 3 B). As blood granulocytes are short-lived, the latter finding is consistent with previous conclusions that Gfi-1b is dispensable for HSC maintenance (Hock et al., 2004a; Khandanpour et al., 2010).
Figure 1.
Generation of a conditional Gfi-1b allele in ES cells and mice. (A) A scheme of the Gfi-1b protein domains illustrating the targeted regions is shown above the restriction maps. LoxP-flanked regions include coding exons for the entire N-terminal protein (ATG-ZF2). The restriction maps show the genomic Gfi-1b locus (top; coding exons in red and noncoding exons in black), the targeting vector (second), the targeted locus after Flp-mediated deletion of NeoR (third), and the excised locus (fourth). The bottom shows a constitutively deleted Gfi-1b allele (KO) generated by Cre-mediated deletion without excision of the NeoR gene. (B–F) Southern blot analyses of Gfi-1b alleles. (B) Documentation of correct targeting at 5′ (digest: BamHI; expected sizes: WT = 12.8 kb, targeted allele = 8.3 kb; probe: 5′ as shown in A). (C) Documentation of correct targeting at 3′ (digest: BamHI/EcoRV; expected sizes: WT = 3 kb, targeted allele = 2 kb; probe: 3′ as shown in A). (D) Documentation of deletion of neoR cassette using Flpe (digest: BamHI; expected sizes: WT = 12.8 kb, floxed-neo+ = 8.3 kb, floxed-Δneo = 6.1 kb; probe: 5′ as shown in A). (E) Documentation of Cre-mediated excision of exons 2–5 in ES cells to generate the constitutive KO allele (A, bottom; digest: BamHI; expected sizes: WT = 12.8 kb; floxed-neo+ = 8.3 kb; KO = 10.5 kb; probe: 5′ as shown in A). (F) Southern blot analysis for genotyping and determination of excision status (digest: BamHI; expected sizes: WT = 12.8 kb, floxed = 6.1 kb, excised = 6.8 kb, KO = 10.5 kb; probe: 5′ as shown in A). (G–I) Gfi-1b genotyping and excision PCR strategies. (G) Scheme of Gfi-1b PCR strategy. (H) Representative genotyping PCR using four primers as shown in G from tail-tip DNA samples (expected bands: WT, 200 bp; floxed, 350 bp; KO, 900 bp). (I) Representative excision PCR using three primers as shown in A from bone marrow cells before (−) and after (+) pIpC treatment (expected bands: WT, 200 bp; floxed, 350 bp; excised, 300 bp; KO, no band).
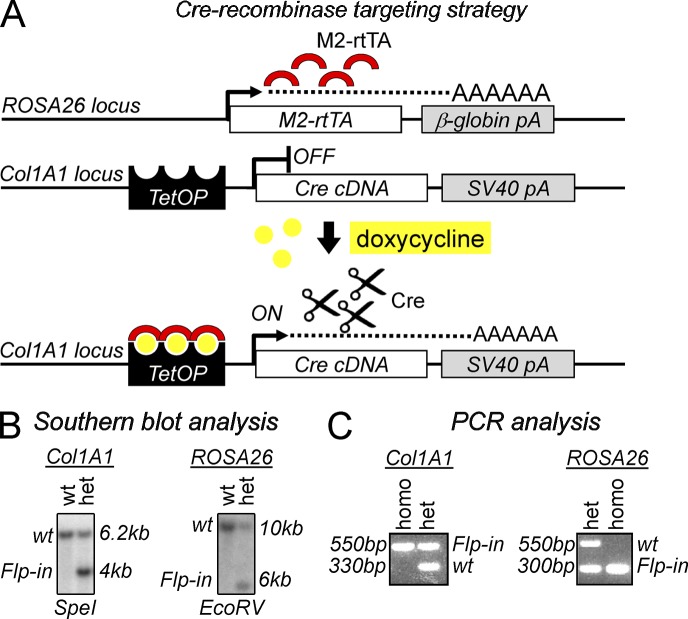
Figure 2.
Generation of mice that permit doxycycline-inducible expression of Cre recombinase (TetO-Cre). (A) Alleles for doxycycline-inducible Cre expression. (top) The M2 reverse tetracycline transactivator (M2-rtTA) is constitutively transcribed off the ROSA26 promoter as described in Beard et al. (2006). (middle) Flp-recombinase–mediated site-specific integration was used as described previously (Beard et al., 2006) to insert the Cre cDNA in the 3′ UTR region of the Collagen (Col) 1A1 locus under the control of a tetracycline-dependent minimal CMV promoter. (bottom) Upon doxycycline treatment (yellow dots), the M2 reverse tetracycline transactivator (red semicircles) binds to the CMV promoter and Cre expression is turned on. pA, poly A; TetOP, tetracycline operator elements fused to CMV minimal promoter. (B) Southern blot analysis confirming integration of the Cre transgene in the Col1A1 locus (left) and M2-rtTA Flp-in the ROSA26 locus (right). ES cells double heterozygous for M2-rtTA and TetOP-Cre were used to generate chimeric animals and germline offspring (not depicted). (C) Representative products after three-primer competitive genotyping PCR for Col1A1 and ROSA26 loci from tail-tip genomic DNA. Col1A1 expected bands: Flp-in, 550 bp; WT, 330 bp. ROSA26 expected bands: Flp-in, 300 bp; WT, 550 bp. het, heterozygous; homo, homozygous.
Figure 3.
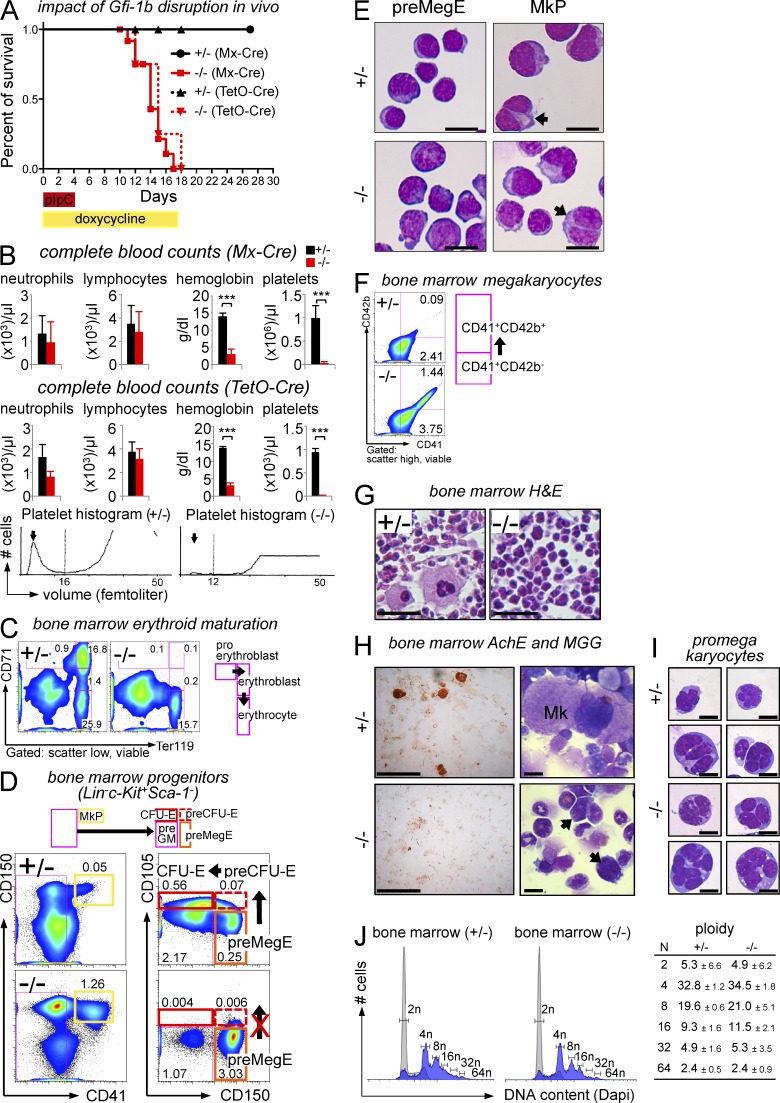
Induced disruption of Gfi-1b in the bone marrow. (A) Kaplan–Meyer survival analysis after conditional Gfi-1b disruption using Mx-Cre (pIpC; +/− n = 15; −/− n = 13) or TetO-Cre (doxycycline; +/− n = 4; −/− n = 4). (B) Automated, complete blood counts after induction of Mx-Cre (top; +/− n = 5; −/− n = 6) or TetO-Cre (middle; days 12–19, +/− n = 4; −/− n = 4). Bottom panels show representative platelet volume histograms. (C) Flow cytometric analysis of bone marrow erythroid maturation (day 13). One representative of five independent experiments is shown. (D) Flow cytometric analysis of bone marrow progenitors (day 13; Pronk et al., 2007); monopotent erythroid progenitors (pre–CFU-E, CFU-E) were undetectable, but bipotent pre-MegEs and monopotent MkPs were increased after Gfi-1b loss. Numbers are frequencies of total bone marrow cells (representative of four independent experiments). (E) Cytospin preparations of flow-sorted pre-MegEs (left) and MkPs (right) stained with MGG. The morphology of both populations (including binucleated MkPs, arrows) was preserved after Gfi-1b disruption. (F) Flow cytometric analysis of bone marrow megakaryocyte maturation (day 15). CD41+CD42b+ megakaryocytic cells were increased ∼15-fold in Gfi-1b−/− bone marrow. Numbers are frequencies of total bone marrow cells (representative of four independent experiments). (G) H&E stain of whole mount bone marrows (day 15) showing the absence of mature megakaryocytes after Gfi-1b loss (−/−, right). (H) Histochemical analysis (left) and MGG stain (right) of total bone marrow cytospin preparations (day 15). After Gfi-1b disruption, no mature megakaryocytes were detected with AchE or MGG stains, but immature multilobulated megakaryocytic cells were abundant (arrows). (I) Cytospin preparations of flow-sorted Gfi-1b+/− (top) and Gfi-1b−/− (bottom three rows) CD41+CD42b+ megakaryocytic cells (MGG) showing high nuclear/cytoplasm ratio, basophilic cytoplasm, faint azurophilic granules, and multilobulated nuclei. Bars: (E, H [right], and I) 10 µm; (G) 50 µm; (H, left) 100 µm. (J) Flow cytometric analysis of bone marrow cell ploidy (day 15). Combined lymphoid plus granulocytic scatter gates were used as internal diploid control (gray histograms), and scatter high gates were used to enrich for megakaryocytic cells (blue histograms). The distribution of cells with high ploidy was preserved after Gfi-1b disruption. Table shows mean ± SD of ploidy data from two independent experiments. Significance was determined with two-tailed Student’s t test: ***, P < 0.001. Error bars denote SDs.
Immunoflow cytometry revealed that differentiated erythroid precursors at and beyond the proerythroblast stage were absent in Gfi-1b−/− bone marrow (Fig. 3 C). Abundant erythroid progenitors were detectable in the spleen after Gfi-1b disruption, but genotyping of sorted splenic, erythroid cells (Ter119+) from Mx-Cre– and TetO-Cre–induced mice revealed that these exclusively harbored unexcised Gfi-1b allele (not depicted). These findings suggested that Cre-mediated Gfi-1b deletion was more efficient in the bone marrow than in the spleen, but terminal erythroid maturation strictly required Gfi-1b at either site. Immunophenotypic progenitor analysis (Pronk et al., 2007) of bone marrow cells demonstrated that the strict requirement for Gfi-1b in the erythroid lineage preceded the proerythroblast stage. Thus, monopotent erythroid progenitors were not detectable after Gfi-1b disruption (pre–CFU-Es and CFU-Es; Fig. 3 D). In contrast, earlier bipotent progenitors of megakaryocytes and erythrocytes (pre-MegEs; Pronk et al., 2007) were markedly increased in Gfi-1b−/− bone marrow (Fig. 3 D, right, orange frames). Cytological analysis of sorted pre-MegEs revealed a proportion of cells with subtle erythroid features (scant cytoplasm, round nucleus, and denser chromatin) in the presence or absence of Gfi-1b (Fig. 3 E, left). Hence, Gfi-1b was not required for some aspects of morphological erythroid maturation at the pre-MegE stage, but in its absence, development did not proceed to the monopotent progenitor stage and beyond.
Immunophenotypic analysis of early megakaryocytic development demonstrated that Gfi-1b−/− bone marrow cells harbored markedly increased numbers of monopotent megakaryocytic progenitors (MkPs; Fig. 3 D, left, yellow frames) with normal, characteristic morphology (Fig. 3 E, right). Further immunophenotypic analysis suggested that development proceeded beyond the progenitor stage with a markedly increased number of cells bearing megakaryocyte markers (CD41+/CD42b+; Fig. 3 F). However, normal megakaryocytes were not detected in Gfi-1b−/− bone marrow sections stained with hematoxylin and eosin (H&E; Fig. 3 G). Histochemical analysis of bone marrow cell cytospin preparations for acetylcholinesterase (AchE) activity, a marker of late megakaryocyte maturation (Fig. 3 H, left), and morphological analysis after May-Grünwald Giemsa (MGG) staining (Fig. 3 H, right) confirmed the absence of mature megakaryocytes. However, the cytospin preparations revealed cells with multilobulated, segmented nuclei (Fig. 3 H, right, bottom, arrows). These were highly enriched after sorting CD41+/CD42b+ cells (Fig. 3 I). They were 15–25 µm in diameter and displayed scant, basophilic cytoplasm with a high nucleus/cytoplasm ratio. Faint azurophilic granules were perceptible (not depicted). The morphology of the Gfi-1b−/− multilobulated cells closely resembles that of normal promegakaryocytes (Long and Williams, 1981), although they were larger (Fig. 3 I). The frequency of cells with promegakaryocyte morphology was increased approximately fivefold in Gfi-1b−/− bone marrow (Gfi-1b+/− 0.4 ± 0.1% and Gfi-1b−/− 1.89 ± 0.4%, n = 3/group, P < 0.005). As endomitosis is completed before the promegakaryocyte stage, in megakaryoblasts (see also Fig. 9; Chang et al., 2007), these data suggested that Gfi-1b was dispensable for polyploidization. Confirming this conclusion, analysis of DNA content of Gfi-1b−/− versus control bone marrow cells revealed no differences in the frequency or distribution of hyperdiploid cells (Fig. 3 J).
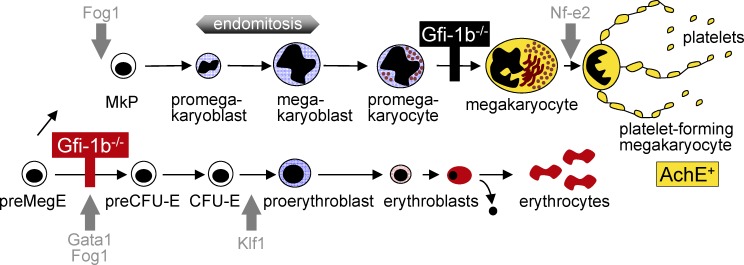
Figure 9.
Schematic representation of Gfi-1b’s role in normal erythropoiesis and thrombopoiesis. The top illustrates the position of the block that follows Gfi-1b disruption in megakaryocytic lineage maturation (black bar). The bottom shows the position of the erythroid lineage block after Gfi-1b loss (red bar). Gray arrows indicate the positions of blocks that follow loss of other lineage regulators.
Together, Gfi-1b loss affects adult megakaryocytic and erythroid development at different stages. In the absence of Gfi-1b, erythroid development is ablated early in progenitors, whereas megakaryocytic maturation proceeds beyond the progenitor stage to the formation of polyploid promegakaryocytes that fail to mature and produce platelets.
Strict requirement for Gfi-1b in erythroid and megakaryocytic colony differentiation
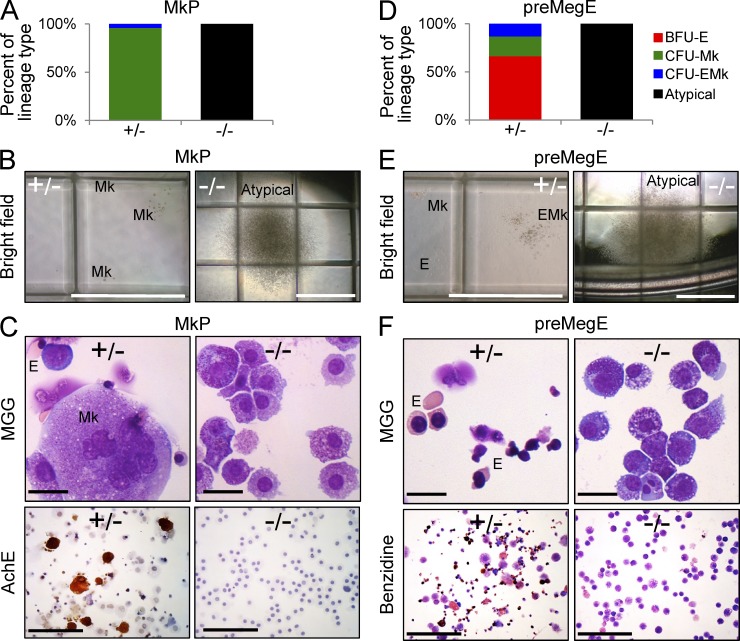
To assess whether Gfi-1b is required for terminal differentiation within erythroid and megakaryocytic lineage cells, we first analyzed colony formation from Gfi-1b−/− or control lineage-committed progenitors in the presence of a cytokine combination that allows for differentiation into all myeloid lineages. As expected (Pronk et al., 2007), sorted control MkPs (Fig. 3 D, yellow frame) predominantly gave rise to colonies containing only megakaryocytes (CFU-Mk) and very few (<10%) colonies containing both erythroid cells and megakaryocytes (CFU-EMk; Fig. 4 A, left). This was judged by colony morphology (Fig. 4 B, left) as well as cellular morphology and AchE activity of cytospin preparations (Fig. 4 C, left). In contrast, Gfi-1b−/− MkPs did not give rise to megakaryocytes or terminal erythroid cells. Instead, they generated atypical, very large colonies that contained immature cells lacking AchE activity (Fig. 4, A–C, right). Thus, Gfi-1b is intrinsically required for megakaryocyte maturation from committed progenitors in vitro.
Figure 4.
Megakaryocytic and erythroid lineage–intrinsic requirements for Gfi-1b in vitro. (A–C) Colony formation from sorted MkPs (days 15–19 after Cre induction; SCF, IL-3, IL-11, GM-CSF, Epo, and Tpo). (A) Distribution of colonies with the indicated morphologies by phase-contrast microscopy (BFU-E, erythroid; CFU-Mk, megakaryocyte; CFU-EMk, erythroid/megakaryocyte; atypical; based on >300 colonies per genotype; three independent experiments scored days 7–10). (B) Microscopic morphologies of typical CFU-Mks and large atypical colonies from Gfi-1b−/− MkPs. (C) Cytospin preparations of pooled colonies stained with MGG (top) and AchE (bottom; representative results of three independent experiments). Gfi-1b−/− MkPs failed to produce typical megakaryocyte colonies but gave rise to large atypical colonies that did not contain mature megakaryocytes. (D–F) Colony formation from sorted bipotent megakaryocyte/erythrocyte progenitors (pre-MegEs; conditions as in A–C). (D) Distribution of colony types (as detailed in A, >300 colonies per genotype, three experiments). (E) Morphologies of typical BFU-Es, CFU-Mks, and CFU-EMks and large, atypical colonies from Gfi1b−/− pre-MegEs. (F) Cytospin preparations of pooled colonies stained with MGG (top) or benzidine (bottom; representative results of three independent experiments). The large atypical colonies from Gfi-1b−/− pre-MegEs did not contain mature erythroid cells (top) or benzidine-reactive (hemoglobin producing) cells (bottom). Bars: (B and E) 1 mm; (C and F, top) 10 µm; (C and F, bottom) 100 µm.
Because monopotent erythroid progenitors were absent after Gfi-1b loss (Fig. 3 D), we tested erythroid differentiation with colony assays from sorted pre-MegEs, a bipotent progenitor population with predominant erythroid output (Fig. 3 D, orange frame). Consistent with previous findings (Pronk et al., 2007), control pre-MegEs gave rise to CFU-Mks as well as bilineage CFU-EMk and a majority of erythroid colonies (BFU-E; Fig. 4, D and E). Cytospin preparations of the colonies confirmed robust terminal erythroid differentiation, including the presence of enucleated red cells and abundant hemoglobin production demonstrated by benzidine reactivity (Fig. 4 F, left). In contrast, Gfi-1b−/− pre-MegEs only gave rise to atypical large colonies (Fig. 4, D and E, right). In cytospin preparations, a larger proportion of cells bore some resemblance to early erythroid cells compared with cells in colonies from MkPs, but hallmarks of terminal erythroid differentiation, such as nuclear condensation or maturation of the cytoplasm, were not appreciated (Fig. 4 F, top right). Consistent with these observations, no benzidine-reactive cells were detected (Fig. 4 F, bottom right). Hence, Gfi-1b is also intrinsically required for erythroid maturation from committed progenitors in vitro.
Gfi-1b disruption results in arrested erythroid and megakaryocytic differentiation in a WT bone marrow microenvironment
To investigate the requirement for Gfi-1b in the presence of a WT microenvironment and without anemia or thrombocytopenia, we transplanted unexcised Gfi-1b conditional bone marrow cells (KO/floxed or WT/floxed; CD45.2) into lethally irradiated recipients (CD45.1) together with WT support bone marrow (CD45.1; Fig. 5 A). After engraftment and recovery of hematopoiesis, we disrupted Gfi-1b by doxycycline administration. As expected in the presence of WT competitor marrow, Gfi-1b disruption did not result in cytopenias (Fig. 5 B). Genotype analysis of DNA from sorted bone marrow populations confirmed that complete excision was achieved (Fig. 5 C). The excised allele was maintained in all control populations bearing a combination of alleles that produced a heterozygous state (Cre → +/−). After biallelic Gfi-1b loss (Cre → −/−), the excised Gfi-1b allele also remained abundant in Gr-1+/Mac-1+ granulocytes, pre-MegEs, and MkPs (Fig. 5 C, second, fourth, and sixth lanes), demonstrating that these populations were maintained in Gfi-1b’s absence. However, the excised allele could not be detected in erythroblasts (Ter119+CD71+), and this population only contained cells derived from the WT competitor marrow (Fig. 5 C, seventh and eighth lanes). These findings suggested striking selection against Gfi-1b loss after, but not at or preceding the pre-MegE stage of erythroid differentiation. Analysis of CD41+CD42b+ megakaryocytic cells also revealed moderate selection for WT Gfi-1b compared with MkPs, but the Gfi-1b−/− allele predominated (Fig. 5 C, 5th, 6th, 9th, and 10th lanes). Analysis of donor or recipient origin with CD45 allelic variants confirmed that MkPs and pre-MegEs were predominantly derived from the conditional donor marrow after Gfi-1b loss, whereas CFU-Es were almost exclusively derived from the competitor marrow after biallelic Gfi-1b disruption (Fig. 5 D). We used a combination of high-resolution microscopy and flow cytometry (Amnis ImageStream MkII system) to study the impact of Gfi-1b on terminal megakaryocytic differentiation in transplant recipients. With this technology, we collected 93 (Gfi-1b+/− transplant recipients) and 195 (Gfi-1b−/− recipients) images of CD41+CD42b+ late megakaryocyte lineage cells and assessed their overall size, nuclear morphology, and CD45.2 expression (Fig. 5, E and F). As expected, the CD45.2-negative cells derived from the competitor marrow included very large, mature megakaryocytes with convoluted nuclei. This was unchanged in CD45.2-positive conditional control cells bearing heterozygous Gfi-1b (Fig. 5, E and F, top). However, after biallelic Gfi-1b loss, CD45.2-positive CD41+CD42b+ cells were invariably small despite the presence of convoluted nuclei (Fig. 5, E and F, bottom). Hence, the early erythroid progenitor arrest and the terminal megakaryocytic differentiation defect after Gfi-1b loss are reproducible in a WT bone marrow microenvironment.
Figure 5.
The megakaryocytic and erythroid defects after loss of Gfi-1b are blood cell autonomous in vivo. (A) Scheme of the experiment. (B) Automated, complete blood counts after 4 wk of doxycycline treatment in Gfi-1b+/− (n = 5) and Gfi-1b−/− (n = 6) posttransplant recipients. Error bars represent SD. (C) Analysis of Gfi-1b gene status by PCR in sorted bone marrow populations in posttransplant recipients. Genomic DNA of sorted Gr1+Mac1+ neutrophils, pre-MegEs, MkPs, CD71+Ter119+ erythroblasts, CD41+CD42+ megakaryocytic cells was analyzed by competitive PCR 7 wk after doxycycline treatment (19 wk after transplant; fl, floxed; ex, excised; see Fig. 1 [G and I]). All populations predominantly contained Gfi-1b−/− cells, except for erythroblasts, which exclusively contained WT cells from competitor marrow (representative of two independent experiments). (D) Flow cytometric analysis of bone marrow progenitors 7 wk after doxycycline treatment in Gfi-1b+/− (top) and Gfi-1b−/− (bottom) posttransplant recipients. CFU-E progenitors were almost all derived from competitor cells (red frame and histogram) in Gfi-1b−/− posttransplant mouse compared with Gfi-1b+/− controls. (E and F) Combination of quantitative imaging and flow cytometric analysis of CD41+CD42+ megakaryocytes in Gfi-1b+/− and Gfi-1b−/− posttransplant recipients, 7 wk after doxycycline treatment. Bone marrow cells were stained with CD45.2 FITC (green pseudocolor), CD42 PE (yellow pseudocolor), CD41 Pacific Blue (purple pseudocolor), and DRAQ5 (DNA; red pseudocolor) and analyzed using Amnis ImageStream MkII. (E) Representative images of competitor cell–derived (comp) CD45.2-negative and conditional cell–derived (cond) CD45.2-positive CD41+CD42+ megakaryocytes in Gfi1b+/− (top) and Gfi1b−/− (bottom) posttransplant recipients. Note that Gfi1b−/− megakaryocytes are dramatically smaller than their WT counterparts. Bars, 10 µm. (F) Quantification of size of competitor cell–derived and conditional cell–derived CD41+CD42+ megakaryocytes in Gfi1b+/− (top) and Gfi1b−/− (middle) posttransplant recipients. Table shows mean ± SD of CD41 and CD42 area scales for a total of 93 and 195 CD41+CD42+ megakaryocytes analyzed from Gfi1b+/− and Gfi1b−/− posttransplant recipients, respectively.
Forced Gfi-1b or Gfi-1 expression similarly rescues erythroid and megakaryocytic colony formation from Gfi-1b−/− progenitors, but Gfi-1b more efficiently promotes proplatelet formation
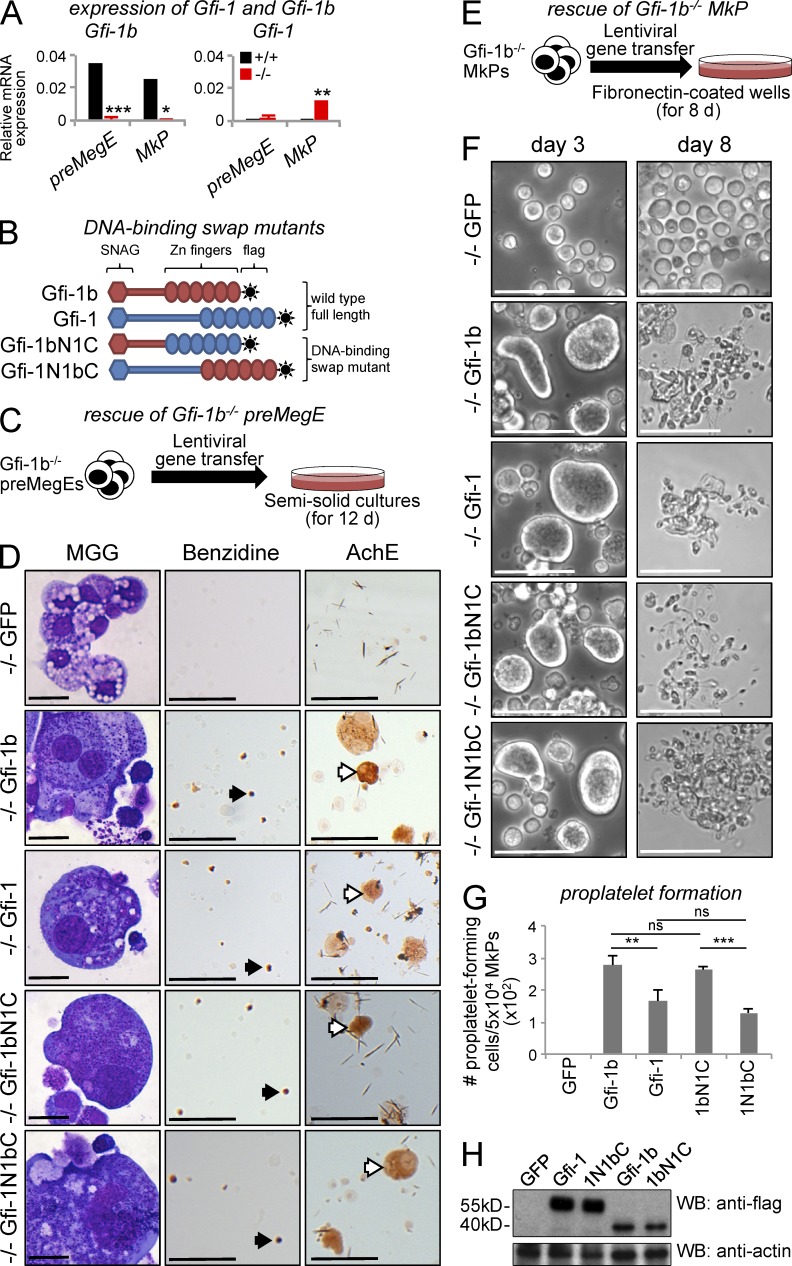
To investigate whether Gfi-1, Gfi-1b’s closely related homologue, could replace it in erythroid and megakaryocytic differentiation, we studied the impact of Gfi-1 and Gfi-1b gene transfer into Gfi-1b−/− progenitors ex vivo (Fig. 6). In most differentiated hematopoietic cells, Gfi-1b and Gfi-1 display mutually exclusive expression patterns (Hock and Orkin, 2006). Gfi-1b has been shown to rescue significant aspects of the phenotype that follows Gfi-1 loss in myeloid cells but not the inner ear defects in Gfi-1−/− mice (Fiolka et al., 2006). It has not been reported whether Gfi-1 can functionally replace Gfi-1b in any setting. As expected from Gfi-1b’s preferential expression in erythroid and megakaryocytic cells (Saleque et al., 2002), WT pre-MegEs and MkPs robustly expressed Gfi-1b (Fig. 6 A, left, black bars) but not Gfi-1 (Fig. 6 A, right, black bars). After Gfi-1b disruption, Gfi-1b expression was abolished in both populations (Fig. 6 A, left, red bars) and the expression of Gfi-1, a direct target of Gfi-1b (Doan et al., 2004; Vassen et al., 2005), was increased (Fig. 6 A, right, red bars). This suggested that Gfi-1 either lacked the potential to promote terminal differentiation of Gfi-1b−/− progenitors or was expressed at insufficient levels.
Figure 6.
Rescue of erythroid and megakaryocytic colony formation and proplatelet formation from Gfi-1b−/− progenitors ex vivo. (A) Gfi-1b (left) and Gfi-1 (right) mRNA expression analysis in Gfi-1b+/+ and Gfi-1b−/− pre-MegE and MkP bone marrow progenitors. mRNA expression relative to β-actin expression is shown as mean ± SD of three independent biological replicates (***, P < 0.001; **, P < 0.005; *, P < 0.05; two-tailed Student’s t test). (B) Scheme of Gfi-1b/Gfi-1 hybrids. In the Gfi-1bN1C mutant (third row), the N terminus of Gfi-1b was fused to the C terminus of Gfi-1. In the Gfi-1N1bC mutant (fourth row), the C terminus of Gfi-1b was fused to the N terminus of Gfi-1. (C) Scheme of the Gfi-1b−/− pre-MegE rescue. (D) Representative cytospin preparations of progenitor colonies on day 12 after lentiviral gene transfer of Gfi-1b−/− pre-MegEs with GFP alone (first row), Gfi-1b with GFP (second row), Gfi-1 with GFP (third row), Gfi-1bN1C with GFP (fourth row), and Gfi-1N1bC with GFP (fifth row). Colonies were stained with MGG (left) and benzidine (middle) and for AchE activity (right). Note the presence of benzidine+ erythrocytes (black arrows) and AchE+ megakaryocytes (white arrows) after transduction of Gfi-1b, Gfi-1, Gfi-1bN1C, and Gfi-1N1bC but not GFP alone. (E) Scheme of the Gfi-1b−/− MkP rescue. (F–H) Analysis of proplatelet formation from Gfi-1b−/− MkPs. (F) Representative phase-contrast micrographs on days 3 (left) and 8 (right) after lentiviral gene transfer of GFP alone (first row), Gfi-1b with GFP (second row), Gfi-1 with GFP (third row), Gfi-1bN1C with GFP (fourth row), and Gfi-1N1bC with GFP (fifth row). Note the formation of proplatelet-forming megakaryocytes in all conditions but GFP alone. Bars: (D, left) 10 µm; (D, middle and right) 100 µm; (F) 50 µm. (G) Quantification of proplatelet formation 8 d after infection of Gfi-1b−/− MkPs. Note that the efficiency of Gfi-1b and Gfi-1bN1C in giving rise to proplatelet is comparable and significantly higher than that of Gfi-1 or Gfi-1N1bC. Data are shown as means ± SD of the number of proplatelet-forming megakaryocytes in three independent experiments (***, P < 0.001; **, P < 0.005; two-tailed Student’s t test). (H) Western blot analysis using M2 anti-flag antibody (top) in day 8 megakaryocytic cultures derived from infected Gfi-1b−/− MkPs. Expression level of β-actin (bottom, same blot after stripping) was used as a control for loading.
We examined the impact of lentiviral vector–mediated gene transfer of C-terminally FLAG-tagged Gfi-1b, Gfi-1, and two hybrids on colony formation from Gfi-1b−/− pre-MegEs (Fig. 6, B and C). Although progenitors infected with a control vector expressing GFP alone did not give rise to erythroid colonies or megakaryocyte-containing colonies, such colonies did form after infection with Gfi-1b or Gfi-1, as well as mutant-expressing vectors. The presence of erythroid cells and megakaryocytes was confirmed in MGG-stained cytospin colony preparations (Fig. 6 D, left), as well as by demonstrating benzidine reactivity (Fig. 6 D, middle) and AchE reactivity (Fig. 6 D, right). These data suggest that Gfi-1 and Gfi-1b exhibit a similar capacity to rescue erythroid differentiation and early megakaryocyte differentiation from Gfi-1b−/− pre-MegEs.
To assess terminal megakaryocyte maturation, we analyzed proplatelet formation from Gfi-1b−/− MkPs in culture with stem cell factor and thrombopoietin on fibronectin-coated dishes. (Fig. 6, E–H). After infection with a GFP control vector alone, Gfi-1b−/− MkPs did not increase in size, become adherent, or form proplatelets (Fig. 6, F [top] and G). In contrast, expression of Gfi-1b and Gfi-1 was associated with cell adhesion and size increase, noticeable between days 2 and 5 (Fig. 6 F, left, second and third panel from top), extrusion of proplatelet processes, and even release of some platelets (approximately days 7–8; Fig. 6 F, right, second and third panel from top). However, the rescue efficiency measured by the number of megakaryocyte-forming proplatelets was significantly reduced with Gfi-1 compared with Gfi-1b in three independent experiments (Fig. 6 G).
Finally, we studied two DNA-binding domain swap mutants. These did not exhibit differences in rescuing erythroid and megakaryocytic colonies from pre-MegEs (Fig. 6 D). Likewise, the first hybrid, in which the zinc finger domain of Gfi-1b was replaced by the zinc fingers of Gfi-1 (Fig. 6 B, third row), was as efficient as Gfi-1b in promoting proplatelet formation from MkPs (Fig. 6, F and G). However, the second hybrid, in which the N-terminal portion of Gfi-1b was replaced by the N-terminal portion of Gfi-1 (Fig. 6 B, bottom), was significantly less efficient than Gfi-1b and was indistinguishable from Gfi-1 in promoting proplatelet formation (Fig. 6, F and G). The differences in rescue efficiency were not a result of reduced protein expression (Fig. 6 H). Thus, the N-terminal portion of Gfi-1b confers a higher efficiency in promoting proplatelet formation, whereas the zinc finger domains of both factors rescue terminal megakaryocyte maturation with equivalent efficiency.
Abnormal but partially preserved erythroid and megakaryocytic lineage gene expression in Gfi-1b−/− progenitors
To shed light on the molecular impact of Gfi-1b loss, we first analyzed the expression of erythroid and megakaryocytic lineage candidate genes by real-time RT-PCR of mRNA from sorted MkPs and pre-MegEs. In accordance with the known lineage biases of MkPs and pre-MegEs (Fig. 4, A and D; Pronk et al., 2007), MkPs expressed megakaryocytic lineage–specific genes at higher levels than pre-MegEs (Fig. 7 A), and pre-MegEs expressed erythroid lineage–specific genes at higher levels than MkPs (Fig. 7 B).
Figure 7.
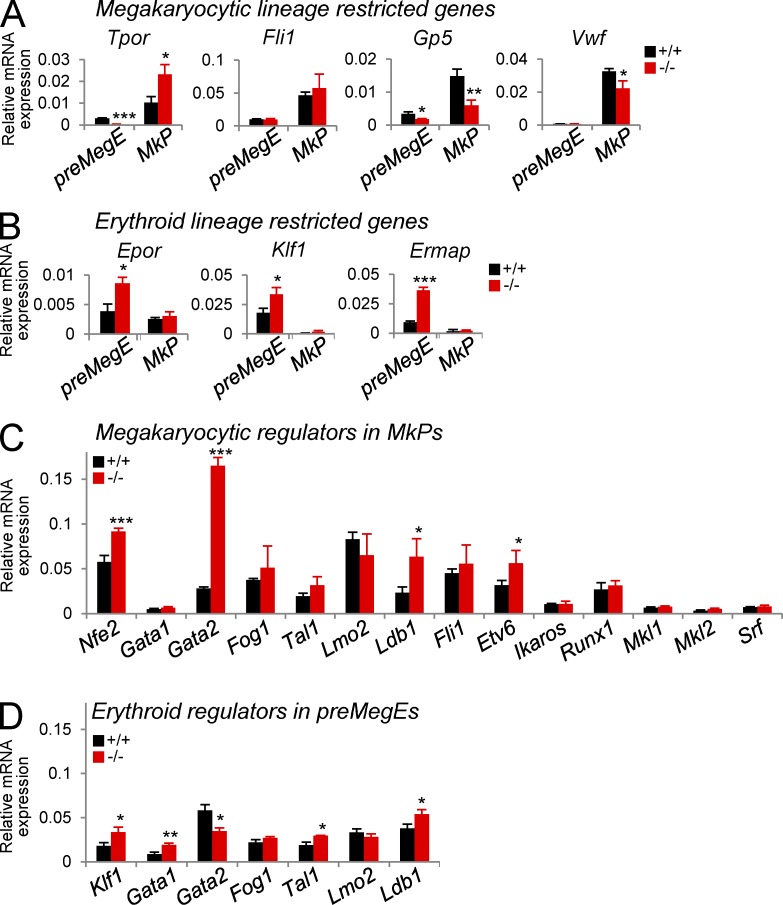
Pre-MegEs and MkPs partially preserve their molecular program after Gfi-1b loss. (A–D) Analysis of gene expression in sorted Gfi-1b+/+ and Gfi-1b−/− pre-MegEs and MkPs. (A) Analysis of megakaryocytic lineage–restricted genes (Tpor, thrombopoietin receptor; Fli1; Gp5, platelet glycoprotein V; Vwf, Von Willebrand factor). (B) Analysis of erythroid lineage–restricted genes (Epor, erythropoietin receptor; Klf1, [Eklf]; Ermap, erythroid membrane-associated protein). (C) Analysis of megakaryocytic transcriptional regulators in MkPs (Nfe2, Gata1, Gata2, Fog1, Tal1 [Scl], Lmo2, Ldb1, Fli1, Etv6 [Tel], Ikaros, Runx1 [Aml1], Mkl1, Mkl2, and Srf). (D) Analysis of erythroid transcriptional regulators in pre-MegEs (Klf1 [Eklf], Gata1, Gata2, Fog1 [Zfpm1], Tal1 [Scl], Lmo2, and Ldb1). mRNA expression relative to β-actin expression is shown as mean ± SD of three independent biological replicates (***, P < 0.001; **, P < 0.005; *, P < 0.05; two-tailed Student’s t test).
After Gfi-1b loss in MkPs, the expression levels of megakaryocyte lineage genes remained higher than in pre-MegEs, but some were significantly altered. Specifically, the mRNA levels of Von Willebrand factor (1.5-fold) and Gp5 (2.5-fold) were decreased in Gfi-1b’s absence, whereas the levels of Fli1 were unchanged, and the levels of the thrombopoietin receptor mRNA (2.2-fold) were markedly increased. Thus, Gfi-1b−/− MkPs did not lose their molecular megakaryocytic identity. Consistent with this conclusion, Gfi-1b−/− MkPs expressed normal or increased levels of 14 transcriptional regulators that have been implicated in the maintenance of normal platelet counts (Fig. 7 C).
In Gfi-1b−/− pre-MegEs, erythroid lineage–specific genes remained expressed at higher levels than in MkPs (Fig. 7 B). The mRNA expression levels of the erythropoietin receptor (approximately twofold), Klf1 (approximately twofold), and the erythroid membrane-associated protein (Ermap; fourfold) were significantly increased after Gfi-1b loss. Therefore, despite the absent potential of Gfi-1b−/− pre-MegEs to give rise to mature erythroid cells, their erythroid molecular program was not extinguished. Accordingly, the expression of six erythroid regulators, Klf1, Gata1, Fog1, Tal1/Scl, Lmo2, and Ldb1, was either unchanged or increased in the absence of Gfi-1b. Gata2 expression was significantly reduced in pre-MegEs, in agreement with the prediction of a proposed regulatory triad between Gfi-1b, Gfi-1, and Gata2 (Moignard et al., 2013). However, in Gfi-1b−/− MkPs, Gata2 expression was altered in the opposite direction (Fig. 7 C). Regardless, lentiviral expression of Gata2 failed to rescue erythroid differentiation from Gfi-1b−/− pre-MegEs (not depicted). Together, the erythroid maturation arrest that follows Gfi-1b loss is unlikely to be the result of expression loss of a previously identified lineage-restricted transcriptional regulator.
Gfi1b functions as a master transcriptional repressor of the erythroid and megakaryocytic lineages
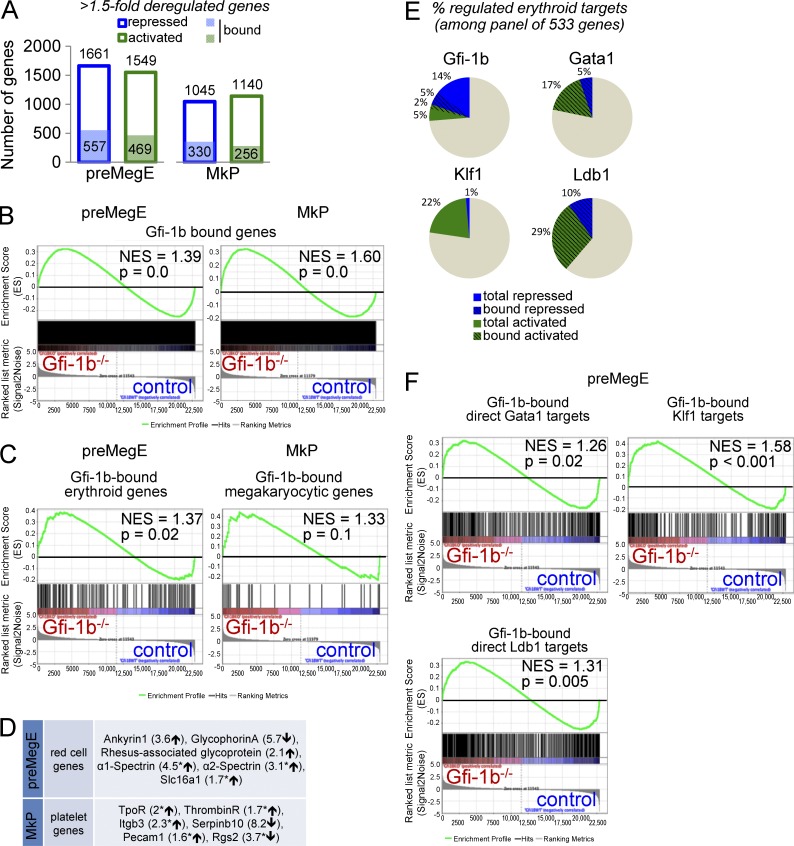
To assess Gfi-1b’s genome-wide impact on gene expression, we compared mRNA profiles in control and Gfi-1b−/− progenitors with microarrays (Fig. 8 A and Tables S1 and S2). A threshold of a 1.5-fold expression change after Gfi-1b disruption defined 3,210 differentially expressed genes in pre-MegEs and 2,185 in MkPs, about half up- and down-regulated in either population. To gauge Gfi-1b’s impact at genes it occupies, we used a published dataset of genomic sites that bind Gfi-1b in the murine hematopoietic progenitor cell line HPC-7 (Wilson et al., 2010); 4,812 genes with peaks between −10 kb from the transcriptional start site and 1 kb after the gene end were defined as bound (Fig. 8 A and Table S3). Among direct Gfi-1b targets, 54.3% (pre-MegEs; Fig. 8 A, left) and 56.3% (MkPs, Fig. 8 A, right) were up-regulated after Gfi-1b loss. Gene set enrichment analysis (GSEA) showed that the increase of up-regulated Gfi-1b targets in Gfi-1b−/− pre-MegEs and Gfi-1b−/− MkPs was highly statistically significant (Fig. 8 B). Therefore, as expected (Zweidler-Mckay et al., 1996; Tong et al., 1998; Saleque et al., 2007), the predominant impact of the local presence of Gfi-1b is repression of transcription.
Figure 8.
Genome-wide analysis of Gfi-1b’s impact on megakaryocytic and erythroid gene expression in progenitors. (A) Affymetrix Mouse GeneChip 1.0 ST microarray analysis of control and Gfi-1b−/− pre-MegEs (left; gene list in Table S1) and MkPs (right; gene list in Table S2). Shown are numbers of genes with 1.5-fold expression changes after Gfi-1b disruption (blue frame: repressed genes; green frames: activated genes). Numbers of direct Gfi-1b targets are shown in filled blue and green boxes (peaks defined as −10 kb from transcription start site to 1 kb from gene end, based on a published ChIP-seq gene set [Wilson et al., 2010]; see Table S3). (B) GSEA of control and Gfi-1b−/− pre-MegEs (left) and MkPs (right) with Gfi-1b–bound gene set (see Table S3). (C) GSEA of control and Gfi-1b−/− pre-MegEs (left) and MkPs (right) with Gfi-1b–bound erythroid gene set (left; 155 Gfi-1b–bound/533 literature-defined genes [Li et al., 2013]; see Table S3) or Gfi-1b–bound megakaryocytic gene set (right; 31 Gfi-1b–bound/57 literature-defined genes [Chen et al., 2007]; see Table S3). (D) Examples of erythroid and megakaryocytic lineage genes that are deregulated after loss of Gfi-1b in pre-MegEs and MkPs. Asterisks indicate Gfi-1b–bound genes (Table S3). (E) Pie charts for comparison of the ratio of repressed and activated literature-defined erythroid genes (Li et al., 2013) by Gfi-1b (top left; Table S1), Gata1 (top right; Yu et al., 2009), Klf1 (bottom left; Tallack et al., 2012), and Ldb1 (bottom right; Li et al., 2013). Dashed lines indicate bound targets. (F) GSEA of control and Gfi-1b−/− pre-MegEs with Gfi-1b–bound, regulated direct Gata1 targets (top left; 355 Gfi-1b–bound/791 Gata1 targets [Yu et al., 2009]; see Table S3), Gfi-1b–bound regulated Klf1 targets (top right; 242 Gfi-1b–bound/813 Klf1-regulated genes [Tallack et al., 2012]; see Table S3), and Gfi-1b–bound, regulated direct Ldb1 targets (bottom; 553 Gfi-1b–bound/1,206 Ldb1 targets [Li et al., 2013]; see Table S3). NESs and nominal p-values are shown for all GSEAs.
To more specifically probe Gfi-1b’s impact on erythroid and megakaryocytic gene programs, we used established gene panels (Chen et al., 2007; Li et al., 2013). 29% (155/533) of the erythroid genes and 55.3% (31/57) of the megakaryocytic genes were bound by Gfi-1b (Table S3). Gfi-1b loss resulted in significant enrichment of up-regulated direct erythroid target genes in pre-MegEs (Fig. 8 C, left) and up-regulated megakaryocytic direct targets genes in MkPs (Fig. 8 C, right). Gfi-1b loss also altered the expression of many important erythroid and megakaryocytic genes that were not bound (Fig. 8 D and Tables S1 and S2).
Finally, we examined the impact of Gfi-1b loss on known targets of other erythroid lineage transcription factors. Recently, genome-wide studies have defined gene sets regulated by Gata1 (Yu et al., 2009), Klf1 (Tallack et al., 2012), and Ldb1 (Li et al., 2013). Superimposing these gene sets on the defined erythroid gene panel showed that all three factors regulate a large fraction of erythroid genes and predominantly increase their expression (Fig. 8 E and Table S3). Gfi-1b also regulates a sizable fraction of the panel genes, but it stands out in that it represses more than two thirds of its erythroid targets (Fig. 8 E). Notably, a high proportion of the targets of Gata1 (44.8%), Ldb1 (45.8%), and Klf1 (29.7%) were occupied by Gfi-1b (Table S3; Wilson et al., 2010). After Gfi-1b loss in the erythroid lineage (pre-MegEs), the Gfi-1b–bound target genes of all three factors were significantly up-regulated (Fig. 8 F). Together, our genomic analysis establishes Gfi-1b as a master transcriptional repressor that regulates a large number of erythroid and megakaryocytic genes in combination with other prominent regulators of these lineages. Its continuous presence is required to restrain the activity of a wide spectrum of genes during adult erythropoiesis and thrombopoiesis.
DISCUSSION
Gfi-1b in the adult bone marrow
In this study we identify strict, lineage-intrinsic requirements for continuous Gfi-1b expression in adult erythropoiesis and thrombopoiesis. After Gfi-1b loss, no red cells, megakaryocytes, or platelets were produced in the bone marrow, whereas progenitors were maintained at increased frequencies. The Gfi-1b−/− progenitors exclusively gave rise to undifferentiated, abnormally large colonies ex vivo. Yet their identity was partially preserved as they retained morphological and molecular traits of the erythroid and megakaryocytic lineages. The requirement for Gfi-1b expression was evident at earlier stages in erythroid lineage than in the megakaryocytic lineage. Although Gfi-1b−/− progenitors initiated erythroid differentiation and expressed many erythroid genes, they did not progress to the fully committed, monopotent erythroid progenitor stage. In contrast, megakaryocytic differentiation progressed beyond the progenitor stage in the absence of Gfi-1b, arresting only after nuclear polyploidization, before cytoplasmic maturation. These Gfi-1b−/− phenotypes were reproduced in WT transplant recipients, in the presence of normal bone marrow and in the absence of anemia and thrombocytopenia.
Our results contradict some findings of another study that examined the consequences of adult Gfi-1b disruption (Khandanpour et al., 2010). These authors reported only moderate anemia without changes in the types of progenitor colonies generated from the bone marrow, including erythrocyte- and megakaryocyte-containing colonies. We were able to reproduce those findings using less efficient regimens for Gfi-1b disruption. A small minority of unexcised cells was sufficient to selectively expand and exclusively maintain late erythropoiesis and megakaryopoiesis (not depicted). Consistent with this, our transplant experiments demonstrate that strong selection against Gfi-1b loss occurs only late, after the pre-MegE and MkP stages (Fig. 5, C and D). Khandanpour et al. (2010) only reported the excision status of HSCs, and it remained unresolved whether late erythroid and megakaryocytic cells indeed lacked Gfi-1b. Notably, we observed comparably severe anemia and thrombocytopenia after Cre-mediated recombination with our doxycycline-inducible TetO-Cre and with Mx-Cre. Thus, the more severe phenotype associated with complete excision was not the result of increased interferon-associated effects (Essers et al., 2009). We conclude that the erythroid and megakaryocytic phenotypes that follow adult Gfi-1b disruption are as severe as those demonstrated in Gfi-1b KO embryos (Saleque et al., 2002).
Unique role for Gfi-1b in megakaryocytic development
Gfi-1b’s role in thrombopoiesis is unique. Similar to our findings, severe thrombocytopenia follows loss of Nf-e2 (Shivdasani et al., 1995), Fog1 (Tsang et al., 1998), and Ldb1 (Li et al., 2010). However, Nf-e2 loss was associated with a later block at the stage of proplatelet formation, and AchE-positive megakaryocytes were abundantly formed (Lecine et al., 1998). In contrast, loss of Fog1 resulted in an earlier block with an absence of MkPs (Mancini et al., 2012). The stage of the requirement of Ldb1 within thrombopoiesis is not fully defined because of its essential role in HSC maintenance (Li et al., 2011). Similarly, Lmo2 and Gata2 are critical for HSC function and megakaryopoiesis but have yet to be studied with lineage-specific KO systems (Warren et al., 1994; Huang et al., 2009). Finally, absence of Gata1, Fli-1, Scl1, Tel/Etv6, Runx1, Mkl1, Mkl2, and Srf was associated with much less severe thrombocytopenia and compatible with the development of mature, if abnormal, megakaryocytes (Tijssen and Ghevaert, 2013). In addition, none of the above regulators exhibited reduced mRNA expression in Gfi-1b−/− MkPs (Fig. 7 C). However, it is highly likely that many individual megakaryocytic genes are jointly regulated by Gfi-1b and the known lineage-restricted regulators. Consistent with this notion, our genomic analysis revealed that significant numbers of Gata1 or Ldb1 targets that are bound by Gfi-1b are up-regulated after Gfi-1b disruption in monopotent MkPs (Fig. 8 F). To our knowledge, an arrest at the promegakaryocyte stage is unprecedented. Thus, Gfi-1b marks a novel entry point for illuminating this important, unexplored stage of thrombopoiesis, when nuclear maturation is largely complete and the cytoplasm still immature. Very recently, mutations of Gfi-1b have been found in two families with platelet-related bleeding disorders (Stevenson et al., 2013; Monteferrario et al., 2014), illustrating the high clinical relevance of understanding Gfi-1b’s molecular function in thrombopoiesis.
Gfi-1b is an essential, major player in the network of erythroid regulators
In the erythroid lineage, the loss of function phenotype associated with Gfi-1b bears striking similarities with findings after the disruption of three other critical lineage-restricted regulators, a complete block of red blood cell production which does not entirely extinguish the early erythroid program (Fig. 9, bottom). In the absence of Klf1, erythroid development was arrested only slightly later, after the formation of fully committed erythroid progenitors (Pilon et al., 2008). Adult disruption of Gata1 or its cofactor Fog1 was followed by an arrest at the same immunophenotypic progenitor stage as Gfi-1b loss (Fig. 9; Mancini et al., 2012). However, we demonstrate that Gfi-1b is dispensable for the expression of these three regulators. Likewise, Gfi-1b was not required for the expression of any of the major components of the pentameric complexes (Ldb1, Scl/Tal1, Lmo2, E2A, and Gata1) that associate with Gata1 to activate a subset of its target genes (Fig. 8 F and not depicted; Soler et al., 2010; Li et al., 2013). In agreement with the preserved or increased expression of these erythroid regulators, GSEA demonstrated significant up-regulation of target genes of Ldb1, Gata1, and Klf1 after Gfi-1b loss (Fig. 8 F and not depicted). By superimposing previously generated datasets (Yu et al., 2009; Wilson et al., 2010; Li et al., 2013), we found that Gfi-1b binds a remarkably high proportion of the direct targets of Gata1 and Ldb1 (44.8% and 45.8%, respectively). As transcription factor binding dynamically changes during erythroid differentiation (May et al., 2013), our indirect comparison, using data from cell lines that are not entirely matched in their differentiation stage, likely underestimates the true number of shared occupied genes. Yet GSEAs revealed that loss of Gfi-1b was associated with a significant increase in up-regulated genes occupied together with Gata1 or Ldb1 (Fig. 8 F). We speculate that the combined abnormal expression of a large number of lineage-associated genes in Gfi-1b’s absence prevents the execution of the complex programs required for the progression of erythroid maturation. The similarities in loss of function phenotypes associated with Gfi-1b, Gata1, and other major erythroid regulators are likely related to the large overlap in their target genes. Regardless, our data provide the first evidence for a broad, direct role of Gfi-1b in the regulation of a wide spectrum of erythroid genes.
Gfi-1b as lineage-restricted master repressor of transcription
Previous studies have established that Gfi-1b functions as a transcriptional repressor (Zweidler-Mckay et al., 1996; Tong et al., 1998). It physically interacts with and locally recruits chromatin-modifying enzymes that generate histone modifications associated with reduced transcription, including Lsd1, G9a, Hdac1, and Hdac2 (McGhee et al., 2003; Duan et al., 2005; Vassen et al., 2006; Saleque et al., 2007). Consistent with these insights into its molecular role, the majority of genes bound by Gfi-1b exhibited increased expression after its disruption in the erythroid and megakaryocytic lineages (Fig. 8 A). In contrast, Klf1 and the Ldb1-assocated pentameric complex predominantly activate gene expression (Soler et al., 2010; Tallack et al., 2012), whereas Gata1 activates and represses about half of its targets in unbiased genome-wide studies (Yu et al., 2009; Kerenyi and Orkin, 2010). The mechanism of Gata1’s repressive activity remains incompletely understood. At some Gata1 target genes, repression involves recruitment of the NuRD complex by Gata1’s cofactor Fog1, but at other Gata1 targets, Fog and NuRD mediate gene activation (Miccio et al., 2010). The presence of the Ldb1-associated pentameric complex strongly predicts activation of Gata1 target genes (Tripic et al., 2009; Yu et al., 2009). Gfi-1b copurifies with Gata1 in biochemical assays, raising the possibility that a physical interaction with Gfi-1b confers repressive activity to Gata1 (Rodriguez et al., 2005; Yu et al., 2009). Consistent with this notion, we demonstrated that a significant number of Gata1 target genes were up-regulated in Gfi-1b−/− progenitors (Fig. 8 F). However, Gfi-1b’s presence only weakly distinguished Gata1-repressed and -activated genes (not depicted). Moreover, Gfi-1b loss significantly correlated with the increased expression of the target genes it shares with Ldb1. Thus, Gfi-1b’s repressive activity is not limited to genes that are repressed by Gata1. The emerging picture is that the expression levels of lineage-restricted genes are determined by the balance of the activity of several multiprotein complexes organized by lineage-restricted transcription factors. Gfi-1b is the hallmark component of the first erythroid/megakaryocyte lineage–restricted complex with predominant repressive activity.
A potential role for Gfi-1b in gene activation might be inferred from our finding that a considerable minority of Gfi-1b–bound genes exhibit reduced expression after Gfi-1b loss (Fig. 8 A). We cannot exclude the possibility that Gfi-1b has unrecognized functions in enhancing gene expression. Although highly speculative, it is conceivable, for example, that the potential interaction with Gata1 may allow Gfi-1b to indirectly recruit factors with a positive impact on gene expression, such as Fog1/NuRD or CBP (Blobel et al., 1998; Tripic et al., 2009). Alternatively, in light of Gfi-1b’s large target gene spectrum, such ostensibly activated genes may be subject to indirect (trans) effects overriding the local repressive impact of Gfi-1b. We note that to date direct evidence for a role of Gfi-1b in activating genes has not been reported. In contrast, in vitro assays, the spectrum of molecules interacting with it, and the predominant impact on gene expression reported here all support its function as a transcriptional repressor.
Further investigations are needed to fully define Gfi-1b’s interplay with the other, more extensively studied lineage-restricted regulators in establishing and fine-tuning the expression levels of many megakaryocytic and erythroid genes. A complete understanding of the transcriptional programs of these vitally important lineages will require a detailed study of the dynamism of the transcription factor occupancy and activities as differentiation proceeds, and it will need to capture the influence of long-range interactions.
MATERIALS AND METHODS
Mice.
Mice were housed at Massachusetts General Hospital in a specific pathogen–free environment. All animal experiments were performed after approval and according to the guidelines and supervision of the Massachusetts General Hospital Subcommittee on Research Animal Care. B6.SJL (ref: 002014) recipient mice and Mx-Cre mice were purchased from The Jackson Laboratory. The R26-M2rtTA::TetOP-Cre mice (TetO-Cre) will be available from The Jackson Laboratory (JAX Stock# 021025). All animals were genotyped by Southern blot or by PCR using the HotStarTaq DNA polymerase (QIAGEN).
Construction of the conditional Gfi-1b targeting vector.
A 9.8-kb Gfi-1b genomic BsaB1–Sal1 fragment including exons 2–6 (Saleque et al., 2002) was subcloned into pBluescript II KS (pBS) vector. A selectable neomycin cassette containing the 5′ LoxP site and flanked by two FRT sites (Liu et al., 2003) was inserted into the Nde1 site of pBS–Gfi-1b. To introduce the 3′ LoxP site, an adapter containing the 5′ Hind3–EcoRV site (primers: sense, 5′-AAGCTTGATATCTAATATAACTTCGTATAATGTATGCTATACGAAGTTATTAG-3′; and antisense, 5′-CTAATAACTTCGTATAGCATACATTATACGAAGTTATATTAGATATCAAGCTT-3′) was inserted into the Swa1 site (between exons 5 and 6 of pBS–Gfi-1b). After sequencing, the vector was linearized with NotI for subsequent electroporation in embryonic stem (ES) cells (see below).
Generation of Gfi-1b–targeted ES cells and mutant mice.
V6.5 (129SvJae × C57BL/6; Eggan et al., 2001) ES cells were cultured on irradiated MEFs generated from DR4 embryos (Tucker et al., 1997) as described previously (Qin et al., 2010). Targeting vectors were introduced into ES cells by electroporation and selected with 400 µg/ml G418. Correctly targeted neomycin-positive ES cells were identified by Southern blot analysis (Fig. 1, A–C). Thereafter, the neomycin resistance cassette was removed by electroporation of the Flpe-expressing pCAGGS vector (Buchholz et al., 1998; Rodríguez et al., 2000), and subsequent puromycin (1 µg/ml) selection (Fig. 1, A and D) leading to the generation of heterozygous Gfi-1bwt/floxed (Gfi-1bwt/fl) ES cells. To create the Gfi-1b–null (Gfi-1bwt/ko) allele (Fig. 1 A, bottom), a plasmid expressing a GFP-Cre fusion protein (pBS500) was introduced by electroporation in targeted neomycin-positive ES cells (Gagneten et al., 1997). After 48 h, GFP-Cre–positive cells were FACS sorted, expanded in culture, and confirmed by Southern blot analysis (Fig. 1 E). Selected Gfi-1bwt/flox and Gfi-1bwt/ko ES cell clones were karyotyped, injected into C57BL/6 blastocysts, and transferred into pseudopregnant foster mothers to generate chimeras. Male chimeras were bred to C57BL/6 female mice for germ line transmission. Homozygous conditional Gfi-1b KO mice (Gfi-1bfl/fl) were generated and had normal viability, fertility, and blood parameters.
Generation of tetracycline-inducible Cre expression transgenic mice.
Tetracycline-inducible Cre expression (M2-rtTA; TetO-Cre) transgenic animals (Fig. 2) were generated from ES cells after targeting the Cre recombinase cDNA (pTurbo-Cre; GenBank accession no. AF334827.1) downstream of the collagen 1A1 locus under the control of a tetracycline-dependent CMV minimal promoter as previously described previously (Beard et al., 2006; Foudi et al., 2009).
Cre-mediated recombination in vivo.
For TetO-Cre–mediated gene recombination, mice harboring M2-rtTA and TetO-Cre as well as targeted Gfi-1b alleles (age 4–8 wk) were subjected to daily doxycycline injections (D9891; Sigma-Aldrich; 10 µg/g mouse weight; diluted 1 mg/ml in PBS) and doxycycline-supplemented drinking water (2 mg/ml doxycycline, 10 mg/ml sucrose) for up to 4 wk. For Mx-Cre–mediated gene recombination, a total of three intraperitoneal injections (at 8.5 µg/g mouse bodyweight) of pIpC (GE Healthcare) were administered every other day.
Bone marrow transplantation.
Competitive bone marrow transplantation was performed by intravenous injection of 4 × 106 total bone marrow cells from control or conditional Gfi-1b KO mice (CD45.2 positive) together with 2 × 106 competitor bone marrow cells (B6.SJL, CD45.1 positive) into B6.SJL recipients that had received 12 Gy of radiation as a split dose as described previously (Hock et al., 2004b).
Complete blood counts.
Analysis was performed using a VetScan MS5 hematology system (Abaxis).
Flow cytometry.
Antibody conjugates were purchased from BD or eBioscience, unless otherwise indicated. Data were collected using either a FACSAria II cell sorter or an LSR II (BD) and analyzed with FlowJo (Tree Star). Dead cells were excluded using 1 µg/ml propidium iodide; doublets were excluded using forward and side scatter width parameters. Antibodies used for bone marrow pre-MegE and MkPs (adapted from Pronk et al. [2007]) are as follows: lineage markers (CD4 [RM4-5], CD8 [53-6], CD3e [145-2C11], B220 [RA3-6B2], Gr-1 [RB6-8C5], CD127 [IL-7Ra, A7R34], and Ter119 [all PE-Cy5]), CD117 (c-Kit, 2B8, APC–eFluor 780), Sca-1 (D7, PE-Cy7), FcgRII/III (2.4G2, PE), CD150 (Slamf1, TC15-12F12.2, APC; BioLegend), CD41 (GPIIb, MWReg30, FITC), and CD105 (Endoglin, MJ7/18, eFluor 450). Antibodies used for bone marrow erythroid precursors (stain performed without red blood cell lysis) were rat anti–mouse CD71 (TFRC, RI7 217.1.4, PE) and rat anti–mouse Ter-119 (Ly76, TER-119, FITC). Antibodies used for bone marrow megakaryocytic precursors were rat anti–mouse CD41 (GPIIb, MWReg30, eFluor 450) and rat anti–mouse CD42b (GPIbalpha, Xia.G5, PE; emfret ANALYTICS). For megakaryocyte ploidy analysis, bone marrow cells were fixed in ice-cold 70% ethanol overnight and stained with 1 µg/ml DAPI (D1306; Invitrogen) for 30 min at room temperature in DAPI/Triton X-100 staining solution (0.1% Triton X-100 in PBS and 1 µg/ml DAPI) and protected from direct light exposure. Thereafter, cells were collected on an Aria II flow cytometer (BD) and analyzed using Kaluza analysis software (Beckman Coulter).
Quantitative imaging flow cytometry.
To assess the morphology and size of CD41+CD42+ megakaryocytes in vivo in transplanted animals, we used an Amnis ImageStream MkII imaging flow cytometer (EMD Millipore), which combines high-resolution microscopy and flow cytometry. In brief, bone marrow cells from Gfi-1b+/− and Gfi-1b−/− posttransplant recipients (7 wk after doxycycline) were immunostained for megakaryocytes as described above, together with a rat anti-CD45.2 FITC antibody (to distinguish conditional KO from competitor cells) and DRAQ5 (to label DNA). Data were collected on the Amnis imaging flow cytometer (105 events/file, 13 files/genotype were merged) and analyzed using IDEAS software (version 6.0). Cellular debris was excluded by gating on DRAQ5-positive events first, followed by CD41-positive/CD42-positive events. After excluding false-positive cellular aggregates, single CD45.2-positive and CD45.2-negative CD41+CD42+ cells were tagged, and their morphology was assessed. Tagged cells were then back-gated on the area of CD41 versus the area of CD42 bivariate plots for size quantification. Data were collected using a 40× objective (66 mm/s, 10-µm core diameter) and five lasers (405 nm, 20 mW; 488 nm, 50 mW; 561 nm, 100 mW; 642 nm, 150 mW; 785 nm, 1.56 mW for SSC). Single color controls were acquired without brightfield or side scatter, and spectral compensation was performed after acquisition.
Generation of expression vectors and mutagenesis.
Gfi-1b and Gfi-1 full-length cDNAs were amplified by PCR and modified to incorporate a C-terminal FLAG tag using the following primers: Gfi-1b sense (NotI), 5′-GCTAGCGGCCGCGCCGCCACCATGCCACGGTCCTTTCTAGTG-3′; and antisense (BamHI), 5′-AATTGGATCCTCACTTGTCGTCATCGTCTTTGTAGTCCTTGAGATTGTGTTGACTCTC-3′; and Gfi-1 sense (NotI), 5′-GCTAGCGGCCGCGCCGCCACCATGCCGCGCTCATTCCTGGTC-3′; and antisense (BamHI), 5′-AATTGGATCCTCACTTGTCGTCATCGTCTTTGTAGTCTTTGAGTCCATGCTGAGTCTC-3′. Two DNA-binding swap mutants were generated by PCR amplification, followed by directional ligation of Gfi-1b N terminus to Gfi-1 C terminus (Gfi-1bN1C) or by ligation of Gfi-1 N terminus to Gfi-1b C terminus (Gfi-1N1bC). Primers were as follows: for Gfi-1b N terminus sense (NotI), 5′-GCTAGCGGCCGCGCCGCCACCATGCCACGGTCCTTTCTAGTG-3′; and antisense, 5′-AGTGTCCATGCCTGGAGAGTA-3′; for Gfi-1b C terminus sense, 5′-TACCACTGTGTCAAGTGCAAC-3′; and antisense (BamHI), 5′-AATTGGATCCTCACTTGTCGTCATCGTCTTTGTAGTCCTTGAGATTGTGTTGACTCTC-3′; for Gfi-1 N terminus sense (NotI), 5′-GCTAGCGGCCGCGCCGCCACCATGCCGCGCTCATTCCTGGTC-3′; and antisense, 5′-GGAGCCGCCGCCCAGCAGCAG-3′; and for Gfi-1 C terminus sense, 5′-TACAAATGCATCAAATGCAGC-3′. DNA fragments were then cloned into the NotI–BamHI site of Phage2 lentivirus (Sommer et al., 2009) directly upstream of an Ires-GFP. All constructs were sequenced before generating vectors.
Lentiviral supernatant production and infection.
Lentiviral supernatants were prepared using 293T packaging cells as previously described (Qin et al., 2012). Aliquots of concentrated lentiviral supernatants were stored at −80°C until the day of infection. Sorted Gfi-1b−/− pre-MegEs and MkPs were suspended in DMEM, 10% FBS, and 1% penicillin-streptomycin containing 50 ng/ml SCF, 10 ng/ml IL-3, and 20 ng/ml TPO. Cells (105 to 5 × 105 in 500 µl) were incubated with 50 µl (diluted in 500 µl culture medium) concentrated lentiviral supernatant supplemented with 8 µg/ml polybrene (H9268, hexadimethrine bromide; Sigma-Aldrich) and centrifuged at 2,500 RPMs for 90 min at room temperature. Cells were washed in complete DMEM before being put in culture. Infected pre-MegEs were seeded at 103 cells/ml in semisolid medium for CFU-C assay, as described below. Infected MkPs were plated in megakaryocyte differentiation condition as described below.
Megakaryocytic differentiation and proplatelet formation.
Infected Gfi-1b−/− MkPs were suspended in DMEM, 10% FBS, and 1% penicillin-streptomycin containing 25 ng/ml SCF and 10 ng/ml TPO and seeded onto fibronectin-coated (40 µg/ml; EMD Millipore) wells. Cells were cultured at 37°C in 5% CO2 for 8 d and monitored with a fluorescence microscope (DMI4000B; Leica) for the emergence of GFP-positive megakaryocytes and proplatelet-forming megakaryocytes.
CFU-C assays from pre-MegEs and MkPs.
Sorted pre-MegEs and MkPs were seeded at 500 cells/dish of 35-mm colony dishes (STEMCELL Technologies) in Methocult M3234 (STEMCELL Technologies) supplemented with 25 ng/ml SCF, 10 ng/ml IL-3, 20 ng/ml IL-11, 10 ng/ml GM-CSF, 3 U/ml Epo, and 10 ng/ml Tpo. Colonies were scored between 7 and 10 d using an inverted microscope (DMI 4000B; Leica).
Cytology and histochemistry.
For whole bone marrow sections, tissues were fixed in Bouin’s solution and embedded in paraffin. Embedded bone marrow blocks were sliced and stained with H&E for conventional morphological assessment. For cytocentrifugation preparations, bone marrow cells were subjected to red blood lysis in ACK buffer (0.15 mM NH4Cl, 1 mM KHCO3, and 0.1 mM Na2EDTA). Between 2 × 105 and 5 × 105 cells were suspended in 100 µl PBS, loaded onto disposable Cytofunnel chambers and spun down for 4 min at 400 RPM using a CytoSpin III cytocentrifuge (Shandon). For MGG stain, cytocentrifuged samples were stained in May-Grünwald solution (MG500; Sigma-Aldrich) for 2 min and in Giemsa (GS500, 5% solution in water; Sigma-Aldrich) for 12 min, followed by two washes in distilled water for 30 s. For benzidine stain, hematopoietic cells (2 × 105 to 5 × 105) were suspended in 200 µl PBS and mixed with 20 µl O-dianisidine/H2O2 mixture for 8 min at room temperature (9 vol of 0.2% o-Dianisidine [D9143] in 10% glacial acetic acid [Sigma-Aldrich] and 1 vol of 30% H2O2 [H325; Thermo Fisher Scientific]).Thereafter, samples were cytocentrifuged, counterstained with May-Grünwald solution for 10 s, and washed in water. For AchE stain, samples were cytocentrifuged and stained using the AchE rapid staining kit (MBL International) according to the manufacturer’s guidelines. Slides were coverslipped using Permount and analyzed by brightfield microscopy.
RNA extraction and quantitative RT-PCR.
Bone marrow pre-MegE and MkPs from a cohort of n = 3 Gfi-1b+/+ and n = 3 Gfi-1b−/− (15–19 d after doxycycline, number of cells ranging from 2 × 104 to 5 × 104 per sample) were double FACS sorted to achieve >99% purity by FACS and suspended in 500 µl TRIzol (Invitrogen). Total RNA was isolated according to the manufacturer’s recommendations (Invitrogen) and purified using the RNeasy Micro kit (QIAGEN). All RNA samples had their integrity checked using an RNA 6000 Pico kit on a 2100 Bioanalyzer (Agilent Technologies) and RIN numbers ranged from 8 to 10. RT was performed using iScript cDNA synthesis kit (Bio-Rad Laboratories). Quantitative RT-PCR was performed using either Brilliant II SYBR green QPCR master mix (Agilent Technologies) or SYBR green I master (Roche) and run on a Stratagene Mx3005P (Agilent Technologies) or LightCycler 480 (Roche), respectively. All reactions were performed in triplicate, and data were generated using the comparative Ct method using actin-b as reference gene (2−ΔΔCt; ΔΔCt = ΔCtsample − ΔCtreference). Primers were designed to span one intron on the corresponding genomic DNA sequence (Tables S4 and S5).
Microarrays and GSEA.
Microarrays were performed at Partners Center for Personalized Medicine (Boston, MA). In brief, 2 ng total RNA (RIN > 8) from control and Gfi-1b−/− pre-MegEs and MkPs were amplified with an Ovation Pico WTA system (NuGEN), reverse transcribed to cDNA with the WT-Ovation Exon Module, and prepared for fragmentation and labeling using the Encore Biotin Module (NuGEN) according to the manufacturer’s protocol. Amplified cDNA were biotinylated, fragmented, and hybridized to Mouse GeneChip 1.0 ST arrays (Affymetrix). The stained arrays were scanned at 532 nm using a GeneChip Scanner 3000 (Affymetrix). The GeneSifter microarray analysis server (Geospiza) was used to generate lists of deregulated genes with >1.5-fold change in expression (Tables S1 and S2). To test whether defined sets of genes show statistically significant and concordant differences between control and Gfi-1b−/− pre-MegEs and between control and Gfi-1b−/− MkPs, we performed GSEA (Mootha et al., 2003; Subramanian et al., 2005) using previously published datasets (Table S3). Each GSEA was run using “Gene Sets” as permutation type with “1,000 permutations.” A normalized enrichment score (NES) is determined and reflects whether genes within a defined dataset are either randomly distributed (NES value close to 0) or primarily found at the top (NES value > 1) or bottom (NES value < −1) of the ranked gene list. GSEA also provides a false discovery rate corresponding to the probability (q-value) of each NES to be a false positive.
Statistical analysis.
Statistical significance was determined by two-tailed Student’s t test. Kaplan–Meyer survival curves were generated using Prism software (version 6; GraphPad Software). Statistical significance of the survival distributions was determined by Log-rank test (Mantel-Cox).
Online supplemental material.
Tables S1 and S2, included as separate Excel files, show lists of deregulated genes after Gfi-1b loss in pre-MegEs and MkPs, respectively. Table S3, included as a separate Excel file, shows gene sets used in genomics analyses. Tables S4 and S5 list primers used for quantitative RT-PCR and genotyping PCR, respectively. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20131065/DC1.
Supplementary Material
Acknowledgments
We thank Laura Prickett-Rice, Meredith Weglarz, and Kat Folz-Donahue from the Harvard Stem Cell Institute Flow Cytometry core at Massachusetts General Hospital. Denille van Buren provided expert technical assistance. We thank William Vainchenker for helpful advice.
This work was supported by National Institutes of Health (NIH) grant R01CA122726 (to H. Hock), the Federal Share of the Program Income earned by the Massachusetts General Hospital on grant C06 CA599267 (to H. Hock), NIH grant R01HL075735 (to S.H. Orkin), NIH grant R01DK096034-01 (to K. Hochedlinger), and NIH grant S10OD012027 (to F.I. Preffer). H. Hock was the recipient of a bridge grant by the American Society of Hematology. J. Qin was the recipient of NIH grant T32 2012D006265 and the Massachusetts General Hospital Executive Committee on Research (MGH ECOR) Fund for Medical Discovery award. A. Foudi was supported by the Deutsche José Carreras Leukämie-Stiftung e. V. (FIJC-09/ED THOMAS fellowship F 09/01), MGH ECOR Fund for Medical Discovery award, and Lady Tata Memorial Trust award.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AchE
- acetylcholinesterase
- ES
- embryonic stem
- GSEA
- gene set enrichment analysis
- HSC
- hematopoietic stem cell
- MGG
- May-Grünwald Giemsa
- MkP
- megakaryocytic progenitor
- NES
- normalized enrichment score
- pIpC
- polyinosinic:polycytidylic acid
- pre-MegE
- progenitor of megakaryocytes and erythrocytes
References
- Akashi K., Traver D., Miyamoto T., Weissman I.L. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197 10.1038/35004599 [DOI] [PubMed] [Google Scholar]
- Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R. 2006. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 44:23–28 10.1002/gene.20180 [DOI] [PubMed] [Google Scholar]
- Blobel G.A., Nakajima T., Eckner R., Montminy M., Orkin S.H. 1998. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl. Acad. Sci. USA. 95:2061–2066 10.1073/pnas.95.5.2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F., Angrand P.O., Stewart A.F. 1998. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 16:657–662 10.1038/nbt0798-657 [DOI] [PubMed] [Google Scholar]
- Capron C., Lacout C., Lécluse Y., Wagner-Ballon O., Kaushik A.L., Cramer-Bordé E., Sablitzky F., Duménil D., Vainchenker W. 2011. LYL-1 deficiency induces a stress erythropoiesis. Exp. Hematol. 39:629–642 10.1016/j.exphem.2011.02.014 [DOI] [PubMed] [Google Scholar]
- Chagraoui H., Kassouf M., Banerjee S., Goardon N., Clark K., Atzberger A., Pearce A.C., Skoda R.C., Ferguson D.J., Watson S.P., et al. 2011. SCL-mediated regulation of the cell-cycle regulator p21 is critical for murine megakaryopoiesis. Blood. 118:723–735 10.1182/blood-2011-01-328765 [DOI] [PubMed] [Google Scholar]
- Chang Y., Bluteau D., Debili N., Vainchenker W. 2007. From hematopoietic stem cells to platelets. J. Thromb. Haemost. 5:318–327 10.1111/j.1538-7836.2007.02472.x [DOI] [PubMed] [Google Scholar]
- Chen C., Fuhrken P.G., Huang L.T., Apostolidis P., Wang M., Paredes C.J., Miller W.M., Papoutsakis E.T. 2007. A systems-biology analysis of isogenic megakaryocytic and granulocytic cultures identifies new molecular components of megakaryocytic apoptosis. BMC Genomics. 8:384 10.1186/1471-2164-8-384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan L.L., Porter S.D., Duan Z., Flubacher M.M., Montoya D., Tsichlis P.N., Horwitz M., Gilks C.B., Grimes H.L. 2004. Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res. 32:2508–2519 10.1093/nar/gkh570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z., Zarebski A., Montoya-Durango D., Grimes H.L., Horwitz M. 2005. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell. Biol. 25:10338–10351 10.1128/MCB.25.23.10338-10351.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan K., Akutsu H., Loring J., Jackson-Grusby L., Klemm M., Rideout W.M., III, Yanagimachi R., Jaenisch R. 2001. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl. Acad. Sci. USA. 98:6209–6214 10.1073/pnas.101118898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Omari K., Hoosdally S.J., Tuladhar K., Karia D., Hall-Ponselé E., Platonova O., Vyas P., Patient R., Porcher C., Mancini E.J. 2013. Structural basis for LMO2-driven recruitment of the SCL:E47bHLH heterodimer to hematopoietic-specific transcriptional targets. Cell Rep. 4:135–147 10.1016/j.celrep.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., Trumpp A. 2009. IFNα activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Fiolka K., Hertzano R., Vassen L., Zeng H., Hermesh O., Avraham K.B., Dührsen U., Möröy T. 2006. Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep. 7:326–333 10.1038/sj.embor.7400618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foudi A., Hochedlinger K., Van Buren D., Schindler J.W., Jaenisch R., Carey V., Hock H. 2009. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27:84–90 10.1038/nbt.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneten S., Le Y., Miller J., Sauer B. 1997. Brief expression of a GFP cre fusion gene in embryonic stem cells allows rapid retrieval of site-specific genomic deletions. Nucleic Acids Res. 25:3326–3331 10.1093/nar/25.16.3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garçon L., Lacout C., Svinartchouk F., Le Couédic J.P., Villeval J.L., Vainchenker W., Duménil D. 2005. Gfi-1B plays a critical role in terminal differentiation of normal and transformed erythroid progenitor cells. Blood. 105:1448–1455 10.1182/blood-2003-11-4068 [DOI] [PubMed] [Google Scholar]
- Gutiérrez L., Tsukamoto S., Suzuki M., Yamamoto-Mukai H., Yamamoto M., Philipsen S., Ohneda K. 2008. Ablation of Gata1 in adult mice results in aplastic crisis, revealing its essential role in steady-state and stress erythropoiesis. Blood. 111:4375–4385 10.1182/blood-2007-09-115121 [DOI] [PubMed] [Google Scholar]
- Hall M.A., Slater N.J., Begley C.G., Salmon J.M., Van Stekelenburg L.J., McCormack M.P., Jane S.M., Curtis D.J. 2005. Functional but abnormal adult erythropoiesis in the absence of the stem cell leukemia gene. Mol. Cell. Biol. 25:6355–6362 10.1128/MCB.25.15.6355-6362.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattangadi S.M., Wong P., Zhang L., Flygare J., Lodish H.F. 2011. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood. 118:6258–6268 10.1182/blood-2011-07-356006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H., Orkin S.H. 2006. Zinc-finger transcription factor Gfi-1: versatile regulator of lymphocytes, neutrophils and hematopoietic stem cells. Curr. Opin. Hematol. 13:1–6 10.1097/01.moh.0000190111.85284.8f [DOI] [PubMed] [Google Scholar]
- Hock H., Hamblen M.J., Rooke H.M., Schindler J.W., Saleque S., Fujiwara Y., Orkin S.H. 2004a. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 431:1002–1007 10.1038/nature02994 [DOI] [PubMed] [Google Scholar]
- Hock H., Meade E., Medeiros S., Schindler J.W., Valk P.J., Fujiwara Y., Orkin S.H. 2004b. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 18:2336–2341 10.1101/gad.1239604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Dore L.C., Li Z., Orkin S.H., Feng G., Lin S., Crispino J.D. 2009. GATA-2 reinforces megakaryocyte development in the absence of GATA-1. Mol. Cell. Biol. 29:5168–5180 10.1128/MCB.00482-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushansky K. 2008. Historical review: megakaryopoiesis and thrombopoiesis. Blood. 111:981–986 10.1182/blood-2007-05-088500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerenyi M.A., Orkin S.H. 2010. Networking erythropoiesis. J. Exp. Med. 207:2537–2541 10.1084/jem.20102260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandanpour C., Sharif-Askari E., Vassen L., Gaudreau M.C., Zhu J., Paul W.E., Okayama T., Kosan C., Möröy T. 2010. Evidence that growth factor independence 1b regulates dormancy and peripheral blood mobilization of hematopoietic stem cells. Blood. 116:5149–5161 10.1182/blood-2010-04-280305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn R., Schwenk F., Aguet M., Rajewsky K. 1995. Inducible gene targeting in mice. Science. 269:1427–1429 10.1126/science.7660125 [DOI] [PubMed] [Google Scholar]
- Lecine P., Villeval J.L., Vyas P., Swencki B., Xu Y., Shivdasani R.A. 1998. Mice lacking transcription factor NF-E2 provide in vivo validation of the proplatelet model of thrombocytopoiesis and show a platelet production defect that is intrinsic to megakaryocytes. Blood. 92:1608–1616 [PubMed] [Google Scholar]
- Li L., Lee J.Y., Gross J., Song S.H., Dean A., Love P.E. 2010. A requirement for Lim domain binding protein 1 in erythropoiesis. J. Exp. Med. 207:2543–2550 10.1084/jem.20100504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Jothi R., Cui K., Lee J.Y., Cohen T., Gorivodsky M., Tzchori I., Zhao Y., Hayes S.M., Bresnick E.H., et al. 2011. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat. Immunol. 12:129–136 10.1038/ni.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Freudenberg J., Cui K., Dale R., Song S.H., Dean A., Zhao K., Jothi R., Love P.E. 2013. Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation. Blood. 121:4575–4585 10.1182/blood-2013-01-479451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Jenkins N.A., Copeland N.G. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13:476–484 10.1101/gr.749203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M.W., Williams N. 1981. Immature megakaryocytes in the mouse: Morphology and quantitation by acetylcholinesterase staining. Blood. 58:1032–1039 [PubMed] [Google Scholar]
- Machlus K.R., Italiano J.E., Jr 2013. The incredible journey: From megakaryocyte development to platelet formation. J. Cell Biol. 201:785–796 10.1083/jcb.201304054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini E., Sanjuan-Pla A., Luciani L., Moore S., Grover A., Zay A., Rasmussen K.D., Luc S., Bilbao D., O’Carroll D., et al. 2012. FOG-1 and GATA-1 act sequentially to specify definitive megakaryocytic and erythroid progenitors. EMBO J. 31:351–365 10.1038/emboj.2011.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May G., Soneji S., Tipping A.J., Teles J., McGowan S.J., Wu M., Guo Y., Fugazza C., Brown J., Karlsson G., et al. 2013. Dynamic analysis of gene expression and genome-wide transcription factor binding during lineage specification of multipotent progenitors. Cell Stem Cell. 13:754–768 10.1016/j.stem.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack M.P., Hall M.A., Schoenwaelder S.M., Zhao Q., Ellis S., Prentice J.A., Clarke A.J., Slater N.J., Salmon J.M., Jackson S.P., et al. 2006. A critical role for the transcription factor Scl in platelet production during stress thrombopoiesis. Blood. 108:2248–2256 10.1182/blood-2006-02-002188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee L., Bryan J., Elliott L., Grimes H.L., Kazanjian A., Davis J.N., Meyers S. 2003. Gfi-1 attaches to the nuclear matrix, associates with ETO (MTG8) and histone deacetylase proteins, and represses transcription using a TSA-sensitive mechanism. J. Cell. Biochem. 89:1005–1018 10.1002/jcb.10548 [DOI] [PubMed] [Google Scholar]
- Miccio A., Wang Y., Hong W., Gregory G.D., Wang H., Yu X., Choi J.K., Shelat S., Tong W., Poncz M., Blobel G.A. 2010. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J. 29:442–456 10.1038/emboj.2009.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola H.K., Klintman J., Yang H., Hock H., Schlaeger T.M., Fujiwara Y., Orkin S.H. 2003. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature. 421:547–551 10.1038/nature01345 [DOI] [PubMed] [Google Scholar]
- Moignard V., Macaulay I.C., Swiers G., Buettner F., Schütte J., Calero-Nieto F.J., Kinston S., Joshi A., Hannah R., Theis F.J., et al. 2013. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat. Cell Biol. 15:363–372 10.1038/ncb2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteferrario D., Bolar N.A., Marneth A.E., Hebeda K.M., Bergevoet S.M., Veenstra H., Laros-van Gorkom B.A., MacKenzie M.A., Khandanpour C., Botezatu L., et al. 2014. A dominant-negative GFI1B mutation in the gray platelet syndrome. N. Engl. J. Med. 370:245–253 10.1056/NEJMoa1308130 [DOI] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Osawa M., Yamaguchi T., Nakamura Y., Kaneko S., Onodera M., Sawada K., Jegalian A., Wu H., Nakauchi H., Iwama A. 2002. Erythroid expansion mediated by the Gfi-1B zinc finger protein: role in normal hematopoiesis. Blood. 100:2769–2777 10.1182/blood-2002-01-0182 [DOI] [PubMed] [Google Scholar]
- Pevny L., Simon M.C., Robertson E., Klein W.H., Tsai S.F., D’Agati V., Orkin S.H., Costantini F. 1991. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 349:257–260 10.1038/349257a0 [DOI] [PubMed] [Google Scholar]
- Pilon A.M., Arcasoy M.O., Dressman H.K., Vayda S.E., Maksimova Y.D., Sangerman J.I., Gallagher P.G., Bodine D.M. 2008. Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol. Cell. Biol. 28:7394–7401 10.1128/MCB.01087-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher C., Swat W., Rockwell K., Fujiwara Y., Alt F.W., Orkin S.H. 1996. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 86:47–57 10.1016/S0092-8674(00)80076-8 [DOI] [PubMed] [Google Scholar]
- Pronk C.J., Rossi D.J., Månsson R., Attema J.L., Norddahl G.L., Chan C.K., Sigvardsson M., Weissman I.L., Bryder D. 2007. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 1:428–442 10.1016/j.stem.2007.07.005 [DOI] [PubMed] [Google Scholar]
- Qin J., Van Buren D., Huang H.S., Zhong L., Mostoslavsky R., Akbarian S., Hock H. 2010. Chromatin protein L3MBTL1 is dispensable for development and tumor suppression in mice. J. Biol. Chem. 285:27767–27775 10.1074/jbc.M110.115410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Whyte W.A., Anderssen E., Apostolou E., Chen H.H., Akbarian S., Bronson R.T., Hochedlinger K., Ramaswamy S., Young R.A., Hock H. 2012. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell. 11:319–332 10.1016/j.stem.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randrianarison-Huetz V., Laurent B., Bardet V., Blobe G.C., Huetz F., Duménil D. 2010. Gfi-1B controls human erythroid and megakaryocytic differentiation by regulating TGF-β signaling at the bipotent erythro-megakaryocytic progenitor stage. Blood. 115:2784–2795 10.1182/blood-2009-09-241752 [DOI] [PubMed] [Google Scholar]
- Rodríguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- Rodriguez P., Bonte E., Krijgsveld J., Kolodziej K.E., Guyot B., Heck A.J., Vyas P., de Boer E., Grosveld F., Strouboulis J. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 24:2354–2366 10.1038/sj.emboj.7600702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S., Cameron S., Orkin S.H. 2002. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 16:301–306 10.1101/gad.959102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S., Kim J., Rooke H.M., Orkin S.H. 2007. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell. 27:562–572 10.1016/j.molcel.2007.06.039 [DOI] [PubMed] [Google Scholar]
- Schulze H., Shivdasani R.A. 2005. Mechanisms of thrombopoiesis. J. Thromb. Haemost. 3:1717–1724 10.1111/j.1538-7836.2005.01426.x [DOI] [PubMed] [Google Scholar]
- Shivdasani R.A., Rosenblatt M.F., Zucker-Franklin D., Jackson C.W., Hunt P., Saris C.J., Orkin S.H. 1995. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell. 81:695–704 10.1016/0092-8674(95)90531-6 [DOI] [PubMed] [Google Scholar]
- Soler E., Andrieu-Soler C., de Boer E., Bryne J.C., Thongjuea S., Stadhouders R., Palstra R.J., Stevens M., Kockx C., van Ijcken W., et al. 2010. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 24:277–289 10.1101/gad.551810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C.A., Stadtfeld M., Murphy G.J., Hochedlinger K., Kotton D.N., Mostoslavsky G. 2009. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 27:543–549 10.1634/stemcells.2008-1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souroullas G.P., Salmon J.M., Sablitzky F., Curtis D.J., Goodell M.A. 2009. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell. 4:180–186 10.1016/j.stem.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck J., Cohet N., Gonnet C., Sarrazin S., Doubeikovskaia Z., Doubeikovski A., Verger A., Duterque-Coquillaud M., Morle F. 2003. Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol. Cell. Biol. 23:1390–1402 10.1128/MCB.23.4.1390-1402.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck J., Weiss-Gayet M., Gonnet C., Guyot B., Vicat J.M., Morlé F. 2010. Inducible Fli-1 gene deletion in adult mice modifies several myeloid lineage commitment decisions and accelerates proliferation arrest and terminal erythrocytic differentiation. Blood. 116:4795–4805 10.1182/blood-2010-02-270405 [DOI] [PubMed] [Google Scholar]
- Stevenson W.S., Morel-Kopp M.C., Chen Q., Liang H.P., Bromhead C.J., Wright S., Turakulov R., Ng A.P., Roberts A.W., Bahlo M., Ward C.M. 2013. GFI1B mutation causes a bleeding disorder with abnormal platelet function. J. Thromb. Haemost. 11:2039–2047 10.1111/jth.12368 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallack M.R., Magor G.W., Dartigues B., Sun L., Huang S., Fittock J.M., Fry S.V., Glazov E.A., Bailey T.L., Perkins A.C. 2012. Novel roles for KLF1 in erythropoiesis revealed by mRNA-seq. Genome Res. 22:2385–2398 10.1101/gr.135707.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijssen M.R., Ghevaert C. 2013. Transcription factors in late megakaryopoiesis and related platelet disorders. J. Thromb. Haemost. 11:593–604 10.1111/jth.12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong B., Grimes H.L., Yang T.Y., Bear S.E., Qin Z., Du K., El-Deiry W.S., Tsichlis P.N. 1998. The Gfi-1B proto-oncoprotein represses p21WAF1 and inhibits myeloid cell differentiation. Mol. Cell. Biol. 18:2462–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripic T., Deng W., Cheng Y., Zhang Y., Vakoc C.R., Gregory G.D., Hardison R.C., Blobel G.A. 2009. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 113:2191–2201 10.1182/blood-2008-07-169417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang A.P., Fujiwara Y., Hom D.B., Orkin S.H. 1998. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 12:1176–1188 10.1101/gad.12.8.1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker K.L., Wang Y., Dausman J., Jaenisch R. 1997. A transgenic mouse strain expressing four drug-selectable marker genes. Nucleic Acids Res. 25:3745–3746 10.1093/nar/25.18.3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassen L., Fiolka K., Mahlmann S., Möröy T. 2005. Direct transcriptional repression of the genes encoding the zinc-finger proteins Gfi1b and Gfi1 by Gfi1b. Nucleic Acids Res. 33:987–998 10.1093/nar/gki243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassen L., Fiolka K., Möröy T. 2006. Gfi1b alters histone methylation at target gene promoters and sites of γ-satellite containing heterochromatin. EMBO J. 25:2409–2419 10.1038/sj.emboj.7601124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas P., Ault K., Jackson C.W., Orkin S.H., Shivdasani R.A. 1999. Consequences of GATA-1 deficiency in megakaryocytes and platelets. Blood. 93:2867–2875 [PubMed] [Google Scholar]
- Wadman I.A., Osada H., Grütz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145–3157 10.1093/emboj/16.11.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren A.J., Colledge W.H., Carlton M.B., Evans M.J., Smith A.J., Rabbitts T.H. 1994. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 78:45–57 10.1016/0092-8674(94)90571-1 [DOI] [PubMed] [Google Scholar]
- Wilson N.K., Foster S.D., Wang X., Knezevic K., Schütte J., Kaimakis P., Chilarska P.M., Kinston S., Ouwehand W.H., Dzierzak E., et al. 2010. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 7:532–544 10.1016/j.stem.2010.07.016 [DOI] [PubMed] [Google Scholar]
- Yien Y.Y., Bieker J.J. 2013. EKLF/KLF1, a tissue-restricted integrator of transcriptional control, chromatin remodeling, and lineage determination. Mol. Cell. Biol. 33:4–13 10.1128/MCB.01058-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Riva L., Xie H., Schindler Y., Moran T.B., Cheng Y., Yu D., Hardison R., Weiss M.J., Orkin S.H., et al. 2009. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell. 36:682–695 10.1016/j.molcel.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweidler-Mckay P.A., Grimes H.L., Flubacher M.M., Tsichlis P.N. 1996. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 16:4024–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.