Liver biopsies from patients with chronic hepatitis C reveal high IFN-λR1 expression, which can be induced with IFN-α and is associated with elevated ISG expression, the IFN-λ3 minor alleles, and nonresponsiveness to peg-IFN-α and ribavirin therapy.
Abstract
The molecular mechanisms that link IFN-λ3 genotypes to differential induction of interferon (IFN)-stimulated genes (ISGs) in the liver of patients with chronic hepatitis C (CHC) are not known. We measured the expression of IFN-λ and of the specific IFN-λ receptor chain (IFN-λR1) in 122 liver biopsies of patients with CHC and 53 control samples. The IFN-λ3 genotype was not associated with differential expression of IFN-λ, but rather IFN-λR1. In a series of 30 primary human hepatocyte (PHH) samples, IFN-λR1 expression was low but could be induced with IFN-α. IFN-α–induced IFN-λR1 expression was significantly stronger in PHHs carrying the minor IFN-λ3 allele. The analysis of liver biopsies of patients with CHC revealed a strong association of high IFN-λR1 expression with elevated ISG expression, with IFN-λ3 minor alleles, and with nonresponse to pegylated IFN-α and ribavirin. The findings provide a missing link between the IFN-λ3 genotype and the associated phenotype of treatment nonresponse.
Hepatitis C virus (HCV) infects an estimated 130–170 million persons worldwide. Chronic hepatitis C (CHC) can lead to cirrhosis and hepatocellular carcinoma (Lauer and Walker, 2001). The treatment for CHC consists of pegylated IFN (peg-IFN)-α and ribavirin, complemented with the HCV protease inhibitors telaprevir or boceprevir for HCV genotype 1 (Ghany et al., 2011). A substantial proportion of patients do not respond to this treatment. The virological response to peg-IFN-α depends on the degree of activation of the endogenous IFN system in the liver (Chen et al., 2005; Asselah et al., 2008; Sarasin-Filipowicz et al., 2008; Dill et al., 2011). Paradoxically, patients with a strong host response to HCV infection whose immune system reacts with a sustained induction of hundreds of IFN-stimulated genes (ISGs) in the liver, in the following designated preactivated patients, are poor responders to peg-IFN-α treatments. Nonresponse to peg-IFN-α in preactivated patients is most likely the consequence of the permanent up-regulation of USP18, an inhibitor of IFN-α signaling through the Jak–STAT pathways (Malakhova et al., 2006; Sarasin-Filipowicz et al., 2009; Dill et al., 2012).
The molecular mechanisms responsible for sustained ISG induction in the liver are unknown. The presence of a T cell infiltrate in the liver of CHC patients suggests that IFN-γ could stimulate ISG induction. However, several groups have reported that the CD8+ T cell response is dysfunctional in regard to IFN-γ secretion (Thimme et al., 2012). Indeed, gene set enrichment analysis revealed a significant enrichment of IFN-α–induced genes in liver biopsies of preactivated patients with CHC, contrary to the late phase of acute hepatitis C that is characterized by a strong enrichment of IFN-γ–induced ISGs (Dill et al., 2012). However, IFN-αs are probably not the drivers of ISG induction because IFN-α signaling is subject to a strong negative feedback inhibition by USP18 that would prevent long-lasting activation of ISGs (Malakhova et al., 2006; Sarasin-Filipowicz et al., 2009). Interestingly, IFN-β signaling is not affected by USP18 to the same extent as IFN-α, despite the fact that both IFN-α and IFN-β belong to the type I IFN family and bind to and signal through the same receptor, IFNAR (Makowska et al., 2011). IFN-β produced by HCV-infected hepatocytes therefore remains a serious contender for being the driver of ISG expression in CHC. However, neither type I nor type II IFNs have been consistently detected in liver biopsies of preactivated patients with CHC.
The discovery, in 2009, that allelic variants near the IFN-λ3 (IL28B) gene are strongly associated with spontaneous clearance of HCV and with response to treatment with peg-IFN-α and ribavirin provided compelling evidence that the type III IFN system is a key component of the host reaction to the virus (Ge et al., 2009; Suppiah et al., 2009; Tanaka et al., 2009; Thomas et al., 2009; Rauch et al., 2010). The molecular mechanisms that link the allelic variants of the IFN-λ3 gene locus to nonresponse to peg-IFN-α and ribavirin are not fully understood, but the significant association of the minor alleles of the IFN-λ3 polymorphism with the induction of ISGs in the liver demonstrates that the IFN-λ system must be critically involved in regulating the innate immune response to HCV (Honda et al., 2010; Urban et al., 2010). More recently, a novel TT/-G polymorphism near the IFN-λ3 gene has been described that is even more strongly associated with spontaneous clearance and treatment response to peg-IFN-α and ribavirin in CHC (Bibert et al., 2013; Prokunina-Olsson et al., 2013). The frameshift variant -G creates a novel 179-aa open reading frame that was designated IFN-λ4 on the basis of its protein sequence homology with type III IFNs (Prokunina-Olsson et al., 2013). Recombinant IFN-λ4 signals through the IFN-λ receptor and has antiviral activity against HCV and coronaviruses in cell culture (Hamming et al., 2013). It is presently not known whether IFN-λ4 is actually produced and functionally important in the HCV-infected human liver. Interestingly, the newly discovered TT/-G polymorphism impacts IFN-λ production in PBMCs. Treatment of PBMCs from patients with at least one -G allelic variant with poly(I:C) resulted in significant lower induction of IFN-λ3 compared with PBMCs from patients homozygous for the major TT allele (Bibert et al., 2013). How lower amounts of IFN-λ3 can be linked to increased expression of ISGs in the liver or how IFN-λ4 could cause a long-lasting ISG induction is presently unknown, but these exciting new findings further point to an important role of the IFN-λ gene locus in the control of the host response to HCV. We therefore analyzed the expression of IFN-λs and their receptor IFN-λR1 (IL28Rα) in liver biopsies and primary human hepatocytes (PHHs) from patients with different IFN-λ3 genotypes.
We found a very low expression of IFN-λR1 in PHHs and in liver biopsies from patients who were not infected with HCV. In PHHs, IFN-λR1 expression was induced by IFN-α, leading to substantially improved responses of the Jak–STAT signaling pathway to IFN-λ. The magnitude of IFN-α–induced IFN-λR1 expression was significantly associated with the IFN-λ3 genotype. IFN-λR1 expression was significantly higher in liver biopsies from patients with CHC compared with uninfected controls. Within the group of patients with CHC, IFN-λR1 expression significantly correlated with the IFN-λ3 genotype. We conclude that the IFN-λ3 genotype affects hepatic ISG expression not through differential expression of IFN-λ3, but through differential expression of its receptor IFN-λR1. Moreover, ex vivo treatment of liver biopsies from patients with high-level expression of IFN-λR1 showed strong responses to IFN-λ and poor responses to IFN-α, whereas the contrary was found in patients with low-level expression of IFN-λR1. These findings provide a rationale for treating peg-IFN-α nonresponder patients with peg-IFN-λ.
RESULTS
Inducible expression of IFN-λ1, IFN–λ2/3, and IFN-λR1 in human liver and PHHs
The strong association of minor alleles of the IFN-λ3 polymorphism with HCV persistence and poor response to peg-IFN-α and ribavirin treatment provides compelling evidence for an important role of the IFN-λ system in the host reaction to HCV. We therefore measured the expression of IFN-λ1, IFN-λ2/3, and IFN-λR1 by quantitative PCR (qPCR) in liver biopsies. Because of the high sequence similarity of 97%, IFN-λ2 and IFN-λ3 mRNA were quantified together using one primer pair that binds to both IFN-λ2 and IFN-λ3. IFN-λ1 mRNA was detectable in 33 and IFN-λ2/3 mRNA in 24 of 53 CHC samples tested (Fig. 1 A). We could not detect IFN-λ1 or IFN-λ2/3 in 22 control biopsies from patients without CHC. Interestingly, IFN-λ1 and IFN-λ2/3 mRNA were detectable in 3 and in 20 of 31 samples from patients with CHB, respectively (Fig. 1 A).
Figure 1.
Expression of IFN-λ1, IFN-λ2/3, and IFN-λR1 in human liver. (A) The expression of IFN-λ1, IFN-λ2/3, and IFN-λR1 was measured in liver biopsy extracts from CHC (n = 53 for IFN-λ1 and IFN-λ2/3; n = 122 for IFN-λR1), non-CHC (n = 22 for IFN-λ1 and IFN-λ2/3; n = 53 for IFN-λR1), and HBV (n = 31) samples by qPCR. Red lines show the statistical median of the individual data points. Statistical analysis was performed using the nonparametric Mann–Whitney test: ***, P < 0.001. (B) IFN-λR1 mRNA was visualized by ISH in fresh-frozen liver biopsy sections using a specific probe coupled with alkaline phosphatase and then revealed by Fast Red substrate (excitation 530 nm, emission 590 nm). Signals (red dots) are predominantly located in hepatocytes. Some signals are localized close to sinusoids. Nuclei are stained with DAPI (blue). Shown are representative images from biopsies A795, A909, A622, and A606. The IFN-λ3 rs12979860 genotype is indicated below the biopsy number. Pictures were obtained from two CHC patients homozygous for the IFN-λ3 rs12979860 C allele (A795 and A909) and two CHC patients carrying one or two T alleles (A622 and A606). Shown are representative images from three sections for each patient. Bar, 20 µm.
IFN-λR1 mRNA was measured in 53 controls, 122 samples from patients with CHC, and 31 samples from patients with CHB and was detectable in all 206 biopsies (Fig. 1 A). The median expression level of IFN-λR1 mRNA expressed as copies per 40 ng of total RNA was 482.5 in CHC samples, which is significantly higher compared with control samples with a median value of 130.0 (Mann–Whitney test; P < 0.001). IFN-λR1 expression was also strongly induced in CHB samples, with a median expression level of 712 copies per 40 ng of total RNA (Fig. 1 A).
We then used a recently adapted in situ hybridization (ISH) method to identify the cell types with IFN-λR1 expression in human liver biopsies. Most of the specific signals were detected in hepatocytes (Fig. 1 B). We also detected signals near the sinusoids that could not unequivocally be localized in hepatocytes. We conclude that the major source IFN-λR1 receptor mRNA measured by qPCR from liver biopsy extracts are hepatocytes, with possible minor quantitative contributions from nonparenchymal sinusoidal cells.
The induction of IFN-λR1 specifically in patients with CHC is reminiscent of the induction of hundreds of ISGs in a substantial proportion of patients with CHC (Chen et al., 2005; Asselah et al., 2008; Sarasin-Filipowicz et al., 2008). Furthermore, it has been reported that the promoter of IFN-λR1 contains a binding site for the IFN-α signal transducer STAT1 (Yang et al., 2010). We therefore tested whether IFN-λR1 is an ISG in PHHs. Indeed, we found a significant induction of IFN-λR1 both on the transcript and the protein level within hours after IFN-α stimulation (Fig. 2, A and B). The IFN-α–induced up-regulation of IFN-λR1 had important consequences for the responsiveness of PHHs to IFN-λ2. Treatment-naive PHHs showed poor to absent phosphorylation of STAT1 in response to 100 ng/ml IFN-λ2. The response was significantly enhanced after the induction of IFN-λR1 through a pretreatment of PHHs with IFN-α (Fig. 2, C and D).
Figure 2.
IFN-α induces IFN-λR1 expression and thereby enhances responsiveness of PHHs to IFN-λ. (A) PHHs were stimulated with 1,000 IU/ml IFN-α for 0, 2, 4, or 6 h. IFN-λR1 expression was analyzed by qPCR. Values are shown as relative to the housekeeping gene GAPDH. Shown are the results from three independent experiments (mean ± SEM). Statistical analysis was performed with the Student’s t test: ***, P < 0.001. (B) PHHs isolated from two non-CHC donors (#231 and #233) were stimulated with 1,000 IU/ml IFN-α for 4 h, and IFN-λR1 was monitored by immunofluorescence using primary anti–IFN-λR1 polyclonal antibody followed by fluorescein (FITC)-coupled secondary antibody. IFN-λR1 signal is shown in green and nuclear staining (DAPI) in blue. (C) PHHs were left untreated (lane 1) or stimulated with 100 ng/ml IFN-λ2 for 15 min before (lane 2) or after (lane 3) treatment with 1,000 IU/ml IFN-α for 4 h. Activation of the Jak–STAT pathway was analyzed with phospho-STAT1–specific antibodies by Western blot, with actin as loading control. Shown is a representative blot of four independent experiments. (D) PHHs isolated from two non-HCV donors (#231 and #233) were prestimulated or not with 1,000 IU/ml IFN-α for 4 h before being exposed to IFN-λ2 for another 15 min. STAT1 activation was monitored by immunofluorescence using anti–pY-STAT1 monoclonal antibody conjugated to Alexa Fluor 555. Phosphorylated STAT1 signal is shown in red and nuclear staining (DAPI) in yellow. Yellow arrowheads indicate nuclear localization of pY-STAT1. (B and D) Shown are representative images from two stainings for each donor. Bars, 40 µm.
Cellular responses to IFN-λ correlate with IFN-λR1 expression levels
To further define the IFN-λR1 expression levels required for the activation of the Jak–STAT signaling pathway and the induction of ISGs by IFN-λs, we generated several Huh7-derived clones stably transfected with IFN-λR1 and selected three of them with different receptor expression levels, designated LR clones 1, 2, and 3 (Fig. 3, A and B). The parent Huh7 clone used for these experiments has very low IFN-λR1 expression and consequently showed only weak phosphorylation of STAT1 upon IFN-λ2 treatment and absent or only very weak induction of ISGs (Fig. 3, C and D). LR clones 1, 2, and 3 had stepwise increased expression levels of IFN-λR1 and showed corresponding increases in Jak–STAT activation and ISG induction by IFN-λ2 (Fig. 3, C and D). Of note, a significant increase in responses to IFN-λ2 was only observed in clones with IFN-λR1 expression of >0.005 relative to GAPDH. Interestingly, IFN-λ2–induced ISG expression in LR clone 2 and 3 cells increased over time during a 24-h treatment period (Fig. 3 D). This is in contrast to the IFN-α–induced ISG expression that is typically transient because of the well-known refractoriness of IFN-α signal transduction pathways in Huh7 cells and liver cells (Larner et al., 1986; Sarasin-Filipowicz et al., 2009; Makowska et al., 2011). In accordance with these published results, IFN-α induced only transient phosphorylation of STAT1 (Fig. 3 E). Contrariwise, IFN-λ signaling showed no refractoriness but prolonged stimulation of the Jak–STAT pathway in LR clone 3 cells (Fig. 3 E).
Figure 3.
IFN-λR1 expression levels determine the strength of Jak–STAT signaling pathway activation by IFN-λ2. (A and B) Huh7 cells were stably transfected with a plasmid containing IFN-λR1 coding sequence. The overexpression of IFN-λR1 (mean ± SEM) was confirmed by qPCR from two independent experiments and by immunofluorescence using primary anti–IFN-λR1 polyclonal antibody followed by Cy3-coupled secondary antibody. Representative images from two independent experiments are shown. Bar, 20 µm. (C) Huh7 cells and Huh7 LR clones were stimulated with 1,000 IU/ml IFN-α or 100 ng/ml IFN-λ2 for 15 min, and pY-STAT1 signal was analyzed by immunoblotting. Shown is a representative blot from two independent experiments. (D) Huh7 (n = 3) and Huh7 LR clones (n = 3) were stimulated with 1,000 IU/ml IFN-α or 100 ng/ml IFN-λ2 for 0, 8, and 24 h. The expression of RSAD2, ISG15, USP18, and IFI44L was assessed by qPCR. Results are expressed as mean ± SEM and are representative of three independent experiments. (E) LR clone 3 cells were stimulated with IFN-α or IFN-λ2 during the time periods indicated in the graph. Activation of the Jak–STAT pathway was detected with phospho-STAT1 Western blots. Shown are the representative results from two independent experiments. (F) PHHs from three non-CHC donors were prestimulated with 1,000 IU/ml IFN-α before being exposed to 100 ng/ml IFN-λ2 for an additional 15 min. The phosphorylation of STAT1 was then monitored by immunoblotting. Additionally, the expression level of IFN-λR1 was measured by qPCR before and after 4-h stimulation with 1,000 IU/ml IFN-α. The IFN-λ3 genotypes (rs12979860 and rs8099917) for each donor are reported below the IFN-λR1 expression graphs.
The correlation between IFN-λR1 expression and activation level of the Jak–STAT pathway by IFN-λ was not restricted to Huh7 cells and was also found in PHHs (Fig. 3 F). We made use of the observation that IFN-α stimulation induced IFN-λR1 to variable degrees. When these cells were stimulated with IFN-λ2 after incubation with IFN-α for 4 h, the strength of STAT1 phosphorylation correlated with the expression level of IFN-λR1. We conclude that in unstimulated PHHs, the IFN-λR1 expression level is low and IFN-λ–induced activation of the Jak–STAT pathway is weak. Responsiveness to IFN-λ strongly increases after induction of IFN-λR1.
IFN-λR1 expression is significantly associated with the IFN-λ3 genotype
There was considerable variation of IFN-λR1 mRNA expression among the CHC samples (Fig. 1 A). To investigate the influence of IFN-λ3 genotypes on IFN-λR1 expression, we genotyped CHC samples for the rs12979860 and for the rs8099917 single-nucleotide polymorphisms. We found significantly higher IFN-λR1 expression in samples from patients having one or two minor IFN-λ3 alleles (Fig. 4 A). To investigate the mechanisms responsible for the association of the IFN-λ3 genotype with IFN-λR1 expression, we genotyped PHH samples from 30 donors not infected with HCV (Table S1), stimulated them for 4 h with IFN-α, and measured the mRNA expression of IFN-λR1 and two well-characterized ISGs, i.e., STAT1 and RSAD2. In unstimulated PHHs, the expression of all three genes was low, independent of the IFN-λ3 genotype (Fig. 4, B and C). IFN-α treatment resulted in a significant induction of all three genes in all samples. For STAT1 and RSAD2, the magnitude of gene induction was not significantly associated with the IFN-λ3 genotype. Contrariwise, there was a significantly stronger induction of IFN-λR1 in samples having one or two minor IFN-λ3 alleles (Fig. 4, B and C).
Figure 4.
IFN-α–induced up-regulation of IFN-λR1 depends on the IFN-λ3 genotype. (A) The IFN-λR1 expression levels were quantified by qPCR in liver biopsies from CHC patients (rs12979860: CC, n = 39 and CT/TT, n = 81; rs8099917: TT, n = 64 and TG/GG, n = 56). Red lines show the statistical median of the individual data points. Statistical analysis was performed using the Mann–Whitney test: **, P < 0.01. (B and C) PHHs from 30 non-CHC donors were stimulated for 4 h with 1,000 IU/ml IFN-α. IFN-λR1, STAT1, and RSAD2 expression were measured by qPCR. The samples were genotyped for IFN-λ3 minor alleles compared with major alleles rs12979860 (CC, n = 16; CT/TT, n = 14; B) and rs8099917 (TT, n = 19; TG/GG, n = 11; C). Statistical analysis was performed using the Mann–Whitney test: **, P < 0.01; ***, P < 0.001.
IFN-λR1 expression is significantly associated with response to peg-IFN-α plus ribavirin treatment of CHC
To address whether the response to treatment with peg-IFN-α and ribavirin is influenced by the expression level of IFN-λR1 in the liver, we reanalyzed the expression values of liver biopsy samples of patients with CHC for whom we had complete virological response data (Table S2). There were 21 samples from patients who had a sustained virological response defined as the absence of detectable HCV RNA in the serum 6 mo after the end of the treatment. 27 samples were from patients who did not respond to peg-IFN-α and ribavirin and remained HCV RNA positive. The median expression of IFN-λR1 measured as copies per 40 ng of total RNA was 732 in pretreatment biopsy samples from nonresponder patients (Fig. 5). In the responders, the median expression was 514. The difference was statistically significant (Mann–Whitney test, P < 0.05). We conclude that the response to treatment with peg-IFN-α and ribavirin negatively correlates with expression levels of IFN-λR1 in the liver.
Figure 5.
IFN-λR1 expression is significantly associated with response to peg-IFN-α plus ribavirin treatment of CHC. The expression of IFN-λR1 was measured in pretreatment CHC liver biopsy extracts by qPCR (SVR [sustained virological response], n = 21; NR [no response], n = 27). Red lines show the statistical median of the individual data points. Statistical analysis was performed using the nonparametric Mann–Whitney test: *, P < 0.05.
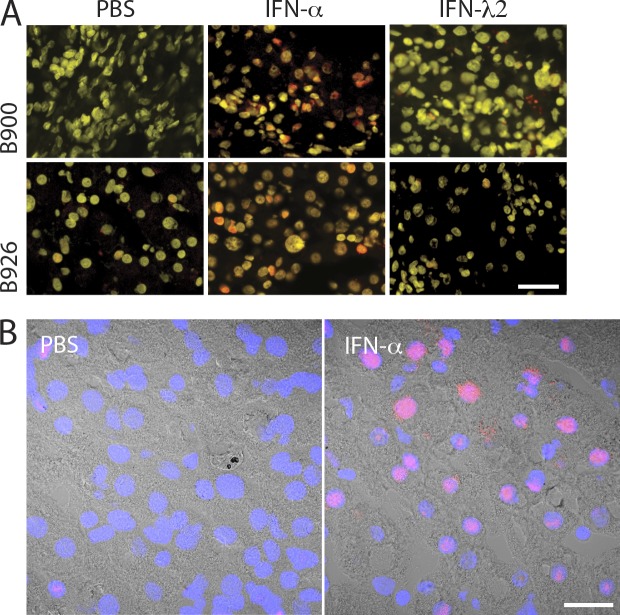
To further address the pharmacological response to IFN-α and IFN-λ, we used an ex vivo treatment protocol using liver biopsy pieces. 2–3-mm-long biopsy cylinder pieces of 1-mm diameter were transferred into test tubes and either left untreated or stimulated with IFN-α or IFN-λ for 15 min. The method was previously used and validated in liver biopsies with different fibrosis stages including cirrhosis (Duong et al., 2004). The activation of the Jak–STAT pathway was assessed by immunostaining of fresh-frozen biopsy material with phospho-STAT1–specific antibodies. For each sample, confocal microscope pictures of three random fields were obtained and used to quantify the number of nuclei with phospho-STAT1 signals. The expression of IFN-λR1 was quantified by qPCR. Two liver biopsies from uninfected patients were used as controls. In these samples, IFN-α induced nuclear phospho-STAT1 signals in most cells, predominantly in hepatocytes, whereas IFN-λ was ineffective (Fig. 6, A and B). The same response pattern was observed in samples from patients with CHC and low expression of the IFN-λR1 (Figs. 7 A and 8). The results were strikingly different in biopsies with high expression levels of IFN-λR1. In many of these samples, nuclear phospho-STAT1 signals were detectable already in the PBS samples, and IFN-α rarely elicited further responses. On the contrary, IFN-λ was able to increase nuclear phospho-STAT1 signals in all nine biopsy samples with high IFN-λR1 (Figs. 7 B and 8). Based on these ex vivo data, we conclude that there is an inverse correlation between responsiveness to IFN-α and IFN-λ in human liver of patients with CHC. Thus, we propose that patients with high-level pretreatment expression of ISGs in the liver, who according to previously published work often respond poorly to IFN-α (Chen et al., 2005; Asselah et al., 2008; Sarasin-Filipowicz et al., 2008; Dill et al., 2011), might indeed have better responses to IFN-λ because of significantly increased expression of IFN-λR1.
Figure 6.
Ex vivo treatment of liver biopsies with IFN-α and IFN-λ. (A) To assess the response to IFN in the absence of HCV infection (positive control for Fig. 7), needle liver biopsies freshly obtained from two non-CHC patients (B900 and B926) were incubated in PBS, IFN-α, or IFN-λ2 for 15 min, and phospho-STAT1 signals were detected by immunofluorescence using anti–pY-STAT1 monoclonal antibody conjugated to Alexa Fluor 555 (positive signals are red). Nuclei were stained with DAPI (in yellow). (B) Liver biopsies from patient B769 were incubated ex vivo in PBS or IFN-α for 15 min, and phospho-STAT1 signals were detected by immunofluorescence using anti–pY-STAT1 monoclonal antibody conjugated to Alexa Fluor 555. Images from fluorescence channels (red for phospho-STAT1 and blue for nuclei) were merged with the corresponding brightfield image. (A and B) Shown are representative images from three stainings for each patient. Bars: (A) 30 µm; (B) 20 µm.
Figure 7.
IFN-α– and IFN-λ–induced nuclear translocation of phospho-STAT1 in ex vivo–treated liver biopsies. (A and B) Needle liver biopsies freshly obtained from CHC patients were incubated in 1,000 IU/ml IFN-α, 100 ng/ml IFN-λ2, or PBS for 15 min at 37°C. The response to IFN-α or IFN-λ was assessed by detection of nuclear translocation of phosphorylated STAT1 by immunofluorescence using anti–pY-STAT1 monoclonal antibody conjugated to Alexa Fluor 555 (positive signals are red). Nuclei were stained with DAPI (in yellow). Sample numbers are indicated to the left to the PBS pictures. (A) Nine liver biopsies with low level expression of IFN-λR1. (B) Nine liver biopsies with high level expression of IFN-λR1. Shown are representative images from three stainings for each patient. Bar, 40 µm.
Figure 8.
Expression of IFN-λR1 correlates negatively with response to IFN-α and positively with response to IFN-λ2. (A) For each CHC patient, three images from each treatment condition (PBS, IFN-α, and IFN-λ2 treated) obtained from confocal microscopy acquisition were used to number cells that are positive for phospho-STAT1. Results are presented as floating bars showing the mean. (B) For each CHC patients, RNAs were isolated from PBS-, IFN-α–, and IFN-λ2–treated samples, and IFN-λR1 expression was quantified by qPCR. The mean of IFN-λR1 copies per 40 ng of total RNA for each CHC patient was then shown. Red lines show the statistical mean of the individual data points. The IFN-λ3 genotypes (rs12979860 and rs8099917) are shown below the sample numbers.
DISCUSSION
The discovery that genetic variants near the IFN-λ3 gene locus are strongly associated with spontaneous clearance of HCV by the host immune system and with response to therapy of CHC with peg-IFN-α and ribavirin was an important milestone in HCV research (Ge et al., 2009; Suppiah et al., 2009; Tanaka et al., 2009; Rauch et al., 2010). IFN-λ (IL28B) genotyping rapidly became a useful tool for clinicians for predicting the likelihood of treatment response of an individual patient, and it became essential for stratifying patients enrolled in clinical treatment trials with peg-IFN-α–containing regimens. The discovery was also important for basic research of host–virus interactions because it identified the previously neglected IFN-λ system as a key component of the host immune answer to HCV. However, the mechanisms that link IFN-λ3 genotypes with spontaneous HCV clearance and with response to peg-IFN-α remained elusive. Important insights came from studies that linked IFN-λ3 genotypes with hepatic ISG expression in CHC (Honda et al., 2010; Urban et al., 2010). Differential activation of the endogenous IFN system in the liver has previously been established as a major determinant of response to peg-IFN-α and ribavirin (Chen et al., 2005; Asselah et al., 2008; Sarasin-Filipowicz et al., 2008). Patients with an activated IFN system are generally poor responders to peg-IFN-α therapy. Therefore, an increased production and secretion of IFN-λ from minor alleles compared with major IFN-λ3 alleles would be an obvious explanation for the association of the minor IFN-λ3 alleles with increased ISG expression in the liver. However, several groups have failed to find increased expression levels of IFN-λ family members in the serum or the liver of patients with minor IFN-λ3 alleles (Honda et al., 2010; Urban et al., 2010; Dill et al., 2011). In the present work, we provide evidence that the link between the IFN-λ3 genotype and hepatic ISG expression is based on differential expression of the IFN-λR1 receptor, and not by differential induction of the IFN-λ ligands. The receptor is detectably expressed in all liver biopsies. The median expression value is significantly higher in samples from patients with CHC, and within the group of CHC samples, there is significantly increased expression in patients carrying the minor IFN-λ3 allele (compared with the major allele) and in patients who were nonresponders to peg-IFN-α and ribavirin therapy (compared with patients who were cured by this treatment). These results are in accordance with previous published results showing an elevated expression of IFNLR1 in patients with CHC infection compared with controls and in Huh7.5 cells infected with HCVcc (cell culture–derived infectious HCV; Dolganiuc et al., 2012; Kohli et al., 2012).
The ligands (IFN-λ1, -2, and -3) were consistently undetectable in liver biopsies from patients not infected with HCV. In CHC patients, IFN-λ1 and -λ2/3 are detected by qPCR in about half of the samples, mostly at low expression levels. In agreement with published work, we did not find a correlation between IFN-λ expression and response to treatment or the IFN-λ3 genotype. We therefore think that we can exclude differential regulation of IFN-λ1 and -λ2/3 as the missing link between the IFN-λ3 genotype and ISG expression. A very interesting alternative explanation has been generated by the recent discovery of a frame shift variant located upstream of the IFN-λ3 gene (Prokunina-Olsson et al., 2013). This variant creates a new open reading frame of 179 aa that has been designated IFN-λ4. Despite considerable sequence diversity between IFN-λ4 and the other members of the IFN-λ family, recombinant IFN-λ4 signals through the IFN-λR to induce an antiviral state against HCV and coronaviruses (Hamming et al., 2013). It is immediately obvious that the exclusive production of IFN-λ4 from the minor IFN-λ3 alleles (more precisely from the ss469415590-ΔG allele) together with the stronger induction of IFN-λR1 in the liver of patients with one or two minor alleles (Fig. 4) provide an attractive explanation for the high level of ISG expression in these patients. For the moment, however, our attempts to detect IFN-λ4 mRNA in liver biopsies failed. Thus, further investigations are needed to clarify whether IFN-λ4 is indeed the cytokine responsible for elevated ISG expression in patients with CHC.
Despite the fact that IFN-λs are not detectable in a substantial percentage of patients with CHC, we think that the IFN-λ system is the most likely candidate for the continuous up-regulation of hundreds of ISGs in preactivated patients. Contrary to IFN-α signaling, IFN-λ signaling does not become refractory during prolonged stimulation (Fig. 3 D; Makowska et al., 2011). Moreover, the strength of IFN-λ–induced activation of the Jak–STAT pathway is regulated to a large degree by the expression of the IFN-λR1 (Figs. 2, 3, 7, and 8). Because IFN-λR1 is an ISG itself, the IFN-λ system can undergo a positive feedback loop that can be maintained as long as there is a minimal amount of IFN-λ ligands to stimulate the expression of IFN-λR1. In this context, our observation that PHHs from non-HCV–infected donors show a differential induction of IFN-λR1 upon IFN-α stimulation according to their IFN-λ3 genotype (Fig. 4, B and C) provides an important missing link between genotype and phenotype. We propose that early during HCV infection the host innate immune system reacts with the induction of type I (IFN-α or IFN-β) and/or III (IFN-λ) IFN. In individuals with one or two minor IFN-λ3 alleles, there is a strong induction of the IFN-λR1. This induction is maintained through a positive feedback loop, and IFN-λR1 remains constitutively induced together with hundreds of other ISGs. In this group of patients, peg-IFN-α is ineffective, most likely because of the permanent induction of the ISG USP18, a strong inhibitor of signaling through the IFN-α receptor (Sarasin-Filipowicz et al., 2009; François-Newton et al., 2011; Dill et al., 2012). On the other side, in patients homozygous for the major IFN-λ3 allele, the induction of IFN-λR1 remains below a critical threshold for the positive feedback loop, and ISG induction fades away. These non-preactivated patients are highly responsive to peg-IFN-α, most likely because IFN-α signaling is not refractory.
Peg-IFN-λ1 is currently undergoing clinical development for the treatment of viral hepatitis. A phase 1b and a phase 2b study demonstrated antiviral activity in patients with CHC and minimal adverse events and hematologic effects (Muir et al., 2010; Muir, A.J., et al. 2012). The 63rd Annual Meeting of the American Association for the Study of Liver Diseases: The Liver Meeting. Abstr. 214). The better tolerability of peg-IFN-λ1 is presumably caused by the restricted tissue distribution of its receptor. Lymphocytes or monocytes are unresponsive to IFN-λ (Dickensheets et al., 2013). The antiviral activity of peg-IFN-λ1 is probably mediated through a direct stimulation of the IFN-λ receptor on hepatocytes. In tissue blots, IFN-λR1 is detectable in the liver (Sheppard et al., 2003), and IFN-λ had antiviral activity and/or activated Jak–STAT signaling in several hepatoma cells lines and PHHs, albeit often with lower efficacy compared with IFN-α (Robek et al., 2005; Doyle et al., 2006; Marcello et al., 2006; Dickensheets et al., 2013). In vivo, the response to IFN-λ in the human liver has not been investigated. Interestingly, in the mouse liver, we and others have found very little IFN-λR1 expression and little to absent Jak–STAT stimulation in the liver in response to systemic application of mouse IFN-λ (Sommereyns et al., 2008; Makowska et al., 2011).
In the present extensive analysis of IFN-λR1 expression in human liver biopsies, we measured rather low expression levels in samples from patients not infected with HCV. Thus, the noninfected human liver has very little response to IFN-λ. This state of relative unresponsiveness could protect the liver from overstimulation by IFN-λ produced during gastrointestinal viral infections and transported via the portal vein into the hepatic circulation.
IFN-λR1 expression is not increased in all CHC patients, but rather predominantly in patients with an induced IFN system in the liver who usually are carriers of the IFN-λ3 minor allele. This differential expression can be explained by the fact that IFN-λR1 is an ISG. The association of IFN-λR1 expression with the IFN-λ3 minor allele has important consequences for the use of peg-IFN-λ1 for the treatment of CHC. Collectively with the results from our ex vivo stimulation experiments (Figs. 7 and 8), the findings provide a rationale for the preferential use of peg-IFN-λ1 in patients with an induced hepatic IFN system who are usually responding poorly to the current standard of care treatment regiments that contain peg-IFN-α. On the negative side, peg-IFN-λ1 could have only limited efficiency in patients with an uninduced endogenous IFN system because of a below-threshold expression of the IFN-λ receptor. A quantification of IFN-λR1 mRNA in liver biopsies could therefore help to select the most promising peg-IFN (peg-IFN-α vs. peg-IFN-λ) to be included in the treatment regimens of individual patients.
MATERIALS AND METHODS
Reagents and antibodies.
Human IFN-α (Roferon-A) was a gift from Roche. IFN-λ2 was purchased from PeproTech, Inc. The following antibodies were used for Western blotting and immunofluorescence staining: phospho-STAT1 and phospho-STAT1 Alexa Fluor 555 conjugated (Cell Signaling Technology), STAT1 (BD), β-actin (Sigma-Aldrich), and IFN-λR1 (ProSci Inc.).
Cell cultures and generation of Huh7 cell clones expressing IFN-λR1.
Huh7 cells were cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin. The coding sequence of IFN-λR1 was amplified from a commercially available IMAGE human cDNA clone (catalog number EHA10001-99865534; Thermo Fisher Scientific) using the primers 5′-TTTTCTAGACGGCAGGAAGGCCATGGC-3′ and 5′-TCTAGACCTGGCCATGTAATGCCCCAAT-3′ and then cloned into a pcDNA3.V5-His vector with XbaI restriction sites. The expression plasmid was transfected into Huh7 cells using FuGENE HD (Roche). Cells were then selected with DMEM culture medium supplemented with 10% FBS and 1 mg/ml G418 (EMD Millipore) to generate stable IFN-λR1–expressing clones.
PHHs were isolated from liver resections from patients at the Strasbourg University Hospitals with approval from the Institutional Review Board and isolated and cultured as described previously (Krieger et al., 2010). The characteristics of the PHH samples are summarized in Table S1.
Patients.
Liver biopsies from CHC and non-CHC patients were obtained in the outpatient clinic of the Division of Gastroenterology and Hepatology, University Hospital Basel. Parts of the biopsy material that were not needed for routine histopathology were used for research purposes after obtaining written informed consent. The use of biopsy material for this project was approved by the Ethikkommission Nordwest- und Zentralschweiz, Basel, Switzerland, protocol number M989/99. Grading and staging of CHC was defined according to the METAVIR classification. Serum HCV RNA was quantified using the COBAS AmpliPrep/COBAS TaqMan HCV Test and the COBAS Amplicor Monitor from Roche. Response to treatment in CHC patients was defined as described previously (Dill et al., 2011). Diagnosis of non-CHC patients was based on clinical, laboratory, and histopathological assessment according to routine clinical practice. CHC and non-CHC patients’ characteristics are summarized in Tables S2 and S3, respectively.
Western blotting.
Protein extracts and immunoblots were performed as described previously (Duong et al., 2004).
RNA extraction and quantitative RT-PCR.
RNA isolation from Huh7 cells was performed with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. DNA digestion was performed with RQ1 RNase-Free DNase (Promega). RNA isolation from PHHs was performed with NucleoSpin RNA II (MACHEREY-NAGEL) according to the manufacturer’s instructions. 1 µg RNA was reverse transcribed with random hexamers and Moloney murine leukemia virus RT (Promega). Real-time qPCR was performed with SYBR green fluorescence (Applied Biosystems) and an ABI 7500 detection system (Applied Biosystems). Primers used for real-time qPCR are listed in Table S4. The IL28A gene shares 97% sequence similarity with the IL28B gene; therefore, we designed primers that specifically recognize both genes. The specificity of the PCR was assessed on 3% agarose gel. For in vitro experiments, gene expression was normalized to human GAPDH expression using the ΔCt method. For analysis in bulk tissues, gene expression was quantified using cloned cDNA (a gift from R. Hartmann, Aarhus University, Aarhus, Denmark) as standard, and results are shown as copies per 40 ng of total RNA. Complete gene expression data from PHH samples are listed in Table S1. Complete gene expression data from CHC patients are listed in Table S2.
Ex vivo treatment of human liver biopsies.
Freshly obtained liver biopsies from CHC patients were incubated with PBS, 1,000 U/ml IFN-α, or 100 ng/ml IFN-λ2 for 15 min at 37°C. Specimens were then embedded in Tissue-Tek OCT (Sakura) and frozen in liquid nitrogen. Cryosections of 8-µm thickness were prepared and used for immunostaining analysis.
Immunofluorescence.
Liver biopsies sections were fixed in freshly prepared 4% formaldehyde for 15 min. After a wash with PBS, they were permeabilized in cold methanol. Background was then removed with Background Buster (Innovex Biosciences, Inc.). Phosphorylated STAT1 was detected using phospho-STAT1–Alexa Fluor 555–conjugated antibody. Sections were mounted with Mount FluorCare DAPI (Carl Roth GmbH).
Adherent cells were incubated with cold methanol for 10 min, washed with TBS-Tween, and incubated in blocking solution (5.5% FBS in TBS-Tween) for 60 min at room temperature. Cells were then washed and incubated with antibodies against phosphorylated STAT1 or IFN-λR1.
Fluorescence ISH.
For the present study, we adapted a highly sensitive and specific ISH system (QuantiGene ViewRNA; Affymetrix) using a custom probe set targeting IFN-λR1 (IL28RA; Affymetrix; GenBank accession no. NM_170743). In brief, 10-µm sections from OCT-embedded liver biopsies were prepared, mounted onto Superfrost Plus Gold glass slides (Thermo Fischer Scientific), and kept at −80°C until use. Upon fixation, washing, and dehydration in ethanol, sections were digested in Protease QF (Affymetrix) and then hybridized with custom-designed QuantiGene ViewRNA probes against target RNA sequences. Bound probes were preamplified and subsequently amplified according to the manufacturer’s instructions. Probe oligonucleotide conjugated to alkaline phosphatase type 1 was added, followed by Fast Red substrate. Finally, slides were counterstained with Meyer’s hematoxylin and embedded with DAPI-containing aqueous mounting medium (Roti-Mount FluorCare DAPI; Roth AG).
Image acquisition, quantification, and data analysis.
Images were acquired using a laser-scanning confocal microscope (LSM710; Carl Zeiss) associated with Zen2 software (Carl Zeiss). For each liver biopsy section, we acquired three random fields. We avoided areas with high lymphocyte concentration and any other areas where the quality of DAPI staining would not allow reliable identification of nuclei.
DNA isolation and rs12979860 and rs8099917 genotyping.
Genomic DNA was isolated from liver biopsies using TRIzol reagent (Invitrogen) and DNeasy Blood & Tissue kit (QIAGEN) according to the manufacturers’ instructions. Genomic DNA from donors listed in Table S1 was obtained from nonparenchymal liver cells using the DNeasy Blood & Tissue kit. rs12979860 and rs8099917 genotyping was performed as described previously (Dill et al., 2011).
Statistical analysis.
Prism4 (GraphPad Software) was used for statistical analysis.
Online supplemental material.
Table S1, included as a separate PDF file, shows PHH donors’ characteristics. Table S2, included as a separate PDF file, shows CHC patients’ characteristics. Table S3, included as a separate PDF file, shows gene expression levels in non-HCV patients. Table S4, included as a separate PDF file, shows primer sequences for qPCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20131557/DC1.
Supplementary Material
Acknowledgments
We thank Prof. P. Bachellier (Service de Chirurgie Générale, Hépatique, Endocrinienne et Transplantation, Hôpitals Universitaires de Strasbourg, France) for providing liver resections for the isolation of PHHs.
This work was supported by Swiss National Science Foundation (SNF) grants 320030_130243 and 310030B_147089 to M.H. Heim and by EU INTERREG-IV-Rhin Supérieur-FEDER-Hepato-Regio-Net 2012, Agence Nationale de Recherche sur le Sida (2012/239, 2012/318, 2013/108), and Laboratoire d’excellence LabEx HEPSYS (Investissement d’Avenir; ANR-10-LAB-28) grants to T.F. Baumert.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CHC
- chronic hepatitis C
- HCV
- hepatitis C virus
- ISG
- IFN-stimulated gene
- ISH
- in situ hybridization
- peg-IFN
- pegylated IFN
- PHH
- primary human hepatocyte
- qPCR
- quantitative PCR
References
- Asselah T., Bieche I., Narguet S., Sabbagh A., Laurendeau I., Ripault M.P., Boyer N., Martinot-Peignoux M., Valla D., Vidaud M., Marcellin P. 2008. Liver gene expression signature to predict response to pegylated interferon plus ribavirin combination therapy in patients with chronic hepatitis C. Gut. 57:516–524 10.1136/gut.2007.128611 [DOI] [PubMed] [Google Scholar]
- Bibert S., Roger T., Calandra T., Bochud M., Cerny A., Semmo N., Duong F.H., Gerlach T., Malinverni R., Moradpour D., et al. ; Swiss Hepatitis C Cohort Study. 2013. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J. Exp. Med. 210:1109–1116 10.1084/jem.20130012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Borozan I., Feld J., Sun J., Tannis L.L., Coltescu C., Heathcote J., Edwards A.M., McGilvray I.D. 2005. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 128:1437–1444 10.1053/j.gastro.2005.01.059 [DOI] [PubMed] [Google Scholar]
- Dickensheets H., Sheikh F., Park O., Gao B., Donnelly R.P. 2013. Interferon-lambda (IFN-λ) induces signal transduction and gene expression in human hepatocytes, but not in lymphocytes or monocytes. J. Leukoc. Biol. 93:377–385 10.1189/jlb.0812395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill M.T., Duong F.H., Vogt J.E., Bibert S., Bochud P.Y., Terracciano L., Papassotiropoulos A., Roth V., Heim M.H. 2011. Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C. Gastroenterology. 140:1021–1031 10.1053/j.gastro.2010.11.039 [DOI] [PubMed] [Google Scholar]
- Dill M.T., Makowska Z., Duong F.H., Merkofer F., Filipowicz M., Baumert T.F., Tornillo L., Terracciano L., Heim M.H. 2012. Interferon-γ–stimulated genes, but not USP18, are expressed in livers of patients with acute hepatitis C. Gastroenterology. 143:777–786 10.1053/j.gastro.2012.05.044 [DOI] [PubMed] [Google Scholar]
- Dolganiuc A., Kodys K., Marshall C., Saha B., Zhang S., Bala S., Szabo G. 2012. Type III interferons, IL-28 and IL-29, are increased in chronic HCV infection and induce myeloid dendritic cell-mediated FoxP3+ regulatory T cells. PLoS ONE. 7:e44915 10.1371/journal.pone.0044915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.E., Schreckhise H., Khuu-Duong K., Henderson K., Rosler R., Storey H., Yao L., Liu H., Barahmand-pour F., Sivakumar P., et al. 2006. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 44:896–906 10.1002/hep.21312 [DOI] [PubMed] [Google Scholar]
- Duong F.H., Filipowicz M., Tripodi M., La Monica N., Heim M.H. 2004. Hepatitis C virus inhibits interferon signaling through up-regulation of protein phosphatase 2A. Gastroenterology. 126:263–277 10.1053/j.gastro.2003.10.076 [DOI] [PubMed] [Google Scholar]
- François-Newton V., Magno de Freitas Almeida G., Payelle-Brogard B., Monneron D., Pichard-Garcia L., Piehler J., Pellegrini S., Uzé G. 2011. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon α response. PLoS ONE. 6:e22200 10.1371/journal.pone.0022200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D., Fellay J., Thompson A.J., Simon J.S., Shianna K.V., Urban T.J., Heinzen E.L., Qiu P., Bertelsen A.H., Muir A.J., et al. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 461:399–401 10.1038/nature08309 [DOI] [PubMed] [Google Scholar]
- Ghany M.G., Nelson D.R., Strader D.B., Thomas D.L., Seeff L.B.; American Association for Study of Liver Diseases. 2011. An update on treatment of genotype 1 chronic hepatitis C virus infection: 2011 practice guideline by the American Association for the Study of Liver Diseases. Hepatology. 54:1433–1444 10.1002/hep.24641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming O.J., Terczyńska-Dyla E., Vieyres G., Dijkman R., Jørgensen S.E., Akhtar H., Siupka P., Pietschmann T., Thiel V., Hartmann R. 2013. Interferon lambda 4 signals via the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J. 32:3055–3065 10.1038/emboj.2013.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M., Sakai A., Yamashita T., Nakamoto Y., Mizukoshi E., Sakai Y., Yamashita T., Nakamura M., Shirasaki T., Horimoto K., et al. ; Hokuriku Liver Study Group. 2010. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 139:499–509 10.1053/j.gastro.2010.04.049 [DOI] [PubMed] [Google Scholar]
- Kohli A., Zhang X., Yang J., Russell R.S., Donnelly R.P., Sheikh F., Sherman A., Young H., Imamichi T., Lempicki R.A., et al. 2012. Distinct and overlapping genomic profiles and antiviral effects of Interferon-λ and -α on HCV-infected and noninfected hepatoma cells. J. Viral Hepat. 19:843–853 10.1111/j.1365-2893.2012.01610.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger S.E., Zeisel M.B., Davis C., Thumann C., Harris H.J., Schnober E.K., Mee C., Soulier E., Royer C., Lambotin M., et al. 2010. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2–CD81–claudin-1 associations. Hepatology. 51:1144–1157 10.1002/hep.23445 [DOI] [PubMed] [Google Scholar]
- Larner A.C., Chaudhuri A., Darnell J.E., Jr 1986. Transcriptional induction by interferon. New protein(s) determine the extent and length of the induction. J. Biol. Chem. 261:453–459 [PubMed] [Google Scholar]
- Lauer G.M., Walker B.D. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41–52 10.1056/NEJM200107053450107 [DOI] [PubMed] [Google Scholar]
- Makowska Z., Duong F.H.T., Trincucci G., Tough D.F., Heim M.H. 2011. Interferon-β and interferon-λ signaling is not affected by interferon-induced refractoriness to interferon-α in vivo. Hepatology. 53:1171–1180 10.1002/hep.24189 [DOI] [PubMed] [Google Scholar]
- Malakhova O.A., Kim K.I., Luo J.K., Zou W., Kumar K.G., Fuchs S.Y., Shuai K., Zhang D.E. 2006. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 25:2358–2367 10.1038/sj.emboj.7601149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello T., Grakoui A., Barba-Spaeth G., Machlin E.S., Kotenko S.V., MacDonald M.R., Rice C.M. 2006. Interferons α and λ inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 131:1887–1898 10.1053/j.gastro.2006.09.052 [DOI] [PubMed] [Google Scholar]
- Muir A.J., Shiffman M.L., Zaman A., Yoffe B., de la Torre A., Flamm S., Gordon S.C., Marotta P., Vierling J.M., Lopez-Talavera J.C., et al. 2010. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology. 52:822–832 10.1002/hep.23743 [DOI] [PubMed] [Google Scholar]
- Prokunina-Olsson L., Muchmore B., Tang W., Pfeiffer R.M., Park H., Dickensheets H., Hergott D., Porter-Gill P., Mumy A., Kohaar I., et al. 2013. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 45:164–171 10.1038/ng.2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A., Kutalik Z., Descombes P., Cai T., Di Iulio J., Mueller T., Bochud M., Battegay M., Bernasconi E., Borovicka J., et al. ; Swiss HIV Cohort Study. 2010. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 138:1338–1345 10.1053/j.gastro.2009.12.056 [DOI] [PubMed] [Google Scholar]
- Robek M.D., Boyd B.S., Chisari F.V. 2005. Lambda interferon inhibits hepatitis B and C virus replication. J. Virol. 79:3851–3854 10.1128/JVI.79.6.3851-3854.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M., Oakeley E.J., Duong F.H., Christen V., Terracciano L., Filipowicz W., Heim M.H. 2008. Interferon signaling and treatment outcome in chronic hepatitis C. Proc. Natl. Acad. Sci. USA. 105:7034–7039 10.1073/pnas.0707882105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M., Wang X., Yan M., Duong F.H., Poli V., Hilton D.J., Zhang D.E., Heim M.H. 2009. Alpha interferon induces long-lasting refractoriness of JAK-STAT signaling in the mouse liver through induction of USP18/UBP43. Mol. Cell. Biol. 29:4841–4851 10.1128/MCB.00224-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C., Roraback J., et al. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63–68 10.1038/ni873 [DOI] [PubMed] [Google Scholar]
- Sommereyns C., Paul S., Staeheli P., Michiels T. 2008. IFN-lambda (IFN-λ) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4:e1000017 10.1371/journal.ppat.1000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppiah V., Moldovan M., Ahlenstiel G., Berg T., Weltman M., Abate M.L., Bassendine M., Spengler U., Dore G.J., Powell E., et al. 2009. IL28B is associated with response to chronic hepatitis C interferon-α and ribavirin therapy. Nat. Genet. 41:1100–1104 10.1038/ng.447 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Nishida N., Sugiyama M., Kurosaki M., Matsuura K., Sakamoto N., Nakagawa M., Korenaga M., Hino K., Hige S., et al. 2009. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat. Genet. 41:1105–1109 10.1038/ng.449 [DOI] [PubMed] [Google Scholar]
- Thimme R., Binder M., Bartenschlager R. 2012. Failure of innate and adaptive immune responses in controlling hepatitis C virus infection. FEMS Microbiol. Rev. 36:663–683 10.1111/j.1574-6976.2011.00319.x [DOI] [PubMed] [Google Scholar]
- Thomas D.L., Thio C.L., Martin M.P., Qi Y., Ge D., O’Huigin C., Kidd J., Kidd K., Khakoo S.I., Alexander G., et al. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 461:798–801 10.1038/nature08463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban T.J., Thompson A.J., Bradrick S.S., Fellay J., Schuppan D., Cronin K.D., Hong L., McKenzie A., Patel K., Shianna K.V., et al. 2010. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 52:1888–1896 10.1002/hep.23912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Luo Y., Wei J., He S. 2010. Integrative genomic analyses on IL28RA, the common receptor of interferon-λ1, -λ2 and -λ3. Int. J. Mol. Med. 25:807–812 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.