Abstract
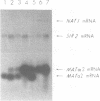
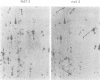
A gene from Saccharomyces cerevisiae has been mapped, cloned, sequenced and shown to encode a catalytic subunit of an N-terminal acetyltransferase. Regions of this gene, NAT1, and the chloramphenicol acetyltransferase genes of bacteria have limited but significant homology. A nat1 null mutant is viable but exhibits a variety of phenotypes, including reduced acetyltransferase activity, derepression of a silent mating type locus (HML) and failure to enter G0. All these phenotypes are identical to those of a previously characterized mutant, ard1. NAT1 and ARD1 are distinct genes that encode proteins with no obvious similarity. Concomitant overexpression of both NAT1 and ARD1 in yeast causes a 20-fold increase in acetyltransferase activity in vitro, whereas overexpression of either NAT1 or ARD1 alone does not raise activity over basal levels. A functional iso-1-cytochrome c protein, which is N-terminally acetylated in a NAT1 strain, is not acetylated in an isogenic nat1 mutant. At least 20 other yeast proteins, including histone H2B, are not N-terminally acetylated in either nat1 or ard1 mutants. These results suggest that NAT1 and ARD1 proteins function together to catalyze the N-terminal acetylation of a subset of yeast proteins.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alfageme C. R., Zweidler A., Mahowald A., Cohen L. H. Histones of Drosophila embryos. Electrophoretic isolation and structural studies. J Biol Chem. 1974 Jun 25;249(12):3729–3736. [PubMed] [Google Scholar]
- Alonso W. R., Nelson D. A. A novel yeast histone deacetylase: partial characterization and development of an activity assay. Biochim Biophys Acta. 1986 Mar 26;866(2-3):161–169. doi: 10.1016/0167-4781(86)90113-2. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Micklem G., Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987 Dec 4;51(5):709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Brandt W. F., Patterson K., von Holt C. The histones of yeast. The isolation and partial structure of the core histones. Eur J Biochem. 1980 Sep;110(1):67–76. doi: 10.1111/j.1432-1033.1980.tb04841.x. [DOI] [PubMed] [Google Scholar]
- Brill S. J., Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988 Jul 29;54(3):403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Campbell J. L. Yeast DNA replication in vitro: initiation and elongation events mimic in vivo processes. Cell. 1982 Nov;31(1):201–213. doi: 10.1016/0092-8674(82)90420-2. [DOI] [PubMed] [Google Scholar]
- Chang T. H., Clark M. W., Lustig A. J., Cusick M. E., Abelson J. RNA11 protein is associated with the yeast spliceosome and is localized in the periphery of the cell nucleus. Mol Cell Biol. 1988 Jun;8(6):2379–2393. doi: 10.1128/mcb.8.6.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberlidge A. G., Isono K. Ribosomal protein modification in Escherichia coli. I. A mutant lacking the N-terminal acetylation of protein S5 exhibits thermosensitivity. J Mol Biol. 1979 Jun 25;131(2):169–189. doi: 10.1016/0022-2836(79)90072-x. [DOI] [PubMed] [Google Scholar]
- Driessen H. P., de Jong W. W., Tesser G. I., Bloemendal H. The mechanism of N-terminal acetylation of proteins. CRC Crit Rev Biochem. 1985;18(4):281–325. doi: 10.3109/10409238509086784. [DOI] [PubMed] [Google Scholar]
- Glembotski C. C. Characterization of the peptide acetyltransferase activity in bovine and rat intermediate pituitaries responsible for the acetylation of beta-endorphin and alpha-melanotropin. J Biol Chem. 1982 Sep 10;257(17):10501–10509. [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy J. M., Klar A. J., Hicks J. B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Feb;6(2):688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G. G., Kronert W. A., Bernstein S. I., Chapman V. M., Smith K. D. Altered turnover of allelic variants of hypoxanthine phosphoribosyltransferase is associated with N-terminal amino acid sequence variation. J Biol Chem. 1988 Jul 5;263(19):9079–9082. [PubMed] [Google Scholar]
- Jörnvall H. Acetylation of Protein N-terminal amino groups structural observations on alpha-amino acetylated proteins. J Theor Biol. 1975 Nov;55(1):1–12. doi: 10.1016/s0022-5193(75)80105-6. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., Fairwell T., Kratofil P., Wills C. Differences in alpha-amino acetylation of isozymes of yeast alcohol dehydrogenase. FEBS Lett. 1980 Feb 25;111(1):214–218. doi: 10.1016/0014-5793(80)80796-4. [DOI] [PubMed] [Google Scholar]
- Kayne P. S., Kim U. J., Han M., Mullen J. R., Yoshizaki F., Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988 Oct 7;55(1):27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Klyce H. R., McLaughlin C. S. Characterization of temperature-sensitive mutants of yeast by a photomicrographic procedure. Exp Cell Res. 1973 Nov;82(1):47–56. doi: 10.1016/0014-4827(73)90243-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Last R. L., Stavenhagen J. B., Woolford J. L., Jr Isolation and characterization of the RNA2, RNA3, and RNA11 genes of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2396–2405. doi: 10.1128/mcb.4.11.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F. J., Lin L. W., Smith J. A. Purification and characterization of an N alpha-acetyltransferase from Saccharomyces cerevisiae. J Biol Chem. 1988 Oct 15;263(29):14948–14955. [PubMed] [Google Scholar]
- Leslie A. G., Moody P. C., Shaw W. V. Structure of chloramphenicol acetyltransferase at 1.75-A resolution. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4133–4137. doi: 10.1073/pnas.85.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Moerschell R. P., Tsunasawa S., Sherman F. Transformation of yeast with synthetic oligonucleotides. Proc Natl Acad Sci U S A. 1988 Jan;85(2):524–528. doi: 10.1073/pnas.85.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Molecular analysis of a cell lineage. Nature. 1983 Apr 21;302(5910):670–676. doi: 10.1038/302670a0. [DOI] [PubMed] [Google Scholar]
- Nelson D. A. Histone acetylation in baker's yeast. Maintenance of the hyperacetylated configuration in log phase protoplasts. J Biol Chem. 1982 Feb 25;257(4):1565–1568. [PubMed] [Google Scholar]
- Nikawa J., Sass P., Wigler M. Cloning and characterization of the low-affinity cyclic AMP phosphodiesterase gene of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Oct;7(10):3629–3636. doi: 10.1128/mcb.7.10.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson B., Flinta C., von Heijne G., Jörnvall H. Structures of N-terminally acetylated proteins. Eur J Biochem. 1985 Nov 4;152(3):523–527. doi: 10.1111/j.1432-1033.1985.tb09227.x. [DOI] [PubMed] [Google Scholar]
- Pestana A., Pitot H. C. Acetylation of nascent polypeptide chains on rat liver polyribosomes in vivo and in vitro. Biochemistry. 1975 Apr 8;14(7):1404–1412. doi: 10.1021/bi00678a010. [DOI] [PubMed] [Google Scholar]
- Rine J., Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987 May;116(1):9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rykowski M. C., Wallis J. W., Choe J., Grunstein M. Histone H2B subtypes are dispensable during the yeast cell cycle. Cell. 1981 Aug;25(2):477–487. doi: 10.1016/0092-8674(81)90066-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Shore D., Squire M., Nasmyth K. A. Characterization of two genes required for the position-effect control of yeast mating-type genes. EMBO J. 1984 Dec 1;3(12):2817–2823. doi: 10.1002/j.1460-2075.1984.tb02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddig M. A., Kinsey J. A., Fincham J. R., Keighren M. Frameshift mutations affecting the N-terminal sequence of Neurospora NADP-specific glutamate dehydrogenase. J Mol Biol. 1980 Feb 25;137(2):125–135. doi: 10.1016/0022-2836(80)90320-4. [DOI] [PubMed] [Google Scholar]
- Smyth D. G., Zakarian S. Selective processing of beta-endorphin in regions of porcine pituitary. Nature. 1980 Dec 11;288(5791):613–615. doi: 10.1038/288613a0. [DOI] [PubMed] [Google Scholar]
- Strathern J. N., Spatola E., McGill C., Hicks J. B. Structure and organization of transposable mating type cassettes in Saccharomyces yeasts. Proc Natl Acad Sci U S A. 1980 May;77(5):2839–2843. doi: 10.1073/pnas.77.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J., Hicks J., Herskowitz I. Control of cell type in yeast by the mating type locus. The alpha 1-alpha 2 hypothesis. J Mol Biol. 1981 Apr 15;147(3):357–372. doi: 10.1016/0022-2836(81)90488-5. [DOI] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Scott J. D., McMullen B., Hurwitz M., Krebs E. G., Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Apr;7(4):1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Travis G. H., Colavito-Shepanski M., Grunstein M. Extensive purification and characterization of chromatin-bound histone acetyltransferase from Saccharomyces cerevisiae. J Biol Chem. 1984 Dec 10;259(23):14406–14412. [PubMed] [Google Scholar]
- Tsunasawa S., Kamitani K., Narita K. Partial purification and properties of the amino-terminal amino acid-acetylating enzyme from hen's oviduct. J Biochem. 1980 Feb;87(2):645–650. doi: 10.1093/oxfordjournals.jbchem.a132789. [DOI] [PubMed] [Google Scholar]
- Tsunasawa S., Stewart J. W., Sherman F. Amino-terminal processing of mutant forms of yeast iso-1-cytochrome c. The specificities of methionine aminopeptidase and acetyltransferase. J Biol Chem. 1985 May 10;260(9):5382–5391. [PubMed] [Google Scholar]
- Vernet T., Dignard D., Thomas D. Y. A family of yeast expression vectors containing the phage f1 intergenic region. Gene. 1987;52(2-3):225–233. doi: 10.1016/0378-1119(87)90049-7. [DOI] [PubMed] [Google Scholar]
- Whiteway M., Freedman R., Van Arsdell S., Szostak J. W., Thorner J. The yeast ARD1 gene product is required for repression of cryptic mating-type information at the HML locus. Mol Cell Biol. 1987 Oct;7(10):3713–3722. doi: 10.1128/mcb.7.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M., Szostak J. W. The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell. 1985 Dec;43(2 Pt 1):483–492. doi: 10.1016/0092-8674(85)90178-3. [DOI] [PubMed] [Google Scholar]
- Yoshida A., Lin M. NH 2 -terminal formylmethionine- and NH 2 -terminal methionine-cleaving enzymes in rabbits. J Biol Chem. 1972 Feb 10;247(3):952–957. [PubMed] [Google Scholar]
- Zweidler A. Resolution of histones by polyacrylamide gel electrophoresis in presence of nonionic detergents. Methods Cell Biol. 1978;17:223–233. [PubMed] [Google Scholar]