Abstract
The ability to perturb individual genes in genome-wide experiments has been instrumental in unraveling cellular and disease properties. Here we introduce, describe the assembly, and demonstrate the use of comprehensive and versatile transcription activator-like effector (TALE) libraries. As a proof of principle, we built an 11-mer library that covers all possible combinations of the nucleotides that determine the TALE-DNA binding specificity. We demonstrate the versatility of the methodology by constructing a constraint library, customized to bind to a known p53 motif. To verify the functionality in assays, we applied the 11-mer library in yeast-one-hybrid screens to discover TALEs that activate human SCN9A and miR-34b respectively. Additionally, we performed a genome-wide screen using the complete 11-mer library to confirm known genes that confer cycloheximide resistance in yeast. Considering the highly modular nature of TALEs and the versatility and ease of constructing these libraries we envision broad implications for high-throughput genomic assays.
Transcription activator-like effectors (TALEs) are proteins secreted by Xanthomonas bacteria to target specific DNA sequences1,2. TALE family members contain a highly conserved and repetitive region (Fig. 1a) and their DNA binding capacities are dependent on the amino acid positions 12 and 13 (i.e. repeat variable diresidue or RVD) of each modular repeat3,4. Recent advances show that TALE proteins consist of a modular DNA-binding domain (DBD) that can be rapidly synthesized de novo5. The ability to complement the DBD with a functional domain permits a wide range of applications that include gene activation, repression, and nucleotide deletion and insertion, in a variety of model organisms and cell types6,7,8,9,10.
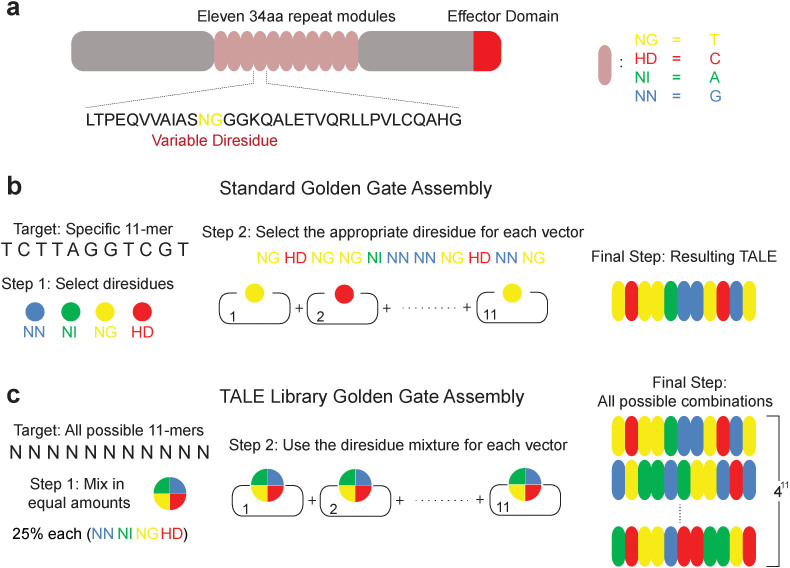
Figure 1. Construction of an 11-mer TALE-VP16 library.
(a) Schematic illustration of a TALE protein depicting the tandem repeat domain and the variable diresidues (RVDs). (b) Schematic illustration of a typical TALE assembling reaction. Corresponding RVDs were chosen for specific nucleotide targets (NI for A, HD for C, NG for T, and NN for G). (c) Schematic illustration of the TALE library assembling. For each position, equal amounts of all four building modules were used, which results in an 11-mer TALE library covering all possible 11-mer DNA targets.
In a parallel domain, high-throughput genome-wide screening experiments continue to provide critical insight into biology and medicine, shedding light on traditional cellular phenotypes, disease networks, and for reverse engineering endogenous pathways. Unfortunately, currently available methods are highly constrained, expensive, and often suffer from non-specific target effects11,12,13,14,15. Traditionally, these assays involve mutations or RNAi and explore the effect of loss of function coupled with selective pressure15,16,17. The complement, or gain-of-function, can be partially accomplished in yeast with introduction of cDNA18. In mammalian cells, a recently developed methodology provides a path for the use of zinc finger19 proteins for genetic screens20,21. This protein-based technology can expand the repertoire of genetic screening in human cells by allowing the delivery of a variety of functional domains. In addition, RNAi-based screening is extensively used for genome-wide screens11,22, and typically relies on shRNA delivery. Finally, the CRISPR-Cas9 system was recently used to perform genetic screens in human cells using genome-scale lentiviral single-guide RNA (sgRNA) libraries23,24.
Here, we introduce novel transcription activator-like effector (TALE) libraries. Our assembly methodology is rapid, modular, and allows precise control of the resulting RVD content and overall DNA binding domain composition. As a proof of principle, we built and verified two libraries: a sequence-constrained library to target a known p53 motif and an 11-mer library that spans all possible 11-nucleotide DNA targets (i.e. resulting to a maximum of 4,194,304 combinations). In contrast, an alternative Golden-Gate assembly system that relies on 432 plasmids assembled using a multi-well plate format, produced a TALE Nuclease (TALEN) library that targets 18,740 exclusively protein-coding gene targets25.
Results
Assembly and verification of the libraries
To assemble our TALE libraries, we modified the Golden Gate protocol (Fig. 1b)5,26. The enabling modification is the use of a predetermined mixture of the four building modules during the first step of the assembly (Fig. 1c). The flanking sequences of the four modules are identical. Therefore, during the enzyme-based digestion and ligase-based ligation reactions these modules have an equal probability in appearing in the final TALE construct (Methods and Supplementary Material). To test the library fidelity we subjected our 11-mer TALE mixture to standard Sanger sequencing using primers flanking the TALE DNA binding domain (Supplementary Table 1, P23 and P24). The sequencing results show that there are 6-nucleotide long repeats, spaced by 102 nucleotides, which have variable signature (Supplementary Fig. 1a). This general profile perfectly matches the TALE tandem repeat structure which contains 102 nucleotides and has RVDs which are 6-nucleotides in length. Importantly the composition of different peaks within these 6-nucleotide noisy elements closely tracks the expected composition. In particular, when equal amounts of all four possible RVDs are used for all positions, the first nucleotide position should consist of 75% A and 25% C (Supplementary Fig. 1b) and indeed the results have the expected sequencing profile (Supplementary Fig. 1c). Similarly, at position 2 of the RVD sequence, we should have only nucleotide A which was confirmed by sequencing (Supplementary Fig. 1c). The sequencing results confirm all positions.
The versatility of the proposed methodology of TALE library construction, perhaps, can be best illustrated by the simplicity in generating specific variants, namely sequence or nucleotide-biased TALE libraries (Supplementary Fig. 2). A sequence-biased TALE library has constrained nucleotides, and is assembled to bind to a specific transcription factor motif, while a nucleotide-biased TALE library has a predetermined ratio of the building modules resulting in desired nucleotide distributions (e.g. GC-rich).
As an example, we assembled a sequence-biased library to target a p53 motif. The p53-responsive element has been identified27 as two copies of the decamer motif 5′-RRRCWWGYYY-3′ (R = A or G, W = A or T, Y = C or T) separated by 0 to 13 random nucleotides. Accordingly, we prepared an 11-mer TALE library which targets 5′-RRRCWWGYYYN-3′ (Methods and Supplementary Material). The validity of this library was again verified by Sanger sequencing, and the results closely match the desired RVD patterns (Supplementary Fig. 3).
TALE library-based yeast one-hybrid assay
We then explored the functional integrity of our complete 11-mer TALE library using a yeast one-hybrid assay (Methods, Yeast One-hybrid Assay). A VP16 activation domain was added to the 11-mer library to enhance the transactivation activities. For the assay, we cloned part of the 5′untranslated region (UTR) and the open reading frame (ORF) of the human SCN9A gene (Supplementary Table 2) in front of an antibiotic resistance gene, Aureobasidin A (Aba) in yeast (bait), and applied our library (prey) (Fig. 2a). We used 50 (15 cm diameter) plates, which resulted to approximately 1 million individual TALE clones (Supplementary Material, Yeast one-hybrid). For assays that require comprehensive library coverage the protocol can be scaled up by increasing the number of plates. The positive clones were confirmed by re-streaking on Aba-containing agar plates (Supplementary Fig. 4). Thirteen of the TALE-VP16 expression plasmids were then rescued and sequenced to extract the RVD sequences (Supplementary Table 3).
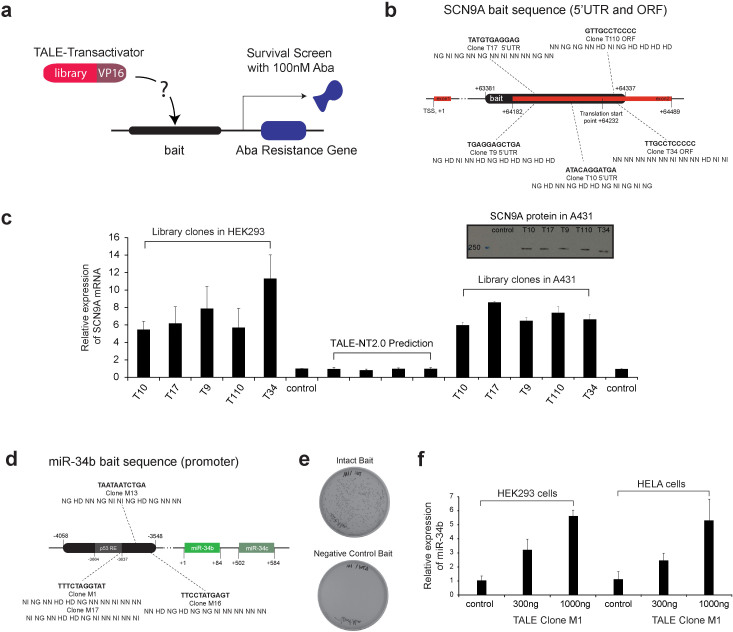
Figure 2. Isolation of TALE-VP16 fusions targeting the human SCN9A gene using the 11-mer TALE-VP16 library and the yeast one-hybrid assay.
(a) Schematic illustration of the yeast one-hybrid assay using the 11-mer TALE-VP16 library. A bait sequence was cloned in front of an antibiotic resistance gene (Aba resistance gene) in yeast. The 11-mer TALE-VP16 library was then transformed into this stable clone and a surviving assay was performed on -Leu plates containing 100 nM Aba. (b) The RVD sequences of isolated TALE-VP16 fusions and their targets within the human SCN9A bait sequence (scale not proportional). TALE-VP16 fusions were shown to bind to both the plus and the minus strands of the SCN9A bait sequence. (c) The isolated TALE-VP16 fusions induced overexpression of endogenous SCN9A in HEK293 cells and A431 cells. The mRNA levels of SCN9A were determined by quantitative RT-PCR. An empty vector (PEF-1) was used as the control. Columns 1-6: All TALE-VP16 fusions were able to effectively induce the overexpression of SCN9A in HEK293 cells (n = 5). Columns 11-16: All TALE-VP16 fusions effectively induced the overexpression of SCN9A in A431 cells (n = 3). Columns 7–10: All 4 TALEs designed according to TALE-NT 2.0 failed to induce the overexpression of SCN9A in HEK293 cells (n = 3). Inlet: Western blot shows that all TALE-VP16 fusions induced the overexpression of SCN9A protein in A431 cells (representative data of two independent experiments). (d) The RVD sequences of isolated TALE-VP16 fusions and their targets within the human miR-34b/c bait sequence (scale not proportional). (e) Confirmation of the binding between isolated TALE-VP16 fusion M1 and its predicted gene target within the human miR-34b/c bait sequence. The isolated clone M1 was predicted to target 5′-TTTCTAGGTAT-3′ within the miR-34b/c bait sequence. The full-length bait sequence (pAbAi-miR-34b/c) or bait with the predicted target site deleted (pAbAi-miR-34b/c (ΔTTTCTAGGTAT)) was stably integrated into yeast cells. TALE-VP16 fusion clone M1 was then transformed into either cell line. Only cells which contained the intact bait sequence survived the 100 nM Aba selection. (f) The isolated TALE-VP16 fusion M1 effectively induced overexpression of miR-34b in a dose-dependent manner in both HEK293 and HeLa cells (n = 3 for both cell lines).
Using TALE-NT 2.0 (TALE Effector Nucleotide Targeter 2.0)28, we were able to confirm that these positive clones are predicted to bind to either the plus or minus strand of three specific locations within the SCN9A bait sequence. Five of the TALE-VP16 clones are illustrated together with their corresponding position (Fig. 2b). To confirm these results, we first generated baits that exclude those predicted DNA target sites. For example, the positive clone T110 induced the expression of Aba resistance gene when the bait sequence is intact (Supplementary Fig. 5a) but it failed to produce surviving clones when the specific DNA target site (5′-GTTGCCTCCCC-3′) was removed (Supplementary Fig. 5b). Subsequently, we cloned these TALE-VP16 fusions into a mammalian expression vector, transiently transfected the plasmids into human embryonic kidney cells (HEK293) and human squamous carcinoma cells (A431), and measured the expression level of SCN9A by quantitative RT-PCR (Methods, Quantitative reverse transcription-PCR). All fusions were able to effectively drive the overexpression of the SCN9A gene in both cell lines (up to 11-fold increase for HEK293 cells and 8.5-fold increase for A431 cells) (Fig. 2c).
In line with these results, the TALE-VP16 fusions also induced the overexpression of SCN9A protein in A431 cells (Fig. 2c, and Supplementary Fig. 6b). Interestingly, we were not able to detect SCN9A protein in HEK293 cell samples, including those transfected with the fusion constructs (Supplementary Fig. 6c). The discrepancy between the mRNA and protein measurements for HEK293 cells may be due to regulatory translational or post-translational mechanisms on the expression of SCN9A29.
We also investigated whether the TALE fusions isolated from our screening were compatible with a virus-based delivery system for potential applications in primary cells. We prepared a TALE T34 recombinant adeno-associated viral vector using the AAV helper-free system (Agilent Technologies). This AAV viral stock was applied to HEK293 cells at a multiplicity of infection (MOI) of 400 and induced the overexpression of the SCN9A gene by 2 fold (Supplementary Fig. 7).
Furthermore, we tested the performance of our TALE-based yeast one-hybrid screening approach for microRNA gene targets. Specifically, we used part of the promoter sequence of the human miR-34b/c gene as the bait sequence (Supplementary Table 2) and isolated four positive clones, which were confirmed by re-streaking on Aba-containing plates (Supplementary Fig. 8). The RVD sequences of these four clones were extracted (Supplementary Table 3) and all are predicted to target the bait (Fig. 2d). Interestingly, two of the clones (M1, M17) target the same sequence within the miR-34b/c promoter (Fig. 2d). Again, we used two methods to confirm the results. We first generated a bait mutant which excludes the predicted target sequence for clone M1 (5′-TTTCTAGGTAT-3′) and prepared the according yeast stable cell line. As expected, in these yeast cells clone M1 failed to induce the expression of Aba resistance gene (Fig. 2e). Furthermore, the TALEM1-VP16 cDNA was cloned into a mammalian expression vector and transiently transfected into HEK293 and HeLa cells. The TALE fusion effectively induced the overexpression of miR-34b in a dose-dependent manner (Fig. 2f).
Our results clearly demonstrate that the complete library can be used to isolate TALEs that bind to a desired DNA sequence. Importantly, we observed that TALE-VP16 can be designed to bind to the minus strand of a target sequence and in addition, can also be designed to target sequences within the ORF. We emphasize that one fundamental difference between our approach and all previous applications of TALEs is that our screening procedure does not rely on TALE-DNA binding prediction algorithms28,30. This is a considerable advantage as available algorithms may not yet fully capture the dynamics of TALE-DNA interactions. Indeed, we designed four 11-mer TALE-VP16 constructs for the same SCN9A bait sequence using the TALE-NT 2.0 program (Supplementary Table 3) and observed that all fusions, as well as combinations, failed to induce the overexpression of SCN9A mRNA levels in HEK293 (Fig. 2c, Supplementary Figs. 7 and 9). Additional insight can be gathered from recovered RVDs (Supplementary Table 3), as none of the isolated positive TALE-VP16 clones contain RVD sequences exactly matching the canonical TALE-DNA binding code. As an example, for the target sequence 5′-GTTGCCTCCCC-3′, instead of the predicted canonical RVD sequence the four isolated clones contained at least one RVD difference. Therefore, the TALE-DNA binding information extracted from our library can be used in the future to refine the TALE-DNA binding algorithms.
TALE library-based benchmark genomic screen in yeast
We next applied our library to screen for TALE-VP16 fusion proteins which confer resistance to cycloheximide in yeast. The underlying mechanisms of cycloheximide resistance are well-studied. For example, in yeast S. cerevisiae, overexpression of ATP-binding cassette (ABC) transporters such as Pdr5p has been shown to contribute to cycloheximide resistance31. In addition, the expression of the PDR5 gene is known to be positively regulated by two homologous zinc finger-containing transcription regulators, Pdr1p and Pdr3p32. We first determined the lowest concentration cycloheximide (0.5 µg/ml) at which the wild-type yeast cells failed to grow (Supplementary Material). We applied the complete TALE-VP16 library and isolated eighteen positive clones which tolerate cycloheximide (e.g., selected clones illustrated in Supplementary Fig. 10). Subsequently, we isolated the TALE-VP16 fusion plasmids and re-transformed them back to the wild-type cells for confirmation, as in the original screen natural mutations of the yeast genome (both gain-of-function33 and loss-of-function34) can artificially increase the resistance to cycloheximide. Five genuine positive clones were confirmed (Fig. 3a), isolated and sequenced to extract the TALE RVD sequences (Supplementary Table 3).
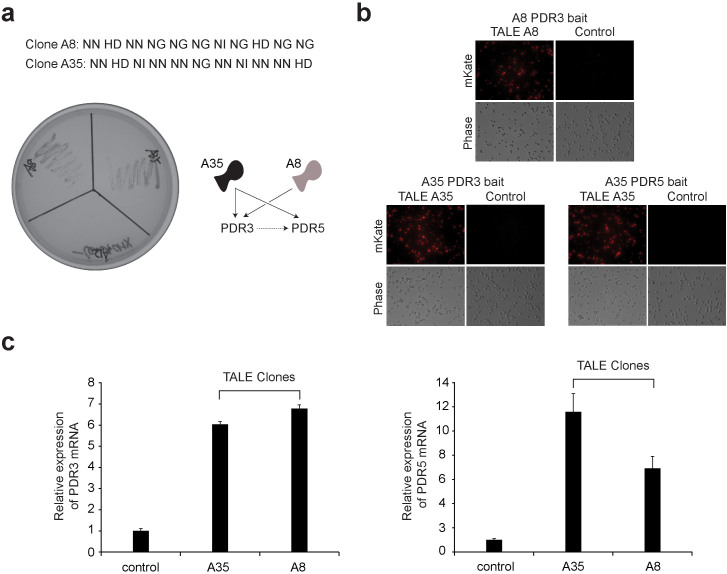
Figure 3. Genetic screen for cycloheximide resistance in yeast using the TALE-VP16 plasmid library.
(a) Confirmation of the genuine positive yeast clones conferring cycloheximide resistance. 18 positive clones were isolated from the cycloheximide resistance screening. Subsequently, these positive TALE-VP16 fusion plasmids were recovered and again re-transformed into the wild-type yeast cells. The transformed cells were then re-streaked onto –Leu plates containing 0.5 µg/ml of cycloheximide. After 3 days, cells transformed with genuine positive clones pGADT7-TALE-A8-VP16 and pGADT7-TALE-A35-VP16 were able to grow robustly. In contrast, cells transformed with the false positive clone pGADT7-TALE-A12-VP16 or the control pGADT7 failed to grow. (b) The isolated TALE-VP16 fusion clones A8 and A35 bind to the promoters of the PDR3 and PDR5 genes. TALE-VP16 fusion clones A8 and A35 were isolated from cycloheximide resistance screening. Both A8 and A35 were predicted to bind to the promoter of PDR3 gene. In addition, A35 was predicted to target the promoter of the PDR5 gene. Four copies of the predicted PDR3/PDR5 promoter targets and their immediate adjacent sequences were cloned in front of a fluorescence reporter gene (mKATE2) in yeast (bait). These yeast stable clones were then transformed with corresponding pGADT7-TALE-A8-VP16, pGADT7-TALE-A35-VP16 or pGADT7 (control). TALE-VP16 fusion clones A35 or A8 potently induced the expression of mKATE2 in yeast cells containing the corresponding baits, while pGADT7 failed to do so (right). (c) The isolated TALE-VP16 fusions A8 and A35 induced overexpression of endogenous PDR3 and PDR5 genes. Wild-type yeast cells were transformed with pGADT7-TALE-A8-VP16, pGADT7-TALE-A35-VP16 or pGADT7 (control). The expression levels of PDR3/PDR5 were measured by quantitative RT-PCR. Both clones were able to effectively induce the overexpression of PDR3 (n = 3) and PDR5 (n = 3).
The sequencing results indicate that two clones (A8, A35) are predicted to target the promoter of the PDR3 gene and, in addition, A35 is also predicted to bind to the promoter of the PDR5 gene (Fig. 3a). Two methods were used to confirm the results. First, we cloned four copies of the predicted PDR3 and PDR5 promoter targets and their immediate adjacent sequences in front of a fluorescence reporter gene (mKate2) in yeast. We then transformed the yeast stable clones with plasmids carrying the TALEs. Indeed both clones induced the expression of mKate2 in cells containing the bait (Fig. 3b). Second, we prepared yeast cells transformed with TALEA8-VP16, TALEA35-VP16 or empty vector and measured the expression levels of PDR3 and PDR5 by quantitative RT-PCR. Again, both clones were able to effectively induce the expression of PDR3 and PDR5 (Fig. 3c).
Discussion
Here, we establish a methodology for the assembly of complete and biased TALE-based libraries. As proof-of-concept, we demonstrate that our methodology can be applied towards designing effective TALEs for any given DNA target (i.e. the yeast-one-hybrid experiments) as well as for genome-wide phenotype screens in yeast.
We show experimentally that the standard TALE design algorithm can fail to predict RVDs that bind to a desired target. Therefore, in the unforeseen cases where computational methods are not successful, the yeast-one-hybrid approach provides an algorithm-independent alternative towards designing TALEs.
A potential pitfall is that instead of the desired bait sequence a TALE may bind to the antibiotic resistance plasmid backbone sequence and induce the expression of the resistance gene. In our case, the negative control experiments (deletion of target) provide proof that the selected TALEs target exclusively the bait (Supplementary Fig. 5 and Fig. 2e). More generally, to avoid false-positives in the TALE-based yeast-one-hybrid assays, we recommend using bait sequences that are sufficiently long, ideally larger than 500 bp (Supplementary Material, Yeast-one-hybrid assay false-positives). We also presume that multiple copies of the targets can be used in cases when the target sequence is short.
An advantage of TALEs compared to other technologies (e.g., RNAi, CRISPR35,36,37) is that the size of the DNA target can be controlled. Increasing the DNA target size will result to superior specificity, albeit at the cost of increasing the library complexity and reducing the coverage in yeast screens. Therefore, a practical strategy towards eliminating off-target effects is to use a relatively short library (e.g., 11-mer) for screening purposes and a posteriori extend the DBDs of the screened TALEs to include flanking target sequences.
An alternative approach towards increasing the size of the targets of the library, without introducing additional experimental burden or increasing the complexity, involves utilizing the RVD properties. For example, it is known that the RVD NN can target both A and G. Therefore, only three modules (i.e. NN, NG and HD) can be used for the assembly of the complete TALE libraries. As an example, a 14-mer TALE library with these three modules will result in a complexity of 314 = 4,782,969. The assembly of larger TALE libraries can be experimentally cumbersome and in some cases practically prohibitive without the use of automated assembly systems.
One additional challenge lies in the fact that there is a certain degree of uncertainty in terms of deciphering the TALE-DNA binding given a RVD sequence, which may complicate the identification of potential target sequences in genome-wide assays. This issue can be resolved experimentally using ChIP-seq38.
In summary, we have successfully established a methodology for the assembly of versatile TALE-based libraries and we demonstrated yeast-one-hybrid and genome-wide screens in yeast cells. We further emphasize that the TALE DBD can be fully customized to target any nucleotide content (e.g., GC-rich areas), nucleotide constraints (e.g., transcription factor motifs), or other modifications (e.g., methylations39,40). For the latter, during the construction of the TALE library, one additional RVD building block can be included in the assembly, namely N*. The star indicates that the amino acids at position 13 of TALE repeats are missing, which has been shown39 to overcome the steric hindrance from the additional methyl moiety present at position 5 of thymine and thus allow efficient recognition of 5-methylated cytosine.
The modularity of the functional domain allows for a range of targeted manipulations including, but not limited to, transactivators, transrepressors, methylases, demethylases, recombinases, and nucleases41. More generally, with TALE libraries we have the ability to introduce multiple rounds of positive and negative screening, and even introduce a combination of positive and/or negative action to study synergistic effects. Finally, the TALE protein coding sequences can be easily isolated and sequenced, which can facilitate the rapid identification of their target genes. To conclude, considering the highly modular nature of TALEs, the ability to control their size and thus fine-tune their specificity, and the versatility and ease of constructing these libraries we foresee a broad range of applications in the future.
Methods
Library construction
A modified protocol based on the Golden Gate Assembly was used to construct the 11-mer TALE-VP16 library. First, a pFUS_A library containing all possible combinations of 10 tandem repeat (RVDs 1-10) and a pFUS_B library containing all combinations of 1 tandem repeat (RVD 11) were prepared as described in Cermak, T. et al.5, except that for each position in a standard 30 µL reaction, 37.5 ng of each of the NN, NI, NG, and HD building blocks were mixed and included. Next, the pFUS_A and pFUS_B plasmid libraries were conjoined using the standard protocol. Similar procedures were used to prepare the p53-responsive element-targeting TALE-VP16 libraries. For each position of the Golden Gate Assembly reactions, the amounts of the four building blocks were adjusted according to the desired nucleotide target. All libraries were then subjected to standard Sanger sequencing using the forward primer P23 and the reverse primer P24 (Supplementary Table 1). The sequencing profiles were then analyzed using FinchTV (Geospiza). For full description, see Supplementary Material.
Yeast one-hybrid assay
The Matchmaker Gold Yeast One-Hybrid Screening System was used (Clontech, #630491). The TALE-VP16 library-based yeast one-hybrid assay was performed according to the protocol (Clontech, protocol #PT4087-1). The positive clones were screened by using SD/-Leu agar medium (Clontech, #630311) containing either 100 ng/ml Aureobasidin A (Clontech, #630466) or 0.5 µg/ml cycloheximide (Sigma-Aldrich, #C7698-1G). Subsequently, the positive clones were cultured in SD/-Leu broth (Clontech, #630310) and the pGADT7-TALE-VP16 plasmids were then prepared using the PrepEase Yeast Plasmid Isolation Kit (Affymetrix, #79220). The plasmids were then rescued into XL10-Gold Ultracompetent cells (Agilent, #200315) and prepared using the Qiagen Plasmid Mini Kit (Qiagen, #12123). For full description, see Supplementary Material.
Prediction of target sequences
All rescued pGADT7-TALE-VP16 plasmids were subjected to standard Sanger sequencing using the forward primer P23 and the reverse primer P24 (Supplementary Table 1), which are flanking sequences of the DNA binding domain of TALE proteins. After extracting the RVD sequences, the TALE potential target sequences were determined by the Target Finder function of TALE-NT 2.028 using the setting “upstream base T”. Since TALE-NT 2.0 only supports sequences which are between 12 and 31 RVDs, for each TALE we conducted four searches by adding one additional RVD (HD or NN or NI or NG) to the end of our RVD sequences. The lowest score from one of these four searches was defined as “the lowest possible score”. The targets were considered as potentially genuine only when the lowest possible score is less than 9, as lower scores indicate higher confidence of the bindings between TALE fusions and their predicted DNA target sequences.
Mammalian cell culture and transfection
All mammalian cells were maintained at 37°C, 100% humidity and 5% CO2. HEK293, A431 and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM, Invitrogen, #11965-1181) supplemented with 10% Fetal Bovine Serum (FBS, Atlanta Biologicals, #S11550), 0.1 mM MEM non-essential amino acids (Invitrogen, #11140-050), 0.045 units/mL of Penicillin and 0.045 units/mL of Streptomycin (Penicillin-Streptomycin liquid, Invitrogen, #15140). For transient transfection, ~300 thousand cells in 1 mL of complete medium were plated into each well of 12-well culture treated plastic plates (Griener Bio-One, #665180) and grown for 16-20 hours. For jetPRIME (Polyplus Transfection, #114-15) transfection, up to 1 µg of the plasmid was added to 75 µL of jetPRIME buffer and 1.75 µL of jetPRIME. Transfection solutions were mixed and incubated at room temperature for 10 min. The transfection mixture was then applied to the cells and mixed with the medium by gently shaking.
Preparation of recombinant AAV vectors
The AAV helper-free system was purchased from Agilent Technologies (#240071). HEK293 cells were seeded on 10-cm cell culture treated petri dishes (Griener Bio-One, #664160). After reaching approximately 70–80% confluence, the cells were transfected with 3.3 µg of AAV-SCN9A-T34, 3.3 µg of pHELPER plasmid and 3.3 µg of pAAV-RC plasmid using 30 µL of jetPRIME. 72 hours post-transfection, the cells were scraped into the growth medium and pelleted by centrifugation at 1000 rpm for 5 min. The supernatant was then removed and the cell pellets were re-suspended in 1 mL of complete growth medium before being subjected to 4 rounds of freeze-thaw cycles between the dry ice-ethanol bath and the 37°C water bath. Subsequently, the cellular debris was collected by centrifugation at 10,000 × g for 10 min. The supernatant (primary recombinant AAV virus stock) was transferred into a fresh tube and stored at −80°C. The QuickTiter AAV Quantitation Kit (Cell Biolabs, #VPK-145-T) was used to determine the titers (genome copy/mL, GC/mL) of the recombinant AAV viral stocks. In brief, any free nucleic acids were first removed and the intact AAV viruses were captured using the QuickTiter AAV Capture Matrix. The captured viruses were then denatured to release the viral nucleic acids, which were subsequently labeled with fluorescence tags. The fluorescent intensities were measured using the FLUOstar Omega plate reader (BMG Labtech) with a 480/520 nm filter set.
Western blot assay
A431 or HEK293 cells were lysed in RIPA buffer (Sigma-Aldrich, #R0278) with protease inhibitors (Sigma-Aldrich, #P8340). Protein concentrations were measured using the Coomassie Plus Bradford assay kit (Thermo Scientific, #23236). For Western blot, 15 µg of whole cell lysates were mixed with loading buffer and boiled for 5 min, and then were run in 4–15% Mini-PROTEAN TGX polyacrylamide gel (Bio-Rad, #456-1083). After transferring, the nitrocellulose membranes were first blotted with 50 mM Tris, 150 mM NaCl and 0.1% Tween-20 (TBS-T) with 5% nonfat milk for 30 min at room temperature. The membranes were then incubated with either anti-SCN9A monoclonal antibody (1:500 dilution in TBS-T, Millipore, #MABN41) or anti-actin monoclonal antibody (1:5,000 dilution in TBS-T, Millipore, #MAB1501) at 4°C overnight. The membranes were then washed with TBS-T three times, each time for 10 min. Subsequently, the membranes were incubated with goat anti-mouse IgG (H+L) secondary antibody (1:10,000 dilution in TBS-T, Thermo Scientific, #31430) at room temperature for 45 min. The membranes were then washed with TBS-T four times, each time for 10 min. The membranes subsequently were incubated with ECL Western blotting substrates (Thermo Scientific, #32106) and then exposed to X-ray films (Sigma-Aldrich, #Z370398).
Quantitative reverse transcription-PCR (rtPCR)
For measurement of mRNAs in mammalian cells, total RNA was extracted using the RNeasy Mini Kit (Qiagen, #74104) 48 hours post transfection. First strand synthesis was performed using the QuantiTect Reverse Transcription Kit (Qiagen, #205311). Quantitative PCR was performed using the KAPA SYBR FAST Universal qPCR Kit (KAPA Biosystems, #KK4601). GAPDH was used for normalization. For measurement of mRNAs in yeast cells, the same protocol was followed, except that total RNA was extracted using the MasterPure Yeast Purification Kit (Epicentre, #MPY80200), and yeast ACT1 was used for normalization. For measurement of miRNAs in mammalian cells, total RNA was extracted using the miRNeasy Micro Kit (Qiagen, #217084). First strand synthesis was performed using the miScript II RT Kit with HiSpec Buffer (Qiagen, #218161). Quantitative PCR was performed using miScript SYBR Green PCR Kit (Qiagen, #218073). RNU6 was used for normalization. The quantitative analysis was performed using the 2−ΔΔCt method. The values of fold-change are reported as mean with standard deviation. For full description, see Supplementary Material.
Fluorescence microscopy
Bait yeast strains containing PDR3/PDR5 promoter target sequences were prepared as described in the yeast two-hybrid section. Subsequently, these yeast stable clones were transformed with corresponding pGADT7-TALE-VP16 plasmids and then grown on SD/-Leu agar medium. Individual positive clones (diameter 2–3 mm) were re-suspended in 1 mL of 0.9% (w/v) NaCl solution and transferred into 12-well culture treated plastic plates. Cells were allowed to fully settle and then imaged using an Olympus IX81 microscope in a Precision Control environmental chamber. The images were captured using a Hamamatsu ORCA-03 Cooled monochrome digital camera. The filter set (Chroma) for mKATE2 is ET560/40x (excitation) and ET630/75m (emission). Data collection and processing was performed using the software package Slidebook 5.0. All images were collected with the same magnification (40×) and exposure times and underwent identical processing.
Author Contributions
Y.L. and L.B. conceived the project and designed the experiments. Y.L. performed the majority of the experiments. K.E. performed TALE-NT2.0 cloning and viral experiments. Y.L. and L.B. analyzed the data. Y.L., K.E., M.Z. and L.B. prepared the manuscript. L.B. supervised the project.
Supplementary Material
Supplementary Material
Acknowledgments
This work was funded by the US National Institutes of Health (NIH) grants GM098984, GM096271, CA17001801, the US National Science Foundation (NSF) grant CBNET-1105524, and the University of Texas at Dallas. We would like to thank S. Baek, M.Guinn, S. Lin and S. Shah for technical support, and the Levene Lab for assistance with Western blot assays.
References
- Zhou J. & Chai J. Plant pathogenic bacterial type III effectors subdue host responses. Curr Opin Microbiol 11, 179–185 (2008). [DOI] [PubMed] [Google Scholar]
- Boch J. & Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 48, 419–436 (2010). [DOI] [PubMed] [Google Scholar]
- Moscou M. J. & Bogdanove A. J. A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501–1501 (2009). [DOI] [PubMed] [Google Scholar]
- Boch J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009). [DOI] [PubMed] [Google Scholar]
- Cermak T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39, e82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q. et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 12, 238–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Zhou R., Kuo Y., Cunniff M. & Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun 3, 968 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J. & Stern D. L. TALE-mediated modulation of transcriptional enhancers in vivo. Nat Methods 10, 762–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 29, 149–153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Liu B., Spalding M. H., Weeks D. P. & Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30, 390–392 (2012). [DOI] [PubMed] [Google Scholar]
- Kaelin W. G. Use and Abuse of RNAi to Study Mammalian Gene Function. Science 337, 421–422 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. D. Assessing the size of gene or RNAi effects in multifactor high-throughput experiments. Pharmacogenomics 11, 199–213 (2010). [DOI] [PubMed] [Google Scholar]
- Cullen B. Enhancing and confirming the specificity of RNAi experiments. Nat Methods 3, 677–681 (2006). [DOI] [PubMed] [Google Scholar]
- Jackson A. L. & Linsley P. S. Noise amidst the silence: off-target effects of siRNAs? Trends Genet 20, 521–524 (2004). [DOI] [PubMed] [Google Scholar]
- Esvelt K. M. & Wang H. H. Genome-scale engineering for systems and synthetic biology. Mol Syst Biol 9, 641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S., Bakal C. & Perrimon N. Genomic Screening with RNAi: Results and Challenges. Annu Rev Biochem 79, 37–64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root D. E., Hacohen N., Hahn W. C., Lander E. S. & Sabatini D. M. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods 3, 715–719 (2006). [DOI] [PubMed] [Google Scholar]
- Jones G. M. et al. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat Methods 5, 239–241 (2008). [DOI] [PubMed] [Google Scholar]
- Beerli R. R., Dreier B. & Barbas C. F. Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci USA 97, 1495–1500 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez B. et al. Modular system for the construction of zinc-finger libraries and proteins. Nat Protoc 5, 791–810 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancafort P., Magnenat L. & Barbas C. F. Scanning the human genome with combinatorial transcription factor libraries. Nat Biotechnol 21, 269–274 (2003). [DOI] [PubMed] [Google Scholar]
- Ute K. et al. Insights into Effective RNAi Gained from Large-Scale siRNA Validation Screening. Oligonucleotides 17, 237–250 (2007). [DOI] [PubMed] [Google Scholar]
- Wang T., Wei J. J., Sabatini D. M. & Lander E. S. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O. et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. et al. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol 31, 251–258 (2013). [DOI] [PubMed] [Google Scholar]
- Li Y., Moore R., Guinn M. & Bleris L. Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Sci Rep 2, 897 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Lane D. & Levine A. J. Surfing the p53 network. Nature 408, 307–310 (2000). [DOI] [PubMed] [Google Scholar]
- Doyle E. L. et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res 40, W117–W122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghouts C. et al. The Intracellular Delivery of a Recombinant Peptide Derived from the Acidic Domain of PIAS3 Inhibits STAT3 Transactivation and Induces Tumor Cell Death. Mol Cancer Res 8, 539–553 (2010). [DOI] [PubMed] [Google Scholar]
- Grau J. et al. Computational Predictions Provide Insights into the Biology of TAL Effector Target Sites. PLoS Comput Biol 9, e1002962 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzi E., Wang M., Leterme S., Van Dyck L. & Goffeau A. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J Biol Chem 269, 2206–2214 (1994). [PubMed] [Google Scholar]
- Katzmann D. J., Burnett P. E., Golin J., Mahe Y. & Moye-Rowley W. S. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol Cell Biol 14, 4653–4661 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourani A., Papajova D., Delahodde A., Jacq C. & Subik J. Clustered amino acid substitutions in the yeast transcription regulator Pdr3p increase pleiotropic drug resistance and identify a new central regulatory domain. Mol Gen Genet 256, 397–405 (1997). [DOI] [PubMed] [Google Scholar]
- Gerlinger U., Gückel R., Hoffmann M., Wolf D. H. & Hilt W. Yeast cycloheximide-resistant crl mutants are proteasome mutants defective in protein degradation. Mol Biol Cell 8, 2487–2499 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31, 822–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey T. S. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet 13, 840–852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton J. et al. Overcoming Transcription Activator-like Effector (TALE) DNA Binding Domain Sensitivity to Cytosine Methylation. J Biol Chem 287, 38427–38432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D. et al. Recognition of methylated DNA by TAL effectors. Cell Res 22, 1502–1504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P. et al. Specific DNA-RNA Hybrid Recognition by TAL Effectors. Cell Rep 2, 707–713 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material