Abstract
The prevalence of toxin/antitoxin (TA) systems in almost all genomes suggests they evolve rapidly. Here we show that an antitoxin from a type V system (GhoS, an endoribonuclease specific for the mRNA of the toxin GhoT) can be converted into a novel toxin (ArT) simply by adding two mutations. In contrast to GhoS, which increases growth, the new toxin ArT decreases growth dramatically in Escherichia coli. Transmission electron microscopy analysis revealed that the nucleoid in ArT-producing cells is concentrated and appears hollow. Whole-transcriptome profiling revealed ArT cleaves 50 additional transcripts, which shows that the endoribonuclease activity of GhoS has been broadened as it was converted to ArT. Furthermore, we evolved an antitoxin for the new toxin ArT from two unrelated antitoxin templates, the protein-based antitoxin MqsA and RNA-based antitoxin ToxI, and showed that the evolved MqsA and ToxI variants are able to counteract the toxicity of ArT. In addition, the de novo TA system was found to increase persistence, a phenotype commonly associated with TA systems. Therefore, toxins and antitoxins from disparate systems can be interconverted.
The ubiquity of toxin/antitoxin (TA) systems in Archaea1 and Bacteria2 appears to be counterintuitive given the cornerstone of Darwinian evolution of increased fitness through natural selection3. TA systems consist of toxic proteins that disrupt cellular processes along with labile antitoxins that counteract this growth inhibition. Without stress, antitoxins mask the toxins and growth is rapid. However, during environmental stress, antitoxins are degraded more rapidly4,5, and toxins are activated. This toxin-mediated reduced growth is therefore beneficial for bacteria for surviving antibiotic stress6,7,8 and for combating phage attack9. Hence, TA systems have evolved to serve physiological needs10.
TA systems can be found in mobile genetic elements (e.g., plasmids) and in chromosomes. The role of plasmid-derived TA systems in stabilizing plasmid copies has been clear since their first discovery11; however, the role of chromosomal-derived TA systems has slowly become more evident over the past three decades. Chromosomal TA systems are involved in tolerance to antibiotics6,7,8, biofilm formation12,13,14, host colonization15, survival16, and pathogenicity17,18. TA systems are also involved in elegant cellular regulation of genes other than their own loci including direct control of another TA system19, selective protein enrichment20,21, and the general stress response4,5.
Given the functional diversities of toxins and antitoxins22, it appears that individual components have been recruited independently from the genetic/proteomic pool to form new TA pairs. For instance, the GhoS antitoxin is homologous to the Cas2 protein from the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated proteins) system23. In the IetS/IetA system, the IetS toxin is highly similar to subtilisin-like serine proteases while the IetA antitoxin is similar to AAA-ATPases24. Mruk and Kobayashi also speculated, based on similarities in genetic structures, biological functions, and regulatory mechanisms, that TA systems and restriction-modification systems may be related25. Furthermore, there are at least 12 toxins/antitoxins acting as endoribonucleases in E. coli23,26, but not all of these are homologs, which means that these toxins/antitoxins must have arisen and assembled at separate times. These observations suggest functional overlap between TA modules and other proteins, and raise the question whether a de novo TA system can be built using extant protein components.
To explore the impact of evolution on TA systems and their physiological role in their host, we set out to test whether one of the most extreme changes in a TA system could occur; i.e., whether an antitoxin could be converted into a toxin. Our hypothesis is that existing components of TA systems are molecularly malleable, and thus, may be used to form novel TA pairs. We found that an antitoxin may be converted into a potent toxin with only two amino acid changes and that two disparate antitoxins (a protein and an RNA) may be created that mask the toxicity of this novel toxin. We refer to this new toxin as ArT (artificial toxin) and the antitoxins as ArA and ArA* (artificial antitoxin).
Results
TA systems are classified as type I when the RNA antitoxin inhibits the toxin mRNA, type II when the protein antitoxin inhibits the toxin protein, type III when the RNA antitoxin inhibits the toxin protein, type IV when the protein antitoxin shields the substrate from the toxin protein, and type V when the protein antitoxin degrades the toxin mRNA27. To synthesize a de novo artificial TA system, we used components from well-studied TA systems of different classes as starting materials: to create the toxin ArT, we started with type V antitoxin GhoS of the E. coli GhoT/GhoS TA system19,23 and to create the antitoxins using two different approaches, we started with type II antitoxin MqsA of the E. coli MqsR/MqsA TA system4,28 and type III antitoxin ToxI of the Erwinia carotovora subspecies atroseptica (also known as Pectobacterium atrosepticum) ToxN/ToxI system29. Error-prone PCR (epPCR) was employed as a means of introducing random mutations into the genes of these disparate components, and the impact on antibiotic persistence was used to assay the impact of the artificial TA module.
Random mutagenesis of ghoS to form artificial toxin ArT
GhoS is an antitoxin endoribonuclease that specifically cleaves the mRNA of toxin GhoT along with 20 mRNAs encoding proteins involved in purine or pyrimidine transport and biosynthesis23. GhoS is not toxic to E. coli and increases growth slightly when overproduced23. To create ArT, GhoS variants were selected based on reduced growth.
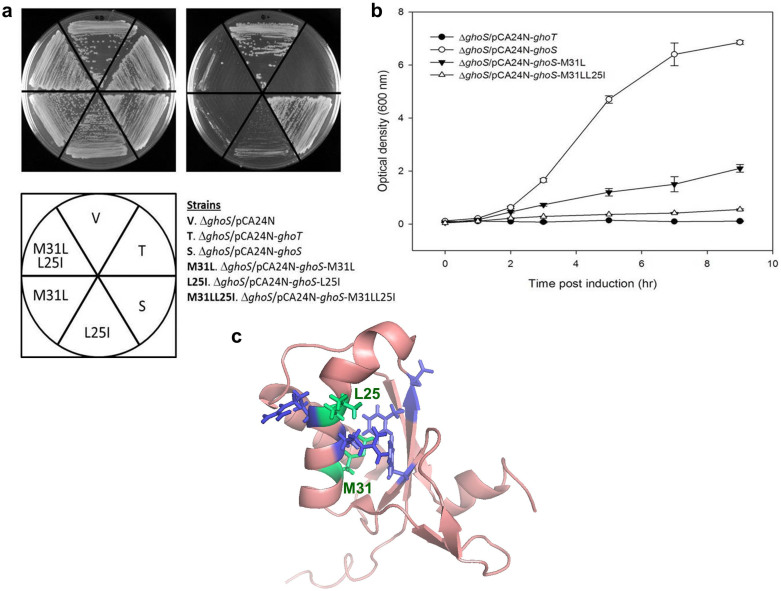
A plasmid library containing ~2 × 105 mutated GhoS variants was generated using epPCR with an error rate of 1.2%. In this plasmid library, all GhoS variants were expressed from an IPTG-inducible T5-lac promoter in pCA24N30. After transforming a ghoS deletion E. coli strain (BW25113 ΔghoS) with this library (to prevent interference by native GhoS), colonies of different size appeared in the absence of IPTG, indicating that the growth of the cells producing these GhoS variants was not uniform and that the basal level of expression of mutated GhoS was sufficient to result in impaired growth. We picked ~2,800 small colonies from this library, and the 10 colonies that grew the slowest on IPTG-containing agar were chosen for sequencing. Although these 10 GhoS variants harbor substitutions at various residues, four residues (D15, M31, E43, and N85) were repeatedly substituted (Fig. S1), and may be important for controlling the activity of GhoS. In particular, the M31L and L25I substitutions alone reduced colony formation of E. coli (Fig. 1a). Growth toxicity was further increased when L25I was introduced into GhoS-M31L to form GhoS-L25I-M31L (termed as ArT hereafter). Colony formation in cells expressing ArT was inhibited (Fig. 1a), and the growth of ArT-expressing strain in LB was reduced to a level comparable to the inhibitory effect of toxin GhoT (Fig. 1b). Therefore, antitoxin GhoS was converted successfully into toxin ArT.
Figure 1. Substitutions M31L and L25I convert GhoS to a toxin.
All plasmids were expressed in E. coli BW25113 ΔghoS. (a) Cells producing either GhoS-M31L, GhoS-L25I, or ArT (M31L + L25I) have reduced growth on LB agar containing 1 mM IPTG (upper right) after 24 h. All strains grew on LB agar without IPTG (upper left). Note that native GhoS does not confer toxicity to cells. The positive control, GhoT, is the membrane toxin from the GhoS/GhoT system. pCA24N denotes empty vector. (b) Reduced growth of cells expressing ArT in liquid LB after the addition of 1 mM IPTG (added at t = 0 h). Error bars indicate s.e.m. (n = 3). (c) Mutations M31L and L25I (green) mapped to the structure of GhoS (pdb: 2llz). Residues in blue denote conserved residues in GhoS.
ArT is independent of GhoT and MqsR and causes ghost-cell formation
We investigated whether the toxicity of ArT depends on either toxin GhoT or toxin MqsR; MqsR, the toxin from the MqsR/MqsA system, activates GhoT by cleaving the mRNA of ghoS19. When ArT was produced in a ghoT-deleted strain (BW25113 ΔghoT), ArT remained toxic (Fig. S2). Moreover, when ArT was produced in an mqsRA deletion strain (BW25113 ΔmqsRA), ArT remained toxic (Fig. S3). Hence, the toxicity of ArT is novel and it is independent of toxins MqsR and GhoT.
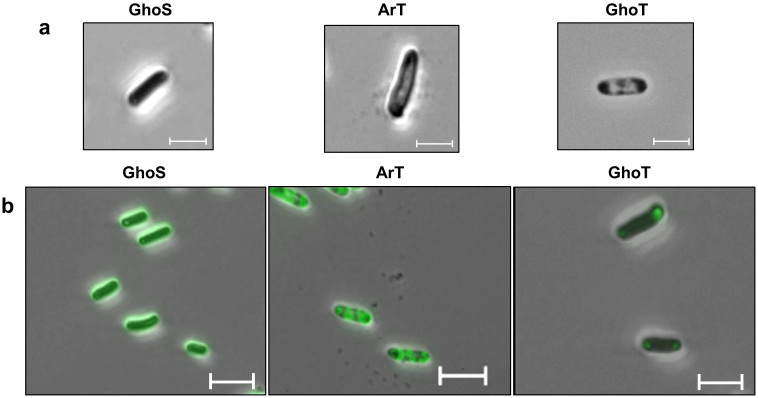
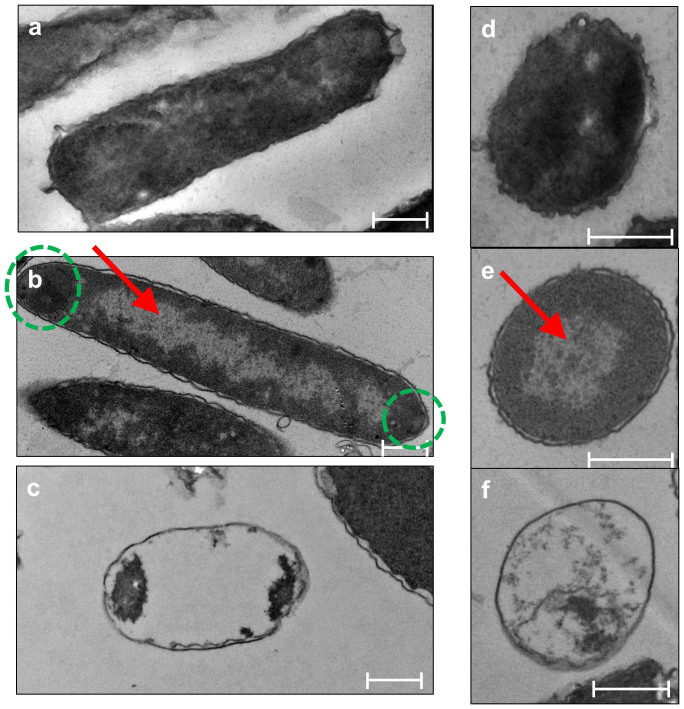
Microscopy was performed to investigate the mechanism of ArT toxicity via morphological changes. Microscopic examination has been used to reveal morphological changes brought about by toxin expression; for example, GhoT-producing cells often appear as “ghost” cells23. Similar to the effects of GhoT, we found that producing ArT causes cells to become “ghost”-like (Fig. 2a). This phenotype was further supported by transmission electron microscopy images of cells producing ArT, in that the center of the cells became translucent (red arrow, Fig. 3b and Fig. 3e), and the surrounding area (cytoplasm) became darker, with the darkest spots at cell poles (green circles, Fig. 3b). Such translucent centers are compact nucleoids that typically appear when growth is suppressed, such as in the event of translation inhibition31. In contrast, true “ghost” cells resemble an empty nest with cytoplasmic regions condensed at the cell poles (Fig. 3c). Unlike cells producing GhoT, cell lysis or breakage was not observed in ArT-producing cells. Using GFP-tagged ArT and fluorescence microscopy, we found that ArT is localized throughout the cells, while GFP-tagged GhoT specifically localized at the cell poles (Fig. 2b). Therefore, ArT causes cells to become “ghost”-like but not to the same extent as GhoT.
Figure 2. ArT-producing cells show “ghost”-like phenotype.
(a) Appearance of cells producing GhoS, ArT, and GhoT under phase contrast at 1,000× magnification (oil immersion). GhoS: BW25113/pCA24N-ghoS, ArT: BW25113/pCA24N-arT, and GhoT: BW25113/pCA24N-ghoT. (b) Localization of GFP-tagged GhoS, ArT, and GhoT using epifluorescence microscopy. Note the “ghost”-like phenotype in cells producing ArT, and the true “ghost” cells in GhoT-expressing cells. GhoS: BW25113/pCA24N-ghoS(GFP+), ArT: BW25113/pCA24N-arT(GFP+), and GhoT: BW25113/pCA24N-ghoT(GFP+). Scale bars denote 5 µm.
Figure 3. TEM images of cells producing GhoS, ArT, and GhoT.
(a) to (c): Longitudinal sections of BW25113/pCA24N-ghoS, BW25113/pCA24N-arT, and BW25113/pCA24N-ghoT, respectively. (d) to (f): Cross-sectional views of BW25113/pCA24N-ghoS, BW25113/pCA24N-arT, and BW25113/pCA24N-ghoT. Scale bars denote 0.5 µm. Red arrow indicates internal translucent areas, and green circles show condensed areas at the poles.
Five catalytic residues are conserved in GhoS (Fig. S1) and its structural homolog, Cas2, from the CRISPR-Cas system, with R28 being the most important residue for the endoribonuclease activity of GhoS23. In ArT, mutations L25I and M31L are located very close to R28 (Fig. 1c), and they may affect the catalytic activity of GhoS. To gain further insights into mechanism of toxicity of ArT, whole-transcriptome studies were conducted for cells producing GhoS and ArT. In comparison to cells harboring an empty plasmid, 20 gene transcripts were degraded upon production of native GhoS23. Among these, four transcripts (purE, purH, purM, and purT) were also degraded in the ArT-producing strain. However, 48 additional transcripts were degraded in cells overproducing ArT compared to those overproducing GhoS. No transcripts were significantly enriched in cells that produce native GhoS, but 19 transcripts were enriched in cells that produce ArT (Supplemental Table 1). These results suggest that ArT has become an endoribonuclease with a broader substrate range.
Random mutagenesis of mqsA to form artificial antitoxin ArA
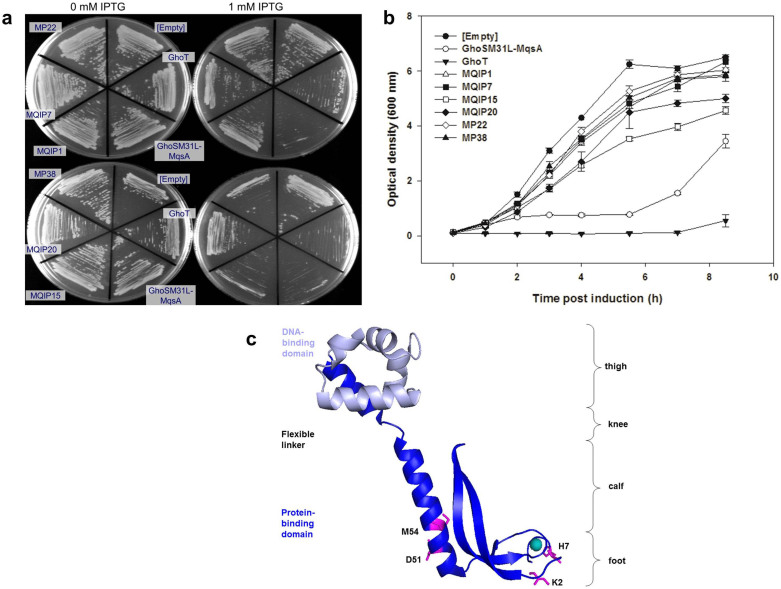
To investigate whether antitoxin activity toward ArT may be readily generated from antitoxins that are not related to ArT, we created MqsA variants via epPCR. MqsA was chosen because the regulation and function of this antitoxin have been well studied4,28,32,33. MqsA has two domains: the toxin protein-binding domain at the N-terminus and the DNA-binding domain at the C-terminus28,33. To prevent interference with the DNA-binding activity of MqsA, we mutagenized only the protein-binding domain at the N-terminus (residues 1 to 76). We introduced a C→T DNA substitution at L81 (silent mutation) to create a HindIII site downstream of the N-terminal domain in order to facilitate mutagenesis of the N-terminal domain. The entire mqsA gene was then placed downstream of ghoS-M31L (in pCA24N-ghoS-M31L), which encodes the toxic GhoS-M31L variant (Fig. 1b). Hence, MqsA variants could be selected based on their ability to reduce the severe growth inhibition of GhoS-M31L with both GhoS-M31L and the MqsA variants produced from the same IPTG-induced promoter. Note that native MqsA is not toxic and does not decrease the toxicity of GhoS-M31L (Fig. 4a and 4b), and that we initiated the experiments to find an antitoxin for the GhoS-M31L toxin variant since we had this variant before we obtained GhoS-L25I-M31L (ArT).
Figure 4. Mutated MqsA variants abolish the toxicity of GhoS-M31L.
All plasmids were expressed in E. coli BW25113 ΔghoS ΔmqsRA ΔKmR using a single plasmid system that produces both the toxin and antitoxin (a) Growth of cells producing GhoS-M31L and different MqsA variants on LB agar with and without IPTG. Note that native MqsA does not abolish the toxicity of GhoS-M31L. (b) Growth of cells expressing GhoS-M31L and different MqsA variants in liquid LB after the addition of 1 mM IPTG (added at t = 0 h). Error bars indicate s.e.m. (n = 3). (c) Residues K2, H7, D51, and M54 (shown as pink sticks) mapped to the structure of MqsA (pdb: 3gn5). These residues were substituted more than once among the six selected MqsA variants. Green sphere denotes zinc ion. Empty: pCA24N, GhoSM31L-MqsA: pCA24N-ghoS-M31L-mqsA, GhoT: pCA24N-ghoT, MQIP1: pCA24N-ghoS-M31L-mqsA-V12A-M54L-K58N-K76Q, MQIP7: pCA24N-ghoS-M31L-mqsA-H7L-P19S-V28A-M54I-K80E-H110Y, MQIP15: pCA24N-ghoS-M31L-mqsA-H7L-I15N-H33L-V75E, MQIP20: pCA24N-ghoS-M31L-mqsA-K2E-MP22-F22L-C37S-D51N-F60L, MP22: pCA24N-ghoS-M31L-mqsA-F22L-C37S-D51N-F60L, MP38: pCA24N-ghoS-M31L-mqsA-K2E-Q8L.
We generated a plasmid library of mutated mqsA using epPCR at a 1.7% error rate, and used this library to transform E. coli with both ghoS and mqsRA deleted (to prevent interference from the native proteins). Among the 4.2 × 108 MqsA variants screened, six MqsA variants were identified as putative antitoxins for GhoS-M31L, as they were able to alleviate the toxicity of GhoS-M31L in the presence of IPTG, and the cells grew as well as the wild-type strain (Fig. 4a and 4b). Four residues (K2, H7, D51, and M54) were mutated in more than one MqsA variant (Fig. S4), which suggests that these mutations were responsible for conferring the improved growth phenotype. We confirmed that in these six MqsA variants, both GhoS-M31L and MqsA variants were co-produced in the cells (Fig. S5). Among these six MqsA variants, we chose MqsA variant MP22 (F22L, C37S, D51N, and F60L, referred to as ArA hereafter) for further analyses.
To investigate whether ArT toxin and ArA antitoxin interact in vivo, we performed a bacterial two-hybrid assay. We cloned arT and arA separately into two plasmids, with each expressing one domain of adenylate cyclase34. If two proteins of interest physically interact, the host forms a blue colony on reporter plates as was found for the positive control (Fig. S6). However, no in vivo interaction was observed for ArT and ArA, as indicated by a lack of blue colonies on reporter plates (Fig. S6).
Random mutagenesis of toxI to form artificial antitoxin ArA*
Given that type II antitoxin MqsA could be converted into a protein that reduces ArT toxicity, we investigated whether we could evolve a second antitoxin for ArT using an RNA-based antitoxin (type III system). We chose ToxI, which is the antitoxin from the ToxIN system29. The RNA-coding region of toxI is made up of 5.5 direct repeats, with each repeat consisting of 36 nt (5′-AGGTGATTTGCTACCTTTAAGTGCAGCTAGAAATTC-3′). We placed toxI under the control of a lac promoter and without a ribosome binding site, and cloned this PLlacO-1::toxI fragment into pCA24N-arT. We generated a plasmid library of mutated toxI using epPCR with a 1% error rate. Similar to the screening strategy used for selecting MqsA variants, we screened for improved growth when ArT was produced along with various ToxI transcripts. Six strains with toxI mutations that completely abolished the toxicity of ArT were isolated (Fig. 5). For each toxI variant, there were 1-6 nucleotide substitutions in the RNA-coding region (Fig. S7). Nucleotide substitutions were found in every repeat, except for the 3rd repeat. Overall, the toxicity of ArT could be readily counteracted by artificially evolved antitoxins generated from an unrelated protein (MqsA) to form the basis of a novel type II TA system and from unrelated RNA (ToxI) to form the basis of a novel type III TA system.
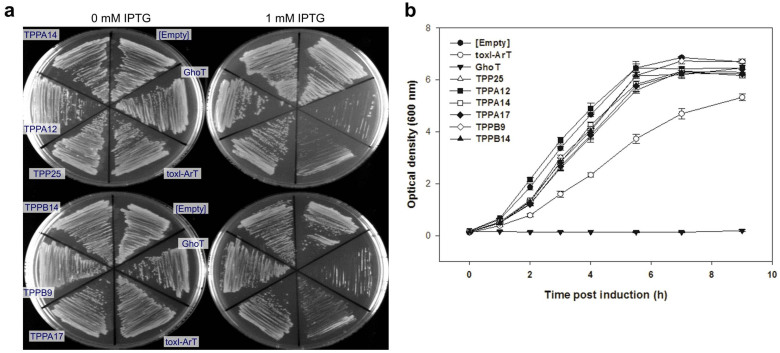
Figure 5. ToxI variants abolished the toxicity of ArT.
All plasmids were expressed in E. coli BW25113 ΔghoS using a single plasmid system that produces both the toxin and antitoxin. (a) Growth of cells producing ArT and different ToxI variants on LB agar with and without IPTG. (b) Growth of cells expressing ArT and different ToxI variants in LB after the addition of 1 mM IPTG (added at t = 0 h). Error bars indicate s.e.m. (n = 3). Empty: pCA24N, toxI-ArT: pCA24N-toxI-arT, GhoT: pCA24N-ghoT, TPP25: pCA24N-toxI-T36C-C73T-A248G-arT, TPPA12: pCA24N-toxI-T9A-T23A-A134T-T167G-T180C-A182T-A245T-arT, TPPA14: pCA24N-toxI-T10A-T116G-A250G-arT, TPPA17: pCA24N-toxI-A34G-A56T-A62T-A182G-T197C-arT, TPPB9: pCA24N-toxI-A1C-A62T-arT, TPPB14: pCA24N-toxI-T188A-arT.
Transcriptional control in the artificial TA system
For type II TA systems, antitoxins typically harbor a DNA-binding domain that allows them to act as transcriptional regulators. For example, MqsA represses mqsRA expression by binding its cognate binding site, the two palindromes in its promoter35. Such transcriptional self-regulation by the antitoxin is canonical for type II TA systems. Hence, we investigated whether the ArT/ArA system could be engineered to be regulated in a similar manner. Given that MqsA binds specific palindromic DNA motif [5′-AACCT (N)3 AGGTT-3′], we cloned the two mqsRA palindromes into the synthetic T5-lac promoter in pCA24N to form PT5-lac-pals (Fig. S8), and tested whether ArA would be able to repress arT expression by binding to the palindromes. To demonstrate arT repression by ArA, we measured arT transcription from PT5-lac-pals using quantitative real-time reverse-transcription PCR (qRT-PCR) upon ArA production from a PARA promoter on a separate plasmid (Fig. S9). In comparison to the empty pBAD plasmid control (arT expression set as 100%), MqsA production via pBAD-mqsA reduced arT expression by 63 ± 18%, while ArA production via pBAD-arA reduced arT expression by 40 ± 15%. Therefore, similar to native MqsA, ArA binds PT5-lac-pals to repress the expression of toxin.
Final constructs
Since transcription from PT5-lac-pals can be controlled by ArA, we subsequently placed ArT, ArA, and ArT+ArA under the control of this promoter. The resulting plasmids are termed as pCA24N-arT (artificial toxin), pCA24N-arA (artificial antitoxin), and pCA24N-arTA (artificial toxin/antitoxin). We re-tested the toxicity of these final constructs in the presence of IPTG, and found that the additional palindromes had minimal effect on the toxicity of ArT, or antitoxicity of ArA (Fig. S10).
Persistence to ampicillin
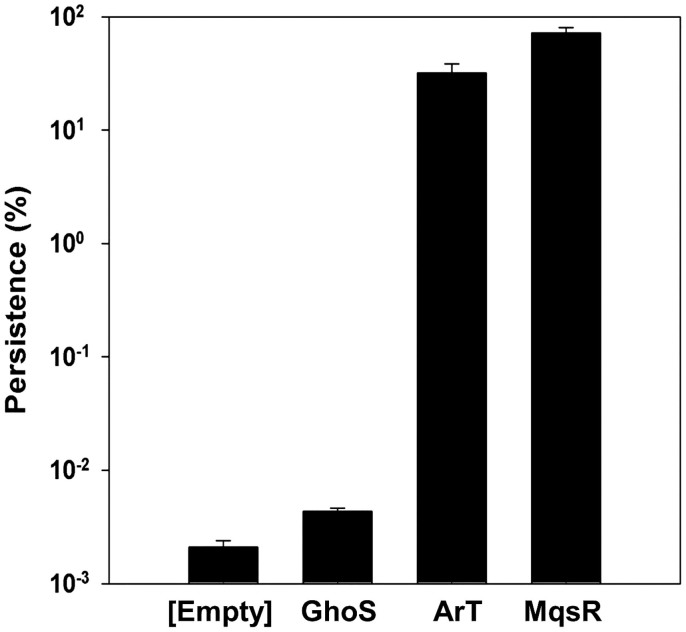
Many toxins, such as MqsR, increase persistence to antibiotics6. Therefore, we measured the persistence level of ArT-expressing cells. In comparison to cells harboring an empty vector, ArT increased persistence to ampicillin by 104 fold (Fig. 6), and this persistence level was only ~2-fold less than MqsR. Note that the persistence level shown by GhoS-expressing cells was not significantly higher than that of the empty vector. In comparison to ArT alone, cells producing both ArT and ArA (with antitoxin ArA masking toxicity of ArT) had 65 ± 20-fold less of an impact on persistence compared to ArT. This shows that ArT is a bona fide toxin, and ArA is an antitoxin that masks the toxicity of ArT.
Figure 6. ArT increases cell persistence to ampicillin.
Persistence of E. coli BW25113 ΔghoS ΔmqsRA ΔKmR harboring different plasmids after ampicillin (100 μg/mL) treatment for 3 h. Empty: pCA24N, GhoS, pCA24N-ghoS, ArT: pCA24N-arT, MqsR: pCA24N-mqsR.
Discussion
In this study, we demonstrate that TA systems evolve readily since we showed that the components of a TA system can be made by evolving existing TA components of different classes. To form a toxin, we converted a native antitoxin to a protein that inhibits growth (ArT) by introducing only two substitutions. To form antitoxins for the novel ArT toxin, we evolved a protein (MqsA to form ArA) as well as evolved a non-coding RNA (ToxI RNA to form ArA*). To form ArA, only four amino acid substitutions were required (Fig. S4), and to form ArA*, as few as one nt change was required (Fig. S7). Therefore, collectively, our results show that TA components are functionally plastic and may be interchanged to generate new systems.
Our results fit well with previous results that have shown that a handful of amino acid substitutions can impact toxin and antitoxin activity. For example, two simultaneous amino acid substitutions (G22S and D291A) abolished the toxicity of HipA (from the HipA/HipB TA system), although this variant still conferred a high persistence phenotype similar to native HipA36. A D83Y substitution also relaxed the interaction specificity between the type II Txe/Axe system that allows the Txe toxin to interact with the YefM antitoxin (from the YoeB/YefM TA system)37. Also, TA systems have been exploited for biotechnological purposes such as the evolution of hemolysin expression-modulating protein (Hha) for biofilm dispersal38.
The five known types of TA systems illustrate the diversity of the molecular mechanisms of TA systems22, especially on how antitoxins counteract the effect of toxins27. Given that we used GhoS (from a type V TA system)23 and MqsA (from a type II TA system)28 as the starting materials for building an artificial TA system, there were two possible mechanisms by which ArA negates the activity of ArT. Firstly, it was possible that ArA sequestered ArT through direct physical interaction, as how MqsA counteracts the activity of MqsR. However, we showed using bacterial two-hybrid assay that ArA does not interact with ArT. Secondly, the toxicity of ArT might be mediated by either MqsR or GhoT, as shown by the type V TA system (MqsR toxin cleaves the ghoS mRNA transcript, and indirectly enriches ghoT mRNA), and ArA production might reduce MqsR and GhoT production. However, the toxicity of ArT is not dependent upon GhoT or MqsR (Fig. S2 and S3), and thus, ArA does not reduce the toxicity of ArT in an MqsR-dependent manner. Overall, these results suggest that TA systems, whether artificial or natural, may exert their effects in a manner that is yet to be fully understood.
One question that has puzzled the TA field is why cells have many TA systems. For example, E. coli has 38 TA systems23,39 and Mycobacterium tuberculosis has 88 TA systems40. Given the plasticity that we demonstrate here in TA systems, it is tempting to ponder whether one of the primary functions of TA systems is to help cells respond to a myriad of stresses. For example, the MqsR/MqsA TA system is activated by oxidative stress4, the RelE/RelB TA system is activated under nutrient starvation conditions41, and we have recently identified a TA system in E. coli that is activated by stress related to exposure to bile acid (Kwan BK and Wood TK, unpublished result). Hence, our results suggest that the cells may co-opt a particular toxin and antitoxin and evolve them to allow the cell to respond to a specific stress.
If our hypothesis is correct, one must consider the source of the building blocks of these new TA systems. Given that the average cell has 3.8 type II chromosomal TA systems27, one likely source for these new TA systems are the existing TA systems found in most genomes. Furthermore, phages are another source of proteins that may be utilized to create new TA systems. Many bacteriophages rely on lytic proteins such as holins and murein hydrolase to lyse the cell42, and these proteins are reminiscent of toxins that disrupt cell membrane and cell wall synthesis such as GhoT8 and PezT43. For example, the cryptic prophages of E. coli now encode at least five TA systems: CbtA/CbeA from CP4-44 prophage44, RelB/RelE45 from Qin prophage, RnlA/RnlB46 from CP4-57 prophage, YpjF/YfjZ47 from CP4-57 prophage, and YkfI/YafW47 from CP4-6 prophage. These five systems may have been created by the host over millions of years by evolving a toxin of the phage, or the phage themselves may have brought an intact TA system. Therefore, TA components may be evolved and their building blocks are readily available.
Methods
Bacterial strains, plasmids, culture conditions, and oligonucleotides
All strains and plasmids used in this study are summarized in Table 1. All strains were grown in lysogeny broth (LB)48 at 37°C, unless indicated otherwise. Kanamycin (50 μg/mL) was used to maintain the Keio strains49, pBS(Kan)-based plasmids50, and pKT25linker-derived plasmids34 (Table 1). Chloramphenicol (30 μg/mL) was used to maintain pCA24N-based plasmids30. Ampicillin (100 μg/mL) was used to maintain pCP2051 and pUT18Clinker-based plasmids34 (Table 1). Mutations L25I and M31L were introduced into pCA24N-ghoS using mutagenic primers to form pCA24N-ghoS-L25I, pCA24N-ghoS-M31L, and pCA24N-arT. E. coli BW25113 ΔghoS ΔmqsRA ΔKmR was created by eliminating the KmR cassette in the BW25113 ΔghoS ΔmqsRA Ω KmR strain19. The mqsR-f and ygiS-r primers were used to verify the KmR deletion. All plasmids were verified by DNA sequencing. All oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA), and primer sequences are listed in Supplemental Table 2.
Table 1. Bacterial strains and plasmids used in this study. KmR, ApR, and CmR denotes kanamycin, ampicillin, and chloramphenicol resistance, respectively.
| Strains or plasmids | Description | Source |
|---|---|---|
| E. coli K-12 | ||
| BTH101 | F−, cya-99 araD139 galE15 galK16 rpsL1 (Str r) hsdR2 mcrA1 mcrB1 | (Gully and Bouveret, 2006) |
| BW25113 | rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | (Baba et al., 2006) |
| BW25113 ΔghoS | BW25113 ΔghoS Ω KmR | (Baba et al., 2006) |
| BW25113 ΔghoS ΔmqsRAΩKmR | BW25113 ΔghoS::frt ΔmqsRA Ω KmR | (Wang et al., 2013) |
| BW25113 ΔghoS ΔmqsRA ΔKmR | BW25113 ΔghoS::frt ΔmqsRA::frt | This study |
| BW25113 ΔghoT | BW25113 ΔghoT Ω KmR | (Baba et al., 2006) |
| BW25113 ΔmqsRA | BW25113 ΔmqsRA ΔKmR | (Kim et al., 2010) |
| Plasmids | ||
| pBAD-mqsA | ApR, pBR322 ori, PARA::mqsA-flag+ | This study |
| pBAD-arA | ApR, pBR322 ori, PARA::arA-flag+ | This study |
| pBAD-myc-His B | ApR, pBR322 ori, PARA::myc-His6+ | Life Tech. |
| pBR-plac | ApR, pBR322 ori, PLlacO-1 | (Guillier and Gottesman, 2006) |
| pBR-plac-toxI | ApR, pBR322 ori, PLlacO-1::toxI+ | This study |
| pBS(Kan)-mqsA | KmR; Plac::mqsA | (Kim et al., 2010) |
| pCA24N | CmR; lacIq | (Kitagawa et al., 2005) |
| pCA24N-arA | CmR; lacIq PT5-lac::arA-flag+ with two mqsRA palindromes between lacO and RBS | This study |
| pCA24N-arT | CmR; lacIq PT5-lac::His6-arT+ with two mqsRA palindromes between lacO and RBS | This study |
| pCA24N-arT-arA | CmR; lacIq PT5-lac::His6-arT+, arA-flag+ with two mqsRA palindromes between lacO and RBS | This study |
| pCA24N-arT (GFP+) | CmR; lacIq PT5-lac::His6-arT-gfp+ | This study |
| pCA24N-ghoS | CmR; lacIq PT5-lac::His6-ghoS+ | (Kitagawa et al., 2005) |
| pCA24N-ghoS(GFP+) | CmR; lacIq PT5-lac::His6-ghoS-gfp+ | (Kitagawa et al., 2005) |
| pCA24N-ghoS-L25I | CmR; lacIq PT5-lac::His6-ghoS+ with TTA (121-123 nt relative to start codon) mutated to ATA | This study |
| pCA24N-ghoS-M31L | CmR; lacIq PT5-lac::His6-ghoS+ with ATG (139-141 nt relative to start codon) mutated to TTG | This study |
| pCA24N-ghoS-M31L-mqsA | CmR; lacIq PT5-lac::His6-ghoS+-M31L-mqsA-flag+ | This study |
| pCA24N-ghoT | CmR; lacIq PT5-lac::His6-ghoT+ | (Kitagawa et al., 2005) |
| pCA24N-ghoT(GFP+) | CmR; lacIq PT5-lac::His6-ghoT-gfp+ | (Kitagawa et al., 2005) |
| pCA24N-mqsR | CmR; lacIq PT5-lac::His6-mqsR+ | (Kitagawa et al., 2005) |
| pCA24N-toxI-arT | CmR; lacIq PT5-lac::His6-arT+, PLlacO-1::toxI+ | This study |
| pCP20 | ApR, CmR; FLP+, λ cI857+, λ pR Repts | (Cherepanov and Wackernagel, 1995) |
| pKT25linker | KmR, p15A ori, Plac::T25+ | (Gully and Bouveret, 2006) |
| pKT25linker-ghoS | KmR, p15A ori, Plac::T25-ghoS+ | This study |
| pKT25linker-arT | KmR, p15A ori, Plac::T25-arT+ | This study |
| pKT25linker-zip | KmR, p15A ori, Plac::T25-zip+ | (Gully and Bouveret, 2006) |
| pUT18Clinker | ApR, ColE1 ori, Plac::T18+ | (Gully and Bouveret, 2006) |
| pUT18Clinker-mqsA | ApR, ColE1 ori, Plac::T18-mqsA+ | This study |
| pUC18Clinker-arA | ApR, ColE1 ori, Plac::T18-arA+ | This study |
| pUT18Clinker-zip | ApR, ColE1 ori, Plac::T18-zip+ | (Gully and Bouveret, 2006) |
arA refers to mqsA with TTC (64-66 nt relative to start codon) mutated to CTC, TGT (109-111 nt relative to start codon) mutated to AGT, GAT (151-153 nt relative to start codon) mutated to AAT, TTT (178-180 nt relative to start codon) mutated to CTT. arT refers to ghoS with TTA (121-123 nt relative to start codon) mutated to ATA, and ATG (139-141 nt relative to start codon) mutated to TTG.
Toxicity tests on solid and liquid media
To investigate the effect of toxic protein production on solid media, at least three independent colonies of each tested strain (harboring different pCA24N-derived plasmids) were streaked on LB-chloramphenicol agar with and without 1 mM IPTG. For growth in liquid media, the OD600 of every culture was monitored every 1 to 2 h. Overnight cultures of each strain were inoculated into 25 mL LB-chloramphenicol to an OD600 of 0.05, grown to an OD600 of 0.1, and 1 mM IPTG was added to induce toxic protein production.
Random mutagenesis of ghoS and toxicity screening
ghoS from plasmid pCA24N-ghoS under the control of the T5-lac promoter was mutated under error-prone conditions (0.5 mM Mn2+ and 5 mM Mg2+)52 using primers pCA24Nf-SH and pCA24Nr-SH. The epPCR product was ligated into the XhoI and HindIII sites of pCA24N, electroporated into E. coli BW25113 ΔghoS, and transformed cells were grown on LB-chloramphenicol agar for 16 h. To screen GhoS variants for increased toxicity (i.e., reduced growth), small colonies (≤0.5 mm in diameter) were picked and transferred onto fresh LB-chloramphenicol agar plates with and without 1 mM IPTG. BW25113 ΔghoS with empty pCA24N, pCA24N-ghoS (native GhoS), and pCA24N-ghoT (GhoT) were tested alongside as the negative and positive controls. Colonies that showed growth on 0 mM IPTG plates, but no growth on 1 mM IPTG plates, were considered as putative positive hits. To confirm the toxicity was due to substituted GhoS, plasmids were isolated from these putative positive clones, verified by DNA sequencing, and re-electroporated into fresh BW25113 ΔghoS cells for further toxicity tests on solid and liquid media.
Random mutagenesis of mqsA and antitoxicity screening
As we wished to mutagenize only the N-terminus (the protein-binding domain, residues 1 to 76) of mqsA (mqsA-N), a C→T silent mutation was introduced to residue L81 of MqsA [from pBS(Kan)-mqsA32] using mutagenic primers mqsA-N-f and mqsA-N-r to create a HindIII site. We then cloned the entire mqsA coding sequence (with the HindIII site) downstream of ghoS-M31L (in pCA24N-ghoS-M31L) using the ArA-f and ArA-r primers (that contain an AccI and BlpI site, respectively). A FLAG tag (DYKDDDDK) was also fused to the C-terminal domain of MqsA via the ArA-r primer. The resulting plasmid is pCA24N-ghoS-M31L-mqsA.
Using the ArA-f and ArA-r primers, mqsA-N was randomly mutated using epPCR as described above. Mutated mqsA-N inserts were digested with AccI and HindIII, and cloned into AccI/HindIII-linearized pCA24N-ghoS-M31L. Ligated product was electroporated into BW25113 ΔghoS ΔmqsRA ΔKmR. To select for reduced growth toxicity, half of the electroporated population was plated directly on LB-chloramphenicol agar with 1 mM IPTG, and variants that gave rise to large colonies (≥0.5 mm in diameter) were chosen for DNA sequencing analysis and retransformation tests. The other half of the population was plated on LB-chloramphenicol agar (without IPTG), and recovered colonies were pooled with fresh LB-chloramphenicol. Approximately 4.2 × 108 cells were then pre-grown in 50 mL LB-chloramphenicol with 1 mM IPTG for 4 h, and aliquots were plated on LB-chloramphenicol agar with 1 mM IPTG. Similarly, large colonies were chosen for DNA sequencing analysis and retransformation tests.
Random mutagenesis of toxI and antitoxicity screening
The template for epPCR of toxI was pCA24N-toxI-arT. This plasmid was created by cloning the PLlacO-1::toxI fragment into the XhoI site of pCA24N-arT via the use of toxIep-f and toxIep-r2 primers. PLlacO-1 is an RBS-less promoter from pBR-plac53. The full-length toxI including its transcriptional terminator (264 bp; Genbank accession: FJ176937) was fused to PLlacO-1 using primers toxI-f and toxI-r.
EpPCR for toxI was performed as described above using primers toxIep-f and toxIep-r2. Mutated toxI inserts were cloned into the XhoI site of pCA24N-arT, and ligated product was used to transform BW25113 ΔghoS via electroporation. Electroporated cells were grown in 1 mL LB-chloramphenicol-kanamycin with 0.1, 0.5, or 1.0 mM IPTG for 16 h, followed by plating on LB-chloramphenicol-kanamycin agar with 1 mM IPTG. The resulting large colonies were chosen for DNA sequencing analysis and retransformation tests.
Autoregulation of TA system
To regulate the expression of ArT and ArA through ArA, a T5-lac promoter containing two mqsRA palindromes was synthesized (GeneArt, Life Technologies), and termed as PT5-lac-pals. In E. coli K-12 MG1655 (Genbank accession number: U00096.2), the mqsRA palindrome 1 is located at position 3166595 bp to 3166609 bp, while mqsRA palindrome 2 is located at position 3166629 bp to 3166639 bp. The PT5-lac-pals cassette was cloned in between lacO2 and the RBS of pCA24N-arT and pCA24N-arT-arA using XhoI and BbvCI. The BbvCI site was pre-introduced into the plasmids using the mutagenic primers pCA24N-BbvCI-f and pCA24N-BbvCI-r. The resulting plasmids were pCA24N-arT and pCA24N-arTA. To construct pCA24N-arA, the NoArT-f and NoArT-r primers were used to amplify the plasmid backbone (without arT) of pCA24N-arTA. The amplified product was allowed to self-ligate using T4 DNA ligase, before being electroporated into BW25113 ΔghoS ΔmqsRA ΔKmR.
TA regulation mediated by ArA
To evaluate the repression of transcription of arT by ArA, arA was cloned into the NcoI and XbaI sites of pBAD-myc-His B (Life Technologies) using primers ArA-NcoI-f2 and ArA-XbaI-r. Ligated product was introduced into BW25113 ΔghoS ΔmqsRA ΔKmR/pCA24N-arT via electroporation. As a positive control, pBAD-mqsA was also constructed in a similar manner, and electroporated into the same strain. For the negative control, pBAD-myc-His B (which expresses only the myc epitope and His tag) was electroporated into the same strain.
Independent cultures of the three strains (BW25113 ΔghoS ΔmqsRA ΔKmR/pCA24N-arT with either pBAD-arA, pBAD-mqsA, or pBAD-myc-His B) were grown in LB-chloramphenicol-ampicillin to an OD600 of 0.5, and 1% l-arabinose was added to induce production of MqsA or ArA. Cells were harvested after 30 min of induction, and total RNA was isolated as described in the following section. qRT-PCR was performed according to manufacturer's instructions (Power SYBR Green RNA-to-CT 1-Step kit, Life Technologies, Carlsbad, CA) using 100 ng of total RNA as the template. Primers were annealed at 60°C, and the housekeeping rrsG (16S rRNA) gene was used to normalize all data. Changes in gene transcripts were calculated using the 2−ΔΔCt formula54.
RNA isolation and whole-transcriptome analysis
Total RNA isolation and DNA microarray analysis were performed as described previously23. RNeasy Mini kit (Qiagen Inc., Valencia, CA) was used to isolate total RNA from E. coli BW25113/pCA24N-arT and BW25113/pCA24N after 1 mM IPTG induction for 90 min. Cells were lysed with 0.1 mm zirconia/silica beads (Biospec, Bartlesville, OK). cDNA synthesis, fragmentation, and hybridization to E. coli GeneChip Genome 2.0 arrays (Affymetrix, P/N 511302) were performed according to previous protocols20. Changes in gene expression were considered significant only when they were 5 fold or more. The gene expression data are accessible through GEO accession number GSE54271.
Microscopy
Strains harboring different pCA24N-based plasmids were grown in LB-chloramphenicol to an OD600 of 0.5, and production of GhoS, ArT, or GhoT was induced by 1 mM IPTG. Cells were collected after 4 h, washed, and resuspended in 0.85% (w/v) NaCl. Cells were then examined at 1,000× magnification using oil immersion under phase contrast and GFP epifluorescence settings on a light microscope (Zeiss Axio Scope.A1, Germany). Microscopy images were processed using ImageJ55.
Transmission electron microscopy (TEM)
Strains (BW25113/pCA24N-ghoS, BW25113/pCA24N-ghoT, and BW25113/pCA24N-arT) were grown in LB-chloramphenicol to an OD600 of 0.5, and 1 mM IPTG was added to induce protein production from plasmids. After 4 h of induction, cells were fixed with 2.5% formaldehyde and 1.5% glutaraldehyde in 100 mM sodium cacodylate buffer (pH 7.4) at 4°C for 12 h, followed by three washes with buffer alone for 5 min each wash. Cells were then fixed with 1% osmium tetroxide for 1 h in the dark, followed by three washes with buffer. After treating the cells with 2% uranyl acetate in the dark for 1 h, cells were sequentially dehydrated in a graded ethanol series (50%, 70%, 85%, 95%, and three times with 100% ethanol, for 5 min each) and 100% acetone (three times for 5 min each). Dehydrated cells were embedded into epoxy resins for at least 12 h, and then sectioned into thin specimens (70 nm thick) using an ultramicrotome (UC6, Leica IL). Specimens were stained with uranyl acetate and lead citrate, before being examined on a FEI Tecnai G2 Spirit BioTwin TEM (Hillsboro, OR) at an accelerating voltage of 120 kV.
Persistence assay
The persistence assay was performed as described previously6 with slight modification. Briefly, each culture was induced with 1 mM IPTG for 2 h at an OD600 of 0.1 (for the strain carrying the empty vector and GhoS-producing strain) or 0.5 (for ArT- or MqsR-producing strains). After adjusting each culture to an OD600 of 1.0, cells were exposed to 100 μg/mL ampicillin for 3 h. Cell viability before and after ampicillin treatment was determined by applying 10 μL drops in serial dilutions56 on LB-chloramphenicol agar, and growing at 37°C for 24 h. Three independent cultures per strain were evaluated.
Bacterial two-hybrid analysis
In vivo protein interactions were assessed using a bacterial two-hybrid approach based on functional reconstitution of adenylate cyclase (CyaA) activity of Bordetella pertussis in an E. coli cya− strain (E. coli BTH101, Table 1). The two catalytic domains of CyaA, T18 and T25, were produced from pUT18Clinker and pKT25linker, respectively. Proteins of interest (GhoS, ArT, MqsA, and ArA) were fused to the C-terminus of either T18 or T25 by cloning their coding sequence into the EcoRI and XhoI sites in the plasmids. The ghoS and arT coding sequence were amplified using primers ghoS-BTH-f and ghoS-BTH-r, while the mqsA and arA coding sequence were amplified using primers mqsA-BTH-f and mqsA-BTH-r. After co-transforming E. coli BTH101 with various combinations of the pUT18Clinker-based and pKT25linker-based plasmids (Table 1), 2 μL of each tested culture was used to streak LB-kanamycin-ampicillin agar containing 40 μg/mL 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) and IPTG at 0, 0.05, or 0.5 mM. Agar plates were incubated at 30°C for 2 days.
Western blot analysis
Strains were grown to an OD600 of 0.6 to 0.9, then 1 mM IPTG was added to induce protein production for 5 h. Cells were centrifuged (13,000 × g, 2 min) and sonicated, and the total proteins were resolved via 18% Tris-tricine-SDS gels. A Western blot was performed with primary antibodies raised against a His tag (Cell Signaling Technology, Danvers, MA) or a FLAG tag (Thermo Scientific, Waltham, MA), and horseradish peroxidase-conjugated goat anti-mouse secondary antibodies (Cell Signaling Technology). Blotted proteins were detected using the chemiluminescence reagents from the SuperSignal West-Pico Chemiluminescence kit (Thermo Scientific).
Author Contributions
T.K.W. conceived the project, and V.W.C.S., H.-Y.C. and B.W.K. performed the experiments. T.K.W. and V.W.C.S. wrote the manuscript, and all authors reviewed the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by the NIH (R01 GM089999). T.K.W. is the Biotechnology Endowed Professor at the Pennsylvania State University. We are grateful for the ASKA and Keio strains provided by the Genome Analysis Project in Japan. We acknowledge the assistance from Missy Hazen (Penn State Microscopy and Cytometry Facility, University Park, PA) in TEM imaging.
References
- Makarova K. S., Wolf Y. I. & Koonin E. V. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 41, 4360–4377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevin E. W. & Barloy-Hubler F. RASTA-Bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 8, R155 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. On the origin of species by means of natural selection. (John Murray, 1859). [Google Scholar]
- Wang X. et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7, 359–366 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Benedik M. J. & Wood T. K. Antitoxin DinJ influences the general stress response through transcript stabilizer CspE. Environ. Microbiol. 14, 669–679 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. & Wood T. K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 391, 209–213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T., Vulić M. & Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8, e1000317 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. Y. et al. Toxin GhoT of the GhoT/GhoS TA system damages the cell membrane to reduce ATP and to reduce growth under stress. Environ. Microbiol., e-pub ahead of print 24 December 2013, 10.1111/1462–2920.12373 (2013). [Google Scholar]
- Pecota D. C. & Wood T. K. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 178, 2044–2050 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. & Wood T. K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 77, 5577–5583 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T. & Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. U. S. A. 80, 4784–4788 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Bedzyk L. A., Thomas S. M., Ye R. W. & Wood T. K. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64, 515–524 (2004). [DOI] [PubMed] [Google Scholar]
- García-Contreras R., Zhang X. S., Kim Y. & Wood T. K. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PLoS One. 3, e2394 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. H., Wang X., Ma Q., Zhang X. S. & Wood T. K. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 191, 1258–1267 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton J. P. & Mulvey M. A. Toxin-antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathog. 8, e1002954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton R., Aggio R. B., Villas-Bôas S. G., Arcus V. L. & Cook G. M. Toxin-antitoxin systems of Mycobacterium smegmatis are essential for cell survival. J. Biol. Chem. 287, 5340–5356 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Walker A. N. & Daines D. A. Toxin-antitoxin loci vapBC-1 and vapXD contribute to survival and virulence in nontypeable Haemophilus influenzae. BMC Microbiol. 12, 263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaine S. et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343, 204–208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ. Microbiol. 15, 1734–1744 (2013).23289863 [Google Scholar]
- González Barrios A. F. et al. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188, 305–316 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai S., Kolodkin-Gal I., Hananya-Meltabashi M., Sacher A. & Engelberg-Kulka H. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genet. 5, e1000390 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner S. J., Poppenberger B. & Rozhon W. Toxin-antitoxin systems: Biology, identification and application. Mob. Genet. Elements 3, e26219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 8, 855–861 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Kiyokawa K., Tanaka K., Moriguchi K. & Suzuki K. Novel toxin-antitoxin system composed of serine protease and AAA-ATPase homologues determines the high level of stability and incompatibility of the tumor-inducing plasmid pTiC58. J. Bacteriol. 191, 4656–4666 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk I. & Kobayashi I. To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. 42, 70–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y. & Inouye M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 9, 779–790 (2011). [DOI] [PubMed] [Google Scholar]
- Goeders N. & Van Melderen L. Toxin-antitoxin systems as multilevel interaction systems. Toxins 6, 304–324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. L. et al. Three dimensional structure of the MqsR:MqsA complex: A novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 5, e1000706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran P. C. et al. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U. S. A. 106, 894–899 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M. et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12, 291–299 (2005). [DOI] [PubMed] [Google Scholar]
- Eltsov M. & Zuber B. Transmission electron microscopy of the bacterial nucleoid. J. Struct. Biol. 156, 246–254 (2006). [DOI] [PubMed] [Google Scholar]
- Kim Y. et al. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 12, 1105–1121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. L., Wood T. K., Peti W. & Page R. Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J. Biol. Chem. 286, 2285–2296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gully D. & Bouveret E. A protein network for phospholipid synthesis uncovered by a variant of the tandem affinity purification method in Escherichia coli. Proteomics 6, 282–293 (2006). [DOI] [PubMed] [Google Scholar]
- Brown B. L., Lord D. M., Grigoriu S., Peti W. & Page R. The Escherichia coli toxin MqsR destabilizes the transcriptional repression complex formed between the antitoxin MqsA and the mqsRA operon promoter. J. Biol. Chem. 288, 1286–1294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch S. B., Henderson T. A. & Hill T. M. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50, 1199–1213 (2003). [DOI] [PubMed] [Google Scholar]
- Połom D., Boss L., Węgrzyn G., Hayes F. & Kędzierska B. Amino acid residues crucial for specificity of toxin-antitoxin interactions in the homologous Axe-Txe and YefM-YoeB complexes. FEBS J. 280, 5906–5918 (2013). [DOI] [PubMed] [Google Scholar]
- Hong S. H., Lee J. & Wood T. K. Engineering global regulator Hha of Escherichia coli to control biofilm dispersal. Microb. Biotechnol. 3, 717–728 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q., Awano N. & Inouye M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol. Microbiol. 79, 109–118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage H. R., Connolly L. E. & Cox J. S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5, e1000767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S. K., Mikkelsen M., Pedersen K. & Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. U. S. A. 98, 14328–14333 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56, 430–481 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutschler H., Gebhardt M., Shoeman R. L. & Meinhart A. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 9, e1001033 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H., Tan Q., Awano N., Wu K. P. & Inouye M. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 84, 979–989 (2012). [DOI] [PubMed] [Google Scholar]
- Gotfredsen M. & Gerdes K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29, 1065–1076 (1998). [DOI] [PubMed] [Google Scholar]
- Koga M., Otsuka Y., Lemire S. & Yonesaki T. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics 187, 123–130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. M. & Shaw K. J. A novel family of Escherichia coli toxin-antitoxin gene pairs. J. Bacteriol. 185, 6600–6608 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. F. & Russell D. W. Molecular cloning: a laboratory manual. 3rd edn, (Cold Spring Harbor Laboratory Press, 2001). [Google Scholar]
- Baba T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canada K. A., Iwashita S., Shim H. & Wood T. K. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J. Bacteriol. 184, 344–349 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P. P. & Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 (1995). [DOI] [PubMed] [Google Scholar]
- Cadwell R. C. & Joyce G. F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2, 28–33 (1992). [DOI] [PubMed] [Google Scholar]
- Guillier M. & Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 59, 231–247 (2006). [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. & Eliceiri K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donegan K., Matyac C., Seidler R. & Porteous A. Evaluation of methods for sampling, recovery, and enumeration of bacteria applied to the phylloplane. Appl. Environ. Microbiol. 57, 51–56 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information