Abstract
Rapid and precise patterning of functional biomaterials is desirable for point-of-care (POC) tissue engineering and diagnostics. However, existing technologies such as dip-pen nanolithography and inkjet printing are currently unsuitable for POC applications due to issues of cost and portability. Here, we report the development of ‘BioPen', a portable tool for continuous, defined and scalable deposition of functional materials with micrometer spatial resolution and nanolitre volumetric resolution. BioPen is based upon the ballpoint pen but with multiple “ink sources” (functional material solutions) and with an apparatus that can be optimized for writing living cells, proteins, nucleic acids, etc. We demonstrate POC detection of human immunodeficiency virus type 1 (HIV-1) nucleic acid by writing on paper with BioPen using “ink” consisting of nucleic acid probes and nucleic acid-modified gold nanoparticles. We also demonstrate POC tissue engineering by writing a continuous pattern of living, functional, interconnected cells with a defined extracellular environment. Because it is simple, accurate, inexpensive and portable, BioPen has broad potential for POC detection of diagnostic biomarkers, and for POC engineering of tissues for a range of healing applications.
Laboratory-based regenerative medicine1,2,3 and diagnostics4,5,6,7,8,9 have progressed significantly over the past decade, in part due to advances in fine control of the deposition of functional materials. The emerging fields of POC tissue engineering and diagnostics, however, are limited by the challenge of precisely controlling the spatial pattern and concentration of nucleic acids10, nanoparticles11, proteins12, and cells13 outside of a laboratory environment. Standard laboratory techniques for patterning (e.g., lithography) have limitations both because functional materials are generally sensitive to environmental conditions, and because these techniques are too cumbersome for POC applications. For instance, conventional lithography techniques such as microcontact printing, capillary micromolding, and microtransfer molding require conditions including hard vacuum, high temperature, ultraviolet irradiation, and strong solvents that are not suitable for POC applications. Maintaining functionality of materials, especially biomaterials, is challenging under these conditions14. Soft lithography techniques are compatible with a broader range of functional materials, but are similarly inadequate for POC applications due to the need for cumbersome laboratory equipment15.
A second class of technologies for patterning functional materials in the laboratory involves writing and printing. The dip-pen uses an “ink”-coated atomic force microscope (AFM) tip to “write” molecules such as nucleic acid with tens of nanometer resolution, and has been used for single biomolecular detection10,16,17,18. However, efforts to scale up to the micrometer scale needed for tissue engineering have not succeeded, and AFM is not yet suitable for POC applications19,20. Technologies to adapt a conventional writing implement to writing of functional materials include a microscale “pencil” for writing of single-walled carbon nanotubes to detect airborne particulates21, but technology to usefully embed biomolecules and living cells into solid “pencil lead” has not yet been developed. A ballpoint pen has been developed to write electrodes for flexible electronics22,23,24, but writing of a functional biomaterial has not yet been achieved.
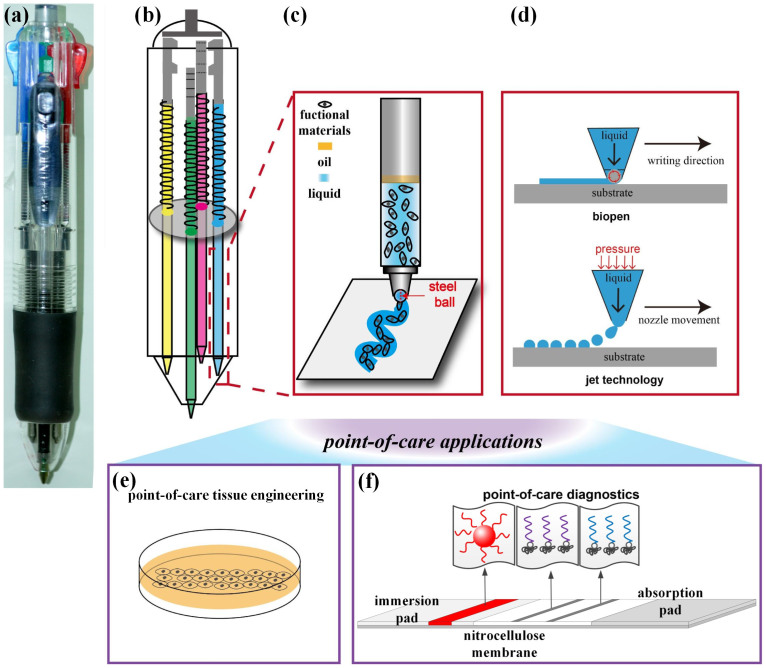
BioPen was developed to enable versatile, portable and precise patterning of functional materials using functional, liquid “ink”. The precursors of several such inks were stored in the four tubular, mechanically-selected cartridge reservoirs of a commercial ballpoint pen. These cartridge reservoirs were emptied, cleaned, and sterilized, filled with functional inks and covered with a layer such as oil to prevent evaporation (Fig. 1a–c).
Figure 1. Schematic of BioPen structure, working principle, and point-of-care applications.
(a) BioPen is constructed by replacing the ink of a commercial ballpoint pen and optimizing conditions for writing specific functional “inks”. (b) One BioPen can house multiple functional cartridges. (c) The suspension of functional materials can be patterned onto a range of substrates by simply writing at a prescribed speed. (d) One advantage of BioPen over inkjet technology is that it lays down continuous rather than discrete streams of ink, enabling superior connectivity. (e)(f) Additional advantages are portability and accuracy, enabling applications including point-of-care tissue engineering and point-of-care diagnostics.
With Biopen, functional materials could be patterned controllably and quantitatively. Line width and mean volume per unit length varied depending upon the ink, the substrate, the diameter of the steel ball that regulated ink flow in a cartridge, and the writing speed, but did not vary with writing pressure. On standard cellulose-fiber printer paper, the width and volume of a written line of rhodamine in phosphate buffered saline (PBS) could be controlled by modulating writing speed and ball size. For a 380 μm ballpoint pen at a writing speed of 4 cm/s, the accuracy of a 329 μm width line was ±3 μm (95% CI) with 103 ± 7 nL/cm (95% CI). Line width increased with ball diameter and decreased with writing speed (Fig. S1). BioPen was also effective and accurate in writing materials over nitrocellulose membranes and glass (Fig. S2a). Accuracy over the entire range of writing speeds tested (2–10 cm/s for 500 μm ball size, providing 88–253 nL/cm, Fig. S2b) was well within the range needed for applications such as POC detection. Surprisingly, accuracy was not affected by writing pressure over the normal physiologic range of writing force. The range of writing forces we measured amongst 10 subjects was 0.6–1.6 N, but spatial and volumetric resolution did not vary significantly over the range of 0.1–3.5 N (Fig. S2c); forces above 3.5 N damaged the paper. A 500 μm ballpoint pen with a writing speed of 6 cm/s produced repeatable lines of width 354 ± 37 μm and volume 112 ± 20 nL/cm (95% CI). BioPen retained all features of a normal ballpoint pen, with user-dependent microscale writing accuracy (Fig. S3).
We next evaluated the degree to which BioPen could deposit specified concentrations of living cells, proteins, and nanoparticles of interest for POC applications. As baseline experiments for comparison to useful functional materials, we studied writing of glass beads and robust cancer cells. Glass beads with diameters of 9–13 μm could be written on a glass substrate with final density directly proportional to the bead concentration within the ink (Fig. S4, initial concentrations of 0.05–5 mg/mL in PBS). Living malignant MCF-7 cells could be written on a Petri dish with cell density proportional to the cell concentration within the cartridge (Fig. S5). Viability of these cells varied slightly with the ball diameter but was always greater than 80% and remained nearly constant after 5 days of culture (Fig. S5).
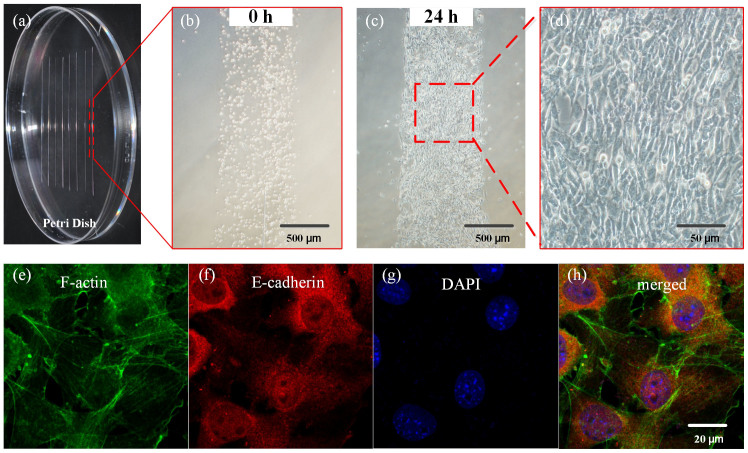
A central challenge with POC tissue engineering applications, and indeed with all bottom-up tissue engineering approaches, is synthesis of tissue elements with long, interconnected networks of active and living cells25. Cell noodles with artificial ECM represent a major success in this area26, but must be synthesized in a laboratory environment and are thus not suitable for POC applications. To demonstrate writing of engineered tissue suitable for POC wound healing applications, lines of fibroblasts embedded in a collagen hydrogel were written on a Petri dish, followed by culture at body temperature (37°C) for 10 min for polymerization. Over the course of several hours, fibroblasts formed remodeled, scar-like scaffolds (Fig. 2) with mature E-cadherin connections, demonstrating POC synthesis of long, interconnected networks of active and living cells. Viability was similar to that seen with cancer cells, demonstrating a continuous tissue element suitable for potential wound healing applications.
Figure 2. Point-of-care tissue engineering.
(a) Writing scar patch assays (3T3 fibroblasts and collagen hydrogel) on a Petri dish. Phase contrast images of cells in one scar patch immediately after writing (b) and after 24 hours of culture (c and d). Fluorescent immunostaining for F-actin (e), E-cadherin (f) and DAPI (g). The merged image (h) shows a mature cellular network with E-cadherin connections.
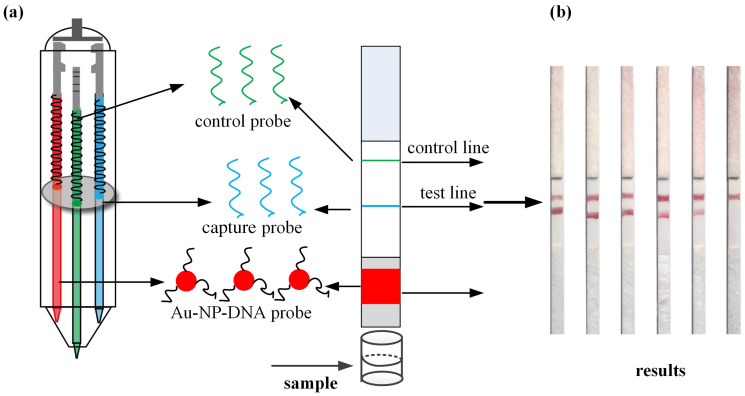
To demonstrate the feasibility and functionality of BioPen for POC diagnostics, we designed ink for detection of nucleic acid associated with the human immunodeficiency virus type 1 (HIV-1) (Fig. 3). Three cartridges were developed: two filled with capture and control probe solutions, respectively, and a third with a gold nanoparticle probe solution. Capture and control probe solutions were nucleic acid-protein complexes in PBS, prepared using biotinylated single strand nucleic acid (24-mer) and streptavidin. Gold nanoparticle probes were a gold nanoparticle-nucleic acid conjugate solution, prepared using gold nanoparticles and thiolated single-strand nucleic acid (24-mer), re-dispersed in an eluent buffer (20 mM Na3PO4, 5% BSA, 0.25% Tween 20, 10% sucrose). Capture and control probe solutions were written on the nitrocellulose membrane of a blank lateral flow test strip as test and control lines. Gold nanoparticle probe solution was written on the conjugate pad. The lateral flow test strip was immersed in 80 μL of SSC buffer (4×) containing a prescribed amount of target or control nucleic acid. As buffer migrated along the lateral flow test strip, clear red band(s) appeared within about 15 min (Fig. 3b). The detection limit was approximately 10 nM. This is comparable to existing disposable nucleic acid biosensors27, but with the advantages associated with a compact, portable pen whose cartridges can be preserved at low temperature for years before use.
Figure 3. Application of BioPen to point-of-care diagnostics.
(a) Fabrication of a lateral flow test strip using BioPen. (b) Rapid detection of HIV-1 nucleic acid at different concentrations: 200 nM, 100 nM, 50 nM, 25 nM, 10 nM of target nucleic acid, and 200 nM of control nucleic acid (from left to right). Au-NP-DNA probe: gold nanoparticle-thiolated oligonucleotide conjugate.
BioPen provides a flexible, precise, quantitative, and inexpensive platform for patterning functional materials at the POC. The technology is sufficiently versatile to allow layers of cells and proteins to be written on various substrates, and is thus useful for POC tissue engineering applications. BioPen further overcomes three challenges encountered when using state-of-the-art inkjet type printing of cells and extracellular matrix proteins. First, BioPen enables a continuous stream of cells and extracellular matrix proteins by directly writing, overcoming the complex synchronization between the movement of nozzle and the flow of liquid (Fig. 1d and Fig. 2). Second, BioPen involves direct contact with the substrate, enabling precise patterning in cases when the substrate deforms or displaces appreciably due to the deposited biomaterial. Third, BioPen can write on curved surfaces, which is challenging for current printing technology.
BioPen is especially promising for inexpensive, POC diagnostics, which we consider to be a critical need. For example, technologies of this character are important given that the regions most heavily afflicted by the HIV have relatively sparse diagnostic medical laboratory facilities and limited trained operators28. Central to this long-term goal is the ability, demonstrated here, to add reagents to lateral flow test strips at a fraction of the cost of standard-of-care gas-driven dispensing modules (such as MatrixTM 1600, Airjet AJQ 3000, and Biojet BJQ 3000 dispensers). In addition to reducing the cost of capital investment by over five orders of magnitude, BioPen reduces the cost of reagents because gas-driven dispensers require pipelines between a reagent reservoir and the tip, while BioPen does not.
The translation of patterned and functional nano- and micro-scale biomaterials into cost-effective tools for solving practical problems has proven a continual challenge29. We are hopeful that BioPen will offer a flexible platform for progress across a number of disciplines.
Methods
Materials
Ink was removed from ballpoint pen cartridges (M&G Stationery Co., Ltd, Shanghai, China) with steel ball diameters of 380 μm, 500 μm and 700 μm. Cartridges were then autoclaved, refilled with a certain volume of functional materials of interest, and stored for further use. In the examples shown, cartridges were then topped with a layer of mineral oil (Sigma-Aldrich, St. Louis, MO, USA). In experiments with rhodamine B or fluorescein isothiocyanate (FITC) (Sigma-Aldrich, St. Louis, MO, USA), fluorophores were diluted to a concentration of 5 μM in phosphate buffered saline (PBS, 1X, pH = 7.2) and pipetted into cartridges. In some experiments, glass beads (Sigma-Aldrich, St. Louis, MO, USA) were added to PBS solution. In others, a cell solution was made. For these, RPMI-1640 culture medium, DMEM (high glucose) culture medium, trypsin, fetal bovine serum (FBS), penicillin and streptomycin were acquired from Thermo Scientific HyClone (Logan, US). MCF-7 breast malignant cells and NIH/3T3 cells were purchased from ATCC (Manassas, US). Printing paper (A4, 80 gsm) was obtained from Double A International Business Co., Ltd (Shanghai, China). Two kinds of nitrocellulose membranes were used: Waterman Protran BA85 and Millipore HFB18002. E-cadherin polyclonal antibody, cy3-conjugated affinipure goat anti-rabbit lgG(H+L) and DAPI were bought from ProteintechTM (Chicago, USA). All the oligonucleotide sequences used in the study were obtained from Sangon Biotechnology Co., Ltd. (Shanghai, China) (Table S1).
Quantitative writing
We developed a method to quantify the volume and width of line when using BioPen to pattern functional materials. First, we quantified the process of using BioPen to write a rhodamine solution on printer paper by imaging the line width using an inverted fluorescence microscope (Olympus X81) and quantifying line width using ImagePro Plus 6 (IPP). The volume was calculated from the reduced weight of water in the cartridge as measured by analytical balance (Sartorius CPA124S) before and after writing. Writing speed was controlled manually by hand and a stopwatch after repeated practice. To minimize error, we measured the reduced weight after writing 100 cm and used a ruler to keep lines straight.
To evaluate the effect of pressure on writing accuracy, we developed a testing system composed of a pen-holder, computer-controlled stage, and a mass loading pallet. A cartridge with ball size of 500 μm and mass 0.01 kg was inserted into the pen-holder, which constrained the pen to move only in the vertical direction. The computer-controlled stage was covered by a sheet of paper, normal to the pen. The sheet of paper was brought into contact with the pen then displaced in a straight line within the plane of the paper at a speed of 6 cm/s. Mass was added to the cartridge in some experiments to increase the contact force from 0.1–3.5 N.
Demonstrations of flexible patterning
To validate the flexibility of BioPen in writing materials into specified patterns, we wrote several patterns on a cellulose printer paper using rhodamine B and FITC “inks” and visualized these under invert florescent microscopy (Olympus X81).
Writing particles
We prepared different concentrations (0.05, 0.5, 5 mg/mL) of glass beads in PBS solution and filled them into three different 700 μm-steel ball cartridges. Then we directly wrote on the surface of Petri dish and imaged using invert phase contrast microscopy (Olympus X81).
Writing tissues
Cells were harvested and cultured using standard procedures described elsewhere30. Briefly, MCF-7 cells were cultured in RMPI-1640 culture medium supplemented with FBS (10%, vol/vol) and penicillin/streptomycin (1%, vol/vol) in humid conditions with 5% CO2. Cells were harvested using trypsin (0.25%, wt/vol) when confluent, and re-suspended in culture medium to reach final concentrations of 0.5, 1, and 2 million cells/mL. We filled these three cell suspensions with different concentrations into three different 700 μm-steel ball cartridges, wrote directly onto the surface of a Petri dish, and imaged using invert phase contrast microscopy (Olympus X81). Cell numbers were quantified from images using IPP. Cell viability was tested directly after writing using a live/dead staining kit, following the manufacturer's instructions (Invitrogen Corporation, Carlsbad, US). Briefly, 4 μL ethidium homodimer (2 mM) and 1 μL calcein AM (4 mM) were added into 2 mL PBS to form a working solution. 30 μL of this working solution was added to cells written by BioPen and incubated at 37°C for 30 min. Finally the cells were imaged using fluorescent microscopy (Olympus X81) and cell viability was analyzed using IPP. All assays were undertaken in a humid environment.
For long time culture, cells were placed directly into incubator after writing. Four hours later, culture medium was added to the Petri dishes. Cell viability was tested again on day 5 using the aforementioned live/dead staining assay.
NIH/3T3 fibroblasts were cultured in DMEM high glucose culture medium and harvested as above. Collagen type I solution (from mouse tails, Shengyou Biotechnological Co., Hangzhou, China) was prepared according to the product instruction. All the reagents and utensils were precooled on ice. Briefly, 40 μL collagen (5 mg/mL in acetic acid) was added to a 1.5 mL tube, followed by adding 2.4 μL NaOH (0.1 M) to the collagen solution with a quick mixing. 4.6 μL PBS (10X) and 60 μL cell suspension (10 million cells per mL) were added to the solution respectively. The cell suspension (with collagen at final concentration of 0.5 mg/mL) was injected into 700 μm-steel ball cartridges. Cells encapsulated in collagen were then written onto the surface of a Petri dish (could be assisted by a pressure controller if necessary) and cultured at 37°C for 10 min before culture medium was added to the Petri dish.
To check the development of a continuous network of cells, we visualized E-cadherin, F-actin and cell nuclei with immunofluorescence according to the manufacturer instructions. Briefly, the cells encapsulated in collagen were fixed with 4% formalin at room temperature for 10 min, then permeabilized with 0.3% Triton X-100 at 4°C for 5 min. 5% BSA (MP Biomedicals, Auckland, New Zealand) was applied at 37°C for 1 hour to block nonspecific recognition. The E-cadherin polyclonal antibody (diluted ration 1:100) was added to the cells and incubated at 4°C overnight. The secondary antibody (diluted ratio 1:100) was added to the cells and incubated at room temperature for 1 hour. Thereafter, F-actin was stained with 100 nM phalloidin (Cytoskeleton, Denver, USA) at room temperature for another 30 min. Finally, DAPI was added to the cells and incubated at room temperature for 10 min. Cells were gently washed with PBS between each step to avoid cross-contamination and to reduce the fluorescent background. The cells were observed using a confocal microscopy (LSM 700, Zeiss).
Detection of nucleic acid
Gold nanoparticles with size of 13 ± 3 nm were prepared according to the reported method with slight modifications31. 2 OD (8.3 nmol) of thiolated oligonucleotide probe was activated32 and added into 15 mL gold nanoparticle solution (~4.3 nM). After standing at 4°C for 16 h, the solution was added with a certain volume of 1% sodium dodecyl sulfate (SDS) to reach a final concentration of 0.01%. 1 h later, 2 M NaCl was added into the solution up to a concentration of 150 mM by four times with 2 h interval. The final solution stood at 4°C for another 24 h and the excessive reagents were removed by centrifugation at 15000 g for 25 min. The supernatant was discarded, and the red pellet was re-dispersed in 1 mL of buffer containing 20 mM Na3PO4, 5% BSA, 0.25% Tween 20 and 10% sucrose. Capture and control probes were previously biotinylated. The capture probe (dry powder, 21.6 nmol) was dissolved in 216 µL of mixing solution containing 165 µL of 2 mg/mL streptavidin PBS solution, 29 µL of PBS and 22 µL of ethanol. The control probe (dry powder, 22.7 nmol) was dissolved in 227 µL of mixing solution containing 173 µL of 2 mg/mL streptavidin PBS solution, 31 µL of PBS and 23 µL of ethanol. The final concentration of capture and control probe solutions were both 100 μM.
Author Contributions
Y.L.H., J.H., T.J.L., G.M.G. and F.X. designed the study and wrote the main manuscript text; Y.L.H. and J.H. carried out the experiments. All authors reviewed the manuscript. Y.L.H. and J.H. contributed equally to this work.
Supplementary Material
Direct writing of functional materials at the point of care
Acknowledgments
This work was financially supported by the National 111 Project of China (B06024), the Major International Joint Research Program of China (11120101002), the National Natural Science Foundation of China (11372243), the International Science & Technology Cooperation Program of China (2013DFG02930), the Key Project of Chinese Ministry of Education (313045), South Wisdom Valley Innovative Research Team Program, International Science & Technology Cooperation Program of China (2013DFG02930), and Fundamental Research Funds for the Central Universities. GMG was also partially supported by the National Institutes of Health (R01HL109505). FX was also partially supported by the China Young 1000-Talent Program and Program for New Century Excellent Talents in University (NCET-12-0437).
References
- Han Y. L. et al. Engineering physical microenvironment for stem cell based regenerative medicine. Drug Discov. Today (2014). 10.1016/j.drudis.2014.01.015. [DOI] [PubMed] [Google Scholar]
- Jain N., Iyer K. V., Kumar A. & Shivashankar G. V. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc. Natl. Acad. Sci. USA 110, 11349–11354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. S. et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 11, 768–774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M., Zhang J., Li Y., Jiang L. & Zhu J.-J. Fabrication of a novel impedance cell sensor based on the polystyrene/polyaniline/Au nanocomposite. Talanta 80, 246–249 (2009). [DOI] [PubMed] [Google Scholar]
- Baumann K. Plant cell biology: Sensing oxygen. Nat. Rev. Mol. Cell Biol. 12, 770–770 (2011). [DOI] [PubMed] [Google Scholar]
- Doyle A. D. & Yamada K. M. Cell biology: Sensing tension. Nature 466, 192–193 (2010). [DOI] [PubMed] [Google Scholar]
- Skinner M. Cell migration: H2O2 sensing: the missing ‘Lynk'. Nat. Rev. Mol. Cell Biol. 13, 2–3 (2012). [DOI] [PubMed] [Google Scholar]
- MacBeath G. & Schreiber S. L. Printing Proteins as Microarrays for High-Throughput Function Determination. Science 289, 1760–1763 (2000). [DOI] [PubMed] [Google Scholar]
- Lockhart D. J. & Winzeler E. A. Genomics, gene expression and DNA arrays. Nature 405, 827–836 (2000). [DOI] [PubMed] [Google Scholar]
- Piner R. D., Zhu J., Xu F., Hong S. & Mirkin C. A. “ Dip-pen” nanolithography. Science 283, 661–663 (1999). [DOI] [PubMed] [Google Scholar]
- Sha B. et al. Potential application of titanium dioxide nanoparticles in the prevention of osteosarcoma and chondrosarcoma recurrence. J. Nanosci. Nanotech. 13, 1208–1211 (2013). [DOI] [PubMed] [Google Scholar]
- Tomizaki K. y., Usui K. & Mihara H. Protein-protein interactions and selection: array-based techniques for screening disease-associated biomarkers in predictive/early diagnosis. FEBS J. 277, 1996–2005 (2010). [DOI] [PubMed] [Google Scholar]
- Guillotin B. & Guillemot F. Cell patterning technologies for organotypic tissue fabrication. Trends Biotechnol. 29, 183–190 (2011). [DOI] [PubMed] [Google Scholar]
- Xia Y. & Whitesides G. M. Soft lithography. Annu. Rev. Mater. Sci. 28, 153–184 (1998). [Google Scholar]
- Cavallini M., Gentili D., Greco P., Valle F. & Biscarini F. Micro- and nanopatterning by lithographically controlled wetting. Nat. Protoc. 7, 1668–1676 (2012). [DOI] [PubMed] [Google Scholar]
- Salaita K., Wang Y. & Mirkin C. A. Applications of dip-pen nanolithography. Nat. Nanotechnol. 2, 145–155 (2007). [DOI] [PubMed] [Google Scholar]
- Vega R. A., Maspoch D., Salaita K. & Mirkin C. A. Nanoarrays of Single Virus Particles. Angew. Chem. Int. Ed. 117, 6167–6169 (2005). [DOI] [PubMed] [Google Scholar]
- Lee K.-B., Park S.-J., Mirkin C. A., Smith J. C. & Mrksich M. Protein Nanoarrays Generated By Dip-Pen Nanolithography. Science 295, 1702–1705 (2002). [DOI] [PubMed] [Google Scholar]
- Gubala V. Harris L. F. Ricco A. J. Tan M. X. & Williams D. E. Point of care diagnostics: status and future. Anal. Chem. 84, 487–515 (2011). [DOI] [PubMed] [Google Scholar]
- Hu J. et al. Advances in paper-based point-of-care diagnostics. Biosens. Bioelectron. 54, 585–597 (2014). [DOI] [PubMed] [Google Scholar]
- Mirica K. A., Weis J. G., Schnorr J. M., Esser B. & Swager T. M. Mechanical Drawing of Gas Sensors on Paper. Angew. Chem. Int. Ed. (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A. et al. Pen-on-Paper Flexible Electronics. Adv. Mater. 23, 3426–3430 (2011). [DOI] [PubMed] [Google Scholar]
- Han Y. L. et al. Benchtop Fabrication of Three-Dimensional Reconfigurable Microfluidic Devices from Paper/Polymer Composite. Lab Chip (2013). [DOI] [PubMed] [Google Scholar]
- Zheng Y., He Z., Gao Y. & Liu J. Direct Desktop Printed-Circuits-on-Paper Flexible Electronics. Sci. Rep. 3 (2013). [Google Scholar]
- Nichol J. W. & Khademhosseini A. Modular tissue engineering: engineering biological tissues from the bottom up. Soft Matter 5, 1312–1319 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. et al. A self-assembly pathway to aligned monodomain gels. Nat. Mater. 9, 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X. et al. Disposable nucleic acid biosensors based on gold nanoparticle probes and lateral flow strip. Anal. Chem. 81, 1660–1668 (2009). [DOI] [PubMed] [Google Scholar]
- Wang S., Xu F. & Demirci U. Advances in developing HIV-1 viral load assays for resource-limited settings. Biotechnol. Adv. 28, 770–781 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesides G. M. Cool, or simple and cheap? Why not both? Lab Chip 13, 11–13 (2013). [DOI] [PubMed] [Google Scholar]
- Han Y. L. et al. Directed self-assembly of microscale hydrogels by electrostatic interaction. Biofabrication 5, 035004 (2013). [DOI] [PubMed] [Google Scholar]
- Hu J. et al. Oligonucleotide-linked gold nanoparticle aggregates for enhanced sensitivity in lateral flow assays. Lab Chip 13, 4352–4357 (2013). [DOI] [PubMed] [Google Scholar]
- Liu J. & Lu Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat. Protoc. 1, 246–252 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Direct writing of functional materials at the point of care