Abstract
Dendritic cells (DCs) are a heterogeneous population within the mononuclear phagocyte system (MPS) that derive from bone marrow precursors. Commitment and specification of hematopoietic progenitors to the DC lineage is critical for the proper induction of both immunity and tolerance. This review summarizes the important cytokines and transcription factors required for differentiation of the DC lineage as well as further diversification into specific DC subsets. We highlight recent advances in the characterization of immediate DC precursors arising from the common myeloid progenitor (CMP). Particular emphasis is placed on the corresponding temporal expression of relevant factors involved in regulating developmental options.
Keywords: Dendritic cell, Lineage commitment, Transcription factor, Flt3-ligand, Common dendritic progenitor
1. Introduction
The immune system comprises a large number of highly differentiated cell types whose functions emerge from coordinated interactions among developmentally heterogeneous cells, and are rarely observed in complete isolation. For example, phagocytic cells such as neutrophils or macrophages best exert their function through interactions with soluble components of the immune system such as antibodies secreted by B cells, or in their absence, mannose-binding lectins or deposition of complement protein C3. Perhaps the most elegant example of coordinated immunity is the generation of high affinity, isotype-switched antibodies capable of neutralizing a pathogen. Such a response requires not only the cell-intrinsic capacity of the B cell, but also linked recognition and activity of a differentiated form of the CD4+ T cell, the T follicular helper cell (Tfh), as well as contributions to T cell priming produced by cells of the phagocyte system called dendritic cells (DC). This three way interaction represents a highly organized system with each cell presenting checkpoints and barriers to improper activation as a means of guarding against autoimmunity. While the molecular mechanisms responsible for the development and diversification of T and B cells are well studied, those responsible for DCs are only beginning to be defined. This review will provide a synopsis of the known cellular and molecular events required for the development of DCs at steady-state.
2. Unique functions and subsets of DC lineages
DCs were discovered nearly 40 years ago by Steinman and Cohn, who identified a “large stellate cell” population among adherent splenocytes on particular glass and plastic surfaces [1]. Beginning with a strictly descriptive analysis, these cells were defined as a distinct lineage, separate from B and T cells as well as granulocytes and macrophages, on the basis of several criteria. Initially, their “dendritic” morphology and clear phagocytic capacity set them apart from lymphocytes. However, it was their profound capacity for stimulating T cells in a mixed lymphocyte reaction (MLR) that prompted the notion that these cells could play a unique role in directing adaptive immunity [2]. Additional hallmarks emerged from flow cytometric analyses that identified a distinct set of cell-surface markers separating DCs from macrophages and other myeloid lineages. Subsequent studies have demonstrated that, in addition to stimulating the immune response, DCs also possess a capacity to impose a tolerogenic state on T cells under appropriate conditions [3;4].
The first two decades of work on DCs largely considered this cell a homogenous population. In 1992, the first clear-cut example of subset differentiation within DCs was put forth, based upon the expression of the murine CD8-alpha marker. Clearly distinct from developing or recirculating thymic T cells, this subset represented a relatively small population within the thymus, and was thought to derive from an intrathymic lymphoid-restricted precursor rather than from bone marrow precursors [5;6]. As a byproduct of differences in identification and separation methods, the demarcation of various subsets has since been somewhat inconsistent [7]. Currently, the field considers four major subsets of lymphoid-resident DCs. In addition to the CD8-alpha positive DC, two other populations of conventional DCs (cDC) are defined by the presence or absence of the cell-surface molecule CD4 [8]. These two populations show a high degree of similarity with respect to function, development, and gene expression, and for simplicity will be grouped together and further referred to as CD4+ cDCs in this review [9]. A fourth population of plasmacytoid DCs (pDC) is defined by the expression of several distinguishing markers including B220, Siglec-H, and Bst2 [10-12]. Human counterparts for each of these DC subtypes have been identified, underscoring the conservation of DC lineage diversification across species, presumably owing to their important functional specialization [13-15]. As discussed briefly below, monocyte-derived cells sharing many features with DCs arise under inflammatory rather than steady-state conditions, although little is currently known regarding the transcriptional and molecular basis of their differentiation [16;17].
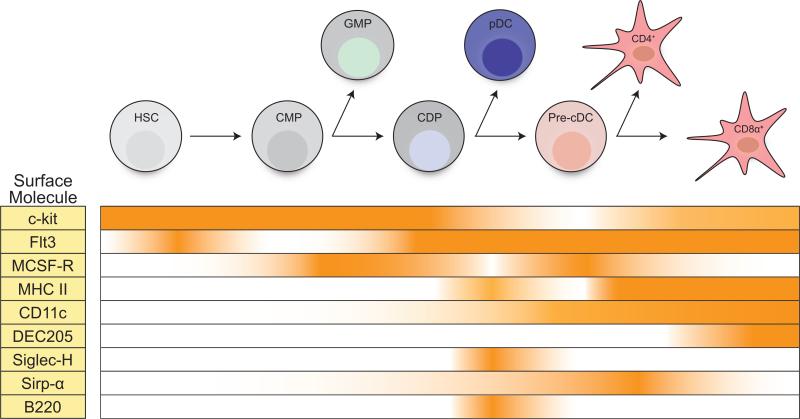
All subsets express high levels of the complement receptor CD11c, and also display a variety of surface receptors endowing them with the properties of growth factor responsiveness, efficient phagocytosis, and antigen presentation. Figure 1 outlines the expression of some of these key surface proteins as DC subsets differentiate from hematopoietic precursors (discussed further below). While they are convenient markers in the murine system, there is no known function of the CD4 and CD8-alpha molecules in the execution of specific functional processes. In addition, these two markers are among the few that do not show conservation with subsets of DCs in the human system.
Figure 1. The temporal expression of genes in DCs and their precursors.
The expression of key surface markers used for the identification of mature dendritic cells (DCs) and their bone marrow precursors is depicted. In addition, the expression of important growth factors required for the development of DC subsets is shown.
Beyond differences in cell-surface receptors as the basis for defining distinct subsets, non-overlapping functions for these populations are beginning to emerge. For example, pDCs appear to be a major source of type I interferon during viral infections. Recent models in which pDCs are selectively eliminated highlight their contributions in some, but not all, settings of infections by pathogens [18;19]. The cDC subsets appear to be primarily concerned with priming adaptive immune responses, particularly those of T cells, although important interactions with other immune cells, such as B cells and NK cells, have also been described [20-22].
Several years after their initial identification, a study by den Haan and colleagues indicated that one potentially unique function of the CD8+ DC is its capacity to cross-present exogenous antigens on MHC class I [23]. As a result, it was hypothesized that these cells are uniquely capable of cross-priming CD8+ T cells, a process first described in 1976 and now implicated as a critical response in viral and anti-tumor immunity [24-26]. By virtue of this unique function, these cells could provide an efficient mechanism to induce CD8+ T cells against pathogens that fail to infect DCs directly, and therefore evade classical mechanisms of inducing CTL (cytotoxic T lymphocyte) responses. In support of this role, CD8+ DCs are now recognized to reside in the T cell zones of the spleen, and are thought to traffic from areas of pathogen trapping into areas of T cell presentation [27;28]. Other studies have also proposed that these cells may provide functions independent of cross-presentation, such as the production of essential cytokines in response to various pathogens [29;30].
CD4+ cDCs for the most part have been considered poor cross-presenters in comparison to their CD8+ counterparts. These cells are largely considered to activate CD4+ T cells, rather than having any specialized role in the induction of CTLs [31]. However, the classical paradigm of functionally segregating cDC subsets based on the efficiency and quality of MHC class I and II presentation is not without controversy. For example, a recent study suggests that the distinction between CD8+ and CD4+ DCs may instead lie in the inability of the former to process antigens for MHC class II efficiently; and when controlled for antigen dose and delivery, both subsets are capable of efficient cross-presentation [32]. In fact, under certain inflammatory conditions, the monocyte-derived DC can present exogenous antigens more efficiently than either of the lymphoid-tissue resident cDCs [33;34]. Collectively, these seemingly conflicting observations may be explained by the type of antigen and the context in which it is being studied. Clearly, additional studies in which DC subsets are selectively ablated are required to substantiate the various competing claims.
3. Accurately defining the DC lineage
As discussed above, the criteria to mark the DC lineage has relied on a constantly evolving rubric, beginning with the purely descriptive analyses of Steinman and colleagues in the 1970's to a more functional and phenotypic characterization at present. Yet such seeming clarity has not resolved the topic of the exact contribution of DCs during an immune response, in part because of the shared expression of many surface markers between DCs and other myeloid cells. In fact, some of the seminal studies over the last decade that relied on genetic or antibody-based depletion of specific myeloid populations also affected several other cell types [35]. For instance, two independent reports have claimed that basophils, and not DCs, are critical for mediating T helper type 2 responses (Th2) [36;37]. However, these reports were disputed by another group who demonstrated that the antibody-based depletion of basophils in the previous studies also removed a key DC population, which actually appears to be necessary for initiating Th2 immunity [38]. More recently, a report claiming the importance of beta-catenin signaling in intestinal DCs to gut tolerance was based upon the selective deletion of this molecule in only DCs, a specious claim given there was also complete deletion in a sizeable fraction of macrophages [39]. These ambiguous results are the byproduct of a strict reliance on cell-surface markers to segregate myeloid populations. Instead, when considering any lineage, especially one as heterogeneous and plastic as DCs, more rigorous criteria need to be applied for its functional characterization in various contexts. Some authors have suggested defining DCs based on their anatomical location, origin from dedicated precursors, and antigen presenting properties in addition to phenotypic markers [40]. However, all of these measures can be inconsistent depending on the physiological context or method of measurement. Therefore, we suggest that the most succinct and reliable definition of the DC lineage is based on the expression of key lineage-specifying transcription factors which are unequivocally required for their development in all settings. Parsing these transcriptional networks will help resolve developmental and functional controversies that currently exist within the field.
4. The developmental origin of DCs
An issue with particular relevance to the transcriptional basis of DC development is their derivation from bone marrow progenitors. It is now clearly established that DCs are short lived and are continuously repopulated in the periphery, both within lymphoid and non-lymphoid tissues, from progenitors that arise in the bone marrow [41;42]. For some period of time, there was confusion about the origin of DCs, based primarily on the initial classification of immune lineages as broadly derived either from a common lymphoid-restricted progenitor (CLP), or from a common myeloid-restricted progenitor (CMP) [43;44]. Whereas other cells of the immune system, such as T cells, B cells or neutrophils, are derived strictly from the CLP or from the CMP, it is now clear that all DC subsets can be derived from either population [45-47]. This unexpected finding overturned the existing paradigm that CD8+ DCs were of lymphoid while CD4+ DCs were of myeloid origin [5;48]. Despite the ability of CLPs to generate DCs upon adoptive transfer into irradiated hosts, the mechanisms and cell intermediates through which this occurs is unclear. Moreover, recent lineage tracing studies by Schlenner et al. indicate that the contribution of this pathway to the steady-state DC compartment is minimal [49]. Therefore, our focus will be on the CMP-derived pathway of DC development.
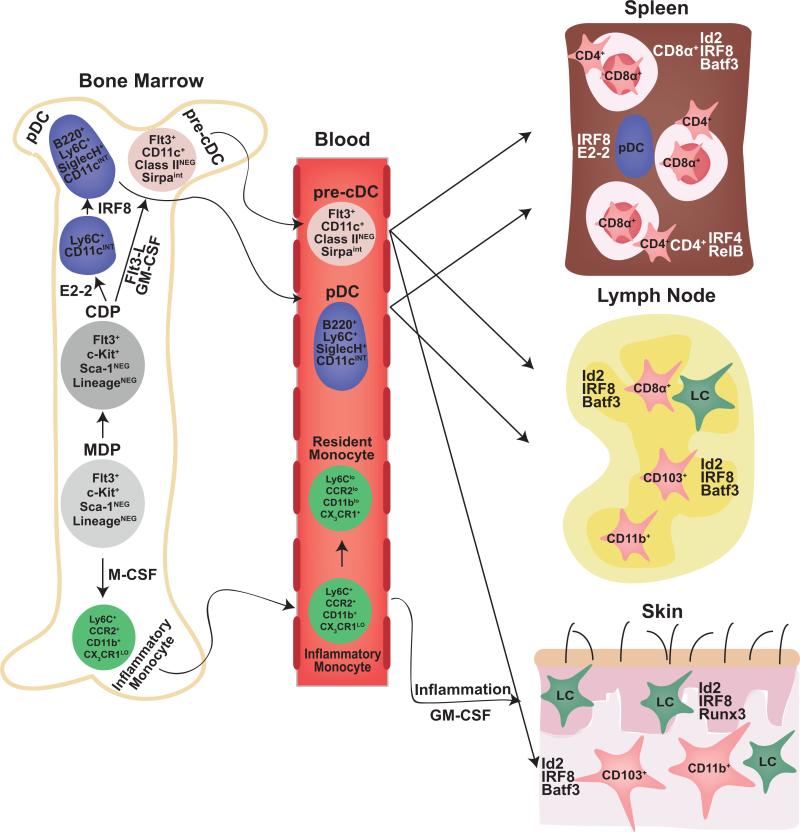
Fogg and colleagues identified the first precursor downstream of the CMP that still retained DC potential, termed the macrophage-dendritic cell precursor (MDP) [50]. As its name would indicate, the MDP strictly has the potential to generate macrophages and DCs whereas alternate myeloid lineages proceed through the granulocyte-macrophage precursor (GMP) [44]. Soon thereafter, two groups reported the identification of a purely DC-restricted bone marrow progenitor called the common dendritic cell progenitor (CDP), which is efficiently able to generate all DC subsets at a clonal level [51;52]. The CDP was shown to originate from the MDP following the loss of monocyte lineage potential [53]. Similar to the MDP, the CDP expresses relatively high levels of macrophage colony-stimulating factor 1 receptor (M-CSFR) and FMS-like tyrosine kinase 3-ligand receptor (Flt3), but lower levels of the stem cell factor receptor (c-Kit). Presently it is thought that both the MDP and the CDP derive exclusively from the CMP in vivo, strengthening the argument that DCs are minimally derived from lymphoid precursors. Finally, the terminal steps in diversification of DC subsets appear to begin at the CDP stage. As outlined in Figure 2, CDPs seem to be the immediate precursor of both pre-pDCs and pre-cDCs, which are cells restricted to their respective lineages but not yet fully mature [54;55]. pDCs complete their maturation in the bone marrow and circulate through the blood to secondary lymphoid organs. In contrast, pre-cDCs seed lymphoid and non-lymphoid tissues via the blood where-upon they complete their differentiation into either the CD8+ or CD4+ cDC subsets.
Figure 2. Stages of DC development.
DCs derive from bone marrow–resident hematopoietic progenitors. Following differentiation of the hematopoietic stem cell (HSC) into the common myeloid progenitor (CMP), alternate myeloid fates are extinguished in the subsequent macrophage-dendritic progenitor (MDP). Commitment to the DC lineage occurs at the next stage termed the common dendritic progenitor (CDP). The CDP has the potential to develop into both plasmacytoid dendritic cells (pDC) and conventional dendritic cells (cDC). This choice occurs as the CDP transitions into either of the intermediates for each lineage: the pre-pDC (not shown) or the pre-cDC. pDCs complete their maturation in the bone marrow and then circulate to secondary lymphoid organs through the blood. In contrast, pre-cDCs seed both lymphoid and non-lymphoid organs and undergo one final fate choice into either CD8+ or CD4+ DCs; or CD103+ or CD11b+ equivalents in the periphery, respectively. The developmental similarity between lymphoid-organ and peripheral DC subsets has been shown using mice deficient in the transcription factors Id2, IRF8, and Batf3. Additionally, monocytes can give rise to DCs in inflammatory settings although the transcriptional mechanism for this process is poorly characterized.
5. Extracellular cues supporting DC development
A number of extracellular cues induce the progression through the progenitor stages described above, mainly by activating several cytokine receptors. The major cytokine receptors influencing DC development are M-CSFR, granulocyte-macrophage colony-stimulating factor receptor (GM-CSFR) and Flt3, which are all expressed variably on early hematopoietic precursors but remain highly expressed on committed DC progenitors (Figure 1).
The importance of these cytokines in the differentiation of DCs was first established by early studies demonstrating the ability of GM-CSF, either alone or in combination with IL-4, to drive the formation of DCs from mouse bone marrow or human peripheral monocytes [48;56]. The unique potency of these cells for antigen presentation, combined with their expression of CD11c and MHC class II, confirmed their identity as bona fide DCs and provided a widely used model system for in vitro studies. Therefore, it was surprising when both GM-CSF- and GM-CSFR-deficient mice showed a largely unperturbed DC compartment, leading to the conclusions that GM-CSF is dispensable or redundant in steady-state DC development [57;58]. The recent identification of TNF-alpha- and iNOS-producing DCs under inflammatory conditions, and their dependence on GM-CSF, points to the relevance of this cytokine in infectious settings and likely explains its ability to induce DC development in vitro [32;59;60].
Nonetheless, these early results suggested that other cytokines were likely responsible for the steady-state development of DCs in vivo. In 1996, McKenna and colleagues reported that the administration of exogenous Flt3-ligand (Flt3L) potently induces the expansion of all DC subsets in vivo [61]. This cytokine was then also shown to support the development of all subsets in vitro [62]. Moreover, forced expression of Flt3 (receptor for Flt3L), or its downstream effector molecule STAT3, in progenitors committed to alternative lineages can re-direct their development into DCs [63]. The requirement for Flt3L in vivo was confirmed by the greatly reduced numbers of DCs in Flt3L-deficient mice and Flt3-deficient mice [64;65]. Interestingly, the absence of Ftl3L presented with a stronger DC phenotype than the absence of Flt3, suggesting that the ligand may interact with a hitherto undefined receptor. These findings have been reinforced by subsequent studies on STAT3-null mice, which appear to phenocopy Flt3L deficiency [66]. Recent evidence suggests that the activation of STAT3 is mediated by mTOR (mammalian target of rapamycin) signaling, and accordingly, chemical inhibition of mTOR with rapamycin perturbs the development of DCs in vitro [67].
While Flt3L appears to be the dominant cytokine controlling steady state DC development, GMCSF may contribute, as mice deficient in both Flt3L and GM-CSF have lower numbers of DCs than either single cytokine deficiency alone [57]. The precise degree of redundancy is still a matter of ongoing work. Much in the same manner as differential localization within the bone marrow has been proposed to regulate B cell development through various stages by distinct actions of cytokines and cell-surface proteins on stroma, a similar combinatorial regulation of DCs has recently been proposed [68;69]. First, as noted above, there is a compound defect in mice deficient in both Flt3L and GM-CSF, suggesting contributions of both cytokines to DC development. In addition, different cytokines appear to have varying effects on the development of specific DC subsets. For instance, GM-CSF favors the development of cDCs while inhibiting the development of pDCs, through a mechanism dependent upon activation of STAT5 [70]. In contrast, culturing bone marrow cells in M-CSF in combination with Flt3L has the opposite effect, favoring pDC development [51]. It is conceivable that specific stromal niches within the bone marrow could support distinct DC developmental outcomes through differential cytokine production. Indirect evidence supporting this model comes from clonal studies of the CDP, which noted that some cells within the population already appear to be biased toward either pDCs or cDCs [52]. The generation of unique, non-overlapping reporter systems for each of the abovementioned cytokines will greatly aid in the resolution of this outstanding question of niche-dependent effects.
6. Transcription factors control DC commitment and specification
Generally, cell fate specification occurs through the actions of transcription factors which may be induced or inhibited by initiating extracellular cues. Recently, a number of transcription factors have been identified that control commitment, specification, and survival of DCs. In this review, we will group these factors into two major categories. The first category consists of transcription factors that are required for the development of early DC progenitors, which for the most part influence all DC subsets. As one might expect with factors influencing such early decisions, the loss of these genes often affects a number of additional immune lineages besides DCs. The second category is comprised of more lineage-restricted transcription factors, which regulate later stages of DC fate decisions. Deficiency of these factors usually results in more specific defects within the DC lineage. It is worth keeping in mind that the expression of some factors from the first category is not extinguished as the steps of commitment and specification unfold, and therefore may influence both progenitors and mature cell types in distinct ways. Importantly, unique developmental outcomes may emerge from transcriptional programs initiated by broadly expressed factors of the first category through the induction and then interaction with factors from the second.
6.1 Factors regulating early DC progenitor development
The landmark discovery of the indispensable role of the zinc finger transcription factor Ikaros in the development of all lymphoid lineages also represents the initial identification of a factor necessary for DC development. Specifically, Ikaros-null mice lack pDCs and CD4+ cDCs, and mice harboring a dominant-negative form exhibit a complete absence of all DC subsets [71;72]. Unexpectedly, Ikaros-null mice retain a residual population of CD8+ cDCs, although the function or origin of these cells has not been examined. The apparent mechanism by which Ikaros controls DC development is through its regulation of a large panel of receptors and transcription factors. Although classically considered to be required only for the priming of lymphoid lineages, gene expression analysis of the earliest cell stages of hematopoietic development revealed that the loss of Ikaros affects the expression of large gene sets associated with both lymphoid and myeloid lineages [73]. For example, the expression of Flt3, IL-7 receptor, Notch1, and the transcription factor Mef2c are all controlled by Ikaros, suggesting that it acts globally on many aspects of lineage specification [74].
Another well-known transcription factor influencing the early development of DCs is the ETS factor PU.1. Recently, PU.1 was unequivocally shown to be necessary for DC development, resolving earlier studies that resulted in conflicting conclusions [75;76]. Mice with a conditional deletion of PU.1 within the hematopoietic compartment fail to develop all DC subsets, but also show defects in other myeloid lineages including but not limited to macrophages and neutrophils [77]. Similar to Ikaros, PU.1 appears to control the induction of a broad range of genes necessary for development of many hematopoietic lineages. In particular, PU.1 controls the early expression of Flt3, GM-CSFR, and M-CSFR on progenitors, although only the absence of Flt3 was confirmed on DC precursors [78;79]. Notably, forced expression of Flt3 in PU.1-deficient progenitor cells did not rescue DC development, reinforcing the notion that PU.1 regulates other important programs aside from the signaling downstream of Flt3.
Finally, Gfi1, a known transcriptional repressor, appears to play a role in the development of all DC subsets. Gfi1-null mice have a global reduction in lymphoid-organ DC compartments, albeit not as severe as either Ikaros- or PU.1-deficient mice [80]. Analysis of Flt3+ DC progenitors revealed a cell-autonomous role for Gfi1 in the activation of STAT3, suggesting that it may act downstream of Ikaros or PU.1. One proposed mechanism is through its ability to inhibit PIAS3 (protein inhibitor of activated STAT3), a specific inhibitor of STAT3 [81]. In this model, loss of Gfi1 leads to the maintenance of PIAS3 activity and thus a reduction in transcriptional events mediated by STAT3. Additionally, studies in human monocytes have shown that Gfi1 can bind to a large number of myeloid gene loci such as JAK3, IL-8, and the CEBP family of transcription factors, indicating a potential role beyond its effect on STAT3 [82]. Interestingly, surviving Gfi1-deficient DCs closely resemble their counterparts from mice deficient in members of the NF-kappaB signaling cascade, suggesting that these two factors may converge on a common developmental pathway (see below) [83]. The dependence of DC development on both PU.1 and Gfi1 is in stark contrast to B cell ontogeny, during which these two factors act in opposition [84]. Whether Ikaros, PU.1, and Gfi1 act in concert or independently in DC development remains to be determined.
6.2 Factors regulating late DC progenitor development
Whereas the factors discussed above appear to be necessary during the HSC to CDP transition, particularly for the proper expression of key cytokine receptors, several transcription factors exert control over developmental options after commitment to the DC lineage has already occurred. These factors are expressed beginning in the CDP stage, and thus specify commitment into subsets or act on survival or proliferation programs in immediate precursors. For the most part, the factors acting in this realm of DC development have been evaluated for their actions on lymphoid resident DCs. However, it is clear that they likely affect the development of corresponding non-lymphoid DCs as well by mediating similar genetic programs in both compartments (Figure 2) [85;86].
The first transcription factor identified to affect a particular subset was RelB, a member of the NF-kappaB family of transcription factors, which also consists of RelA (p65), c-Rel, p50 and p52. In contrast to RelA and c-Rel, RelB selectively forms a heterodimer with p52 or the precursor form, p100 [87]. Expression of RelB is induced in the pre-cDC and then preferentially maintained in CD4+ DCs. Consequently, mice deficient in RelB show a significant decrease in the CD4+ population of cDCs, a defect determined to be cell intrinsic, while CD8+ cDCs and pDCs appear to be unaffected [88;89].
Although studies investigating the mechanism by which RelB controls DC differentiation are lacking, one gene suggested to be a target of RelB is interferon regulatory factor 4 (IRF4), whose deletion also results in a large reduction in the number of CD4+ DCs [90]. IRF4, one of nine IRF family members, contains an N-terminal DNA-binding domain (DBD) and a C-terminal IRF-associated domain (IAD). IRF4 is known to interact with a number of other transcription factors, including ETS factors and AP1 family members. In different settings, these interactions are critical in determining the DNA-binding specificity and functional nature of the IAD, either as an activator or repressor of transcription [91-93]. However, whether these interactions occur within the context of DCs remain to be defined, although PU.1 (ETS) and Batf3 (AP1) are attractive candidates.
Of the IRF family members, the member most functionally related to IRF4 is IRF8. IRF8 has also been demonstrated to play a critical role in DC differentiation. IRF8 deficiency leads to a complete loss of both CD8+ cDCs and pDCs, and mice lacking both IRF4 and IRF8 exhibit essentially a compound phenotype, with a severe reduction in the total number of DCs and an absence of all but a few CD4+ and double negative cDCs [94;95]. This result suggests that these factors may provide an overlapping function in certain subsets, but for most DCs, these two factors display essentially non-overlapping patterns of expression. It is not yet known what signals are controlled by the IRF proteins, or what signals or transcription factors lead to the induction of IRF4 or IRF8 in their respective subsets.
In contrast to the selective expression of IRFs within DC subsets, Batf3 is expressed in both CD8+ and CD4+ cDCs, but loss of Batf3 leads to an abrogation in the development of only one of these subsets, the CD8+ DC [96]. Batf3 is a member of the AP1 family of transcription factors that forms an obligate heterodimer with cJun or other Jun paralogs. However, unlike other AP1 members, Batf3 lacks a C-terminal extension that would provide a transcriptional activation domain, and instead consists only of a basic DNA-binding motif followed by a dimer-forming leucine zipper region [97;98]. Batf3 is most similar to a related factor Batf, which was recently found to be necessary for Th17 and Tfh differentiation as well as for isotype-switching in B cells [99;100]. These recent findings reverse the earlier notion that these minimal AP1 family members, Batf3 and Batf, were simply dominant-negative analogs of Fos, and instead suggest that these factors form unique heterodimers with Jun that posses distinct transcriptional activities. In the case of Batf3, this activity presumably includes the activation of genes selectively expressed by CD8+ DCs.
Mice deficient in Batf3 have normal numbers of precursors, including CDPs and pre-cDCs [85], and appear deficient in only the most terminal stage of CD8+ DC maturation. This final step is defined by the induction of genes important for the function of this DC subset, including Langerin and CD103. However, at present the immediate transcriptional targets of Batf3 are unknown.
The factors discussed above appear to regulate terminal steps of commitment to particular DC lineages. In contrast, two additional transcription factors appear to regulate the survival of terminally committed precursors or mature cell types. The transcription factor Rbp-j, a critical mediator of the Notch signaling pathway, was recently found to be required for the survival of CD4+ DCs in vivo [101]. Mice deficient in Rbp-j lack more than 50% of their CD4+ DCs. Detailed analysis revealed that commitment to this lineage appears to be unaffected since the population of splenic pre-cDCs in these mice is normal. Rather, higher levels of Annexin-V staining and BrdU incorporation in surviving CD4+ DCs suggest increased apoptosis of the terminal cell stage.
Similarly, the zinc finger transcription factor Bcl6 appears to regulate the survival of cDC precursors [102]. Bcl6 is a BTB (bric-a-brac, tramtrack, broad) -domain containing repressor that also has important actions in Tfh differentiation and exerts a repressive action on plasma cell genes within germinal center B cells [103-105]. In DCs, Bcl6 deficiency causes a severe reduction in both types of cDCs. While in vitro Flt3L experiments suggest a more pronounced effect on CD8+ DC equivalents, the in vivo results indicate a larger effect on CD4+ DC frequency. In contrast to Rbp-j deficiency, Bcl6-null mice do appear to exhibit increased Annexin V staining in cDC precursors and accordingly, express markers of apoptosis such as p53.
7. The balance between E2-2 and Id2 determines the choice between cDC and pDC fates
A family of transcription factors known as class I basic helix loop helix (bHLH) factors plays an important role in the differentiation and survival of a number of lymphocyte populations [106]. Class I bHLH proteins, also referred to as E proteins, comprise a family of four members: E12, E47, E2-2, and HEB. These proteins function either as homodimers or heterodimers between family members, and bind to a conserved DNA motif referred to as an E-box. The DNA-binding activity of these heterodimers can be interrupted by formation of a complex between E proteins and members of the inhibitor of differentiation (Id) HLH protein family, which lack the necessary DNA-binding motif of the basic region. Of the four members of the Id protein family, Id2 and Id3 appear to be the major inhibitors of E2 protein activity during lymphocyte development.
The first indication of the importance of E proteins in DC development arose from the observation that over-expression of Id2 or Id3 inhibits the development of pDCs in vitro [107]. This was extended by the finding that Id2 expression is induced in vitro in response to GM-CSF, and that Id2 is required for in vivo development of CD8+ DCs, but not other subsets [108]. Surprisingly, the frequency of pDCs is increased in these mice suggesting that Id2 may function to divert cDC precursors away from the pDC fate. This hypothesis is supported by the finding that DCs developing in Id2-deficient mice also show de-repression of many genes normally associated with B cells as well as pDCs, as they share a large common gene signature [109].
A major breakthrough in the understanding of DC subset development resulted from the discovery that E2-2 is required for the development of pDCs in vivo [54]. These findings also helped to establish the identity of a pDC-restricted precursor: a CD11c+ Ly6C+ SiglecH- cell termed the pre-pDC. Mechanistically, E2-2 regulates a large pDC gene program including the direct regulation of other key transcription factors associated with pDC development, including IRF8, Bcl11a, and Spi-B. Interestingly, E2-2 also appears to repress genes associated with cDC subsets [110]. This was shown using an inducible Cre-deletion system whereby E2-2 was deleted in mature pDCs. Terminal deletion of E2-2 results in the induction of a number of cDC-associated genes including the re-expression of Id2. However, only a small subset of cDC genes appears to be affected. Therefore, it is not clear whether the emergence of these genes represents a conversion of pDCs into bona fide cDCs, or instead into the recently reported alternative CD8+ CX3CR1+ DC lineage that appears to be more closely related to pDCs [111]. In any event, de-repression of Id2 caused by the loss of E2-2 expression now highlights an important regulatory circuit that controls the switch between pDCs and cDCs. In this circuit, induction of Id2 decreases E2-2 expression, and E2-2 activity suppresses Id2 expression. This arrangement represents a classical “flip-flop” circuit based upon mutual antagonism of alternately expressed factors. Presently, it is unknown what signals might control the induction of alternate states of this circuit.
8. Gene expression analysis of the CDP points to “pDC-priming.”
Elaborating the transcriptional mechanism of T cell development was greatly aided by the identification of distinct cellular stages through which T cells progress. For example, the fate choice between T and NK cell lineages in the thymus was known to take place during the CLP to pro-T cell transition and to be dependent on Notch1 [112]. Careful gene expression analysis of intermediate cell stages allowed three groups to identify the downstream factor Bcl11b as a key mediator of this specification process [113-115] .
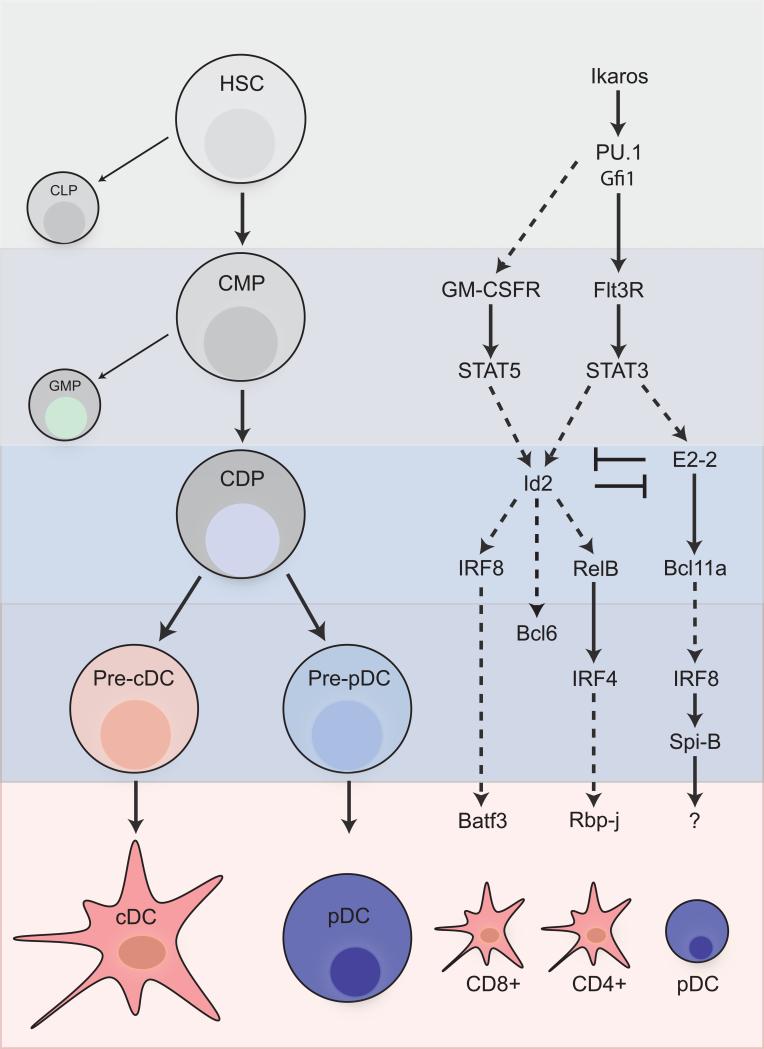
In contrast to T cell development, the specific intermediate stages of DC development have only recently been identified, as discussed above. In addition, the difficulty in identifying and isolating these progenitors, as well as their relative paucity in the bone marrow, has made them difficult to study. As a result, the relationships between the transcription factors that regulate DC development and the progenitor stages at which they act are still unclear. One plausible arrangement of the transcription factor network with respect to cellular differentiation is shown in Figure 3. In this scheme, we have diagrammed the potential relationships between key transcription factors, some of which were suggested in the original reports.
Figure 3. Transcription factor networks control DC development.
A number of transcription factors regulate the commitment and survival of dendritic cells at various stages of development. However, the coordinated action of these factors with respect to one another as well as to relevant progenitor stages has not been well defined. One plausible network of these factors is depicted alongside the cellular stage in which they likely exert their functions. Solid arrows indicate connections that have been proven or suggested within original reports. Dotted arrows indicate hypothesized relationships.
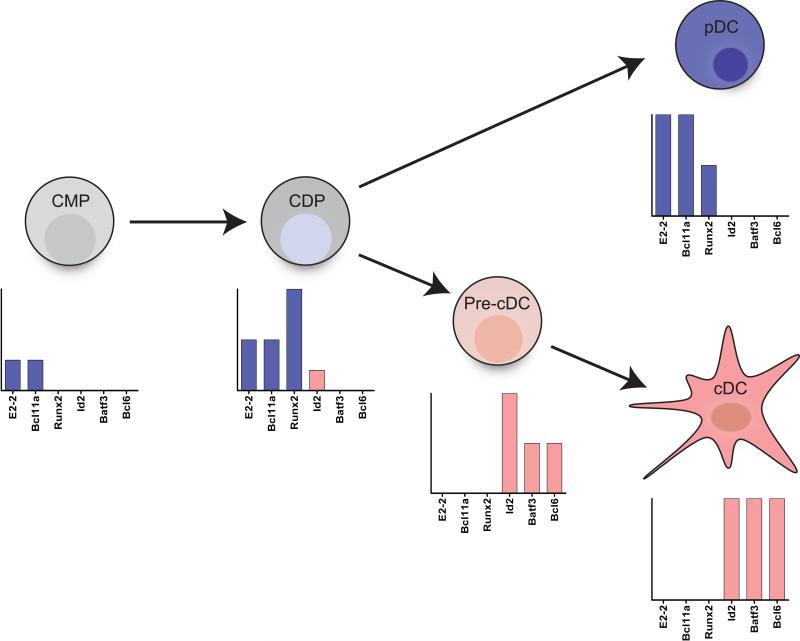
We propose that the CDP represents the key stage at which the choice between the pDC and cDC lineage is made. The CDP is the natural candidate for this choice, since it has extinguished its ability to form alternate lineages, but still retains the ability to generate all DC subsets. Recent studies, as well as unpublished studies from our lab, have provided expression profile data for each sequential stage of DC differentiation in vivo including the CDP [116]. Strikingly, analysis of transcription factors expressed in CDPs reveals that a number of pDC factors, including E2-2, Bcl11a, and Runx2, are already expressed at moderate levels (Figure 4). In contrast, factors associated with the cDC lineage, such as Id2, Batf3, and Bcl6, are expressed either at very low levels or not at all. One interpretation of this data is that CDPs may be set to “default” into the pDC lineage unless a trigger is received to induce Id2. Id2 becomes highly expressed at the precDC stage where-upon pDC-associated genes are lost. Such a default pathway could be likened to the strong skewing of naïve T cells towards the Th2 phenotype, which is mediated by an auto-activation of Gata-3 [117].
Figure 4. CDPs are “pDC-primed.”.
CDPs represent the last common precursor for both cDCs and pDCs, and thus a likely stage of specification into either lineage. This decision is dependent on a mutually antagonist balance between the transcription factors E2-2 and Id2. Gene expression analysis of progenitor populations reveals that CDPs express a number of pDC-specific transcription factors. Shown is the expression of pDC factors (E2-2, Bcl11a, and Runx2) in comparison to cDC factors (Id2, Batf3, and Bcl6). Expression of pDC factors and the lack of expression of cDC factors in the CDP points to a “default” pDC pathway during DC differentiation. Re-direction into the cDC lineage requires the induction of Id2 and repression of pDC factors in the pre-cDC.
9. Concluding remarks
The transcriptional networks regulating the diversification of myeloid lineages are currently being elucidated. Arguably, DCs remain the most enigmatic and developmentally uncharacterized cell type within this system. However, recent studies have defined key steps in the DC differentiation pathway by identifying restricted bone marrow precursors. We hope that these advances will encourage the current transition of the field from a relatively descriptive state to one in which molecular mechanisms are emphasized. This shift in approach is needed to clarify confusion regarding the precise role of the DC lineage, and particularly specific DC subsets, in settings of infection and tolerance. For example, our understanding of the functional differences between pDCs and cDC subsets has greatly improved since the identification of transcription factors which are required for their development, such as E2-2 and Batf3, respectively. The deletion of either gene leads to a model in which the exact function of the relevant cell can be interrogated.
Clearly, many questions regarding the transcriptional regulation of DC fate decisions still remain unanswered. The basis for the initial commitment to the DC lineage or its split from the monocyte pathway downstream of the MDP is unknown. Similarly, the mechanisms regulating the transition from the cDC precursor into either the CD4+ or CD8+ subset are also unclear. Another key issue is whether the combinatorial action of cytokines within distinct stromal niches plays any role in influencing these fate choices. Answers to these outstanding questions will allow for the arrangement of the current transcription factor network into a resolvable structure that can be used for manipulation of DCs in clinical settings.
Acknowledgements
We are grateful to the primary authors of the work cited in this review and to members of the Murphy lab for exciting and helpful discussions on the topic.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J Exp. Med. 1973;137:1142–1162. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinman RM, Witmer MD. Lymphoid dendritic cells are potent stimulators of the primary mixed leukocyte reaction in mice. Proc. Natl. Acad. Sci U. S A. 1978;75:5132–5136. doi: 10.1073/pnas.75.10.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohnmacht C, et al. Constitutive ablation of dendritic cells breaks self-tolerance of CD4 T cells and results in spontaneous fatal autoimmunity. J Exp. Med. 2009;206:549–559. doi: 10.1084/jem.20082394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnberg T, et al. Lack of conventional dendritic cells is compatible with normal development and T cell homeostasis, but causes myeloid proliferative syndrome. Immunity. 2008;29:986–997. doi: 10.1016/j.immuni.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Vremec D, et al. The surface phenotype of dendritic cells purified from mouse thymus and spleen: investigation of the CD8 expression by a subpopulation of dendritic cells. J. Exp. Med. 1992;176:47–58. doi: 10.1084/jem.176.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu L, Li CL, Shortman K. Thymic dendritic cell precursors: relationship to the T lymphocyte lineage and phenotype of the dendritic cell progeny. J. Exp. Med. 1996;184:903–911. doi: 10.1084/jem.184.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naik SH. Demystifying the development of dendritic cell subtypes, a little. Immunology and Cell Biology. 2008;86:439–452. doi: 10.1038/icb.2008.28. [DOI] [PubMed] [Google Scholar]
- 8.Vremec D, Pooley J, Hochrein H, Wu L, Shortman K, shortman CD4 and CD8 expression by dendritic cell subtypes in mouse thymus and spleen. J. Immunol. 2000;164:2978–2986. doi: 10.4049/jimmunol.164.6.2978. [DOI] [PubMed] [Google Scholar]
- 9.Edwards AD, Chaussabel D, Tomlinson S, Schulz O, Sher A, Sousa CRE. Relationships among murine CD11c(high) dendritic cell subsets as revealed by baseline gene expression patterns. J. Immunol. 2003;171:47–60. doi: 10.4049/jimmunol.171.1.47. [DOI] [PubMed] [Google Scholar]
- 10.Blasius AL, Giurisato E, Cella M, Schreiber RD, Shaw AS, Colonna M. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol. 2006;177:3260–3265. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, et al. Characterization of Siglec-H as a novel endocytic receptor expressed on murine plasmacytoid dendritic cell precursors. Blood. 2006;107:3600–3608. doi: 10.1182/blood-2005-09-3842. [DOI] [PubMed] [Google Scholar]
- 12.Grouard G, Rissoan MC, Filgueira L, Durand I, Banchereau J, Liu YJ. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J. Exp. Med. 1997;185:1101–1111. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crozat K, et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J Exp. Med. 2010;207:1283–1292. doi: 10.1084/jem.20100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulin LF, et al. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J Exp. Med. 2010;207:1261–1271. doi: 10.1084/jem.20092618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachem A, et al. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J Exp. Med. 2010;207:1273–1281. doi: 10.1084/jem.20100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Randolph GJ, Inaba K, Robbiani DF, Steinman RM, Muller WA. Differentiation of phagocytic monocytes into lymph node dendritic cells in vivo. Immunity. 1999;11:753–761. doi: 10.1016/s1074-7613(00)80149-1. [DOI] [PubMed] [Google Scholar]
- 17.Randolph GJ, Beaulieu S, Lebecque S, Steinman RM, Muller WA. Differentiation of monocytes into dendritic cells in a model of transendothelial trafficking. Science. 1998;282:480–483. doi: 10.1126/science.282.5388.480. [DOI] [PubMed] [Google Scholar]
- 18.Swiecki M, Gilfillan S, Vermi W, Wang Y, Colonna M. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity. 2010;33:955–966. doi: 10.1016/j.immuni.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swiecki M, Colonna M. Unraveling the functions of plasmacytoid dendritic cells during viral infections, autoimmunity, and tolerance. Immunol Rev. 2010;234:142–162. doi: 10.1111/j.0105-2896.2009.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sapoznikov A, Pewzner-Jung Y, Kalchenko V, Krauthgamer R, Shachar I, Jung S. Perivascular clusters of dendritic cells provide critical survival signals to B cells in bone marrow niches. Nat Immunol. 2008;9:388–395. doi: 10.1038/ni1571. [DOI] [PubMed] [Google Scholar]
- 21.McCartney S, et al. Distinct and complementary functions of MDA5 and TLR3 in poly(I:C)-mediated activation of mouse NK cells. J Exp. Med. 2009;206:2967–2976. doi: 10.1084/jem.20091181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyake T, et al. Poly I:C-induced activation of NK cells by CD8 alpha+ dendritic cells via the IPS-1 and TRIF-dependent pathways. J Immunol. 2009;183:2522–2528. doi: 10.4049/jimmunol.0901500. [DOI] [PubMed] [Google Scholar]
- 23.den Haan JM, Lehar SM, Bevan MJ. CD8(+) but not CD8(−) dendritic cells cross-prime cytotoxic T cells in vivo. J Exp. Med. 2000;192:1685–1696. doi: 10.1084/jem.192.12.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bevan MJ. Cross-priming for a secondary cytotoxic response to minor H antigens with H-2 congenic cells which do not cross-react in the cytotoxic assay. J Exp. Med. 1976;143:1283–1288. doi: 10.1084/jem.143.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sigal LJ, Crotty S, Andino R, Rock KL. Cytotoxic T-cell immunity to virus-infected non-haematopoietic cells requires presentation of exogenous antigen. Nature. 1999;398:77–80. doi: 10.1038/18038. [DOI] [PubMed] [Google Scholar]
- 26.Ochsenbein AF, et al. Roles of tumour localization, second signals and cross priming in cytotoxic T-cell induction. Nature. 2001;411:1058–1064. doi: 10.1038/35082583. [DOI] [PubMed] [Google Scholar]
- 27.Idoyaga J, Suda N, Suda K, Park CG, Steinman RM. Antibody to Langerin/CD207 localizes large numbers of CD8alpha+ dendritic cells to the marginal zone of mouse spleen. Proc. Natl. Acad. Sci U. S A. 2009;106:1524–1529. doi: 10.1073/pnas.0812247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu CH, Miyake Y, Kaise H, Kitamura H, Ohara O, Tanaka M. Novel Subset of CD8 alpha(+) Dendritic Cells Localized in the Marginal Zone Is Responsible for Tolerance to Cell-Associated Antigens. J. Immunol. 2009;182:4127–4136. doi: 10.4049/jimmunol.0803364. [DOI] [PubMed] [Google Scholar]
- 29.Reis e Sousa C, et al. In vivo microbial stimulation induces rapid CD40 ligand-independent production of interleukin 12 by dendritic cells and their redistribution to T cell areas [see comments] J. Exp. Med. 1997;186:1819–1829. doi: 10.1084/jem.186.11.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hochrein H, Shortman K, Vremec D, Scott B, Hertzog P, O'Keeffe M. Differential production of IL-12, IFN-alpha, and IFN-gamma by mouse dendritic cell subsets. J Immunol. 2001;166:5448–5455. doi: 10.4049/jimmunol.166.9.5448. [DOI] [PubMed] [Google Scholar]
- 31.Dudziak D, et al. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- 32.Kamphorst AO, Guermonprez P, Dudziak D, Nussenzweig MC. Route of antigen uptake differentially impacts presentation by dendritic cells and activated monocytes. J Immunol. 2010;185:3426–3435. doi: 10.4049/jimmunol.1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheong C, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 2010;143:416–429. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano H, et al. Blood-derived inflammatory dendritic cells in lymph nodes stimulate acute T helper type 1 immune responses. Nat. 2009;10:394–402. doi: 10.1038/ni.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hume DA. Macrophages as APC and the dendritic cell myth. J Immunol. 2008;181:5829–5835. doi: 10.4049/jimmunol.181.9.5829. [DOI] [PubMed] [Google Scholar]
- 36.Yoshimoto T, et al. Basophils contribute to T(H)2-IgE responses in vivo via IL-4 production and presentation of peptide-MHC class II complexes to CD4+ T cells. Nat Immunol. 2009;10:706–712. doi: 10.1038/ni.1737. [DOI] [PubMed] [Google Scholar]
- 37.Sokol CL, Chu NQ, Yu S, Nish SA, Laufer TM, Medzhitov R. Basophils function as antigen-presenting cells for an allergen-induced T helper type 2 response. Nat Immunol. 2009;10:713–720. doi: 10.1038/ni.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammad H, et al. Inflammatory dendritic cells--not basophils--are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J Exp. Med. 2010;207:2097–2111. doi: 10.1084/jem.20101563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manicassamy S, et al. Activation of beta-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science. 2010;329:849–853. doi: 10.1126/science.1188510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geissmann F, Gordon S, Hume DA, Mowat AM, Randolph GJ. Unravelling mononuclear phagocyte heterogeneity. Nat Rev Immunol. 2010;10:453–460. doi: 10.1038/nri2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu K, Waskow C, Liu X, Yao K, Hoh J, Nussenzweig M. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat Immunol. 2007;8:578–583. doi: 10.1038/ni1462. [DOI] [PubMed] [Google Scholar]
- 42.Kamath AT, et al. The development, maturation, and turnover rate of mouse spleen dendritic cell populations. J. Immunol. 2000;165:6762–6770. doi: 10.4049/jimmunol.165.12.6762. [DOI] [PubMed] [Google Scholar]
- 43.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 44.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 45.Manz MG, Traver D, Miyamoto T, Weissman IL, Akashi K. Dendritic cell potentials of early lymphoid and myeloid progenitors. Blood. 2001;97:3333–3341. doi: 10.1182/blood.v97.11.3333. [DOI] [PubMed] [Google Scholar]
- 46.Manz MG, et al. Dendritic cell development from common myeloid progenitors. Ann. N. Y. Acad. Sci. 2001;938:167–173. doi: 10.1111/j.1749-6632.2001.tb03586.x. [DOI] [PubMed] [Google Scholar]
- 47.Traver D, et al. Development of CD8alpha-positive dendritic cells from a common myeloid progenitor. Science. 2000;290:2152–2154. doi: 10.1126/science.290.5499.2152. [DOI] [PubMed] [Google Scholar]
- 48.Inaba K, et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp. Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schlenner SM, et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 50.Fogg DK, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 51.Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3(+) M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 52.Naik SH, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 53.Liu K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cisse B, et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naik SH, et al. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat Immunol. 2006;7:663–671. doi: 10.1038/ni1340. [DOI] [PubMed] [Google Scholar]
- 56.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp. Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kingston D, Schmid MA, Onai N, Obata-Onai A, Baumjohann D, Manz MG. The concerted action of GM-CSF and Flt3-ligand on in vivo dendritic cell homeostasis. Blood. 2009 doi: 10.1182/blood-2009-02-206318. [DOI] [PubMed] [Google Scholar]
- 58.Vremec D, Lieschke GJ, Dunn AR, Robb L, Metcalf D, Shortman K. The influence of granulocyte/macrophage colony-stimulating factor on dendritic cell levels in mouse lymphoid organs. Eur. J Immunol. 1997;27:40–44. doi: 10.1002/eji.1830270107. [DOI] [PubMed] [Google Scholar]
- 59.Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- 60.Xu YK, Zhan YF, Lew AM, Naik SH, Kershaw MH. Differential development of murine dendritic cells by GM-CSF versus flt3 ligand has implications for inflammation and trafficking. J. Immunol. 2007;179:7577–7584. doi: 10.4049/jimmunol.179.11.7577. [DOI] [PubMed] [Google Scholar]
- 61.Maraskovsky E, et al. Dramatic numerical increase of functionally mature dendritic cells in FLT3 ligand-treated mice. Adv. Exp. Med. Biol. 1997;417:33–40. doi: 10.1007/978-1-4757-9966-8_6. [DOI] [PubMed] [Google Scholar]
- 62.Brasel K, De Smedt T, Smith JL, Maliszewski CR. Generation of murine dendritic cells from flt3-ligand-supplemented bone marrow cultures. Blood. 2000;96:3029–3039. [PubMed] [Google Scholar]
- 63.Onai N, Obata-Onai A, Tussiwand R, Lanzavecchia A, Manz MG. Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon-producing and dendritic cell development. J Exp. Med. 2006;203:227–238. doi: 10.1084/jem.20051645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McKenna HJ, et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- 65.Waskow C, et al. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat Immunol. 2008;9:676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laouar Y, Welte T, Fu XY, Flavell RA. STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity. 2003;19:903–912. doi: 10.1016/s1074-7613(03)00332-7. [DOI] [PubMed] [Google Scholar]
- 67.Sathaliyawala T, et al. Mammalian target of rapamycin controls dendritic cell development downstream of Flt3 ligand signaling. Immunity. 2010;33:597–606. doi: 10.1016/j.immuni.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tokoyoda K, Egawa T, Sugiyama T, Choi BI, Nagasawa T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 2004;20:707–718. doi: 10.1016/j.immuni.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 69.Schmid MA, Kingston D, Boddupalli S, Manz MG. Instructive cytokine signals in dendritic cell lineage commitment. Immunol Rev. 2010;234:32–44. doi: 10.1111/j.0105-2896.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- 70.Esashi E, Wang YH, Perng O, Qin XF, Liu YJ, Watowich SS. The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity. 2008;28:509–520. doi: 10.1016/j.immuni.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu L, Nichogiannopoulou A, Shortman K, Georgopoulos K. Cell-autonomous defects in dendritic cell populations of Ikaros mutant mice point to a developmental relationship with the lymphoid lineage. Immunity. 1997;7:483–492. doi: 10.1016/s1074-7613(00)80370-2. [DOI] [PubMed] [Google Scholar]
- 72.Allman D, et al. Ikaros is required for plasmacytoid dendritic cell differentiation. Blood. 2006;108:4025–4034. doi: 10.1182/blood-2006-03-007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoshida T, Ng SY, Georgopoulos K. Awakening lineage potential by Ikaros-mediated transcriptional priming. Curr. Opin. Immunol. 2010;22:154–160. doi: 10.1016/j.coi.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guerriero A, Langmuir PB, Spain LM, Scott EW. PU.1 is required for myeloid-derived but not lymphoid-derived dendritic cells. Blood. 2000;95:879–885. [PubMed] [Google Scholar]
- 76.Anderson KL, Perkin H, Surh CD, Venturini S, Maki RA, Torbett BE. Transcription factor PU.1 is necessary for development of thymic and myeloid progenitor-derived dendritic cells. J Immunol. 2000;164:1855–1861. doi: 10.4049/jimmunol.164.4.1855. [DOI] [PubMed] [Google Scholar]
- 77.Carotta S, et al. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 78.Zhang DE, Hetherington CJ, Chen HM, Tenen DG. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol. 1994;14:373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hohaus S, Petrovick MS, Voso MT, Sun Z, Zhang DE, Tenen DG. PU.1 (Spi-1) and C/EBP alpha regulate expression of the granulocyte-macrophage colony-stimulating factor receptor alpha gene. Mol Cell Biol. 1995;15:5830–5845. doi: 10.1128/mcb.15.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rathinam C, et al. The transcriptional repressor Gfi1 controls STAT3-dependent dendritic cell development and function. Immunity. 2005;22:717–728. doi: 10.1016/j.immuni.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 81.Rodel B, et al. The zinc finger protein Gfi-1 can enhance STAT3 signaling by interacting with the STAT3 inhibitor PIAS3. EMBO J. 2000;19:5845–5855. doi: 10.1093/emboj/19.21.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duan Z, Horwitz M. Targets of the transcriptional repressor oncoprotein Gfi-1. Proc. Natl. Acad. Sci U. S A. 2003;100:5932–5937. doi: 10.1073/pnas.1031694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kobayashi T, et al. TRAF6 is a critical factor for dendritic cell maturation and development. Immunity. 2003;19:353–363. doi: 10.1016/s1074-7613(03)00230-9. [DOI] [PubMed] [Google Scholar]
- 84.Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Edelson BT, et al. Peripheral CD103(+) dendritic cells form a unified subset developmentally related to CD8 alpha(+) conventional dendritic cells. J. Exp. Med. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ginhoux F, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp. Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 88.Wu L, D'Amico A, Winkel KD, Suter M, Lo D, Shortman K. RelB is essential for the development of myeloid-related CD8alpha- dendritic cells but not of lymphoid-related CD8alpha+ dendritic cells. Immunity. 1998;9:839–847. doi: 10.1016/s1074-7613(00)80649-4. [DOI] [PubMed] [Google Scholar]
- 89.Burkly L, et al. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 1995;373:531–536. doi: 10.1038/373531a0. [DOI] [PubMed] [Google Scholar]
- 90.Suzuki S, et al. Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha- dendritic cell development. Proc. Natl. Acad. Sci U. S. A. 2004;101:8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 92.Brass AL, Kehrli E, Eisenbeis CF, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 93.Biswas PS, Bhagat G, Pernis AB. IRF4 and its regulators: evolving insights into the pathogenesis of inflammatory arthritis? Immunol Rev. 2010;233:79–96. doi: 10.1111/j.0105-2896.2009.00864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tamura T, et al. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 95.Schiavoni G, et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J Exp. Med. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iacobelli M, Wachsman W, McGuire KL. Repression of IL-2 promoter activity by the novel basic leucine zipper p21SNFT protein. J Immunol. 2000;165:860–868. doi: 10.4049/jimmunol.165.2.860. [DOI] [PubMed] [Google Scholar]
- 98.Bower KE, Fritz JM, McGuire KL. Transcriptional repression of MMP-1 by p21SNFT and reduced in vitro invasiveness of hepatocarcinoma cells. Oncogene. 2004;23:8805–8814. doi: 10.1038/sj.onc.1208109. [DOI] [PubMed] [Google Scholar]
- 99.Schraml BU, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Betz BC, et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 2010;207:933–942. doi: 10.1084/jem.20091548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp. Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ohtsuka H, et al. Bcl6 is required for the development of mouse CD4+ and CD8alpha+ dendritic cells. J Immunol. 2011;186:255–263. doi: 10.4049/jimmunol.0903714. [DOI] [PubMed] [Google Scholar]
- 103.Johnston RJ, et al. Bcl6 and Blimp-1 Are Reciprocal and Antagonistic Regulators of T Follicular Helper Cell Differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Niu H, Cattoretti G, Dalla-Favera R. BCL6 Controls the Expression of the B7-1/CD80 Costimulatory Receptor in Germinal Center B Cells. J. Exp. Med. 2003;198:211–221. doi: 10.1084/jem.20021395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nurieva RI, et al. Bcl6 Mediates the Development of T Follicular Helper Cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 107.Spits H, Couwenberg F, Bakker AQ, Weijer K, Uittenbogaart CH. Id2 and Id3 inhibit development of CD34(+) stem cells into predendritic cell (pre-DC)2 but not into pre-DC1. Evidence for a lymphoid origin of pre-DC2. J Exp. Med. 2000;192:1775–1784. doi: 10.1084/jem.192.12.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hacker C, et al. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat Immunol. 2003;4:380–386. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- 109.Robbins SH, et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 2008;9:R17. doi: 10.1186/gb-2008-9-1-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ghosh HS, Cisse B, Bunin A, Lewis KL, Reizis B. Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 2010;33:905–916. doi: 10.1016/j.immuni.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bar-On L, et al. CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc. Natl. Acad. Sci U. S A. 2010;107:14745–14750. doi: 10.1073/pnas.1001562107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pui JC, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 113.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li P, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ikawa T, et al. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 116.Felker P, et al. TGF-beta1 accelerates dendritic cell differentiation from common dendritic cell progenitors and directs subset specification toward conventional dendritic cells. J Immunol. 2010;185:5326–5335. doi: 10.4049/jimmunol.0903950. [DOI] [PubMed] [Google Scholar]
- 117.Ouyang W, et al. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]