Abstract
A sensor capable of continuously measuring specific molecules in the bloodstream in vivo would give clinicians a valuable window into patients’ health and their response to therapeutics. Such technology would enable truly personalized medicine, wherein therapeutic agents could be tailored with optimal doses for each patient to maximize efficacy and minimize side effects. Unfortunately, continuous, real-time measurement is currently only possible for a handful of targets, such as glucose, lactose, and oxygen, and the few existing platforms for continuous measurement are not generalizable for the monitoring of other analytes, such as small-molecule therapeutics. In response, we have developed a real-time biosensor capable of continuously tracking a wide range of circulating drugs in living subjects. Our microfluidic electrochemical detector for in vivo continuous monitoring (MEDIC) requires no exogenous reagents, operates at room temperature, and can be reconfigured to measure different target molecules by exchanging probes in a modular manner. To demonstrate the system's versatility, we measured therapeutic in vivo concentrations of doxorubicin (a chemotherapeutic) and kanamycin (an antibiotic) in live rats and in human whole blood for several hours with high sensitivity and specificity at sub-minute temporal resolution. Importantly, we show that MEDIC can also obtain pharmacokineticparameters for individual animals in real-time. Accordingly, just as continuous glucose monitoring technology is currently revolutionizing diabetes care, we believe MEDIC could be a powerful enabler for personalized medicine by ensuring delivery of optimal drug doses for individual patients based on direct detection of physiological parameters.
INTRODUCTION
The paradigm of personalized medicine holds the promise to revolutionize healthcare by delivering “the right drug, at the right dose, and at the right time”(1). Toward the realization of this potential, the past decade has witnessed significant advancements in the development of biosensors capable of sensitive and specific detection of small-molecule, nucleic acid and protein targets (2, 3). A limitation of most these sensors, however, is that they generally only perform single time-point measurements. This prevents their use for continuous, real-time monitoring of molecular analytes, an ability that could be of considerable medical value. Indeed, continuous, real-time molecular monitoring is available for only a handful of analytes that produce readily measureable signals from analyte-specific reactions (e.g., detection of glucose through glucose oxidase activity) (4, 5). A universal architecture that can continuously measure in vivo concentrations of a wide range of circulating biomolecules would enable many potentially transformative applications in medicine; for example, continuous monitoring of cardiac markers (e.g., troponin) could predict an oncoming heart attack (6), measurements of chemokines could provide early warning for infection or autoimmune flare-ups (7) and real-time tracking of chemotherapeutic agents could facilitate administration of a therapeutic dose that is continuously optimized for maximal efficacy and minimal side effects for a specific cancer patient (8).
The design of such a sensor poses extraordinary technological challenges (9–11). First, the sensor must operate continuously, without sample preparation, batch processing or addition of exogenous reagents. Second, it must achieve sufficient sensitivity, selectivity and dynamic range, and demonstrate the ability to resolve changes in analyte concentrations at physiological time-scales. Finally, it must be resistant to fouling even after prolonged exposure to whole blood and other complex samples, and remain stable with high signal-to-noise ratios (SNR) while operating in this environment for extended periods of time. To date, no platform has satisfactorily addressed these challenges, and there remains an urgent need for a generalizable approach to continuous in vivo detection of clinically relevant target molecules in real-time.
In response, we have developed the microfluidic electrochemical detector for in vivo continuous monitoring (MEDIC)—a biosensor platform that can be readily reconfigured to continuously measure a diverse array of biomolecules in real-time. As proof of concept, we have used MEDIC to measure in vivo concentrations of doxorubicin (DOX), a widely-used chemotherapeutic, in human whole blood and in live rats (Fig. 1A). We chose DOX because it exhibits substantial, clinically meaningful changes in pharmacokinetics across populations, and even over the course of treatment for a single individual (12). By simply exchanging the probes in our MEDIC chip, we were also able to achieve real-time in vivo measurement of kanamycin, an antibiotic, demonstrating the inherent modularity of this platform.
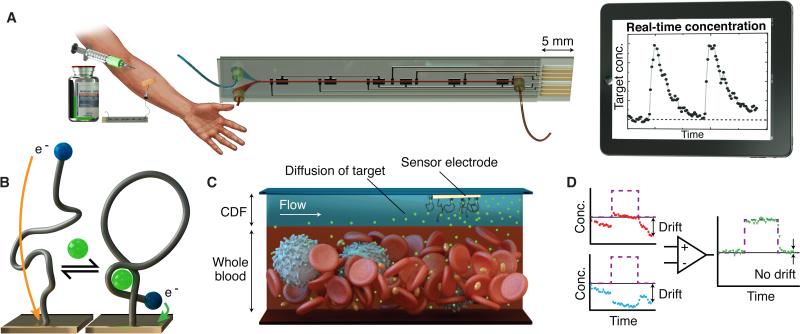
Figure 1. MEDIC overview.
MEDIC achieves real-time quantitative measurement of specific molecules in the blood of living animals. (A) The envisioned set-up, where the MEDIC chip is connected to the patient’s bloodstream to measure drug pharmacokinetics. (B) The aptamer probe is tethered to the gold electrode. Binding of target (green) induces a reversible conformational change in the probe, increasing the rate of electron transfer between an electrochemical redox reporter (blue) and a microfabricated electrode, yielding a measurable current change, shown in (A) as a function of time. (C) The continuous diffusion filter (CDF),formed by vertically stacked laminar flow of buffer (blue) and blood (red), as shown in the microfluidic device in (A), permits access to the target molecules while selectively excluding blood-borne interferents. (D) Signal-on (red) and signal-off (blue) both exhibit significant drift in response to a pulse of target (purple). Kinetic differential measurement (KDM, green) improves accuracy of real-time current measurements by minimizing drift and enhancing SNR.
The MEDIC system overcomes the long-standing limitations of real-time in vivo detection. By supporting measurement of a range of drugs and biomarkers, we expect MEDIC to elucidate individual and variable patient pharmacokinetics and, when translated to the clinic, enable personalized and adaptable dosing for individual patients to drive optimal disease treatment.
RESULTS
MEDIC design and function
MEDIC integrates multiple technological advances to overcome the challenges that have previously thwarted development of biosensors for continuous in vivo molecular detection. The central element of MEDIC is the electrochemical, aptamer-based sensor (13). This is a conformation-changing aptamer probe that is covalently attached via one terminus to an integrated electrode within the MEDIC device and modified at the other terminus with a redox reporter (Fig. 1B). Upon binding to its target molecule, the probe undergoes a conformational rearrangement that modulates the redox current and generates an electrochemical signal. Since the conformational change is reversible, our probe enables continuous, sensitive, label-free detection with rapid kinetics (see below). Importantly, detection is highly specific—only binding of the target triggers this conformation change, while non-specific binding of interferents does not generate an electrochemical signal.
Within the MEDIC chip, the aptamer probes are protected by a continuous-flow diffusion filter (CDF), which prevents blood cells and other high-molecular-weight interferents from physically occluding the sensor surface during operation (Fig. 1C). Finally, these are integrated with an electrochemical kinetic differential measurement (KDM) technique that self-corrects for signal drift and enhances the SNR (Fig. 1D), enabling stable and quantitative detection over four hours of continuous operation.
Aptamer probes are sensitive and specific in space and time
For our initial tests, we designed a DOX-specific aptamer probe using a previously described DNA aptamer (14). We conjugated its 3’ end to a methylene blue (MB) reporter and its 5’ end to an alkane thiol for attachment to gold working electrodes within the MEDIC chip microchannel (Fig. S1). Target binding induces a conformational change in the aptamer that modulates electron transfer between MB and the electrode (Fig. 1B) (13). This modulation is expected to produce a readily measurable change in current at the MB reduction peak when the sensor is interrogated using square-wave voltammetry (SWV). The relative change in current (the signal gain) thus provides a direct measurement of DOX concentration.
To characterize the DOX probe's sensitivity, we fitted the probe's signal gain to a Langmuir isotherm, and obtained an apparent equilibrium dissociation constant (Kd) of 824 ± 18 nM (mean ± SEM) (Fig. 2A). The sensor achieved a limit of detection (LOD) in buffer of 10 nM, with a dynamic range of 0.1–10 μM, spanning the drug's therapeutic range (15). Importantly, the sensor did not respond to 1,000-fold greater concentrations of ifosfamide (Ifex), mesna, mitomycin-C (MTC), dacarbazine (DTIC), or cisplatin (CDDP) (Fig. 2B)—agents commonly administered with DOX. The sensor also exhibited rapid kinetic response; after a 5 min pulse of 600 nM DOX, we measured kon = 3.0 ± 0.35 μM-1·min-1 and koff = 1.35 ± 0.05 min1 (means ± SEM). The probe therefore reached 90% saturation within 45 s and then returned to within 10% of baseline by 100 s (Fig. 2C). Because the alpha-phase plasma clearance time of DOX in humans is between 6 and 26 min (16), this temporal resolution is sufficient for clinical applications.
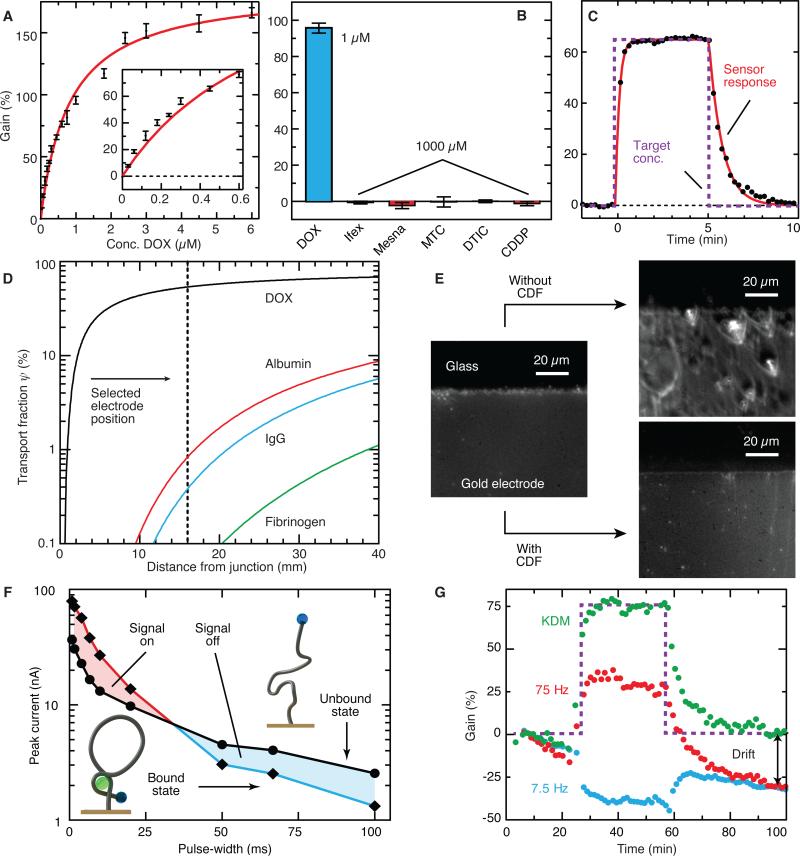
Figure 2. Testing the core components of MEDIC.
(A to C) DOX aptamer sensor characterization. (A) Dose-response of the DOX aptamer probe to characterize limit of detection and dynamic range (mean ± SD, n = 20). (B) Demonstration of probe specificity based on lack of response to other chemotherapeutic agents (mean ± SD, n = 15). (C) Temporal response to DOX.(D) CDF design and performance. Numerical simulations of transport fraction for DOX and control proteins crossing the CDF. (E) Optical micrographs showed fouling in whole blood, as evidenced by the illuminated biomass collected on the electrode surface.. (F) Mechanism and effectiveness of KDM. SVW pulse width is varied and peak current is recorded in the unbound and DOX-bound states (data are means, n = 6). In the unbound state (black line), MB is slowly reduced, yielding a gradual decay in current. Conversely, MB is rapidly reduced in the target-bound state (colored line). We can thus invert the response from “signal-on” to “signal-off” by changing the interrogation frequency. (G) The response to a DOX pulse (purple) at 75 and 7.5 Hz without KDM (red and blue lines, respectively). Applying KDM (green) minimized drift and increased signal amplitude.
The continuous diffusion filter prevents sensor fouling in human whole blood
Although aptamer-based electrochemical sensors perform well when challenged in undiluted blood serum (17), real-time measurement in whole blood has been thwarted to date by fouling from blood cells and high-molecular-weight clotting factors. We circumvented this problem with the CDF—a flowing, liquid-phase filter that exploits differences in diffusive transport (18). We established the CDF within the MEDIC chip by forming a vertically stacked laminar stream of isotonic buffer between the blood sample and electrodes (Fig. 1C).
Because of its high diffusivity (1 × 10−9 m2/s) (19), DOX rapidly crossed the CDF, while larger, lower-diffusivity interferents, such as albumin, immunoglobulin, and fibrinogen, cannot reach the sensor surface during their transit time through the MEDIC chip. To optimize CDF performance, we modeled transport of DOX and several potential interferents, including human serum albumin (HSA), immunoglobulin G (IgG), fibrinogen, and red blood cells. To do so, we created a finite-element model of the MEDIC channel to solve for velocity and concentration distributions. A CDF 125-μm-high and with a total flow-rate of 1 ml/h yielded a Reynolds number of ~0.1, enabling laminar flow stacking throughout the device. This is equivalent to a flow-rate of 0.75 ml/h for blood; under these conditions, sampling over the course of a full day would draw on only ~0.3% of the average human's blood volume. We verified the flow pattern via confocal microscopy (Fig. S2A), observing consistent results across multiple flow-rate ratios (Fig. S2B). To achieve optimal filtering performance, we adjusted CDF height and analyte residence time by controlling the total flow and ratio of the two flow-rates. We calculated average concentrations of DOX and interferents at the sensor surface (Csurf) as a function of position along the channel and normalized these to their input concentration (C0) to yield the transport fraction, ψ = Csurf /C0 (Fig. S3). The optimal CDF yielded a transport fraction that was high for DOX (ψ = 54%) and lower for interferents, including albumin, IgG, and fibrinogen (ψ < 1%) (Fig. 2D). Sensor placement was optimal at 16 mm from the buffer-blood junction. Imaging before and after 20 min in flowing whole human blood confirmed that the CDF effectively prevented fouling of the sensor surface (Fig. 2E).
Kinetic differential measurement eliminates drift during continuous blood monitoring
Accurate, long-term measurement requires maintaining sufficient SNR with minimal drift. Differential measurement strategies can counteract drift, but these typically require a physically separate reference probe to obtain a target-independent, common background signal that can be subtracted from the sensor signal. The reproducible fabrication of such a perfectly matched background-correction sensor is often challenging, rendering this approach problematic. To surmount this, we have developed KDM, which employs a single probe to obtain both measurements. KDM exploits the difference in charge transfer kinetics between the target-bound and unbound states of the probe. In the absence of target, the unbound probe is unfolded and expanded; this results in a slow electron transfer rate, because the electrochemical reporter approaches the electrode surface only rarely (Fig. 2F) (20). The target-bound probe, in contrast, supports rapid electron transfer because it constrains the reporter near the electrode surface. We interrogated the probe at multiple SWV frequencies by varying the pulse-width () (Fig. S4) to observe the rate of MB reduction. At short pulse-widths, we observed higher current in the bound compared to the unbound state (Fig. 2F), as the faster kinetics allow more rapid reduction of MB. However this rapid reduction results in depletion of MB, such that with longer pulse-widths, we observed lower current in the bound state (Fig. 2F). This difference in electron transfer rate allowed us to obtain a “signal-on” response when we interrogated probes using high-frequency SWV, and a “signal-off” response when we interrogated at lower frequencies (Fig. S5A). Critically, these two output signals exhibited matching responses to the background (Fig. S5B), thus we can differentially combine them to reduce drift. In response to a 2-μM pulse of DOX in whole blood over 90 min, KDM reduced the drift from ~30% to < 2% (Fig. 2G). Moreover, because the two output signals differed in polarity, taking their difference yielded >300% improvement in SNR (Fig. S5C).
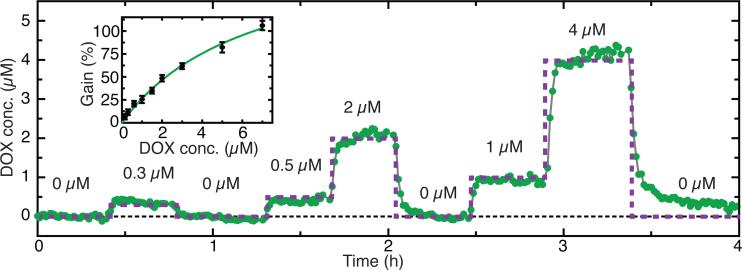
By combining the aptamer probes with these CDF and KDM techniques, MEDIC can achieve continuous, sensitive measurement of absolute analyte concentrations in a flowing stream of whole human blood for extended periods of operation (Fig. 3). To demonstrate this, we constructed a standard curve to relate concentration and differential gain (Fig. 3, inset). After pulsing DOX at 0.3, 0.5, 1, 2, and 4 μM in whole human blood, MEDIC tracked concentrations with a stable baseline for 4 hours. During this time, the measured concentration differed from the known concentration on average by 0.06 μM, verifying its ability to quantify unknown DOX concentrations continuously and in real-time.
Figure 3. Real-time measurement of DOX in vitro in human whole blood.
Continuous, real-time measurement of DOX in human whole blood (green) relative to actual concentrations (purple) over the course of 4 hours. Blood flow rate = 0.75 ml/h, Buffer flow rate = 0.25 ml/h. Inset shows a standard curve relating KDM signal to DOX concentration. Data are means ± SD (n = 20).
Real-time pharmacokinetic monitoring of multiple analytes in live animals
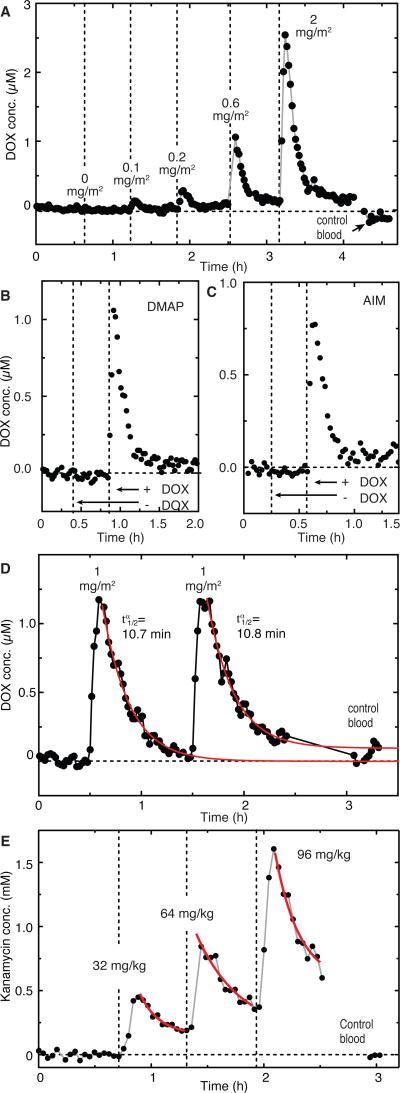
Having shown that MEDIC can reliably operate in human whole blood in vitro, we next performed real-time DOX measurements using live rats. Briefly, we injected DOX intravenously into anesthetized Sprague-Dawley rats while continuously drawing blood into the chip using an indwelling catheter. We dosed a rat with increasing DOX concentrations (0–2 mg/m2) over 4.5 hours (Fig. 4A). Vehicle-only injection (negative control) yielded no observable signal change. In contrast, the lowest DOX dose (0.1 mg/m2) resulted in a peak in vivo concentration of 0.13 μM, while doses of 0.2, 0.6, and 2 mg/m2 resulted in peak concentrations of 0.3, 1.0, and 2.5 μM, respectively—a therapeutically relevant range for human dosing (15). A final measurement made on blood collected prior to dosing confirmed minimal drift during the experiment (0.1 μM) (Fig. 4A). Importantly, we observed a signal in rats injected with the chemotherapeutic drug cocktails DMAP (dacarbazine, mitomycin-C, DOX, and cisplatin) and AIM (DOX, ifosfamide, and mesna) when these cocktails contained DOX, but not when DOX was excluded (Fig. 4B, C).
Figure 4. Real-time measurement of drugs in the blood of live rats.
(A) MEDIC measurement of changing in vivo DOX concentrations over several hours. (B and C) Sensor specificity demonstrated in vivo in rats. We subjected rats to chemotherapy cocktails DMAP (B)and AIM (C) in the absence or presence of DOX. (D) Peak concentrations and after two intravenous injections of DOX into a rat. (E) MEDIC analysis with the aptamer probe substituted to enable continuous real-time measurement of kanamycin. Control blood contains no drug. Measurements were performed at the end of the experiment by switching the blood draw from the animal to a tube containing a sample of the animal's blood taken at the beginning of the experiment, prior to injection of the drug. The gap between these measurements represents the time required to transition the intake tube from the animal to this control sample. Each graph corresponds to a single animal, and each data point corresponds to a single KDM signal.
This capability to continuously measure in vivo DOX concentrations enabled us to obtain key pharmacokinetic parameters for individual animals in real-time. For example, after administering two DOX injections (1 mg/m2) an hour apart to the same rat, we reproducibly measured equivalent peak concentrations (1.17 and 1.16 μM) and alpha-phase half-life values ( = 10.7 ± 0.5 and 10.8 ± 1 min; mean ± SEM) (Fig. 4D). These values are in agreement with previously published values for intravenously injected DOX in rats (21). Highlighting the potential importance of such “personalized” pharmacokinetics, we observed substantial variability in when we performed these measurements in rats (Fig. S6). Indeed, this variability held even when we normalized the dose for each animal by body surface area, the current gold standard for DOX dosing (Pearson correlation coefficient = -0.73).
MEDIC can be readily reconfigured to measure different target molecules by exchanging its aptamer probe in a modular manner. As an exemplar, we exchanged our DOX probe for one that supports continuous detection of the antibiotic kanamycin (22). After obtaining a calibration curve as described above (Fig. S7), we serially injected rats with kanamycin doses of 32, 64, and 96 mg/kg, followed by a baseline control. We observed peak concentrations of 0.45, 0.85, and 1.6 mM, respectively (Fig. 4E), and measured of 7.2, 10, and 9 minutes, in agreement with previously reported values for kanamycin (23). Unlike DOX, however, kanamycin concentrations did not return to baseline because much of the clearance of this drug occurs in the beta phase ( ≈ 2-3 h) (24), which is considerably longer than our dosing interval (40 min).
DISCUSSION
We present a real-time biosensor in a miniaturized, disposable chip format that can continuously measure the concentration of a wide range of circulating molecules in living animals with excellent precision and sub-minute temporal resolution. We achieved direct measurement of therapeutic concentrations of DOX in human whole blood, even when co-administered with a 1000-fold excess of other chemotherapeutic agents. This illustrates that the MEDIC device is highly sensitive and specific. Experiments in rodents demonstrated that MEDIC is effective at discriminating individual differences in drug pharmacokinetics. Variability in pharmacokinetic parameters has been reported for DOX in humans (25), which can result in dramatically different patient outcomes (26). This offers strong evidence for this platform's potential clinical utility in designing patient-appropriate dosing regimens. Finally, the modular architecture of MEDIC means that it can be adapted to a wide range of target molecules simply by exchanging the aptamer probes, as demonstrated by our follow-up real-time measurements of kanamycin in live rats.
MEDIC's real-time measurement capabilities could establish a new paradigm in therapeutic drug monitoring, with immediate applications in acute care. As reported in the literature (27), drug dosing strategies based on pharmacokinetic models using physiological parameters such as sex, mass or body surface area often fail to capture clinically important inter-and intra-patient variability. This may in part account for the inconsistencies often observed in response to therapy, wherein patients receiving similar doses of doxorubicin, for example, exhibit markedly different toxic and therapeutic responses (12, 28). Accordingly, individualized drug dosing based on therapeutic drug monitoring has led to considerable improvements in patient outcomes, especially for drugs with narrow therapeutic ranges (29). Unfortunately, current monitoring strategies use methods such as chromatography, immunoassays or mass spectrometry to measure drug levels, requiring significant sample handling and processing that can delay availability of results for days (30). This lag-time is a long-standing problem in acute clinical settings such as emergency medicine, surgery and treatment of infectious disease, where immediate response to rapidly-changing conditions is critical (31). By tracking patient pharmacokinetics in real-time, MEDIC could give clinicians the capacity to provide individualized and optimal dose control at the point of care.
Although we have demonstrated a potential solution to a long-standing problem, there are several limitations in this study. First, MEDIC was configured to measure the pharmacologically active free form of the drug in whole blood. This is valuable for monitoring a range of drugs including chemotherapeutics, anti-infective agents, immunosuppressants, and anti-convulsants (32). However, for many drugs, it is important to measure their metabolites or drug-protein complexes. In order to detect these molecules, it will be important to develop appropriate aptamer probes that can specifically distinguish and measure the free drug, metabolites, and complexes. Fortunately, aptamers have been discovered for a broad spectrum of biological molecules (33), and by using advanced selection techniques, aptamers with exquisite specificity to closely-related analogs can be generated (34). Second, we have presently demonstrated continuous MEDIC operation with stable performance over multiple hours. Although we believe this range of continuous operation is sufficient for many acute clinical applications, the capacity for much longer-term operation (e.g., days and weeks) could prove highly valuable in chronic care applications, and improved device architecture to support substantially longer operation is a subject of ongoing work.
Although we tested the performance of the device with whole human blood, we have not yet tested MEDIC directly connected to human patients. Such transition will require extensive testing to satisfy regulatory requirements. Specifically, we will perform comparative studies to determine the absolute accuracy of our MEDIC system with respect to standard reference values over time, and ascertain the frequency with which the disposable sensor must be replaced. One advantage of our device from a regulatory perspective is that since it achieves analysis with minimal blood volumes, we do not anticipate the need to recirculate blood back to patients after testing. We initially envision the use of MEDIC as a replacement for standard periodic measurement in hospitals and other clinical settings, and it will therefore be essential for us to demonstrate improved patient outcomes and to make the system easy to use for nurses and other health workers. In addition, we will strive to achieve a cost advantage relative to existing measurement strategies, which will be favorable from a reimbursement perspective. More generally, the entry of continuous glucose monitoring (CGM) technology into the clinical market over the past decade offers an invaluable precedent, and will be used as a model for us to translate our technology as well.
We also foresee numerous opportunities for extending the capabilities of MEDIC; for example, simultaneously measuring not only the drug being administered but also the biomarkers indicative of the patient's response to that drug by integrating multiple probes into one device (35). This would facilitate a closed feedback loop between the drug and the patient's response, which can then be adjusted to continuously deliver an optimal therapeutic dose in real-time. For example, optimal dosing of the immunosuppressant cyclosporine for patients undergoing organ transplantation is notoriously difficult because of inter- and intra-patient variability (36). Appropriate dosing is crucial because the effects of the drug are proportional to its concentration (37), but cyclosporine dose has also been correlated with levels of pro-inflammatory chemokines such as CXCL9 (38, 39), which are associated with nephropathy, acute graft rejection, and graft failure (40). Thus, the capacity to continuously measure both CXCL9 and cyclosporine using MEDIC could enable continuous, closed-loop delivery of optimal cyclosporine doses to enhance therapeutic outcome for individual patients in real-time (41, 42). Importantly, such adaptive dosing technology could also enable the expanded use of drugs with narrow therapeutic indices. In this way, just as real-time continuous glucose sensors have revolutionized diabetes care and contributed to the ongoing development of an “artificial pancreas” via closed-loop control of insulin (43), we believe MEDIC could offer a general framework for achieving closed-loop control of numerous different in vivo biological processes, thereby yielding more effective treatments for a broad range of medical conditions.
MATERIALS AND METHODS
Study design
Our study consisted of three major components: in vitro characterization with buffers, human whole blood testing and live animal testing. The objective of our study was to demonstrate proof of concept capability to monitor drugs directly from the blood of living subjects in real-time. Characterization in buffer was required to demonstrate function of the aptamer probe, including its sensitivity, specificity and temporal response. Human whole blood samples were utilized to assess the capacity of the CDF and KDM to support real-time measurement of target for a period of at least four hours, and compared to measurement without these components. Our Animal Study protocol, In Vivo Small Molecule Detection, was approved by the UCSB IACUC and assigned protocol number 824. Measurements were taken once animal surgeries were stabilized, and concluded with euthanasia of each animal. Blood samples and individual animals were selected randomly for each experiment. Blinding was not applicable to this study, and none of the experiments were blinded. Replication conditions for each experiment are defined and described in the Results section.
Device fabrication
Two borofloat glass wafers (top and bottom; Mark Optics Inc), each 10 cm in diameter and 500 μm thick, were cleaned by sequential immersion in acetone, isopropanol and deionized water (DI). Gold working, counter and reference electrodes were then patterned onto the top wafer at 8-mm intervals photolithographically and electron-beam evaporated (VES 2550, Temescal) to a thickness of 200 nm on 20-nm titanium adhesion layers. Subsequently, a CNC mill (Flashcut CNC) with 1.1-mm diamond bit (Triple Ripple, Abrasive Technology) was used to drill fluidic vias (two inlets, one outlet) in the top wafer. The top and bottom wafers were then diced (DISCO) into device pieces with dimensions of 58 mm × 11 mm and 53 mm × 11 mm, respectively.
The 500-μm-wide top (buffer) and bottom (blood) flow channels were both cut from a 250-μm-thick PDMS sheet (BISCO Silicones, Rogers Corporation) using a cutting plotter (CE5000-60, Graphtec). The buffer channel was first bonded to the diced electrode glass substrate via 10-s corona-ozone treatment (BD-20AC, Electro-Technic Products, Inc) The blood channel was bonded to the buffer channel, and then to the bottom glass substrate, each with the same ozone treatment. Alignment was performed under a standard inverted microscope via an x – y – z – θ stage (Newport Corporation) and vacuum chuck. Fluidic port connectors (Labsmith) were glued onto the device with 5-minute epoxy (ITW Devcon), readying the chip for probe immobilization.
Fluidic instrumentation
All flow to the device was controlled via syringe pump (PhD 2000, Harvard Apparatus). The MEDIC input port was connected to an intravenous cannula for animal studies, or multiport valve (Upchurch Scientific) for in vitro studies. The valve was used to select samples bearing different target concentrations. A 3-ml syringe loaded with 1x SSC was placed in a pump and connected to the buffer port on the MEDIC device via a 30-cm length of Tygon (Saint-Gobain Performance Plastics) tubing, with 2.29-mm outer diameter (OD) and 0.76-mm inner diameter (ID). The MEDIC device output port was connected to a primed but otherwise empty 10 ml ‘waste’ syringe placed in a second pump, via 1/16-inch OD and 0.5-mm ID Teflon tubing (Thomson Instrument Company). To monitor flow-rates in real-time, a flow meter (Mitos Flow Rate Sensor, Dolomite Microfluidics) was employed in-line between the output port and syringe pump. The buffer layer was established by engaging the buffer pump at 0.25 ml/h. Simultaneously, sample was continuously drawn into the device by engaging the waste pump at 1 ml/h.
Aptamer probe preparation and immobilization
The aptamer probes were synthesized by Biosearch Technologies with the following sequences: DOX-28: 5’-HS-(CH2)6-ACC ATC TGT GTA AGG GGT AAG GGG TGG T-MB-3’; kanamycin: 5’-HS-(CH2)6-GGG ACT TGG TTT AGG TAA TGA GTC CC-MB-3’. DOX-28 is a truncated form of the Wochner 41-nucleotide anthracycline aptamer (14). We selected a region of the aptamer suspected to support a hairpin structure, with an apparent active binding site in the base. We engineered the stem to enable effective signal transduction through stabilization of the folded state. The kanamycin probe was adapted from (22). Each probe was thiolated at the 5’-end to facilitate self-assembly on the gold working electrodes, and conjugated with an MB redox label at the 3’-end to enable target binding-induced charge transfer modulation. Gold electrode cleaning and probe immobilization methods were adapted from (17).
To measure aptamer affinity, a dose-response curve was obtained by subjecting the probe to DOX (Sigma-Aldrich) concentrations ranging from 50 nM to 10 μM in 1× SSC at a flow rate of 1 mL/h. At each concentration, the signal was permitted to equilibrate and then ten subsequent readings were obtained from two separate electrodes and averaged as the reported values. The same procedure was applied for specificity characterization, where DOX was injected at 1 μM while off-target reagents MTC, Ifex, Mesna, DTIC or CDDP (Sigma-Aldrich) were injected at 1 mM concentrations. Kinetic measurements were performed in a reaction-limited regime by flowing target-free buffer at 10 mL/h for 5 minutes to obtain a stable baseline, followed by a 5-minute 1 μM DOX pulse, and then back to buffer. Voltammograms were taken in immediate succession to provide maximal temporal resolution. Dose-response curves were obtained by fitting data to a one-site binding model, and kinetic-response curves were obtained by fitting data to a one-phase association and dissociation model (GraphPad Prism version 5.00C for Mac OS X, GraphPad Software).
LOD was calculated as the concentration that gives a signal three baseline standard deviations (sbaseline) above zero:
To determine the dynamic range, the maximum detectable concentration (MDC) was calculated as the concentration where the average is three standard deviations less than maximum signal. Here is the average standard deviation of the four largest concentrations used:
Voltammetry
Electrochemical measurements were conducted with an electrochemical analyzer (730B, CH Instruments). The chip was connected via an 8-pin card-edge connector (Molex) and subjected to SWV scans. Working electrodes were scanned in continuous succession with an average scan period of 8 seconds. To compare the potential difference between the patterned reference electrode and a standard Ag/AgCl electrode, the channel was submerged into a flask which contained buffer and an Ag/AgCl rod electrode with porous Teflon tip (CH Instruments) was utilized. MB redox peak was typically observed at approximately -260 mV (with respect to Ag/AgCl reference), or approximately -110 mV (with respect to the pattered reference electrode). A potential range of -50 mV to -450 mV (with respect to Ag/AgCl reference) was scanned to reliably capture the entire MB reduction peak. In order to determine the optimal frequency for KDM, frequency-sweep scans were performed from 5–800 Hz, with amplitude of 25 mV. A custom peak-fitting script was created to fit the SWV measurements with a Gaussian curve on a hyperbolic baseline. Peak currents were then normalized to a baseline peak current to generate signal gain. Unless otherwise noted, all reported gains were obtained via KDM with the difference divided by the average of gains from 7.5 Hz and 75 Hz signals. To demonstrate sensing with a handheld electrochemical analyzer, we used the PalmSens EmStat2 (Palm Instruments BV). We observed a comparable effective signal; to account for instrument differences, KDM was performed with constituent frequencies of 150 and 5 Hz, and a separate dose-response curve was obtained.
CDF characterization
COMSOL Multiphysics V4.2a was used to simulate target and non-target diffusion in the microchannel in three dimensions via laminar flow and transport of diluted species modules. To simplify the simulation geometry, we focused on the straight portion of the channel from the junction of the two vertically stacked flows to the outlet region. Based on an expected operational sample-to-buffer throughput ratio of 3:1, the domain depths were assigned to 400 μm and 100 μm, respectively. Input flow-rates were defined at 0.75 and 0.25 ml/h for buffer and sample, respectively, with open pressure defined at the output. Buffer was represented as water, while the blood sample was approximated as water with density increased to 1060 kg/m3 and viscosity increased to 4 mPa·s. A diffusion constant of 1 ×10−9 m2/s was assumed for DOX (19). The diffusivities of HSA, IgG, and fibrinogen—blood protein interferents associated with biofouling—were estimated as 5 ×10−11, 4 ×10−11, and 2 ×10−11 m2/s, respectively (44–46). Analytes were supplied in the sample stream at a concentration of 1 μM at the sample input and convected according to the prior-solved velocity field, and reported concentrations were normalized by this value for the respective species. Solving for the velocity field and then the convection diffusion equation yields the concentration field throughout the channel. An x-y cross-section was taken at the top of the channel in the plane of the electrodes (Fig. S3A), scaled at 20% in x with respect to y. This cross-section confirmed an expected transport fraction in excess of ~50% of input at a point 20 mm from the junction, with parabolic isolines. Examination of y-z sections along the channel (Fig. S3B) showed diffusion of DOX from the target-rich flow stream to the initially pure buffer stream.
All confocal imaging data were obtained via Fluoview 1000 MPE (Olympus) with a 10× objective. To obtain unobstructed views of the channel, we fabricated devices exclusively for imaging that were identical to normal devices save the absence of electrodes. These were mounted upside-down on a microscope slide clipped to the microscope stage in order to prevent the connectors from occluding the view of the channel. With the device mounted upside-down, the buffer and blood inputs were switched to enable normal operation with blood on the bottom and buffer on the top. The motorized stage enabled digital encoding of location data. The x−y origin was set at the center of the channel junction for a given device. All location data were thus derived from the stage readout with respect to junction center. All image depth data were likewise derived from the readout of the motorized nosepiece actuating in the z-axis. To image the buffer and blood streams, we respectively added fluorescein and rhodamine WT fluorescent dyes (Bright Dyes) to each at a 1:10,000 dilution from stock. The rhodamine and fluorescein dyes were excited with 559-nm and 473-nm lasers, respectively. To negate channel cross-talk, all images were collected via sequential illumination. To demonstrate flow stacking in the absence of diffusion, we increased the flow-rates of the buffer and blood streams 10-fold to 2.5 and 7.5 ml/h, respectively, since small-molecule dye diffusion was readily observed under normal operating conditions.
KDM characterization
The rate of charge transfer from our redox-labeled probe is a function of the applied voltage, probe flexibility, and redox reporter location. At a fixed voltage,
where F is Faraday's constant, [MB] is the concentration of methylene blue, [leucoMB] is the concentration of reduced leucomethylene blue, and k1 and k-1 are the forward and reverse reaction rates, respectively. In the unbound probe conformation, the kinetics were slow and MB was only gradually reduced to leucoMB. However, in the bound confirmation, the kinetics were fast and MB was reduced rapidly.
The KDM signal (d) was obtained by taking the difference of the signal-on (Gon) and signal-off (Goff) and dividing by the average as:
Optimal KDM was obtained by selecting Gon and Goff at frequencies with well-matched drift. This was determined empirically by examining drift over a range of frequencies.
We defined SNR as:
where is the average signal in the presence of target and starget is the standard deviation.
Human whole blood
We obtained male human whole blood with sodium citrate anticoagulant (Bioreclamation, LLC) within approximately one week of the date it was drawn from the donor. Whole blood was refrigerated at 4 °C until use. Prior to use, blood was warmed to room temperature and then passed through a 40-μm pore cell-strainer (ThermoFisher Scientific) to remove or break up any large aggregates while letting blood constituents pass through. Concentrated target drug was then doped to the desired dose. Whole blood was otherwise unmodified.
Live animal studies
All rats used in this work were Sprague-Dawley males (n = 9) from Charles River Laboratories. To safely mount the device and catheters, rats were sedated throughout the duration of the experiment via isoflurane gas (1–4%) and ketamine/xylazine (56.25 mg/kg and 7.5 mg/kg respectively). Animals were placed dorsal side down on a heating pad. The right jugular vein was isolated and catheterized (11 cm long, 0.64 mm OD, 0.3 mm ID; Dow Corning) to provide an intravenous injection port. We performed an initial injection of 0.4 ml heparin solution (1000 U/ml USP), followed by periodic injections of 0.1 ml every 40 minutes to prevent clot formation in the catheters. The left jugular vein was isolated and cannulated (Insyte Autoguard Shielded IV Catheter, BD). A 0.3-ml pre-drug injection control blood sample was taken and set aside for the duration of the experiment. The end of a 10-cm length of Tygon tubing (2.29-mm OD and 0.76-mm ID) was inserted into the cannula to connect the intravenous blood source to the blood port of the MEDIC device. The remaining fluidic instrumentation was configured as described in the prior section. Target concentrations were adjusted for a constant 0.3 ml injection volume, and injected via the input catheter in a 30-second bolus at >30 min intervals. Upon completion of each experiment, subjects were euthanized via intravenous Euthasol (Virbac Animal Health) injection. The MEDIC input tube was then redirected to the control blood to obtain post-experiment readings of zero-target blood samples. Observed concentrations were derived from a dose-response calibration curve obtained a priori (Fig. S7). Dosing was normalized to the animal's body surface area.
Supplementary Material
Acknowledgements
We thank A. Yang and J. Sommerson for insightful discussions.
Funding: Supported by NIH (R01A1076899), Army Research Office (W911NF-09-D-001 and W911NF-10-2-0114) and the Garland Initiative. We are grateful for the support of the UCSB NNIN Nanofabrication Facility and UCSB MCDB Microscopy Facility, supported through the NCRR Shared Instrument Grant #1S10RR022585-01A1.
Footnotes
Author contributions: B.S.F. and H.T.S. conceived the experiments. B.S.F. and D.A.H. designed the device, probes, KDM, and performed in vitro tests with buffer, cross-reactivity studies, human whole blood studies, and animal studies. B.S.F performed confocal measurement studies. D.M., K.P., and N.W.performed the animal surgeries. B.S.F, D.A.H., K.H., K.P., D.M., T.K., R.J.W., and H.T.S.discussed the results and performed analysis. A.J.B. created the custom peak fitting software.T.K. created the animal study protocol. B.S.F., D.A.H., M.E., K.W.P., and H.T.S. co-wrote and edited the manuscript.
Competing interests: Patent application filed pertaining to results: U.S. Provisional Patent Application No. 61/784,130 “In-Vivo Monitoring of Molecular Targets” (B.S.F., D.A.H., and H.T.S. are listed as inventors of this application).
REFERENCES AND NOTES
- 1.Hamburg MA, Collins FS. The path to personalized medicine. The New England journal of medicine. 2010;363:301–4. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 2.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nature biotechnology. 2010;28:595–9. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patolsky F, Zheng G, Lieber CM. Fabrication of silicon nanowire devices for ultrasensitive, label-free, real-time detection of biological and chemical species. Nature protocols. 2006;1:1711–24. doi: 10.1038/nprot.2006.227. [DOI] [PubMed] [Google Scholar]
- 4.Baker DA, Gough DA. A Continuous, Implantable Lactate Sensor. Analytical Chemistry. 1995;67:1536–1540. [Google Scholar]
- 5.Wilson GS, Hu Y. Enzyme-based biosensors for in vivo measurements. Chemical reviews. 2000;100:2693–704. doi: 10.1021/cr990003y. [DOI] [PubMed] [Google Scholar]
- 6.Keller T, Zeller T, Peetz D, Tzikas S, Roth A, Czyz E, Bickel C, Baldus S, Warnholtz A, Fröhlich M, Sinning CR, Eleftheriadis MS, Wild PS, Schnabel RB, Lubos E, Jachmann N, Genth-Zotz S, Post F, Nicaud V, Tiret L, Lackner KJ, Münzel TF, Blankenberg S. Sensitive troponin I assay in early diagnosis of acute myocardial infarction. The New England journal of medicine. 2009;361:868–77. doi: 10.1056/NEJMoa0903515. [DOI] [PubMed] [Google Scholar]
- 7.Bauer JW, Petri M, Batliwalla FM, Koeuth T, Wilson J, Slattery C, Panoskaltsis-Mortari A, Gregersen PK, Behrens TW, Baechler EC. Interferon-regulated chemokines as biomarkers of systemic lupus erythematosus disease activity: a validation study. Arthritis and rheumatism. 2009;60:3098–107. doi: 10.1002/art.24803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, Dummer R, McMahon M, Stuart DD. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013:1–6. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Microfluidic diagnostic technologies for global public health. Nature. 2006;442:412–8. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 10.Gaster RS, Hall DA, Nielsen CH, Osterfeld SJ, Yu H, Mach KE, Wilson RJ, Murmann B, Liao JC, Gambhir SS, Wang SX. Matrix-insensitive protein assays push the limits of biosensors in medicine. Nature medicine. 2009;15:1327–32. doi: 10.1038/nm.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plaxco KW, Soh HT. Switch-based biosensors: a new approach towards real-time, in vivo molecular detection. Trends in biotechnology. 2010;29:1–5. doi: 10.1016/j.tibtech.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desoize B, Robert J. Individual dose adaptation of anticancer drugs. European journal of cancer. 1994;30A:844–51. doi: 10.1016/0959-8049(94)90304-2. [DOI] [PubMed] [Google Scholar]
- 13.Xiao Y, Lubin A. a., Heeger AJ, Plaxco KW. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angewandte Chemie. 2005;44:5592–5595. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 14.Wochner A, Menger M, Orgel D, Cech B, Rimmele M, a Erdmann V, Glökler J. A DNA aptamer with high affinity and specificity for therapeutic anthracyclines. Analytical biochemistry. 2008;373:34–42. doi: 10.1016/j.ab.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 15.Greene RF, Collins JM, Jenkins JF, Speyer JL, Myers CE. Plasma pharmacokinetics of adriamycin and adriamycinol: implications for the design of in vitro experiments and treatment protocols. Cancer research. 1983;43:3417–21. [PubMed] [Google Scholar]
- 16.Camaggi C, Comparsi R, Strocchi E. Epirubicin and doxorubicin comparative metabolism and pharmacokinetics. Cancer chemotherapy. 1988:221–228. doi: 10.1007/BF00262774. [DOI] [PubMed] [Google Scholar]
- 17.Swensen JS, Xiao Y, Ferguson BS, Lubin AA, Lai RY, Heeger AJ, Plaxco KW, Soh HT. Continuous, real-time monitoring of cocaine in undiluted blood serum via a microfluidic, electrochemical aptamer-based sensor. Journal of the American Chemical Society. 2009;131:4262–6. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brody JP, Yager P. Diffusion-based extraction in a microfabricated device. Sensors and Actuators A: Physical. 1997;58:13–18. [Google Scholar]
- 19.Eikenberry S. A tumor cord model for doxorubicin delivery and dose optimization in solid tumors. Theoretical biology & medical modelling. 2009;6:16. doi: 10.1186/1742-4682-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uzawa T, Cheng RR, White RJ, Makarov DE, Plaxco KW. A mechanistic study of electron transfer from the distal termini of electrode-bound, single-stranded DNAs. Journal of the American Chemical Society. 2010;132:16120–6. doi: 10.1021/ja106345d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueda Y, Munechika K, Kikukawa A, Kanoh Y, Yamanouchi K, Yokoyama K. Comparison of efficacy, toxicity and pharmacokinetics of free adriamycin and adriamycin linked to oxidized dextran in rats. Chemical & pharmaceutical bulletin. 1989;37:1639–41. doi: 10.1248/cpb.37.1639. [DOI] [PubMed] [Google Scholar]
- 22.Rowe AA, Miller EA, Plaxco KW. Reagentless measurement of aminoglycoside antibiotics in blood serum via an electrochemical, ribonucleic acid aptamer-based biosensor. Analytical chemistry. 2010;82:7090–5. doi: 10.1021/ac101491d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Nazawi MH, Homeida AM. Comparative Pharmacokinetic Studies of Kanamycin in Camels, Sheep and Goats. Scientific Journal of King Faisal University (Basic and Applied Sciences) 2008;10:105–113. [Google Scholar]
- 24.McGregor D. Kirk-Othmer Encyclopedia of Chemical Technology. John Wiley & Sons, Inc.; 2000. [Google Scholar]
- 25.a Dobbs N, Twelves CJ, Gillies H, a James C, Harper PG, Rubens RD. Gender affects doxorubicin pharmacokinetics in patients with normal liver biochemistry. Cancer chemotherapy and pharmacology. 1995;36:473–6. doi: 10.1007/BF00685796. [DOI] [PubMed] [Google Scholar]
- 26.Elis A, Lishner M, Walker S, Atias D, Korenberg A, Koren G. Doxorubicin in lymphoma: association between pharmacokinetic variability and clinical response. Therapeutic drug monitoring. 2010;32:50–2. doi: 10.1097/FTD.0b013e3181c3a16d. [DOI] [PubMed] [Google Scholar]
- 27.Khalil F, Läer S. Physiologically based pharmacokinetic modeling: methodology, applications, and limitations with a focus on its role in pediatric drug development. Journal of biomedicine & biotechnology. 2011;2011:907461. doi: 10.1155/2011/907461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bach DM, Straseski JA, Clarke W. Therapeutic drug monitoring in cancer chemotherapy. Bioanalysis. 2010;2:863–79. doi: 10.4155/bio.10.48. [DOI] [PubMed] [Google Scholar]
- 29.Evans WE, V Relling M, Rodman JH, Crom WR, Boyett JM, Pui CH. Conventional compared with individualized chemotherapy for childhood acute lymphoblastic leukemia. The New England journal of medicine. 1998;338:499–505. doi: 10.1056/NEJM199802193380803. [DOI] [PubMed] [Google Scholar]
- 30.DiFrancesco R, Rosenkranz S, Mukherjee AL, Demeter LM, Jiang H, DiCenzo R, Dykes C, Rinehart A, Albrecht M, Morse GD. Quality assessment for therapeutic drug monitoring in AIDS Clinical Trials Group (ACTG 5146): a multicenter clinical trial. Therapeutic drug monitoring. 2010;32:458–66. doi: 10.1097/FTD.0b013e3181e4427a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyer A, Gruson D, Bouchet S, Clouzeau B, Hoang-Nam B, Vargas F, Gilles H, Molimard M, Rogues A-M, Moore N. Aminoglycosides in septic shock: an overview, with specific consideration given to their nephrotoxic risk. Drug safety: an international journal of medical toxicology and drug experience. 2013;36:217–30. doi: 10.1007/s40264-013-0031-0. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez D, Schmidt S, Derendorf H. Importance of relating efficacy measures to unbound drug concentrations for anti-infective agents. Clinical microbiology reviews. 2013;26:274–88. doi: 10.1128/CMR.00092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nature reviews. Drug discovery. 2010;9:537–50. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science (New York, N.Y.) 1994;263:1425–9. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 35.Pavlovic E, Lai RY, Wu TT, Ferguson BS, Sun R, Plaxco KW, Soh HT. Microfluidic Device Architecture for Electrochemical Patterning and Detection of Multiple DNA Sequences. Langmuir: the ACS journal of surfaces and colloids. 2008;24:1102–7. doi: 10.1021/la702681c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang CY, Schneider HG, Levvey B, Mitchell L, Snell GI. Tacrolimus Level Variability Is a Novel Measure Associated with Increased Acute Rejection in Lung Transplant (LTx) Recipients. The Journal of Heart and Lung Transplantation. 2013;32:S170. [Google Scholar]
- 37.Halloran PF. Immunosuppressive drugs for kidney transplantation. The New England journal of medicine. 2004;351:2715–29. doi: 10.1056/NEJMra033540. [DOI] [PubMed] [Google Scholar]
- 38.V Naidu B, Krishnadasan B, Byrne K, Farr AL, Rosengart M, Verrier ED, Mulligan MS. Regulation of chemokine expression by cyclosporine A in alveolar macrophages exposed to hypoxia and reoxygenation. The Annals of thoracic surgery. 2002;74:899–905. doi: 10.1016/s0003-4975(02)03746-3. discussion 905. [DOI] [PubMed] [Google Scholar]
- 39.Linfert D, Chowdhry T, Rabb H. Lymphocytes and ischemia-reperfusion injury. Transplantation reviews (Orlando, Fla.) 2009;23:1–10. doi: 10.1016/j.trre.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rotondi M, Netti GS, Lazzeri E, Stallone G, Bertoni E, Chiovato L, Grandaliano G, Gesualdo L, Salvadori M, Schena FP, Romagnani P, Serio M. High pretransplant serum levels of CXCL9 are associated with increased risk of acute rejection and graft failure in kidney graft recipients. Transplant international: official journal of the European Society for Organ Transplantation. 2010;23:465–75. doi: 10.1111/j.1432-2277.2009.01006.x. [DOI] [PubMed] [Google Scholar]
- 41.Böhler T, Nolting J, Kamar N, Gurragchaa P, Reisener K, Glander P, Neumayer H-H, Budde K, Klupp J. Validation of immunological biomarkers for the pharmacodynamic monitoring of immunosuppressive drugs in humans. Therapeutic drug monitoring. 2007;29:77–86. doi: 10.1097/FTD.0b013e318030a40b. [DOI] [PubMed] [Google Scholar]
- 42.Corris PA, Kirby JA. A role for cytokine measurement in therapeutic monitoring of immunosuppressive drugs following lung transplantation. Clinical and experimental immunology. 2005;139:176–8. doi: 10.1111/j.1365-2249.2005.02711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El-Khatib FH, Russell SJ, Nathan DM, Sutherlin RG, Damiano ER. A bihormonal closed-loop artificial pancreas for type 1 diabetes. Science translational medicine. 2010;2:27ra27. doi: 10.1126/scitranslmed.3000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrade JD, Hlady V. Protein adsorption and materials biocompatibility: a tutorial review and suggested hypotheses. Advances in Polymer Science. 1986;79:1–63. [Google Scholar]
- 45.Koutsopoulos S, Unsworth LD, Nagai Y, Zhang S. Controlled release of functional proteins through designer self-assembling peptide nanofiber hydrogel scaffold. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4623–8. doi: 10.1073/pnas.0807506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahuja AS, Hendee WR, Carson PL. Transport phenomena in laminar flow of blood. Physics in medicine and biology. 1978;23:928–36. doi: 10.1088/0031-9155/23/5/009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.