Abstract
Streptococcus mutans is a cariogenic oral pathogen whose virulence is determined largely by its membrane composition. The signal recognition particle (SRP) protein-targeting pathway plays a pivotal role in membrane biogenesis. S. mutans SRP pathway mutants demonstrate growth defects, cannot contend with environmental stress, and exhibit multiple changes in membrane composition. This study sought to define a role for ylxM, which in S. mutans and numerous other bacteria resides directly upstream of the ffh gene, encoding a major functional element of the bacterial SRP. YlxM was observed as a produced protein in S. mutans. Its predicted helix-turn-helix motif suggested that it has a role as a transcriptional regulator of components within the SRP pathway; however, no evidence of transcriptional regulation was found. Instead, capture enzyme-linked immunosorbent assay (ELISA), affinity chromatography, and bio-layer interferometry (BLI) demonstrated that S. mutans YlxM interacts with the SRP components Ffh and small cytoplasmic RNA (scRNA) but not with the SRP receptor FtsY. In the absence of FtsY, YlxM increased the GTP hydrolysis activity of Ffh alone and in complex with scRNA. However, in the presence of FtsY, YlxM caused an overall diminution of net GTPase activity. Thus, YlxM appears to modulate GTP hydrolysis, a process necessary for proper recycling of SRP pathway components. The presence of YlxM conferred a significant competitive growth advantage under nonstress and acid stress conditions when wild-type and ylxM mutant strains were cultured together. Our results identify YlxM as a component of the S. mutans SRP and suggest a regulatory function affecting GTPase activity.
INTRODUCTION
In bacteria, approximately 25% to 30% of proteins are translocated to the membrane or beyond (1). Streptococcus mutans, the acidogenic and cariogenic Gram-positive pathogen implicated in human tooth decay, is no exception. Its ability to take up and ferment dietary carbohydrates, quorum-sensing and signal transduction mechanisms, competence pathways, and resilience to environmental stressors are in large part due to the composition of its membrane (2).
The signal recognition particle (SRP) pathway is a cotranslational translocation pathway conserved in all kingdoms of life that is involved in proper biogenesis of membrane and secreted proteins. In bacteria, the SRP pathway consists of a minimum of three universally conserved components: Ffh, scRNA, and FtsY. Ffh has GTPase activity and forms a stable complex with a 7S or 4.5S small cytoplasmic RNA (scRNA) to form the SRP. The established view is that the SRP binds to nascent polypeptide chains containing a hydrophobic signal sequence as they emerge from the ribosome exit tunnel (ribosome nascent-chain, or RNC, complex). The SRP-RNC complex next interacts with the membrane-bound SRP receptor (SR), termed FtsY in bacteria, also a GTPase. Upon interaction of the SRP with FtsY, there is mutual stimulation of the GTPase activities of Ffh and FtsY, bound GTP molecules are converted to GDP, the RNC is transferred to the SecYEG translocase, and translation continues with translocation to insert the polypeptide into or through the membrane (reviewed in reference 3).
Most of what we know about the bacterial SRP comes from studies of Escherichia coli, where interactions of pathway components have been characterized. Ffh and scRNA interact strongly to form the SRP (4, 5). Functional activities of Ffh are ascribed to its M and NG domains. The M domain interacts with the scRNA and nascent polypeptides containing a hydrophobic SRP signal sequence (6–8). Ffh and FtsY interact with one another through their highly similar NG domains. In each, the N domain is followed by a GTPase G domain (9, 10). Efficient membrane protein targeting requires that the targeting apparatus minimizes nonproductive cargo release, ineffective Ffh-FtsY interactions, and premature disruption of the RNC-Ffh-FtsY complex due to futile GTPase activity. Defects in GTPase activity lead to protein translocation deficiencies (11).
S. mutans displays several attributes that make studies of its protein translocation systems of interest. An unexpected finding was that S. mutans survives in the absence of an SRP, in stark contrast to findings for Escherichia coli and Bacillus subtilis (12–14). Although SRP pathway mutants of S. mutans are viable, they are sensitive to environmental stress (15). The closely related species Streptococcus pyogenes can also survive disruption of the SRP but with diminished virulence (16). Another distinguishing feature of S. mutans is that it and most other Gram-positive organisms contain two paralogs of the Alb3/OxaI/YidC family of membrane-localized protein insertases (YidC1 and YidC2 in S. mutans). This is also in contrast to what exists in E. coli and other Gram-negative organisms that contain a single YidC (15). S. mutans YidC2 can substitute for the Saccharomyces cerevisiae mitochondrial homolog OxaI to mediate cotranslational translocation in the absence of an SRP pathway, a property lacking in E. coli YidC (17).
In S. mutans, ffh is contained within a five-gene operon named sat (secretion and acid tolerance) (Fig. 1A) (18, 19). The functions of the other genes within this genetic locus are unknown. ylxM is present not only in S. mutans but also in all other streptococci, where it is located immediately upstream of ffh. In fact, the presence of ylxM (see Fig. S1 in the supplemental material) and its location upstream of ffh (Fig. S2) are highly conserved among Gram-positive bacteria. The distribution of ylxM is not limited to Gram-positive organisms. The genomes of the Gram-negative coccus Veillonella parvula and the nonfirmicute Gram-negative organisms Fusobacterium nucleatum and Thermovirga lienii also contain ylxM. ylxM is almost always located in immediate proximity to genes encoding SRP components, either ffh or ftsY (20) and sometimes both (Fig. S2). The fact that ylxM is linked with other SRP components in so many species across a range of phyla suggests that it may play a role within the pathway.
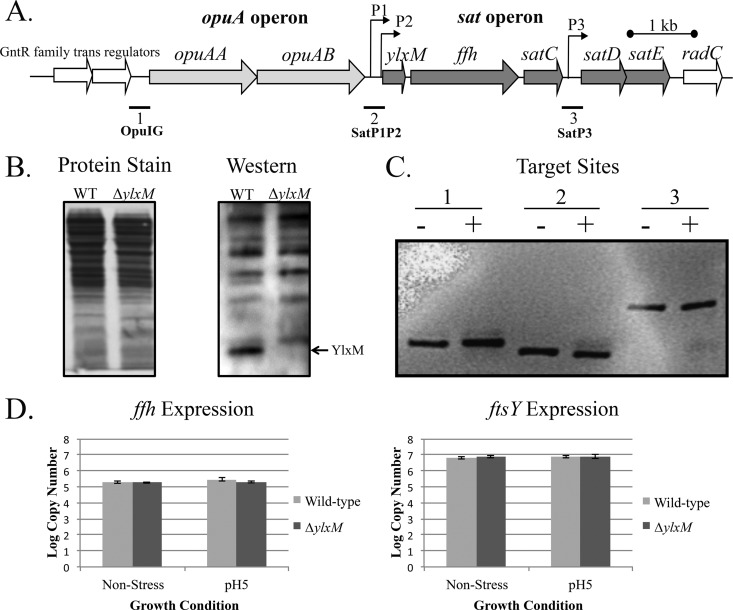
FIG 1.
Genetic locus and expression of ylxM in S. mutans. (A) Schematic representation of the 5-gene S. mutans sat operon and its genetic context. The locations of previously identified promoters (19) are indicated by arrows. The locations of target regions 1, 2, and 3 used in electrophoretic mobility shift assays are indicated in bold. OpuIG, opu operon intergenic region (target region 1); SatP1P2, sat operon promoter 1 (P1) and promoter 2 regions (target region 2); SatP3, sat operon promoter 3 region (target region 3). (B) Colloidal gold total protein staining and Western blot analysis of YlxM in whole-cell lysates from the S. mutans wild type and the ΔylxM mutant. (C) The binding of YlxM to potential regulatory sites within or upstream of the sat operon was evaluated by electrophoretic mobility shift assay. DNA mobility was evaluated in the presence (+) and absence (−) of YlxM. Target regulatory sites were as follows: 1, opu operon intergenic region; 2, sat operon promoter 1 and promoter 2 regions; 3, sat operon promoter 3 region (as shown in panel A). (D) S. mutans ffh and ftsY expression was evaluated by qPCR in wild-type and ΔylxM mutant strains. Cultures were grown under nonstress (pH 7.0) or acid stress (pH 5.0) conditions.
While the existence of ylxM has been recognized for some time, it has not been widely studied and no function has yet been established (20–23). First reported in Mycoplasma mycoides, it was predicted to encode a 13-kDa protein with a helix-turn-helix (HTH) domain that might act as a transcriptional regulator of SRP components (20). Subsequently, an S. mutans strain lacking ylxM demonstrated a modest growth defect in comparison to the growth of the parental strain (19). Later, recombinant S. pyogenes YlxM was crystallized and the presence of an HTH structure was confirmed, suggesting that the protein binds DNA, although no functional assays were performed (23). A more recent study reported that deletion of ylxM led to increased detection of Ffh in B. subtilis cell lysates by Western blotting, although no increase in ffh message level was observed (24). Our initial experiments revealed no evidence for YlxM as a transcriptional regulator; hence, we speculated that YlxM might serve a role within the SRP pathway itself. Here we present our results that identify interactions of S. mutans YlxM with Ffh and scRNA and demonstrate that YlxM functions to modulate the GTPase activity of SRP pathway components.
MATERIALS AND METHODS
Bacterial expression strains.
S. mutans ylxM was cloned into pET151D-TOPO using primers PC178F (CACCATGGAGATCGAAAAAACCAATCG) and PC178R (TTAGTCTCTATTATCAATAGTCGTC) with strain UA159 genomic DNA as the template. The plasmid was used to transform chemically competent E. coli BL21 Star(DE3) cells (Invitrogen, Carlsbad, CA). Protein expression was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 37°C. The Ffh expression strain was constructed using primers AH2F (TTTTCTGCAGTATATTGTGATCATAGCAATCCTT) and AH2R2 (TTTTTTTTTGCTCAGCTTTGTATTTTATTGGGAAGACCATTA). The PCR product was cloned into pET15b and used to transform chemically competent E. coli BL21(DE3) cells (Invitrogen, Carlsbad, CA). Expression was induced overnight at room temperature with 0.2 mM IPTG. ftsY was cloned and expressed similarly to ffh, except that 0.5 mM IPTG was used for induction for 4 h at 37°C. Primers for ftsY amplification were AH1F2 (TTTTTATGGATCCTATGGGTTTATTTAATCGCTTATTTGGT) and AH1R2 (TTTTCTGCAGTATATTGTGATCATAGCAATCCTT). To complement the ylxM deletion in S. mutans, ylxM was amplified by PCR using strain NG8 chromosomal DNA as the template and primers MW36F (CTGGCGTCGACATGCATCATCACCATCACCATATGGAGATCGAAAAAACCAATC) and MW36R (CTGGCGGATCCTTAGTCTCTATTATCAATAGTCG) and cloned into pVPT-NT7 (25). The recombinant expression construct was used to transform strain BK140 (19) to generate the strain denoted herein as +YlxM.
Purification of proteins.
Recombinant proteins were purified using metal ion affinity chromatography. YlxM and FtsY were purified from whole-cell lysates using HisPur cobalt resin (Thermo Scientific, Rockford, IL). Most Ffh produced in E. coli was insoluble; therefore, protein was extracted from inclusion bodies using the BugBuster protein extraction reagent (Novagen, Darmstadt Germany) before purification on nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen, Hilden, Germany) under denaturing conditions in the presence of 8 M urea. Ffh preparations included RNase to prevent copurification of scRNA. Ffh was refolded by dialysis into renaturation buffer (200 mM NaCl, 2 mM MgCl2, 50 mM glycine, 10% glycerol, pH 7.6). Proper refolding was confirmed by circular dichroism (data not shown), reconstitution of GTPase activity (see Fig. 4), and interaction with FtsY (see Fig. S5 in the supplemental material). All proteins were dialyzed into HM buffer (10 mM HEPES, 10 mM MgCl2, pH 8.0) before use.
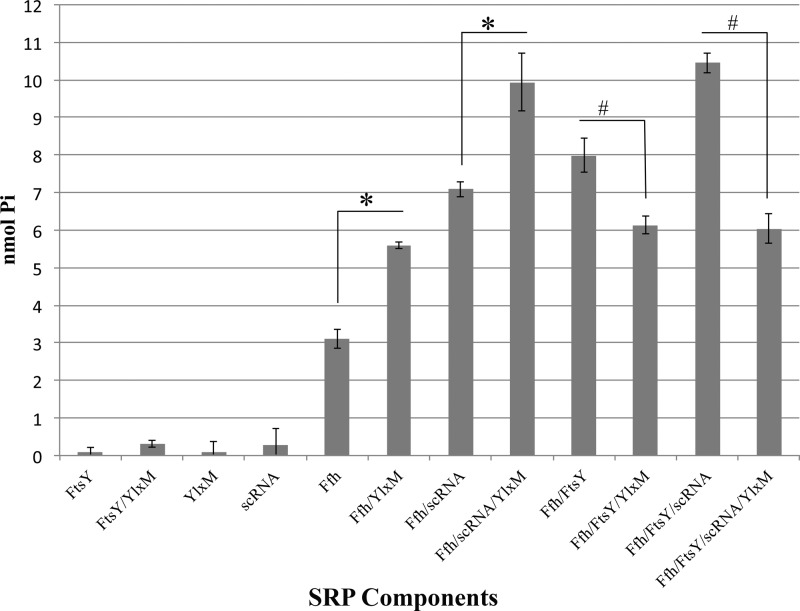
FIG 4.
YlxM affects the GTPase activity of SRP components. An endpoint colorimetric assay was used to detect the amount of inorganic phosphate (Pi) released upon hydrolysis of GTP to GDP after 1 h by Ffh and/or FtsY in the presence of YlxM and/or scRNA. Assays were performed in triplicate. Significant differences were determined by two-way ANOVA. * indicates a significant increase in activity, and # indicates a significant decrease in activity (P < 0.01).
Anti-YlxM, Ffh, and FtsY antisera.
Polyclonal rabbit antisera were generated against purified recombinant YlxM, Ffh, and FtsY (Lampire Biological Laboratories, Pipersville, PA). The anti-FtsY antiserum was exhaustively adsorbed with purified Ffh to eliminate cross-reactive antibodies. The anti-Ffh antiserum did not cross-react with FtsY, so no cross-adsorption was necessary. Anti-YlxM antibodies were affinity purified using purified YlxM covalently coupled to Sepharose beads as described below. The specificities of the Ffh, FtsY, and YlxM antisera and levels of background reactivity are shown in Fig. S3 in the supplemental material.
Western blot detection of YlxM in S. mutans.
Whole-cell lysates of exponential-phase cultures were prepared from S. mutans strain UA159 and a corresponding ΔylxM strain (kindly provided by Robert Quivey, University of Rochester) by glass bead breakage in a Mini-Beadbeater 8 apparatus (BioSpec Products, Inc., Bartlesville, OK). Cell lysates were electrophoresed through 12% sodium dodecyl sulfate (SDS)-polyacrylamide gels, transblotted onto Immobilon polyvinylidene difluoride (PVDF) membranes (Sigma-Aldrich, St. Louis, MO), reacted with anti-YlxM affinity-purified antibodies followed by horseradish peroxidase-labeled anti-rabbit IgG, and developed using the enhanced-chemiluminescence (ECL) Western blotting system (GE Healthcare) and Hyperfilm (GE Healthcare) according to the manufacturer's instructions. As a loading control, total protein staining was performed using colloidal gold (Bio-Rad, Hercules, CA).
qPCR.
To measure the levels of ffh and ftsY mRNA, real-time quantitative reverse transcriptase PCR (RT-qPCR) was performed with S. mutans RNA prepared as described elsewhere (26). Briefly, RNA was purified from UA159 and the ΔylxM mutant and was grown to mid-exponential phase under nonstress (pH 7) or acid stress (pH 5) conditions. cDNA was synthesized using 200 ng of total RNA and a SuperScript III first-strand synthesis system (Invitrogen, Carlsbad, CA). Target-specific primers used for real-time qPCRs were as follows: RTffhFWD (AGGGTTGAGCGGTGCTAATA), RTffhRVS (GTTTCATAGCAAATTCGCCG), RTylxMFWD (TCTGACTACGTCGTCCGC), and RTylxMRVS (TCAATATGGAAATCTTGTTCTG). Copy numbers of target genes were determined using the iQ SYBR green supermix (Bio-Rad) in an iCycler iQ real-time PCR detection system (Bio-Rad). Cycling conditions were as follows: 1 cycle of 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All expression data were normalized to the copy number of 16S rRNA in each sample. qPCR data were derived from two separate experiments. Each experiment consisted of triplicate PCRs from duplicate RT reactions. Student's t test was used to assess statistical significance.
EMSAs.
To determine if YlxM binds to regulatory sequences in the vicinity of ffh, potential target regions within and upstream of the sat operon were PCR amplified with 5′-end-biotinylated primers. These included three promoters previously identified within the sat operon and an intergenic region within the upstream opu operon (Fig. 1A) that was identified as a potential target of regulation using the Inverted Repeats Finder and RibEx (19, 27, 28). Target site 1 was the upstream opu intergenic region. An ∼330-bp probe was generated with primers MW17F (GGAGATGACTTTGCTTTTTCAC) and MW17R (TGGTGAGATGTTTTATTTCAAGTT). Target site 2 included the promoter 1 and 2 regions of the sat operon. An ∼150-bp probe was created using primers MW18F (GAAGGTTGCATCTTGGACAAG) and MW18R (AAGCGCATTCATTCGATTGGTT). Target site 3 was promoter 3 of the sat operon. An ∼200-bp probe was made using primers MW19F (CCATCATTGTCATTATCGTAACT) and MW19R (GATCGCCAATAATAGCGATGTA). Binding reactions were performed with 20 fmol of biotinylated target DNA mixed with 26 pmol recombinant YlxM (rYlxM) in electrophoretic mobility shift assay (EMSA) binding buffer (10 mM Tris, 50 mM KCl, 10 mM dithiothreitol [DTT], pH 7.5) for 30 min prior to electrophoresis on a nondenaturing 6% polyacrylamide gel. Samples were electroblotted onto PVDF and developed using the LightShift EMSA kit with appropriate internal controls (Thermo Scientific, Waltham, MA).
Protein interaction studies. (i) Capture ELISA.
Ninety-six-well polystyrene plates (Costar, Corning, NY) were coated with 200 ng of purified recombinant YlxM or maltose binding protein (MBP) as a negative control. Wells were blocked with PBST (phosphate-buffered saline containing 0.3% Tween 20). Twofold serial dilutions of Ffh, beginning at 200 ng, were added to the wells and incubated for 2 h at 37°C. Unbound proteins were removed by washing them with PBST. Bound Ffh was detected with specific rabbit antisera, followed by horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG and development with o-phenylenediamine.
(ii) Affinity capture on YlxM-Sepharose.
Four milligrams of purified, recombinant YlxM was covalently coupled to 1 ml cyanogen bromide-activated Sepharose 4B (Sigma-Aldrich) beads according to the manufacturer's instructions. Unreacted sites were blocked with ethanolamine. S. mutans cells harvested from a 100-ml stationary-phase culture were lysed, and the whole-cell lysate was reacted with the YlxM matrix or a no-protein bead-only negative-control matrix for 16 h at 4°C on a rotator. The Sepharose matrix was placed in a column, and unbound proteins were removed by extensive washing with PBS. Bound proteins were eluted using 0.2 M glycine-HCl (pH 2.5) and were immediately neutralized by collection in 100 mM Tris, 0.2% sodium azide (pH 8.0). The unbound proteins, wash, and elution fractions were separated by electrophoresis through 4-to-20%-gradient SDS-polyacrylamide gels (Bio-Rad, Hercules, CA), transferred to PVDF, and visualized by Western blotting with rabbit polyclonal antisera against Ffh, FtsY, affinity-purified anti-YlxM antibodies, or a commercially available murine anti-6His antiserum (Invitrogen).
BLI experiments.
The scRNA DNA was PCR amplified from S. mutans strain UA159 using primers MW28F (TAATACGACTCACTATAGGGGGAGCAACAGCTTTGCGT) and PC62R (AGAGCACACAGCTTACATC). Underlined bases indicate the T7 promoter region required for in vitro transcription (IVT). SRP RNA was transcribed using the MEGAshortscript T7 IVT kit (Ambion, Carlsbad, CA). The IVT scRNA was biotinylated using the RNA 3′-end biotinylation kit from Pierce (Thermo Scientific, Waltham, MA). Bio-layer interferometry (BLI) experiments were performed on an Octet QKe instrument (FortéBio, Menlo Park, CA). Streptavidin-coated sensor tips were wetted in 1× kinetics buffer (1 mM phosphate, 15 mM NaCl, 0.002% Tween 20, 0.005% sodium azide, 0.1 mg/ml bovine serum albumin [BSA], pH 7.4) prior to exposure to the biotinylated scRNA. After incubation with 25 μg/ml of scRNA, unreacted streptavidin molecules on the tips were quenched with 50 μg/ml biocytin. The loaded tips were equilibrated in 1× HM buffer and then reacted with purified Ffh or YlxM at 1 μM concentrations. Binding was measured over 15 min, followed by a 30-min dissociation period in HM buffer without protein. Controls included a sample lacking protein (buffer only) and a sample with no scRNA on the sensor. Affinity calculations were performed using FortéBio Octet data analysis software. The following fit parameters were utilized: Savitzky-Golay filtering, a 1:1 binding model, and global fitting. Only data with an R2 of >0.90 were included to ensure quality. The BLI measurements were repeated three times for Ffh and five times for YlxM.
GTP hydrolysis assays.
Recombinant Ffh and FtsY were assayed for GTPase activity individually and in combination with S. mutans scRNA and/or recombinant YlxM. An endpoint colorimetric assay was used to measure hydrolysis in solution, as reflected by the amount of inorganic phosphate (Pi) released by GTP hydrolysis, as described previously (29–31). Assays containing a 360 nM final concentration (each) of Ffh, FtsY, and/or YlxM and 2 mM GTP were brought to a final volume of 150 μl in HM buffer and incubated at 30°C for 1 h. scRNA was included where indicated in the figures at a concentration of 13.3 μM. Following incubation, SDS was added to a final concentration of 6% to stop GTP hydrolysis. Ascorbic acid and ammonium molybdate were added to 6 and 1%, respectively, and the mixture was incubated for 5 min. Sodium citrate, sodium (meta)arsenite, and acetic acid were then added to a final concentration of 1% and a final reaction volume of 1.05 ml. The absorbance of each reaction mixture was measured at 850 nm. A standard curve of inorganic phosphate was linear from 2 to 150 nmol of Pi and used to determine the amount of Pi released from each assay. Substrate-only controls lacking protein and reaction components and a time zero control with 6%-SDS-denatured proteins were used to correct for nonspecific hydrolysis and background. Experiments were performed in triplicate. Statistically significant differences between groups were determined by two-way analysis of variance (ANOVA) using GraphPad Prism 5 (La Jolla, CA). Nonmatching parameters and Bonferroni posttests were used to determine differences among the groups.
Competition assays.
To determine if ylxM confers a competitive advantage in S. mutans, ylxM-positive (wild-type-NG8 [WT-NG8]) and ΔylxM (with a kanamycin resistance cassette in place of ylxM) strains were grown separately to mid-exponential phase in Todd-Hewitt broth (ThermoFisher, Waltham, MA) supplemented with 0.3% yeast extract (THYE broth), pH 7.0, at 37°C. Cell numbers were normalized by determining the optical density at 600 nm (OD600), and the two strains were mixed in a 1:1 ratio, placed in fresh THYE (pH 7.0 or pH 5.0, i.e., under nonstress or acid stress conditions, respectively) at 37°C and allowed to grow to stationary phase before serial dilution and plating onto THYE agar (pH 7.0 or pH 5.0) in triplicate. Total numbers of CFU were determined, and plates were replica plated onto THYE agar plates with 500 μg/ml kanamycin to determine the number of colonies that were wild type versus lacking ylxM. Similar experiments were also performed using the complemented ylxM mutant (+YlxM) strain, as well as other SRP mutants (Δffh, ΔftsY, and ΔscRNA gene strains) in the presence of appropriate antibiotics (15). Each experiment was plated in triplicate and performed three times. Numbers from all plates were averaged and are reported as percentages of the total number of colonies. Student's t test was used to determine significance.
PTS assay.
As an indirect measurement of membrane protein insertion, the ability of S. mutans cells to take up sorbitol by the sorbitol phosphoenolpyruvate-sugar phosphotransferase system (PTS) was evaluated as described elsewhere (32) using wild-type, ΔylxM, Δffh, and +YlxM strains grown in defined media containing 1% sorbitol.
RESULTS
YlxM is produced as a detectable protein in S. mutans.
To begin our studies, we confirmed that ylxM is expressed and produced as a protein in S. mutans. Affinity-purified rabbit anti-YlxM antibodies were used to identify YlxM contained in S. mutans cellular lysates by Western blotting. A 13-kDa band, consistent with the predicted size of YlxM, was identified in whole-cell lysates prepared from the wild-type strain but not in samples prepared from a ΔylxM mutant strain (Fig. 1B).
YlxM is not a transcriptional regulator of ffh or ftsY in S. mutans.
We evaluated known and putative regulatory regions within and near the sat operon that might serve as potential targets of YlxM binding but found no evidence to suggest any such activity when electrophoretic mobility shift assays were performed (Fig. 1C). In addition, we used quantitative real-time PCR to evaluate the effect of the presence or absence of ylxM on the transcription of ffh and ftsY under both nonstress and acid stress growth conditions. No significant differences in ffh or ftsY expression were detected when wild-type and ΔylxM mutant strains were compared (Fig. 1D). Neither ffh nor ftsY expression was influenced by acid stress. Furthermore, in contrast to a previous report on B. subtilis (24), neither Ffh nor FtsY protein production appeared to be substantially altered in the absence of ylxM (see Fig. S4 in the supplemental material).
YlxM interacts with Ffh but not FtsY.
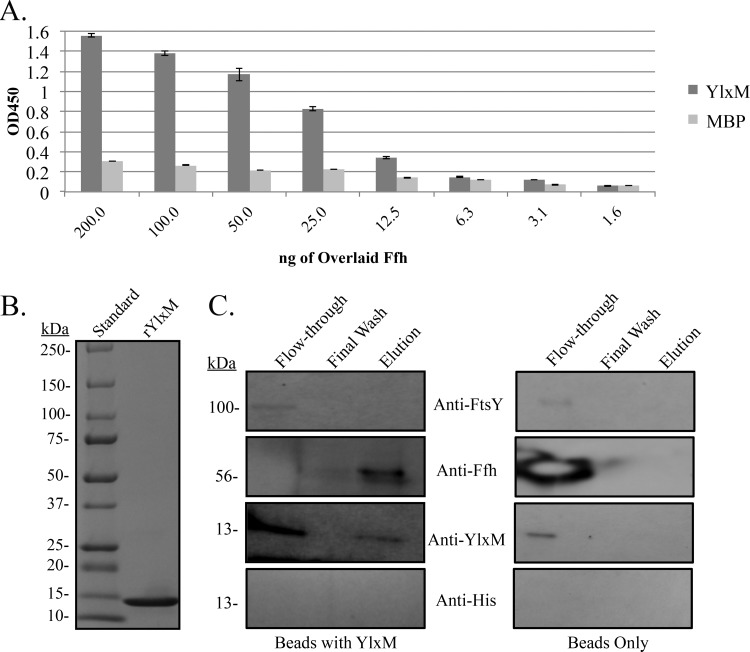
Since YlxM did not appear to function as a transcriptional regulator, we next evaluated whether it might serve a role within the SRP pathway. To determine whether YlxM interacts with known SRP components, 6-histidine-tagged recombinant proteins were purified. A capture ELISA demonstrated a YlxM-Ffh interaction (Fig. 2A). In contrast, YlxM did not interact with FtsY (see Fig. S5 in the supplemental material), which shares homologous domains with Ffh, indicating that the YlxM-Ffh interaction is specific. To determine whether the interaction of YlxM with Ffh could also occur within the context of other streptococcal proteins, purified YlxM (Fig. 2B) was covalently coupled to a Sepharose matrix, and an S. mutans cellular lysate was applied to the column. FtsY, Ffh, and YlxM were all present in the cellular lysate and detectable in the column flowthrough (Fig. 2C). In the case of Ffh, the high level of reactivity with the anti-Ffh antiserum caused quenching of the detection reagent in the flowthrough fraction. Western blot analysis also demonstrated the presence of a 56-kDa Ffh band, but not FtsY, in the elution fraction (Fig. 2C). In addition, a 13-kDa band was detected with anti-YlxM antibodies in the elution fraction. This band was not recognized by an anti-His antibody, indicating that it represented the untagged, native S. mutans protein derived from the cell lysate rather than the His-tagged recombinant protein used to make the column. Hence, S. mutans Ffh and YlxM were both captured and eluted from a purified rYlxM affinity column. No SRP proteins were captured by the bead-only negative control (Fig. 2C).
FIG 2.
Identification of a YlxM and Ffh interaction. (A) ELISAs showing immobilized YlxM (or MBP as a negative control) incubated with serial dilutions of Ffh. Bound protein was detected with monospecific polyclonal rabbit anti-Ffh antisera. (B) Coomassie blue stain of purified recombinant YlxM (rYlxM) that was covalently coupled to Sepharose to generate the affinity matrix. (C) Western blot analysis of S. mutans whole-cell lysates applied to the YlxM affinity matrix (Beads with YlxM) or a bead-only control. Captured proteins were eluted as described in Materials and Methods and subjected to SDS-polyacrylamide gel electrophoresis, electroblotted onto a PVDF membrane, and reacted with polyclonal rabbit anti-Ffh, -FtsY, -YlxM, or -6His antibodies.
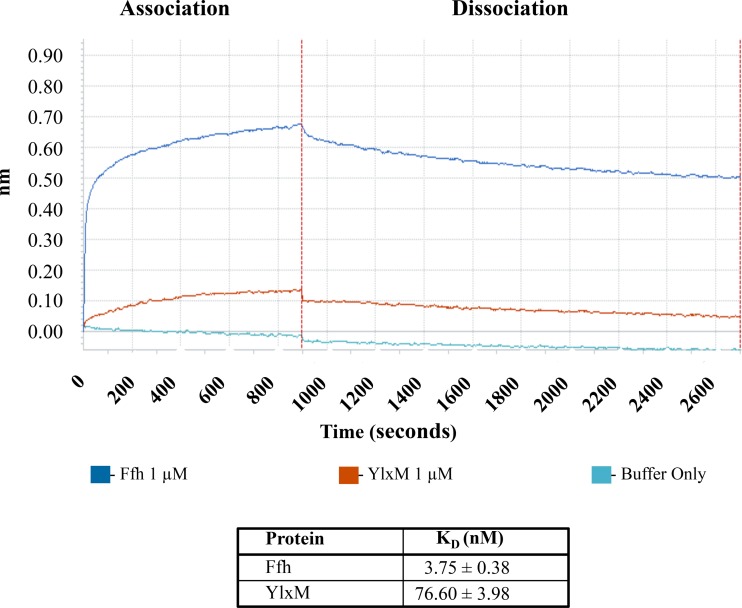
S. mutans YlxM associates with the 4.5S scRNA.
Given that YlxM contains an HTH motif suggestive of a nucleic acid binding function, we also tested whether it is capable of interacting with the SRP scRNA. The S. mutans scRNA was transcribed in vitro and biotinylated before being immobilized on streptavidin-coated sensor tips. Binding of Ffh and YlxM to the immobilized scRNA was evaluated by bio-layer interferometry (BLI) (Fig. 3). As expected, Ffh bound to the immobilized scRNA. The calculated equilibrium dissociation constant of this interaction was 3.75 ± 0.38 nM and is consistent with previously published values obtained using other methods (4, 5). YlxM also bound to the immobilized scRNA. Because YlxM is substantially smaller than Ffh, 13 kDa compared to 56 kDa, the optical thickness measured at the biosensor tip was less. In the case of YlxM, the calculated dissociation constant was 76.60 ± 3.98 nM.
FIG 3.
Evaluation of the interactions of S. mutans Ffh and YlxM with scRNA by bio-layer interferometry. Association and dissociation curves of Ffh or YlxM, with scRNA coated on the sensor tip, are shown. Calculated equilibrium dissociation constants ± standard errors of the mean are shown in the table.
The GTPase activity of SRP components is modulated by YlxM.
Ffh and FtsY are GTPases that form a heterodimer and mutually stimulate one another, leading to the hydrolysis of GTP to GDP. This hydrolysis is fundamental to the recycling of SRP pathway components and reflects their functional activities. Ffh exhibited intrinsic GTPase activity in the absence of additional components (Fig. 4), consistent with previous reports on Streptococcus pneumoniae, Mycoplasma mycoides, and Mycobacterium tuberculosis (5, 33, 34). The GTPase activity of Ffh was significantly increased in the presence of scRNA or FtsY, with maximal activity measured in the presence of all three components. The GTPase activity of Ffh alone, as well as that of Ffh/scRNA, was also significantly increased upon addition of YlxM. However, when YlxM was added to Ffh or Ffh/scRNA in the presence of FtsY, there was a significant decrease in measured GTPase activity.
YlxM confers a competitive growth advantage to S. mutans.
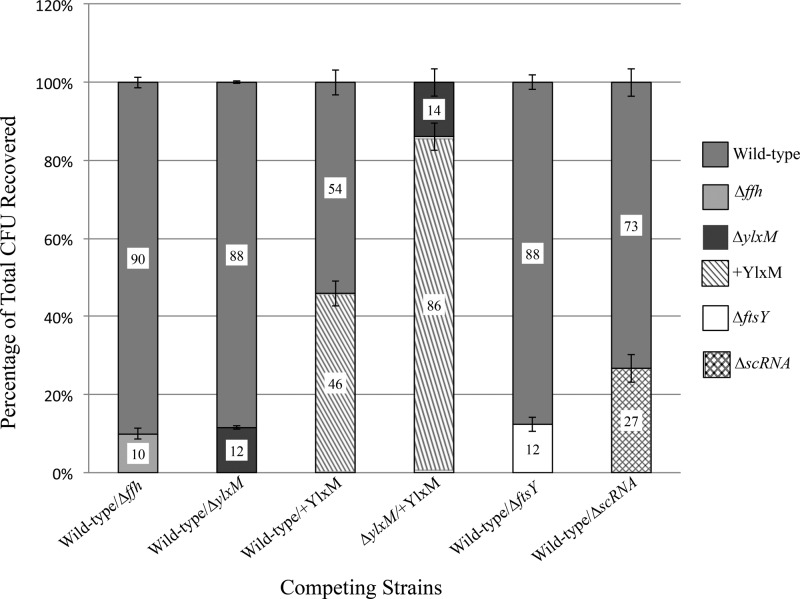
The high degree of evolutionary conservation of ylxM within genetic loci containing ffh and/or ftsY suggests that it confers a selective advantage to organisms that contain it. To test the fitness of wild-type S. mutans relative to that of a ΔylxM mutant, the two strains were mixed at a 1:1 ratio and cultured to stationary phase prior to being plated. The ΔylxM mutant was clearly outcompeted by the parental strain (Fig. 5). At the end of the incubation period, almost 90% of the recovered organisms grown under nonstress conditions represented the wild type. Under acid stress conditions, over 90% of the recovered organisms were wild type (see Fig. S6 in the supplemental material). The presence of the kanamycin marker used to identify the ylxM mutant has no measurable effect on the growth characteristics of S. mutans (data not shown). Complementing the ΔylxM strain with YlxM expressed from a plasmid restored the fitness of the strain. We also tested mutants lacking other known SRP components to assess their competitive ability. Like the ΔylxM strain, the Δffh and ΔftsY strains represented approximately 10% of the recovered cells when grown in competition with the wild type, whereas the ΔscRNA gene strain represented approximately 30% of the recovered cells (Fig. 5).
FIG 5.
YlxM confers a competitive advantage to S. mutans. Wild-type and SRP mutant strains were mixed at a 1:1 ratio and grown to stationary phase before being plated onto THYE agar. The experiments were performed in triplicate, and the data are reported as the percentage of total colonies of each strain recovered. Significant differences between the recovery of the wild type and that of the SRP mutant strains were detected as follows: for the Δffh strain, P was <0.01; for the ΔylxM strain, P was <0.01; for the ΔftsY strain, P was <0.01; and for the ΔscRNA gene strain, P was equal to 0.02. There was no significant difference in wild-type values compared to +YlxM values (P = 0.34).
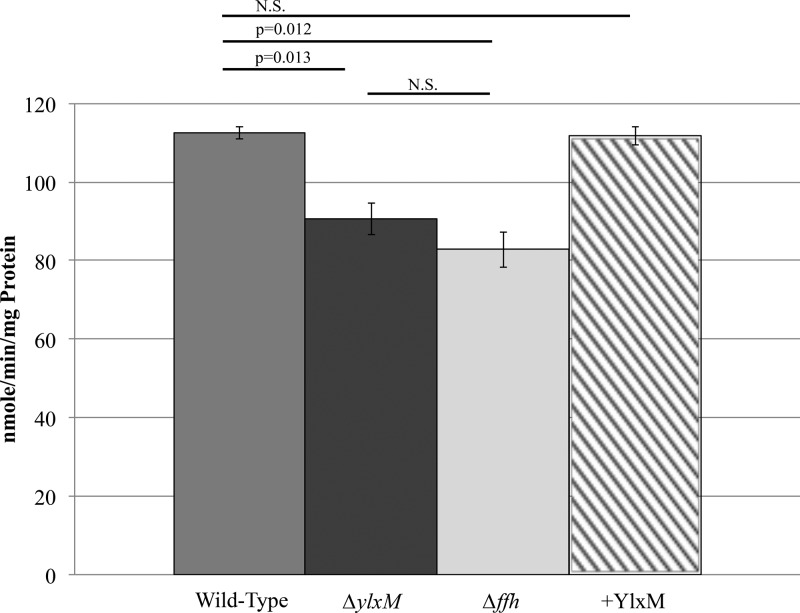
YlxM affects membrane protein insertion in S. mutans.
To evaluate whether YlxM affects the efficiency of membrane protein insertion in S. mutans, we measured sorbitol uptake via the sorbitol phosphoenolpyruvate-sugar phosphotransferase system (PTS) as an indirect measure. Uptake of sorbitol, unlike that of other sugars that can be taken up by more than one of the 14 known PTSs in S. mutans, is mediated by way of a single PTS via the membrane protein EIIC (35–37). Sorbitol uptake was impaired in the ylxM mutant and was restored by complementation of YlxM by plasmid-based expression (Fig. 6), which is similar to results seen with the ffh mutant. This suggests that YlxM contributes to the efficiency of SRP-mediated membrane protein insertion. Not surprisingly, sorbitol uptake was not completely eliminated in the ffh or ylxM mutant strains since S. mutans possesses YidC2, which can support cotranslational membrane protein insertion in the absence of an SRP (17). As with our previous results in which simultaneous deletion of ffh and yidC2 was lethal (15), double deletion of ylxM and yidC2 proved to be fatal (see Fig. S7 in the supplemental material). This suggests that S. mutans can survive deletion of yidC2 only if the SRP is fully operational. In contrast, while more impaired than the ylxM single mutant, a ylxM yidC1 double deletion mutant was still viable (Fig. S7). This is consistent with the viability of an ffh yidC1 double deletion mutant (15).
FIG 6.
Sorbitol uptake in S. mutans is influenced by YlxM. Wild-type, ΔylxM, Δffh, and +YlxM strains were assayed for sorbitol PTS activity as described in Materials and Methods. Each bar represents the average of results from three replicates. Shown are P values as determined by Student's t test. N.S. (not significant), P > 0.05.
DISCUSSION
Prior literature on YlxM suggested that it functions as a transcriptional regulator of SRP components (20, 23). We found that this is not the case in S. mutans. Instead, YlxM interacted with Ffh and the scRNA and modulated the net GTPase activity of pathway components. This activity suggests a functional role for YlxM within the SRP pathway.
The bacterial SRP machinery has been extensively characterized in E. coli, where the pathway is subject to complex spatial and temporal regulation through a series of defined checkpoints (reviewed in references 3 and 38 to 41). These checkpoints involve highly choreographed interactions with a number of external factors, including signal peptides, ribosome nascent-chain complexes, membrane lipids, and the SecYEG translocon (38, 42–45). Fewer studies have been performed with Gram-positive organisms. Although there are apparent similarities in SRP machineries across species, unique characteristics have been found as well, highlighting the need for further study among different bacteria. Reports on E. coli described a role for the scRNA and its interaction with the M domain of Ffh in the formation of Ffh/FtsY complexes and subsequent GTPase activity of SRP pathway components (4, 43, 44). However, studies of Streptococcus pneumoniae and Mycoplasma mycoides reported that the scRNA-Ffh interaction is not an absolute requirement for Ffh/FtsY complex formation nor for GTPase activity (5, 34). Thus, it has been postulated that scRNA may have a regulatory role that is not as significant in some organisms as it is in E. coli (5, 34). In our competitive-fitness assays, we found that a lack of scRNA was less detrimental than a loss of YlxM, Ffh, or FtsY when the bacteria were grown under nonstress conditions; however, under acid stress conditions, the scRNA mutant was almost as impaired as the ylxM, ffh, and ftsY mutants. In S. mutans, the SRP pathway can be eliminated altogether and viability maintained, albeit with severe consequences (15). In this organism, deletion of ffh, ftsY, or the scRNA gene results in a pronounced growth defect and sensitivity to acid and osmotic stress, whereas elimination of ffh also confers sensitivity to oxidative stress.
While bacterial SRP is composed minimally of Ffh, FtsY, and scRNA, it should not be surprising to identify the presence of additional accessory proteins. In mammalian systems, the SRP pathway is much more complex. Proteins such as SRP68/72, SRP9/14, and SRP19 all contribute to the regulation of the pathway in ways that are not completely understood (reviewed in reference 3). It has been predicted that as-yet-unknown proteins may work in concert with the scRNA to regulate bacterial Ffh/FtsY activity (46, 47). It appears that YlxM represents such an accessory factor. In ELISAs, S. mutans YlxM and Ffh interacted with one another without the need for additional proteins or cofactors. In addition, Ffh from S. mutans cell lysates was captured using a recombinant YlxM affinity column. Bio-layer interferometry experiments with in vitro-transcribed S. mutans scRNA confirmed a high-affinity interaction with Ffh, and a lower-affinity interaction of scRNA with YlxM in the absence of Ffh was also identified. In B. subtilis, the histone-like protein HBsu complexes with the Alu domain of its larger 7S scRNA and is presumed to function as part of the SRP, since HBsu mutants show reduced translocation efficiency of a model substrate (48, 49). While the S. mutans genome contains a gene encoding a protein similar to HBsu (the nucleoid-associated protein HU), S. mutans scRNA does not contain an Alu domain. The interaction of YlxM with the scRNA therefore likely occurs via a mechanism different from that of HBsu. As stated above, YlxM contains a predicted HTH domain, and such a structure has been clearly identified in YlxM from S. pyogenes (23). Although traditionally thought of as a DNA binding element, the HTH domain of Ffh has been shown to interact with the scRNA (6). It will be important in future structural studies to delineate how YlxM interacts with the scRNA and determine whether its HTH domain is critical for this binding.
The fact that YlxM interacted with both of the minimally conserved components of the SRP suggested that it contributes a functional role to the pathway. The GTPase activities of Ffh and FtsY are necessary for recycling of pathway components, and measurement of GTPase is a common measure of SRP pathway activity (3, 29, 40, 42, 50). We therefore performed GTPase activity assays on purified recombinant S. mutans proteins and confirmed that, as in other organisms (5, 34, 51), S. mutans Ffh GTPase activity is significantly increased upon the addition of its heterodimer partner FtsY. We also observed intrinsic GTPase activity associated with Ffh alone, a property that has been reported for several bacteria (5, 33, 34, 50). Values from our colorimetric assays were comparable to those published in studies of the chloroplast SRP (29, 30). As expected, the addition of scRNA to Ffh or to Ffh/FtsY resulted in a significant net increase in GTPase activity. The presence of YlxM also significantly enhanced the GTPase activity of Ffh, both alone and in combination with scRNA. However, in the presence of FtsY, for example, when YlxM was added to Ffh/FtsY or to Ffh/FtsY/scRNA, a significant inhibitory effect on net GTPase activity was observed. Thus, YlxM appears to have a bifunctional role, as it can increase or decrease net GTPase activity depending on the presence or absence of other pathway components.
The underlying reason for an SRP pathway-modulating factor such as YlxM is not immediately apparent. Unlike with the elimination of the known SRP pathway components Ffh, scRNA, and FtsY, deletion of the S. mutans gene encoding YlxM does not result in pronounced stress sensitivity, and only a modest effect on growth is observed (19). Still, ylxM is highly conserved in Gram-positive bacteria, and a number of Gram-negative bacteria harbor the gene. Our current results indicate that YlxM is physiologically relevant, as direct competition experiments demonstrated that the presence of ylxM in S. mutans conferred an obvious advantage to the cells. Interestingly, under both nonstress and acid stress conditions, elimination of ylxM was found to be as detrimental to the competitive fitness of S. mutans as was elimination of ffh or ftsY. This suggests that the SRP pathway must be not only operational but also optimally efficient to confer a selective advantage to the bacteria. Use of the S. mutans sorbitol uptake pathway as an indirect measure of proper membrane protein insertion confirmed that YlxM contributes in a meaningful way to the functional activity of the SRP pathway. Furthermore, we found that a ylxM yidC2 double deletion mutant is not viable. This result is similar to results with yidC2 ffh, yidC2 ftsY, and yidC2 scRNA gene double mutants, which also are not viable (15). The lethality of simultaneous elimination of ylxM and yidC2 suggests that less-than-full operational efficiency of the SRP is fatal when YidC2 is not present. Taken together, these results suggest that the improved SRP function in the presence of YlxM is sufficient to warrant its retention in a myriad of bacterial genomes.
YlxM may have a function analogous to that of YlxH, an accessory protein recently found to regulate the activity of FlhF, a protein involved in flagellar biosynthesis in B. subtilis (46). Like Ffh and FtsY, FlhF is a member of the SIMIBI family of GTPases. A similarity between the ylxH flhF and ylxM ffh (ftsY) genes is their frequent juxtaposition. YlxH utilizes its activator helix to promote homodimerization of FlhF. Although YlxM and YlxH share little sequence homology and do not demonstrate any obvious conserved domains, YlxM's measurable influences on the GTPase activities of Ffh and Ffh/FtsY suggest that it too may influence homodimer and/or heterodimer formation or stability, likely in the presence of the scRNA. Such speculation will require in-depth structural studies in the future. The potential for alternative interactions and outcomes of SRP pathway components is beginning to be reported; hence, YlxM's participation in membrane protein insertion and secretion may be unconventional. For example, an alternative SRP pathway model that docks SRP/FtsY complexes at the membrane prior to the recruitment of RNCs has been proposed (52). It is thought that the docked Ffh/FtsY complex may forgo its GTPase activity to prevent premature dissociation of Ffh and FtsY. The presence of a negative regulator of Ffh/FtsY GTPase activity, such as YlxM, may provide an explanation for how such events in a membrane-associated SRP pathway might occur. Another model proposes that FtsY alone recruits RNCs to the membrane and that the interaction of Ffh with nascent polypeptide and FtsY occurs after RNCs are docked at the membrane (53). Even within the conventional pathway, the entire series of events from the initial interaction of the SRP with RNCs through docking and cargo handover and release are not fully understood. As stated by Akopian et al., the increased complexity stemming from the presence of additional factors within the mammalian SRP adds layers of nuance and regulation that remain to be elucidated (3). Our studies reveal a further layer of complexity within the S. mutans SRP pathway and identify YlxM as a contributing functional component. This new information will aid in the design and interpretation of studies of the SRP pathway in the many bacteria that contain it.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nathan Lewis for scientific discussion and critical review of the manuscript and Justin Runac for technical assistance.
This work was supported by NIH grant R01DE08007 to L.J.B. and NIH grants T90DE021990 and F31DE023710 to M.L.W.
Footnotes
Published ahead of print 21 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01465-13.
REFERENCES
- 1.Driessen AJ, Nouwen N. 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77:643–667. 10.1146/annurev.biochem.77.061606.160747 [DOI] [PubMed] [Google Scholar]
- 2.Banas JA. 2004. Virulence properties of Streptococcus mutans. Front. Biosci. 9:1267–1277. 10.2741/1305 [DOI] [PubMed] [Google Scholar]
- 3.Akopian D, Shen K, Zhang X, Shan SO. 2013. Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 82:693–721. 10.1146/annurev-biochem-072711-164732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siu FY, Spanggord RJ, Doudna JA. 2007. SRP RNA provides the physiologically essential GTPase activation function in cotranslational protein targeting. RNA 13:240–250. 10.1261/rna.135407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macao B, Luirink J, Samuelsson T. 1997. Ffh and FtsY in a Mycoplasma mycoides signal-recognition particle pathway: SRP RNA and M domain of Ffh are not required for stimulation of GTPase activity in vitro. Mol. Microbiol. 24:523–534. 10.1046/j.1365-2958.1997.3551729.x [DOI] [PubMed] [Google Scholar]
- 6.Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. 2000. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287:1232–1239. 10.1126/science.287.5456.1232 [DOI] [PubMed] [Google Scholar]
- 7.Keenan RJ, Freymann DM, Walter P, Stroud RM. 1998. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94:181–191. 10.1016/S0092-8674(00)81418-X [DOI] [PubMed] [Google Scholar]
- 8.Janda CY, Li J, Oubridge C, Hernandez H, Robinson CV, Nagai K. 2010. Recognition of a signal peptide by the signal recognition particle. Nature 465:507–510. 10.1038/nature08870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. 2004. Heterodimeric GTPase core of the SRP targeting complex. Science 303:373–377. 10.1126/science.1090827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM. 2004. Substrate twinning activates the signal recognition particle and its receptor. Nature 427:215–221. 10.1038/nature02250 [DOI] [PubMed] [Google Scholar]
- 11.Shan SO, Chandrasekar S, Walter P. 2007. Conformational changes in the GTPase modules of the signal reception particle and its receptor drive initiation of protein translocation. J. Cell Biol. 178:611–620. 10.1083/jcb.200702018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips GJ, Silhavy TJ. 1992. The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359:744–746. 10.1038/359744a0 [DOI] [PubMed] [Google Scholar]
- 13.Ribes V, Romisch K, Giner A, Dobberstein B, Tollervey D. 1990. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell 63:591–600. 10.1016/0092-8674(90)90454-M [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O'Reilly M, O'Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U. S. A. 100:4678–4683. 10.1073/pnas.0730515100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasona A, Crowley PJ, Levesque CM, Mair RW, Cvitkovitch DG, Bleiweis AS, Brady LJ. 2005. Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc. Natl. Acad. Sci. U. S. A. 102:17466–17471. 10.1073/pnas.0508778102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosch JW, Vega LA, Beyer JM, Lin A, Caparon MG. 2008. The signal recognition particle pathway is required for virulence in Streptococcus pyogenes. Infect. Immun. 76:2612–2619. 10.1128/IAI.00239-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funes S, Hasona A, Bauerschmitt H, Grubbauer C, Kauff F, Collins R, Crowley PJ, Palmer SR, Brady LJ, Herrmann JM. 2009. Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc. Natl. Acad. Sci. U. S. A. 106:6656–6661. 10.1073/pnas.0809951106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutierrez JA, Crowley P J, Cvitkovitch DG, Brady LJ, Hamilton IR, Hillman JD, Bleiweis AS. 1999. Streptococcus mutans ffh, a gene encoding a homologue of the 54 kDa subunit of the signal recognition particle, is involved in resistance to acid stress. Microbiology 145:357–366. 10.1099/13500872-145-2-357 [DOI] [PubMed] [Google Scholar]
- 19.Kremer BH, van der Kraan M, Crowley PJ, Hamilton IR, Brady LJ, Bleiweis AS. 2001. Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J. Bacteriol. 183:2543–2552. 10.1128/JB.183.8.2543-2552.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samuelsson T, Macao B, Bolske G. 1997. A 13-kDa protein with a helix-turn-helix motif is encoded by bacterial operons related to the SRP pathway. Biochem. Biophys. Res. Commun. 231:839–843. 10.1006/bbrc.1997.6199 [DOI] [PubMed] [Google Scholar]
- 21.Honda K, Nakamura K, Nishiguchi M, Yamane K. 1993. Cloning and characterization of a Bacillus subtilis gene encoding a homolog of the 54-kilodalton subunit of mammalian signal recognition particle and Escherichia coli Ffh. J. Bacteriol. 175:4885–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez JA, Crowley PJ, Brown DP, Hillman JD, Youngman P, Bleiweis AS. 1996. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 178:4166–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oganesyan V, Pufan R, DeGiovanni A, Yokota H, Kim R, Kim SH. 2004. Structure of the putative DNA-binding protein SP_1288 from Streptococcus pyogenes. Acta Crystallogr. D Biol. Crystallogr. 60:1266–1271. 10.1107/S0907444904009394 [DOI] [PubMed] [Google Scholar]
- 24.Zanen G, Antelmann H, Meima R, Jongbloed JD, Kolkman M, Hecker M, van Dijl JM, Quax WJ. 2006. Proteomic dissection of potential signal recognition particle dependence in protein secretion by Bacillus subtilis. Proteomics 6:3636–3648. 10.1002/pmic.200500560 [DOI] [PubMed] [Google Scholar]
- 25.Zhou M, Fives-Taylor P, Wu H. 2008. The utility of affinity-tags for detection of a streptococcal protein from a variety of streptococcal species. J. Microbiol. Methods 72:249–256. 10.1016/j.mimet.2007.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn SJ, Wen ZT, Burne RA. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 74:1631–1642. 10.1128/IAI.74.3.1631-1642.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abreu-Goodger C, Merino E. 2005. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 33:W690–W692. 10.1093/nar/gki445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. 2004. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 14:1861–1869. 10.1101/gr.2542904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis NE, Marty NJ, Kathir KM, Rajalingam D, Kight AD, Daily A, Kumar TK, Henry RL, Goforth RL. 2010. A dynamic cpSRP43-Albino3 interaction mediates translocase regulation of chloroplast signal recognition particle (cpSRP)-targeting components. J. Biol. Chem. 285:34220–34230. 10.1074/jbc.M110.160093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goforth RL, Peterson EC, Yuan J, Moore MJ, Kight AD, Lohse MB, Sakon J, Henry RL. 2004. Regulation of the GTPase cycle in post-translational signal recognition particle-based protein targeting involves cpSRP43. J. Biol. Chem. 279:43077–43084. 10.1074/jbc.M401600200 [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez-Romo P, Sanchez-Nieto S, Gavilanes-Ruiz M. 1992. A modified colorimetric method for the determination of orthophosphate in the presence of high ATP concentrations. Anal. Biochem. 200:235–238. 10.1016/0003-2697(92)90458-J [DOI] [PubMed] [Google Scholar]
- 32.Moye ZD, Zeng L, Burne RA. 2014. Modification of gene expression and virulence traits in Streptococcus mutans in response to carbohydrate availability. Appl. Environ. Microbiol. 80:972–985. 10.1128/AEM.03579-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palaniyandi K, Veerasamy M, Narayanan S. 2012. Characterization of Ffh of Mycobacterium tuberculosis and its interaction with 4.5S RNA. Microbiol. Res. 167:520–525. 10.1016/j.micres.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 34.Zheng F, Zook C, Campo L, Henault M, Watson H, Wang QM, Peng SB. 2002. Identification and characterization of Streptococcus pneumoniae Ffh, a homologue of SRP54 subunit of mammalian signal recognition particle. Biochem. Biophys. Res. Commun. 292:601–608. 10.1006/bbrc.2002.6694 [DOI] [PubMed] [Google Scholar]
- 35.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434–14439. 10.1073/pnas.172501299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ajdic D, Pham VTT. 2007. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J. Bacteriol. 189:5049–5059. 10.1128/JB.00338-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vadeboncoeur C, Pelletier M. 1997. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol. Rev. 19:187–207. 10.1111/j.1574-6976.1997.tb00297.x [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Rashid R, Wang K, Shan SO. 2010. Sequential checkpoints govern substrate selection during cotranslational protein targeting. Science 328:757–760. 10.1126/science.1186743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Schaffitzel C, Ban N, Shan SO. 2009. Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc. Natl. Acad. Sci. U. S. A. 106:1754–1759. 10.1073/pnas.0808573106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grudnik P, Bange G, Sinning I. 2009. Protein targeting by the signal recognition particle. Biol. Chem. 390:775–782. 10.1515/BC.2009.102 [DOI] [PubMed] [Google Scholar]
- 41.Saraogi I, Shan SO. 2011. Molecular mechanism of co-translational protein targeting by the signal recognition particle. Traffic 12:535–542. 10.1111/j.1600-0854.2011.01171.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akopian D, Dalal K, Shen K, Duong F, Shan SO. 2013. SecYEG activates GTPases to drive the completion of cotranslational protein targeting. J. Cell Biol. 200:397–405. 10.1083/jcb.201208045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peluso P, Herschlag D, Nock S, Freymann DM, Johnson AE, Walter P. 2000. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science 288:1640–1643. 10.1126/science.288.5471.1640 [DOI] [PubMed] [Google Scholar]
- 44.Peluso P, Shan SO, Nock S, Herschlag D, Walter P. 2001. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry 40:15224–15233. 10.1021/bi011639y [DOI] [PubMed] [Google Scholar]
- 45.Lam VQ, Akopian D, Rome M, Henningsen D, Shan SO. 2010. Lipid activation of the signal recognition particle receptor provides spatial coordination of protein targeting. J. Cell Biol. 190:623–635. 10.1083/jcb.201004129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bange G, Kummerer N, Grudnik P, Lindner R, Petzold G, Kressler D, Hurt E, Wild K, Sinning I. 2011. Structural basis for the molecular evolution of SRP-GTPase activation by protein. Nat. Struct. Mol. Biol. 18:1376–1380. 10.1038/nsmb.2141 [DOI] [PubMed] [Google Scholar]
- 47.Bange G, Sinning I. 2013. SIMIBI twins in protein targeting and localization. Nat. Struct. Mol. Biol. 20:776–780. 10.1038/nsmb.2605 [DOI] [PubMed] [Google Scholar]
- 48.Nakamura K, Yahagi S, Yamazaki T, Yamane K. 1999. Bacillus subtilis histone-like protein, HBsu, is an integral component of a SRP-like particle that can bind the Alu domain of small cytoplasmic RNA. J. Biol. Chem. 274:13569–13576. 10.1074/jbc.274.19.13569 [DOI] [PubMed] [Google Scholar]
- 49.Yamazaki T, Yahagi S, Nakamura K, Yamane K. 1999. Depletion of Bacillus subtilis histone-like protein, HBsu, causes defective protein translocation and induces upregulation of small cytoplasmic RNA. Biochem. Biophys. Res. Commun. 258:211–214. 10.1006/bbrc.1999.0615 [DOI] [PubMed] [Google Scholar]
- 50.Samuelsson T, Olsson M. 1993. GTPase activity of a bacterial SRP-like complex. Nucleic Acids Res. 21:847–853. 10.1093/nar/21.4.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller JD, Bernstein HD, Walter P. 1994. Interaction of E. coli Ffh/4.5S ribonucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature 367:657–659. 10.1038/367657a0 [DOI] [PubMed] [Google Scholar]
- 52.Braig D, Mircheva M, Sachelaru I, van der Sluis EO, Sturm L, Beckmann R, Koch HG. 2011. Signal sequence-independent SRP-SR complex formation at the membrane suggests an alternative targeting pathway within the SRP cycle. Mol. Biol. Cell 22:2309–2323. 10.1091/mbc.E11-02-0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bibi E. 2012. Is there a twist in the Escherichia coli signal recognition particle pathway? Trends Biochem. Sci. 37:1–6. 10.1016/j.tibs.2011.09.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.