Abstract
The Bacillus subtilis spoVAEa and spoVAF genes are expressed in developing spores as members of the spoVA operon, which encodes proteins essential for the uptake and release of dipicolinic acid (DPA) during spore formation and germination. SpoVAF is likely an integral inner spore membrane protein and exhibits sequence identity to A subunits of the spore's nutrient germinant receptors (GRs), while SpoVAEa is a soluble protein with no obvious signals to allow its passage across a membrane. However, like SpoVAD, SpoVAEa is present on the outer surface of the spore's inner membrane, as SpoVAEa was accessible to an external biotinylation agent in spores and SpoVAEa disappeared in parallel with SpoVAD during proteinase K treatment of germinated spores. SpoVAEa and SpoVAD were also distributed similarly in fractions of disrupted dormant spores. Unlike spoVAD, spoVAEa is absent from the genomes of some spore-forming members of the Bacillales and Clostridiales orders, although SpoVAEa's amino acid sequence is conserved in species containing spoVAEa. B. subtilis strains lacking SpoVAF or SpoVAEa and SpoVAF sporulated normally, and the spores had normal DPA levels. Spores lacking SpoVAF or SpoVAEa and SpoVAF also germinated normally with non-GR-dependent germinants but more slowly than wild-type spores with GR-dependent germinants, and this germination defect was complemented by ectopic expression of the missing proteins.
INTRODUCTION
The spoVA operon of Bacillus subtilis is expressed only in the developing spore during sporulation and encodes seven proteins, with the genes in the order spoVAA, -B, -C, -D, -Eb, -Ea, and -F. At least five of these SpoVA proteins, SpoVAA, -B, -C, -D, and -Eb, are necessary for normal B. subtilis spore formation (1, 2). These proteins are essential for the uptake of dipicolinic acid (DPA) into the dormant spore (3–7); in turn, DPA is essential for spore stability, as DPA-less B. subtilis spores germinate spontaneously after their release from the sporangium and during spore purification (8). At least some SpoVA proteins are also essential for DPA release in spore germination, and one SpoVA protein, SpoVAD, binds DPA specifically, with this binding essential for DPA uptake in sporulation (6). SpoVAD is a soluble protein and has no obvious signal peptide or membrane-spanning or anchor sequences. However, assessment of SpoVAD's location and accessibility in spores indicates that this protein is on the outer surface of the spore's inner membrane (IM) (4, 9, 10).
In contrast to SpoVAD, SpoVAA, -B, -C, -Eb, and -F appear to be integral membrane proteins on the basis of predictions from primary sequences and in some cases the localization of proteins expressed in growing bacteria (3, 11, 12). Although SpoVAF exhibits significant sequence identity to the A subunits of the spore's nutrient germinant receptors (GRs) (2), SpoVAA, -B, -C, -Eb, and -Ea exhibit no significant identity to known proteins. However, SpoVAEa is also predicted to be a soluble protein. Consequently, in this work, we have examined the location and accessibility of SpoVAEa in B. subtilis spores compared to those of SpoVAD; the number of SpoVAEa molecules in spores; the role of this protein, as well as SpoVAF, in DPA uptake in sporulation and DPA release during spore germination; and the presence of a spoVAEa gene in other spore-forming members of the orders Bacillales and Clostridiales.
MATERIALS AND METHODS
Preparation of purified SpoVAEa and generation and purification of antiserum.
The spoVAEa gene was amplified by PCR with genomic DNA from B. subtilis strain PS832, a prototrophic 168 strain derivative, as the template. The 5′ primer introduced a NotI site, and the 3′ primer introduced a KpnI site. Deletion of a predicted destabilizing loop corresponding to residues 124 to 135 was accomplished by overlap PCR (6), giving ΔspoVAEa. The resulting PCR products, ΔspoVAEa and the intact spoVAEa gene, were cloned into a modified pET15b vector containing a His6 tag and a tobacco etch virus (TEV) protease cleavage site (13). The ΔSpoVAEa protein (residues 2 [Δ124-135] to 203) and intact SpoVAEa were expressed in Escherichia coli BL21 Star(DE3) (Invitrogen, Grand Island, NY) initially by growth at 37°C in Luria broth (LB) (14) plus ampicillin (100 μg/ml) and then by induction with 1 mM isopropyl-β-d-thiogalactopyranoside at an optical density at 600 nm (OD600) of 0.8 and subsequent growth at 21°C for 16 h. Both SpoVAEa proteins were soluble and were purified by Ni2+-nitrilotriacetic acid affinity chromatography under native conditions, followed by TEV protease cleavage of the His6 tag and cation-exchange chromatography (SD200; GE Healthcare, Piscataway, NJ) (6, 13).
The purified ΔSpoVAEa protein was dialyzed against phosphate-buffered saline (PBS; 50 mM sodium phosphate, 150 mM NaCl, pH 7.2), adjusted to a concentration of 1 mg/ml in PBS, and supplied for polyclonal antibody production in rabbits (Pocono Rabbit Farm and Laboratory, Canadensis, PA). The antibody was detected in a bleed 2 months after its initial injection and was affinity purified with Pierce AminoLink Plus Immobilization kit (Pierce, Rockford, IL) in accordance with the manufacturer's instructions. Briefly, 1 mg of purified SpoVAEa protein was added to an AminoLink Plus Resin column (∼2 ml) in 0.1 M sodium citrate–0.05 M sodium carbonate coupling buffer (pH 10) and mixed by rocking at room temperature for 4 h, resulting in the formation of semistable Schiff base bonds. The column was washed with 4 ml of PBS, and reduction with sodium cyanoborohydride overnight at 4°C, followed by quenching with 1 M Tris-HCl (pH 7.4), was performed, resulting in stable secondary amine bonds. The antigen column was washed with 10 ml of 1 M NaCl and equilibrated in PBS, and then 2 ml of antiserum was added. After mixing by rocking for 1 h at room temperature, the column was washed with 8 ml of PBS. Bound antibody was eluted with 2 ml of 0.2 M glycine-HCl (pH 2.5) into a tube containing 100 μl of 1 M Tris-HCl (pH 8.9) to neutralize the glycine buffer. The purified antibody was finally dialyzed at 4°C against PBS and used for Western blot analysis.
B. subtilis strains used and spore preparation and germination.
The B. subtilis strains used in this work are listed in Table 1; they are isogenic derivatives of strain PS832, a laboratory 168 strain, including (i) PS533 (15), which contains plasmid pUB110, providing resistance to kanamycin (10 μg/ml); (ii) PS3411 (4) (termed ↑spoVA), in which the spoVA operon is under the control of the strong forespore-specific promoter of the sspB gene (PsspB) (PsspB-spoVA is turned on at the same time in sporulation as the normal spoVA operon but gives ∼5-fold higher levels of at least SpoVAD); and (iii) PS3406 (16), which lacks most of the spoVA operon and the gene encoding the spore cortex lytic enzyme SleB. PS3406 spores lack DPA, but because of their lack of SleB, these spores are stable and can be isolated.
TABLE 1.
Genotypes, spore phenotypes, and sources of strains used in this study
| B. subtilis strain | Genotype; spore phenotypea | Source or reference |
|---|---|---|
| PS533 | Wild type/pUB110 Kmr | 15 |
| PS832 | Wild-type 168 trp+ | Laboratory stock |
| PS3406 | spoVA sleB; no SpoVA proteins, stable DPA-less spores | 16 |
| PS3411 | PsspB-spoVA; elevated SpoVA protein levels | 4 |
| PS4348 | spoVAEa; Cmr, lacks SpoVAEa and likely SpoVAFa | This work |
| PS4351 | spoVAF; Cmr, lacks SpoVAF | This work |
| PS4361 | spoVAF; Tcr, lacks SpoVAF | This work |
| PS4362 | spoVAEa; Tcr, lacks SpoVAEa and likely SpoVAFa | This work |
| PS4363 | spoVAF amyE::PsspB-spoVAEa; has SpoVAEa but not SpoVAF | This work |
| PS4364 | spoVAEa amyE::PsspB-spoVAEa; has SpoVAEa but not SpoVAF | This work |
| PS4374 | spoVAEa amyE::PspoVA-spoVAEa; has SpoVAEa but not SpoVAF | This work |
| PS4381 | spoVAF amyE::PsspB-spoVAF; has SpoVAFa and SpoVAEa | This work |
| PS4383 | spoVAEa amyE::PsspB-spoVAF; has SpoVAFa but not SpoVAEa | This work |
| PS4390 | spoVAF amyE::PspoVA-spoVAF; has SpoVAFa | This work |
| PS4391 | spoVAEa amyE::PsspB-spoVAEa-F; has SpoVAFa and SpoVAEa | This work |
| PS4392 | spoVAF amyE::PsspB-spoVAEa-F; has SpoVAFa and SpoVAEa | This work |
The presence of SpoVAF is based only on the finding that the spoVAF gene is present in a construct that should yield SpoVAF synthesis; there is no direct proof that SpoVAF is indeed synthesized.
A B. subtilis strain with a deletion in the spoVAEa cistron was constructed as follows. A DNA fragment consisting of bp +2 to −246 relative to the spoVAEa translation start codon was PCR amplified from PS832 chromosomal DNA with primers that inserted BamHI and PstI restriction sites upstream and downstream, respectively, of the translation start site. After digestion with BamHI and PstI, the fragment was cloned between these sites in plasmid pKG57 (10), which is pBluescript II KS with a chloramphenicol resistance (Cmr; 5 μg/ml) cassette (cam), and the recombinant plasmid, pPVE1, was isolated in E. coli DH5α. A DNA fragment consisting of bp −272 to +122 relative to the translation stop codon in spoVAEa was PCR amplified with primers that inserted HindIII and ClaI sites upstream and downstream, respectively, of the translation stop codon. This fragment was digested with HindIII and ClaI and cloned between these sites in plasmid pPVE1, giving plasmid pPVE2 (spoVAEa::cam), which has fragments from either end of spoVAEa flanking the cam cassette. Plasmid pPVE2 was linearized by digestion with ClaI and used to transform B. subtilis strain PS832 to Cmr. PCR of one Cmr colony confirmed the expected chromosomal structure, and this strain was termed PS4348 (spoVAEa mutant).
A B. subtilis strain with a deletion of the spoVF gene was constructed by a two-step strategy that generated a derivative of plasmid pKG57 (10) termed pPVE4 in which upstream and downstream spoVAF fragments flanked a cam gene. In the first step, a DNA fragment consisting of bp +91 to −662 relative to the spoVAF translation start site was PCR amplified, the fragment was cut with BamHI and PstI (sites added in the upstream and downstream PCR primers, respectively), and the resultant fragment was inserted between the BamHI and PstI sites of plasmid pKG57 to create plasmid pPVE3. A PCR consisting of 275 bp upstream and 103 bp downstream of the spoVAF translation stop site was then amplified from PS832 genomic DNA, the fragment was cut with HindIII and ClaI (sites added in upstream and downstream PCR primers, respectively), and the resultant fragment was inserted between the BamHI and ClaI sites in plasmid pPVE3, giving plasmid pPVE4 (ΔspoVAF::cam). The individual steps in this plasmid's construction were confirmed by PCR and restriction enzyme digestion. Plasmid pPVE4 was then used to transform B. subtilis to Cmr, and proper integration of the spoVAF::cam fragment at the spoVAF chromosomal locus was confirmed by PCR, giving strain PS4351 (spoVAF).
B. subtilis strains carrying the spoVAEa or spoVAF genes at the amyE locus under the control of PsspB (4) were constructed as follows. The region from bp −490 to −1 relative to the translation start site of the B. subtilis sspB gene (this region has PsspB, as well as a strong ribosome-binding site [RBS]) was amplified from PS832 DNA with primers with an EcoRI site in the upstream primer and a region of overlap with the spoVAEa translation start site in the downstream primer. The upstream primer for spoVAEa amplification was complementary to the downstream primer for PsspB, and the downstream spoVAEa primer would amplify the complete spoVAEa coding region and a 3′ BamHI site. For overlap PCR, we amplified a product that has PsspB plus a good RBS just upstream of the spoVAEa coding region and between the EcoRI and BamHI sites, with the amplified PsspB promoter and spoVAEa fragments as the template. The overlap PCR product of the expected size was purified, digested with EcoRI and BamHI, and ligated to similarly cut plasmid pDG364 (17), and the recombinant plasmid (pPVE5) was isolated in E. coli DH5α. This plasmid was used to transform B. subtilis strain PS4362 (derived from strain PS4348 with the Cmr cassette changed to a Tcr cassette [18]) to a Cmr Tcr amylase-negative phenotype by a double-crossover event, giving strain PS4364, and the expected genomic structure in the amyE region of this strain was confirmed by PCR. The construction of a B. subtilis strain in which spoVAF expression was under PsspB control was similar to that for strain PS4264 described above, except that the overlap PCR product was flanked by BamHI sites. Plasmid pPVE7 with the spoVAF overlap PCR product was used to transform B. subtilis strain PS4361 (derived from strain PS4351 with the Cmr cassette changed to a Tcr cassette [18]) to a Cmr Tcr amylase-negative phenotype by a double-crossover event, giving strain PS4381, and the expected genomic structure in the amyE region of this strain was confirmed by PCR. Plasmids pPVE5 and pPVE7 were used to transform strains PS4361 and PS4362, respectively, giving strains PS4363 and PS4383.
The spoVAEa-spoVAF genes were also amplified together; the upstream primer for spoVAEa amplification was complementary to the downstream primer for PsspB (plus the strong RBS), and the downstream spoVAF primer would amplify the complete spoVAEa-spoVAF coding region plus a 3′-BamHI site, and the overlap PCR product was flanked by BamHI sites. Plasmid pPVE8 with the PsspB-spoVAEa-spoVAF overlap PCR product was used to transform B. subtilis strains PS4362 and PS4361, respectively, giving strains PS4391 and PS4392.
B. subtilis strains carrying the spoVAEa or spoVAF gene at the amyE locus under the control of the promoter for the spoVA operon (PspoVA) were constructed by a strategy identical to that described above. The region from bp −183 to −1 relative to the translation start site of the B. subtilis spoVAA gene (this is the first gene of the spoVA operon and includes PspoVA) was amplified from PS832 DNA, and this region was overlapped with either the spoVAEa or the spoVAF gene. The overlap amplicon PspoVA-spoVAEa or PspoVA-spoVAF was cloned into plasmid pDG364 (17), and the recombinant plasmid (pPVE6 or pPVE9, respectively) was isolated in E. coli DH5α. This plasmid was transformed into strain PS4362 or PS4361, respectively, giving strain PS4374 or PS4390.
Spores of all B. subtilis strains were prepared at 37°C with 2× Schaeffer's glucose (SG) medium either on agar plates or in liquid medium, and spores were purified as described previously (8, 19). The spores used in this work were free (>98%) of growing or sporulating cells, germinated spores, and cell debris as shown by phase-contrast microscopy.
Nutrient germination of spores was preceded by heat activation (75°C, 30 min) in water, followed by cooling on ice. Germination was in 10 mM l-valine–25 mM K-HEPES buffer (pH 7.4) at 37°C, 10 mM l-asparagine–10 mM d-glucose–10 mM d-fructose–10 mM KCl (AGFK) in 25 mM K-HEPES buffer (pH 7.4) at 37°C, a 1:1 mixture of 60 mM CaCl2 and 60 mM DPA (CaDPA) adjusted to pH 8 with Tris base at 23°C, or 1 mM dodecylamine–25 mM K-HEPES buffer (pH 7.4) at 45°C. In germinations with AGFK, dodecylamine, and l-valine, spores were at an initial OD600 of 0.5, and in CaDPA germination, spores were at an OD600 of 2. Germination was routinely assessed by the presence of 50 μM TbCl3 from the beginning of germination and continuously measuring the release of DPA from spores by its fluorescence with Tb3+ as described previously (l-valine) (20) and in some experiments by (i) adding TbCl3 to 50 μM to aliquots of germination incubations taken at various times and measuring Tb-DPA fluorescence (AGFK and dodecylamine) or (ii) phase microscopic analysis of ∼100 spores either directly or in aliquots of CaDPA-germinating cultures taken at various times and made 0.1 M in EDTA to halt germination.
High-pressure (HP) germination of spores was carried out differently from the germination methods described above and was done essentially as described previously (21), with 1-ml samples of spores at an OD600 of 1 at either 37°C and 150 MPa or 50°C and 550 MPa. Levels of spore germination in samples treated at HP for various hold times were assessed by first centrifuging the treated samples, suspending the pelleted spores in ∼20 μl water, and examining ∼100 spores by phase-contrast microscopy to determine the percentages of spores that had become phase dark and thus had germinated. The spores of the two strains whose HP germination was to be compared were prepared, purified, and HP treated at the same time.
Analysis of the germination of multiple individual spores by differential interference contrast (DIC) microscopy was done as described previously (22, 23). Briefly, heat-activated spores (1 μl; ∼108 spores/ml of water) were spread on the surface of a microscope coverslip that was then dried in a vacuum desiccator for 5 to 10 min. Coverslips were then mounted on and sealed to a microscope sample holder kept at 37°C. The DIC microscope was set such that the polarizer and analyzer were crossed and thus the DIC bias phase was zero. After the addition of preheated (37°C) germinant-buffer solution (10 mM l-valine in 25 mM K-HEPES buffer, pH 7.4) to spores on the coverslips, a digital charge-coupled device camera (16 bit, 1,600 by 1,200 pixels) was used to record the DIC images at a rate of 1 frame/15 s for 60 to 120 min. The images obtained were analyzed with a computation program in Matlab to locate each spore's position and to calculate the average pixel intensity of an area of 20 by 20 pixels that covered the whole individual spore in the DIC image. The DIC image intensity of each individual spore was plotted as a function of the incubation time (with a resolution of 15 s), and the initial intensity at T0 (the first DIC image recorded after the addition of the germinant) was normalized to 1 and the intensity at the end of measurements was normalized to 0. These data allowed the determination of kinetic parameters of the germination of individual spores defined previously (22, 23), including the following: (i) Tlag, the time between germinant addition and initiation of fast CaDPA release; (ii) Trelease, the time of completion of fast CaDPA release; (iii) ΔTrelease, equal to Trelease − Tlag; (iv) Tlys, the time when a spore's DIC image intensity stops decreasing and when spore cortex hydrolysis and core swelling are complete; (v) ΔTlys, equal to Tlys minus Trelease; (vi) Ilag, the relative DIC image intensity at Tlag; (vii) Irelease, the relative DIC image intensity at Trelease.
Analysis of DPA accumulation during the sporulation of various strains was carried out in liquid 2× SG medium at 37°C. Cultures were inoculated to an OD600 of 0.1 from an overnight culture (the inoculation time was defined as time zero), and at various times, 1-ml aliquots were taken, centrifuged in a microcentrifuge, washed twice with 1 ml of water, suspended in 1 ml of water, and boiled for 30 min to extract DPA. The samples were centrifuged after cooling on ice for >15 min, and the supernatant fluid was saved. The DPA in these supernatant fluids was assayed by its fluorescence with Tb3+ as described previously (20).
For analysis of the biotinylation or proteinase K sensitivity of germinated spores, heat-activated spores were germinated with l-valine as described above but without TbCl3 and at an OD600 of 2. After ∼60 min of incubation when germination was >90% complete as determined by phase-contrast microscopy, the spores were harvested by centrifugation and suspended in a small volume, and the remaining dormant spores were removed by centrifugation through 50% HistoDenz as described previously (9).
Analytical procedures.
Dormant spores were decoated to remove some protein from the spore's outer coat, as well as the outer membrane; the cortex layer surrounding the spore's IM and the core was disrupted with lysozyme, followed by mild sonication with glass beads; the lysate obtained was centrifuged at low speed to isolate the integument (I) fraction; and the supernatant fluid was centrifuged at high speed to give the IM (M) fraction and a soluble (S) fraction, all as described previously (24–26). In some cases, the sonicated, lysozyme-ruptured spores (defined as the lysate) were made 1% in SDS and 55 mM in dithiothreitol, and the mixture was stored frozen. Germinated spores were extracted similarly but without prior decoating.
SpoVAD and SpoVAEa, as well as GR subunits and the GerD protein, were detected in the lysate or various fractions of disrupted spores described above by Western blotting with specific antiserum to either SpoVAEa prepared as described above or GerD, SpoVAD, or GR subunits (4, 25, 26). The levels of these proteins in spores were determined by Western blot analyses of spore fractions or lysates, and comparisons of the intensities of protein bands with those of known amounts of the purified protein antigens with ImageJ were all done as described previously (4, 25, 26). The concentrations of the antigens were determined by measuring the OD280 of these proteins and their predicted molar extinction coefficients at this wavelength. In a number of experiments, different amounts of protein from some spore lysates were run on the same Western blot to facilitate the quantitation of specific antigen levels in lysates from spores of different strains.
Assessment of the accessibility of SpoVAD and SpoVAEa in chemically decoated dormant spores and intact germinated spores (isolated as described above) to the biotinylation agent sulfo-N-hydroxysuccinimide-SS-biotin (EZ-Link Sulfo-NHS-SS-Biotin reagent; Pierce) was assayed in both M fractions and lysates as described previously (9). Treatment of intact germinated spores with proteinase K (Sigma Chemical Co., St. Louis, MO), stopping of proteinase K digestion by the addition of the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF), and subsequent Western blot analysis of SpoVAD and SpoVAEa in M fractions or lysates were all done as described previously (9).
RESULTS
Characteristics and distribution of SpoVAEa in spore formers.
The spoVA operon of B. subtilis was originally annotated as containing six genes, spoVAA-spoVAF. However, there was a sequencing error in the spoVAE gene in earlier work, and B. subtilis spoVAE actually comprises two genes now annotated as spoVAEa and spoVAEb. The gene order in the spoVAE region is spoVAD-spoVAEb-spoVAEa-spoVAF. While spoVAEa is present in many members of the order Bacillales, it is less prevalent in those of the order Clostridiales, although the amino acid sequence of SpoVAEa is well conserved in those species that have a spoVAEa gene, with 42% of all residues conserved ≥86% in the 36 species examined (see Fig. S1A and Table S1 in the supplemental material; data not shown). However, the SpoVAEa sequence shows no obvious similarity to other proteins in available databases. Notably, a spoVAF gene is routinely associated with a spoVAEa gene only in Bacillus species and is found only infrequently with a spoVAEa gene in genera such as Alicyclobacillus, Amphibacillus, Brevibacillus, Geobacillus, Lysinibacillus, and others, as well as members of the order Clostridiales (7; data not shown). In contrast, the gene upstream of spoVAEa, spoVAEb, is found in the genomes of all of the spore-forming members of the orders Bacillales and Clostridiales (7), and the sequence of the SpoVAEb protein is also well conserved, with ∼35% of all residues conserved ≥86% in the 26 species examined (see Fig. S1B and Table S2 in the supplemental material).
Like many of the spoVA-encoded proteins, SpoVAEb is predicted to be relatively hydrophobic and is further predicted to have four membrane-spanning domains by multiple prediction programs (data not shown). In contrast, the SpoVAEa protein is predicted to be relatively hydrophilic and its sequence contains no obvious predicted membrane-spanning domains (data not shown). In the latter regard, SpoVAEa is similar to the SpoVAD protein, which also lacks any obvious membrane-spanning or attachment signals, even though SpoVAD has been shown to associate with the spore's IM (4). SpoVAD is also a soluble globular protein and has recently been shown to be on the outer surface of the spore's IM (6, 9). Given that SpoVAEa is a soluble protein, it was thus of interest to determine if this protein is also associated with the spore's IM and, if so, whether the protein is on the outer or the inner surface of this membrane.
Detection, location, and quantitation of SpoVAEa in spores.
In order to detect SpoVAEa in spore extracts, the B. subtilis ΔspoVAEa gene was cloned and the ΔSpoVAEa protein, which lacks amino acids 124 to 135 of the SpoVAEa protein (see Materials and Methods), was overexpressed in E. coli. The protein was soluble and was purified and used to immunize rabbits to generate a polyclonal antiserum against SpoVAEa. The antiserum was then purified and used in a Western blot analysis of various fractions of dormant wild-type and ↑spoVA and spoVA sleB mutant spores (Fig. 1). A band corresponding to the expected size of SpoVAEa was not detected in the material removed from wild-type spores by the decoating procedure used prior to spore lysis and subsequent fractionation (data not shown), as also found previously for SpoVAD (4). However, a band of ∼22 kDa, the expected size of SpoVAEa, was detected in wild-type spore fractions, but this band was absent from spoVA mutant (strain PS3406) spore fractions. The intensity of the putative SpoVAEa band was also significantly higher in the soluble (S) and integument (I) fractions of spores of strain PS3411, in which the spoVA operon is overexpressed ∼5-fold from the strong forespore-specific PsspB promoter (4). SpoVAD was also absent from spoVA sleB mutant spores, and its level was elevated in ↑spoVA mutant spores, as expected (4). Thus, the band detected by the anti-SpoVAEa serum was almost certainly SpoVAEa.
FIG 1.
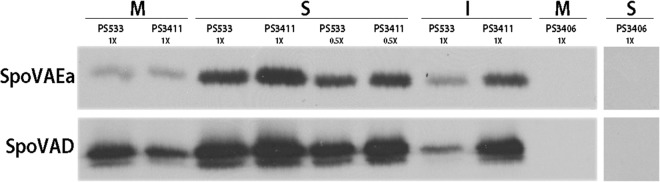
Detection of SpoVAEa and SpoVAD in spores and their distribution in different spore fractions. Spores of B. subtilis strains PS533 (wild type), PS3411 (↑spoVA mutant), and PS3406 (sleB spoVA mutant) were purified and decoated, removing some spore coat protein, as well as the spore outer membrane. The spores were then disrupted and fractionated as described in Materials and Methods, giving the spore IM fraction (M), the soluble fraction (S) consisting of soluble proteins from the spore core, and the integument fraction (I) consisting of insoluble coat protein, partially degraded spore cortex, and IM that is not sheared off the I fraction (9, 25–27). Aliquots of the various fractions from equal numbers of spores (∼108) were then subjected to Western blot analysis with antiserum against SpoVAEa and SpoVAD. All samples were run on the same Western blot, except the S fraction from PS3406 spores. The purified SpoVAD protein migrated identically to the SpoVAD antigen detected in spores, while the SpoVAEa antigen detected in spores ran more slowly than the His-tagged full-length SpoVAEa protein used as a marker (data not shown).
While we expected that most SpoVAEa protein would be associated with the spore's outer layer, the coat, cortex, outer membrane, or IM on the basis of previous localization experiments with intact spores (10), most was actually found in the S fraction, which previous work has shown is derived largely from the spore core (9, 25, 26), with slightly less in the insoluble I fraction that pellets upon low-speed centrifugation of decoated spore lysates (Fig. 1). The I fraction contains the large amount of coat protein not removed from spores by decoating and fragments of spore cortex peptidoglycan, as well as much of the spore's IM that is not sheared off the spore cortex during fractionation (25–27). Similar results were obtained when the same Western blot was probed for SpoVAD (Fig. 1). These results were not unexpected on the basis of recent work that also found much SpoVAD in the spore's soluble fraction and further that much spore IM and integral IM proteins partition with the I fraction (27). Presumably, the hydrophilic SpoVAD and SpoVAEa proteins are only loosely bound to the IM in intact spores and readily dissociate in spore lysates. Analysis of the levels of SpoVAD and SpoVAEa in the various spore fractions gave a value of ∼6,000 molecules of SpoVAD per spore, similar to the value determined recently (27), while the level of SpoVAEa was lower, at ∼750 molecules per spore (Fig. 2).
FIG 2.
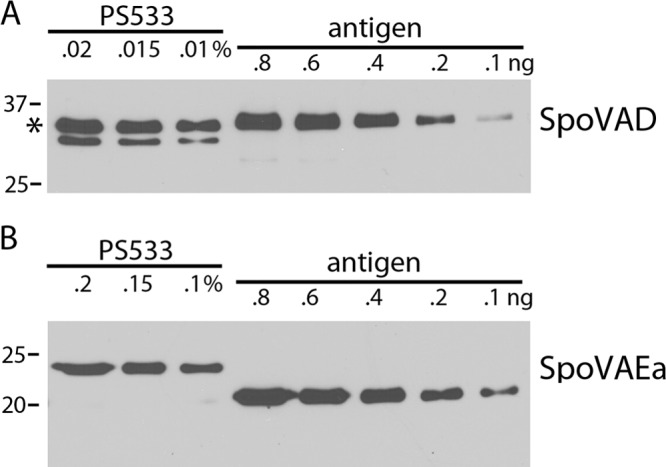
(A, B) Levels of SpoVAD (A) and SpoVAEa (B) in wild-type spores. Various amounts of the total lysate from 8.2 × 108 spores of strain PS533 (wild type) were subjected to Western blot analysis as described in Materials and Methods alongside various amounts of purified SpoVAD or SpoVAEa protein. (A) The intensities of the SpoVAD band in various amounts of the lysate (upper band at 36 kDa [asterisk]) were compared to the intensities of known amounts of purified SpoVAD. (B) The intensities of the SpoVAEa band at 23 kDa in various amounts of lysate were compared to the intensities of various amounts of purified ΔSpoVAEa that ran at ∼20 kDa. The molecular mass of the purified ΔSpoVAEa protein was lower than that of the SpoVAEa protein in the lysate, probably because the purified ΔSpoVAEa protein had an internal deletion, as described in Materials and Methods. Analysis of the various band intensities by ImageJ gave values of 5,800 molecules of SpoVAD/spore and 740 molecules of SpoVAEa/spore. The numbers of SpoVAD and SpoVAEa molecules per spore determined in three independent Western blot analyses were all within ±20% of these values. The values to the left of both panels are molecular sizes in kilodaltons.
Topology of SpoVAEa in spores.
Previous work has shown that SpoVAD is largely, if not completely, on the outer surface of the intact decoated spore's IM, as in germinated spores this protein is accessible to a biotinylation agent, as well as to proteinase K, while a soluble spore core protein is not (9). Analysis of the labeling of the SpoVAEa and SpoVAD proteins by this same biotinylation agent found that the biotinylation of both proteins in dormant and germinated wild-type spores was quite similar, as ∼40% of both proteins was biotinylated in dormant spores, with ∼65% of both proteins biotinylated in germinated spores (Fig. 3A and B). Similar results were obtained when proteins obtained in the M fraction (Fig. 3A and B) or the spore lysate were examined (data not shown). SpoVAE and SpoVAD were also both sensitive to proteinase K in intact germinated spores when proteins in either the M fraction (Fig. 3C) or the spore lysate were examined (data not shown). Importantly, the conditions used for the biotinylation of dormant and germinated spores and the proteinase K treatment of germinated spores have been shown not to biotinylate or digest a soluble spore core protein, in particular, one that is very proteinase K sensitive in spore extracts (9; data not shown). These results together indicate that, like SpoVAD, SpoVAEa is also on the outer surface of the spore's IM.
FIG 3.
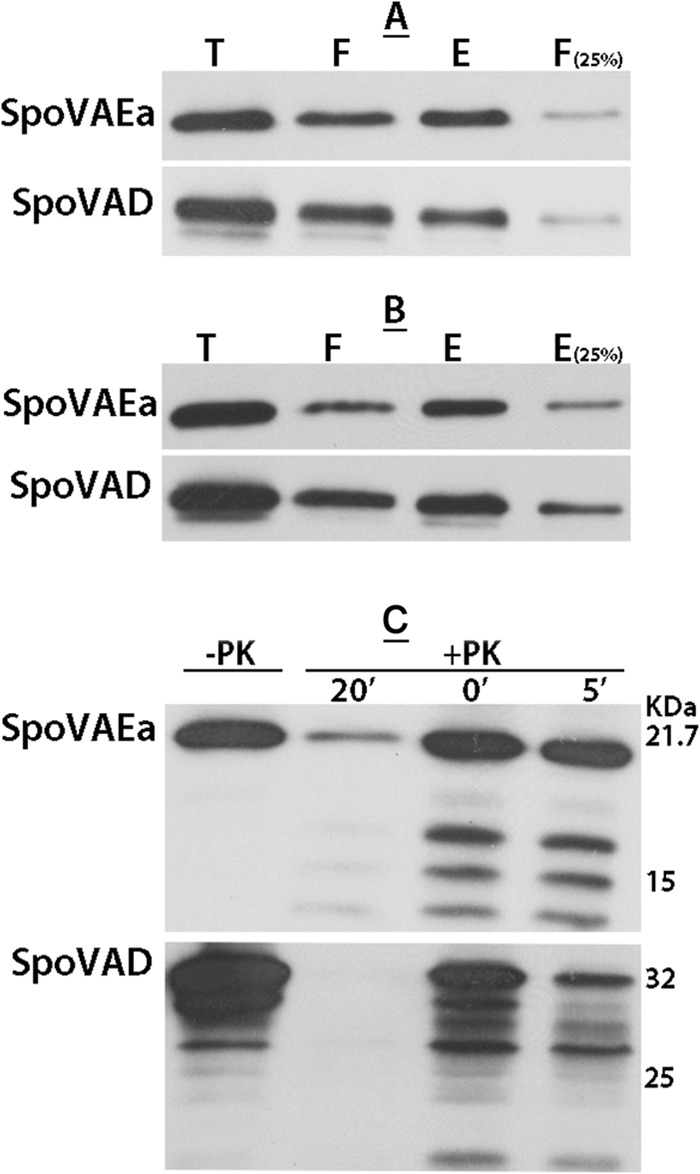
(A to C) Levels of biotinylation of SpoVAEa and SpoVAD in dormant (A) and germinated (B) spores and proteinase K susceptibility of SpoVAEa and SpoVAD in germinated spores (C). Decoated dormant (A) or intact germinated (B) spores of B. subtilis strain PS533 (wild type) were biotinylated, the biotinylation reagent was quenched, spores were lysed, IM fractions were isolated, and proteins were solubilized, giving the total (T) fraction. These proteins were adsorbed to NeutrAvidin beads (Pierce); proteins that did not absorb were termed the flowthrough (F) fraction, and proteins eluted with dithiothreitol were termed the eluted (E) fraction. Aliquots of these three fractions from the same amount of spores, as well as 25% of the amounts of the F fraction (A) and the E fraction (B) were subjected to Western blot analysis with either anti-SpoVAEa or anti-SpoVAD serum. ImageJ analysis of the intensities of the bands showed that the levels of biotinylation of SpoVAEa in dormant and germinated spores were 40 and 65%, respectively, while the levels of SpoVAD biotinylation in dormant and germinated spores were 36 and 65%, respectively. (C) Germinated B. subtilis PS533 (wild-type) spores were treated with proteinase K (25 μg/ml) for various times, and proteinase K was inactivated as described in Materials and Methods immediately after addition or after 5 or 20 min of digestion at 25°C. IMs were then isolated, and equal aliquots of the IM fraction were subjected to Western blot analysis with antiserum against SpoVAEa or SpoVAD. Note that some SpoVAD and SpoVAEa was digested with proteinase K even if PMSF was added immediately after the protease was added, presumably because proteinase K inactivation was not instantaneous.
Lack of essential function of SpoVAEa and SpoVAF in sporulation.
The fact that SpoVAEa is not encoded in the genomes of many spore-forming members of the orders Bacillales and Clostridiales suggests that this protein may not be essential for DPA movement in B. subtilis spore formation and germination, as is also reported to be the case for SpoVAF (2). To test SpoVAEa's function in spore formation in light of evidence that SpoVA proteins are important in DPA uptake by the developing spore, a spoVAEa deletion replacement mutation was integrated into the wild-type B. subtilis chromosome, giving strain PS4348. Analysis of the sporulation of this strain indicated that the strain sporulated normally and accumulated DPA levels in the resultant spores that were within ∼5% of those in the parental wild-type spores and that the rate of DPA accumulation by developing forespores during sporulation of the spoVAEa strain was quite similar to that of the wild-type strain (Fig. 4; data not shown). Spores of the spoVAEa mutant strain lacked SpoVAEa, as expected, but had normal levels of SpoVAD (Fig. 5A). Levels of GerD and the GR subunits GerAA, GerAC, GerBC, and GerKA were also normal in spoVAEa mutant spores when spore lysates were examined by Western blot analysis with various specific antisera (data not shown). Since the mutation generating the spoVAEa strain would be expected to eliminate the expression of spoVAF, we also constructed a spoVAF strain. The latter strain had normal SpoVAD and SpoVAEa levels, as expected (Fig. 5B), and also accumulated DPA during sporulation relatively similarly to the wild-type strain and to essentially wild-type levels in spores (Fig. 4; data not shown).
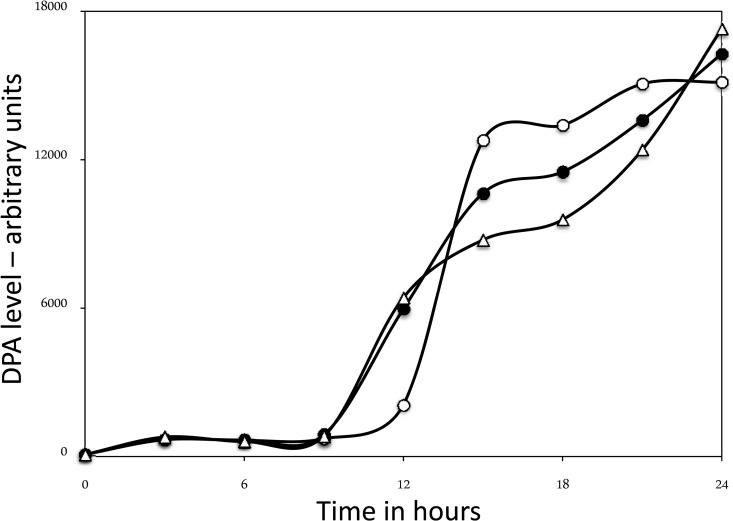
FIG 4.
DPA accumulation during sporulation of wild-type and spoVAEa and spoVAF mutant spores. Spores of strains PS533 (wild type), PS4348 (spoVAEa mutant), and PS4351 (spoVAF mutant) were sporulated in liquid 2× SG medium and 37°C. At various times, samples were taken, washed, and extracted and DPA was analyzed in extracts as described in Materials and Methods. Time zero is the time when cultures were inoculated to an OD600 of ∼0.1. Symbols: ○, wild type; ●, spoVAEa mutant; △, spoVAF mutant.
FIG 5.
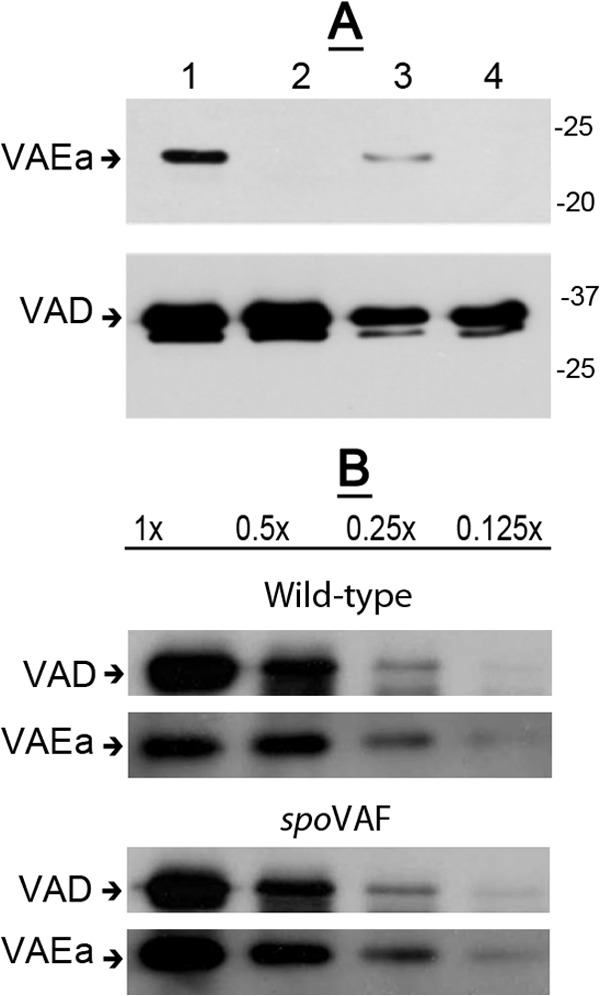
(A, B) Levels of SpoVAD and SpoVAEa in lysates of wild-type and spoVAEa and spoVAF mutant spores. Spores of B. subtilis strains PS533 (wild type) (A, B), PS4348 (spoVAEa mutant) (A), and PS4351 (spoVAF mutant) (B) were decoated, and lysates were prepared as described in Materials and Methods. Various amounts of protein in the lysates from the spores of the two strains (∼5 × 107 spores for analysis of SpoVAEa or ∼5 × 106 spores [each defined as 1×] for analysis of SpoVAD) were analyzed by Western blotting with antiserum against either SpoVAEa or SpoVAD as described in Materials and Methods. The lanes in panel A are as follows: 1, 1× protein from wild-type spores; 2, 1× protein from spoVAEa mutant spores; 3, 0.25× protein from wild-type spores; 4, 0.25× protein from spoVAEa mutant spores. The arrows labeled VAEa and VAD on the left of the panels denote the migration positions of SpoVAEa and SpoVAD, respectively, and the values to the right of panel A are the masses of molecular size marker proteins in kilodaltons.
Germination of spoVAEa and spoVAF mutant spore populations.
While the sporulation, in particular the DPA accumulation, of strains lacking SpoVAEa and/or SpoVAF was normal, there is evidence that SpoVA proteins are also involved in DPA release during spore germination (5). Consequently, it was of interest to examine the germination of spoVAEa and spoVAF mutant spores. Germination of B. subtilis spores can be triggered either by a nutrient germinant such as l-valine or the AGFK mixture via GRs located in the spore's IM or by compounds that do not act via GRs, including CaDPA and dodecylamine (1, 3). The spoVAEa and spoVAF mutant spores germinated essentially identically to wild-type spores with the non-GR-dependent germinants CaDPA and dodecylamine (Fig. 6A and B; data not shown). However, the germination of the spoVAEa and spoVAF mutant spores with the GR-dependent nutrient germinant l-valine was slower than that of wild-type spores (Fig. 6C), with the spoVAEa mutant spores exhibiting the slowest germination. The spoVAEa mutant spores also exhibited slower germination than wild-type spores with AGFK, although spoVAF mutant spores did not (Fig. 6D). These mutant spore germination phenotypes were complemented by the ectopic expression of spoVEAa and spoVAF (see below). The slower valine germination of the spoVAEa and spoVAF mutant spores was also seen when germination was measured solely by phase-contrast microscopy (data not shown). In the latter experiment, it took ∼1 h for wild-type spore germination with l-valine to reach ∼95% completion, while spoVAEa and spoVAF mutant spores took 3 to 4 h to reach ∼95% completion of l-valine germination (data not shown). This overall germination behavior was seen with several independent preparations of spoVAEa and spoVAF mutant spores, and the dependence of the rate of spoVAEa mutant spore germination on the l-valine concentration was essentially identical to that of wild-type spores (data not shown).
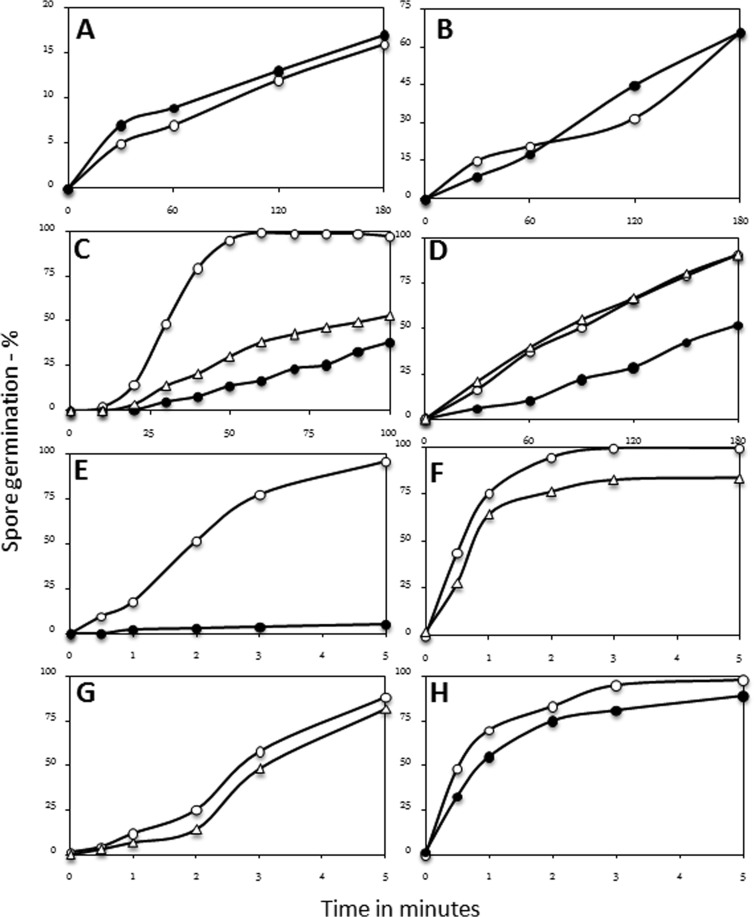
FIG 6.
(A to H) Germination of wild-type and spoVAEa and spoVAF mutant spores with various agents. Wild-type (PS533) and spoVAEa (PS4348) and spoVAF (PS4351) mutant spores were germinated with CaDPA (A), dodecylamine (B), l-valine (C), AGFK (D), HP at 150 MPa (E, G), or HP at 550 MPa (F, H), and spore germination was assessed as described in Materials and Methods. Symbols: ○, wild-type spores; ●, spoVAEa mutant spores; △, spoVAF mutant spores.
The slower germination of spoVAEa mutant spores than wild-type spores with GR-dependent germinants was also seen when germination was triggered by 150 MPa of HP, which also acts via GRs (1, 3), although the germination of the spoVAF mutant spores was only slightly slower than that of wild-type spores (Fig. 6E and G). However, with a HP of 550 MPa, which acts primarily on the SpoVA protein channel and largely in a GR-independent manner (1, 3), the germination of the spores of the two mutant strains was only slightly slower than that of wild-type spores (Fig. 6F and H).
Germination of individual spoVAEa mutant spores.
It was clear from the results described above that SpoVAEa, SpoVAF, or both proteins are important in normal GR-dependent spore germination. However, these experiments measured the germination of spore populations and additional information can be gleaned about the germination process by examining the germination of individual spores (22, 23). In particular, a variety of kinetic germination parameters can be obtained by monitoring the germination of individual spores. Consequently, we compared the l-valine germination of large numbers of wild-type and spoVAEa mutant spores (Fig. 7A to C; Table 2). As expected from analyses of the l-valine germination of spore populations, the germination of individual spoVAEa mutant spores was also slower than that of wild-type spores (Fig. 7A) and a significant percentage of the spoVAEa mutant spores did not germinate in the observation period. Examination of the DIC image intensity curves of large numbers of individual germinating wild-type and spoVAEa mutant spores further allowed the determination of the average values of the germination kinetic parameters Tlag, Trelease, ΔTrelease, and ΔTlys, which were defined in Materials and Methods (Fig. 7B and C; Table 2). Notably, the average Tlag, the time prior to initiation of rapid CaDPA release, of spoVAEa mutant spores was >2-fold longer than that of wild-type spores, and the difference is actually larger than 2-fold, as while 97% of wild-type spores germinated during the observation period, only 74% of the spoVAEa mutant spores did so and these spores thus have Tlag values of >90 min. The spoVAEa mutation also had effects on the average ΔTrelease and ΔTlys values, which are the times for fast CaDPA release and for cortex lysis plus core expansion, respectively (Table 2). However, these effects were relatively small, certainly smaller than the >2-fold effects on the average Tlag, and the effects on the ΔTrelease and ΔTlys values are probably not significant.
FIG 7.
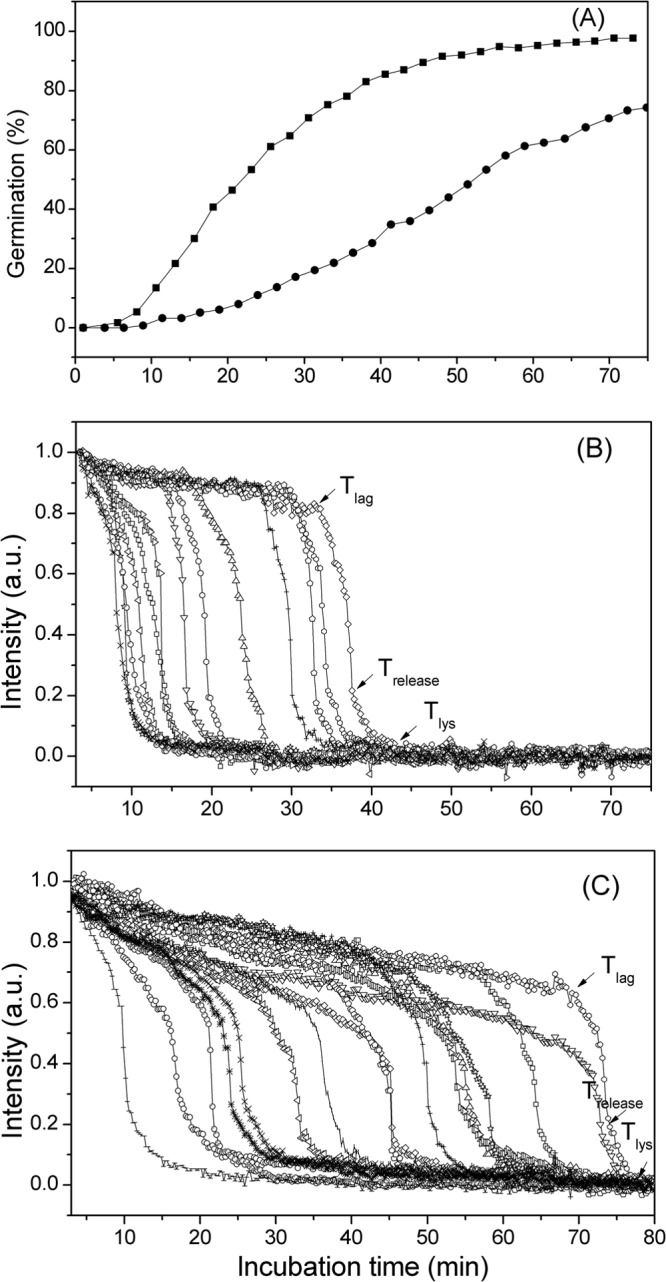
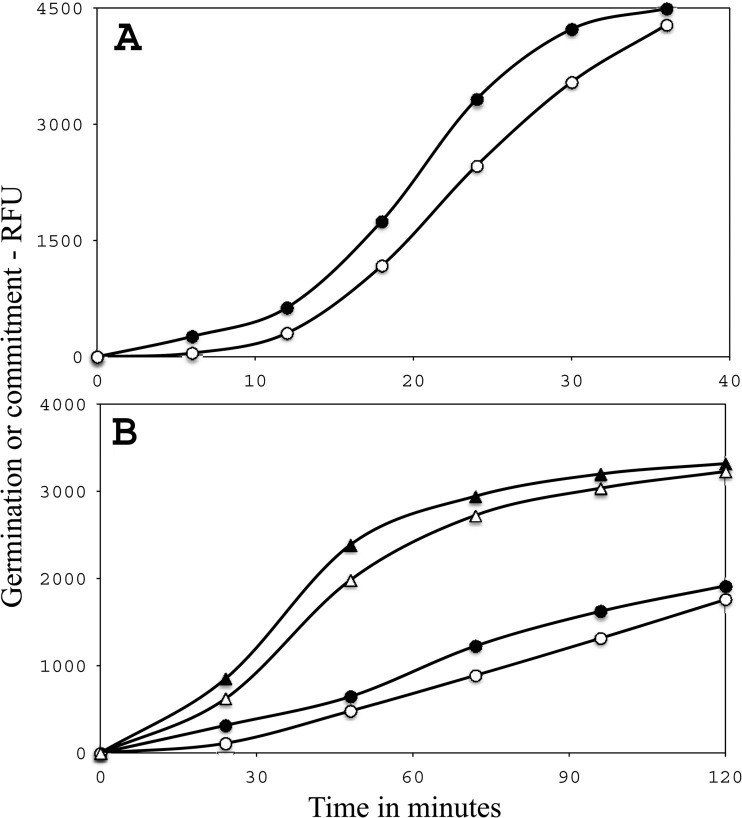
(A to C) Analysis of the l-valine germination of individual wild-type and spoVAEa mutant B. subtilis spores. Heat-inactivated spores of strains PS533 (wild type) and PS4348 (spoVAEa mutant) were germinated with l-valine, the germination of individual spores was monitored by DIC microscopy, and various germination parameters were determined as described in Materials and Methods and illustrated in panels B and C. (A) The percent germination of >300 individual wild-type (■) and spoVAEa mutant (●) spores was monitored for 75 to 80 min. (B, C) The DIC image intensities of 13 to 16 randomly chosen wild-type (B) or spoVAEa mutant (C) spores were monitored for 75 to 80 min. The Tlag, Trelease, and Tlys values of one selected spore are shown in panels B and C. a.u., arbitrary units.
TABLE 2.
Kinetic parameters of the l-valine germination of wild-type and spoVAEa mutant sporesa
| Spores | Tlag (min) | Trelease (min) | ΔTrelease (min) | ΔTlys (min) | Ilag | Irelease |
|---|---|---|---|---|---|---|
| PS533 (wild type) | 21 ± 14b | 25 ± 14 | 3.8 ± 1.2 | 7 ± 4 | 0.81 ± 0.08 | 0.21 ± 0.07 |
| PS4348 (spoVAEa) | 45 ± 18c | 49 ± 19 | 4.9 ± 1.8 | 10 ± 6 | 0.60 ± 0.09 | 0.16 ± 0.07 |
The germination of individual spores with l-valine was analyzed as described in Materials and Methods, and kinetic parameter values were determined as described previously and as indicated in Fig. 7B and C.
Spores were observed for 90 min, and the data are average values and standard deviations from analyses of ≥80 spores that germinated.
This value is a significant underestimate of the Tlag value for the entire spoVAEa mutant spore population, since only 74% of the spores examined germinated during the 90-min observation period while 97% of the wild-type spores did so.
The analyses of multiple individual spores also revealed that because of their longer average Tlag, the spoVAEa mutant spores exhibited a period of a slow decrease in DIC image intensity prior to Tlag that was much longer than that of wild-type spores. As a consequence, the spoVAEa mutant spore Ilag value, the average relative DIC image intensity at Tlag, was much lower than that of wild-type spores, although the average DIC image intensities at Trelease, the Irelease values, were relatively similar for wild-type and spoVAEa mutant spores. However, the precise meaning of the slow decrease in DIC image intensity prior to Tlag for either wild-type or spoVAEa mutant spores is not clear (see Discussion).
Commitment of spoVAEa and spoVAF mutant spores to germination.
The analysis of the germination of multiple individual wild-type and spoVAEa mutant spores noted above indicated that the defect in the mutant spores was their much longer average Tlag values. One event that is known to take place in Tlag is the process of commitment, whereby a spore is committed to continue through germination even if a nutrient germinant is removed (20). Therefore, we examined the kinetics of commitment in l-valine germination of wild-type and spoVAEa and spoVAF mutant spores (Fig. 8A and B). As found previously (20), commitment took place 5 to 15 min before CaDPA release with wild-type spores. However, the average times between commitment and germination were 2- to 3-fold longer for spoVAEa and spoVAF mutant spores, respectively. Given the much slower germination of these two mutant spore types, it thus appears that the defect in these mutant spores is primarily in the period prior to commitment, especially since the length of time between commitment and CaDPA release is larger when germination is slower (20).
FIG 8.
(A, B) Commitment to germination of spores of various strains. Spores of strain PS533 (wild type) (A) and strains PS4348 (spoVAEa mutant) (○, ●) and PS4351 (spoVAF mutant) (△, ▲) (B) were germinated with l-valine, and both commitment (●, ▲) and CaDPA release (○, △) were measured as described in Materials and Methods. Measurement of the times between commitment and CaDPA release for wild-type spores at 6, 12, 28, 24, and 30 min (A) and spoVAEa and spoVAF mutant spores at 24, 48, 72, and 96 min (B) gave average values of ∼5, ∼10, and ∼14 min, respectively. RFU, relative fluorescence units.
Complementation of the germination defect of spoVAEa and spoVAF mutant spores.
The results described above suggested that perhaps both SpoVAEa and SpoVAF are essential for normal DPA release during spore germination by nutrients. To further examine this possibility, we integrated copies of spoVAEa, spoVAF, or both genes under the control of either the spoVA promoter or the very strong forespore-specific PsspB promoter at the amyE locus of the spoVAEa and spoVAF mutant strains and examined the l-valine germination and SpoVAEa levels of spores of the resultant strains (Fig. 9; data not shown). The results showed that ectopic expression of spoVAEa plus spoVAF under PsspB control restored relatively normal germination and ∼5-fold higher-than-normal SpoVAEa levels in spoVAEa or spoVAF mutant spores, while expression of either spoVAEa or spoVAF alone did not (data not shown). Surprisingly, ectopic expression of spoVAEa alone from PsspB gave levels of SpoVAEa in spores that were ∼10-fold higher than when spoVAEa and spoVAF were expressed together from PsspB (data not shown). Ectopic expression of spoVAEa and spoVAF from PspoVA in the spoVAEa mutant strain gave no complementation of the spore germination defect, and Western blot analysis showed that the levels of SpoVAEa in these spores were <5% of the levels in wild-type spore (data not shown). Unfortunately, we have no antiserum against SpoVAF, so the levels of this protein in spores of the strains complemented by spoVAF expression alone or with spoVAEa are not known.
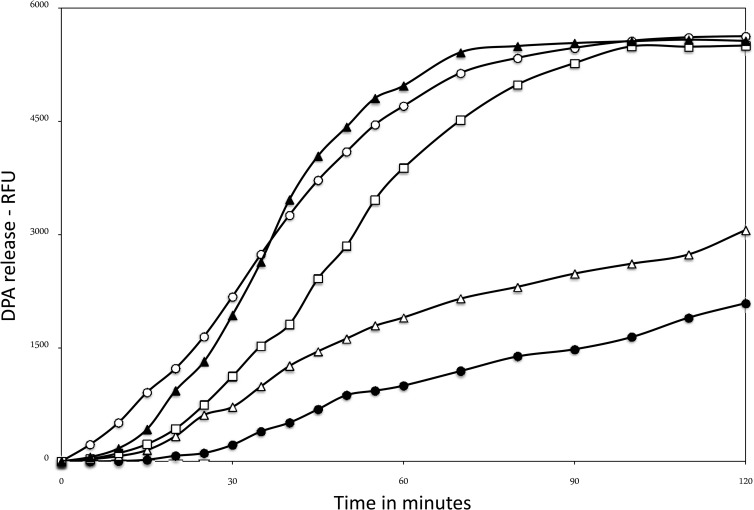
FIG 9.
Nutrient germination of spoVAEa or spoVAF mutant spores with or without ectopic expression of spoVAEa and spoVAF. Heat-activated spores of various strains were germinated with l-valine, and DPA release was measured by determining Tb-DPA fluorescence as described in Materials and Methods. Results are presented in relative fluorescence units (RFU). Numbers of RFU of spores of different strains were corrected for the amounts of DPA in different spore preparations. Symbols: ○, PS832 (wild type); ●, PS4348 (spoVAEa mutant); △, PS4351 (spoVAF mutant); □, PS4391 (spoVAEa PsspB-spoVAEa-spoVAF mutant); ▲, PS4392 (spoVAF PsspB-spoVAEa-spoVAF mutant).
DISCUSSION
There are a number of interesting features of the SpoVAEa protein. First, SpoVAEa is made in the forespore, where the spoVA operon is transcribed, and is present on the outer surface of the spore's IM yet has no obvious signal sequence or transmembrane segments that might direct this hydrophilic protein to the IM's outer surface. In this regard, SpoVAEa behaves similarly to the hydrophilic SpoVAD protein. Second, SpoVAEa distribution in spore fractions is similar to that of the SpoVAD protein, which has been shown to be an IM protein by several methods (4, 10). However, SpoVAEa and SpoVAD are bound only loosely to the IM, since in fractionated spore lysates most of these proteins are found in the supernatant fraction, although SpoVAD and SpoVAEa do not interact directly, as assessed in a pulldown assay (6). This lack of interaction in a pulldown assay has also been seen between SpoVAEa and the auxiliary germination protein GerD (Y.-Q. Li and B. Hao, unpublished data). In contrast to the hydrophilic SpoVAD and SpoVAEa proteins, the other SpoVA proteins are most likely to be integral IM proteins. While the arrangement of the SpoVA proteins in the IM is not known, it seems likely that (i) all of these proteins are in a complex in the IM, with SpoVAD and SpoVAEa on the outer surface; and (ii) this protein complex comprises a channel for CaDPA movement into and out of the developing and germinating spore.
Third, the level of SpoVAEa in spores is much lower than that of SpoVAD. While the reason for this is not clear, the spoVAEa mRNA may be translated more poorly than the spoVAD mRNA, as the RBS for the latter gene is a good match to the optimal RBS for translation of B. subtilis mRNAs, as is the RBS for spoVAEb (28) (Fig. 10). In contrast, the spoVAEa RBS is by no means a perfect match to the optimal RBS, and this is also the case for the spoVAF RBS (Fig. 10).
FIG 10.

Intergenic regions of the spoVA operon showing translational start regions for spoVAD, spoVAEb, and spoVAEa. Triplets in italics are the stop codons for the upstream gene, the translation initiation codon for the downstream gene is in bold, and underlined residues are matches to the optimal B. subtilis RBS (28), which is shown below the sequence along with the optimal spacing between the RBS and the translation initiation codon. Note that the translation stop codon for spoVAEa is within the coding sequence of spoVAF.
The other notable feature of SpoVAEa is that this protein is not essential for DPA uptake in B. subtilis sporulation, as is also the case for SpoVAF, while SpoVAA, -B, -C, -D, and -Eb are essential (2, 4, 6). Notably, not only do several temperature-sensitive spoVAC mutants exhibit alterations in DPA uptake during sporulation at the nonpermissive temperature, but spores of these mutants made at the permissive temperature also exhibit slow DPA release during germination (5, 29). spoVAEa and spoVAF mutant spores do not exhibit any major defect in DPA uptake in sporulation, while spoVAEa mutant spores and, to a slightly lesser extent, spoVAF mutant spores do exhibit germination defects, but only in GR-dependent germination. A possible function of these proteins is thus some role in linking GR activation to the opening of the SpoVA DPA channel, although SpoVAEa and SpoVAF are clearly not essential for the opening of this channel. This lack of an essential role for SpoVAEa and SpoVAF in spore formation and germination is consistent with the absence of the spoVAEa and spoVAF genes in many members of the orders Bacillales and Clostridiales.
Probably the major findings from this overall work are that SpoVAEa, SpoVAF, or both proteins (i) are not essential for normal DPA uptake in sporulation (ii) but are essential for normal DPA release in GR-dependent spore germination, although not in GR-independent germination. The latter finding is consistent with SpoVA proteins being components of a DPA channel in the spore's IM, with a possible function of SpoVAEa being to coordinate communication between GRs and the gating of this DPA channel, as suggested above. The effect of SpoVAEa, SpoVAF, or both proteins appears to be to decrease the Tlag value for GR-dependent germination, while the CaDPA release process itself, as measured by ΔTrelease values, was relatively unaffected by a spoVAEa mutation. The absence of SpoVAEa also lengthens the time between germinant addition and commitment. This was also seen previously when times for commitment and DPA release for spores of several mutants in the spoVAC gene were determined, but this may be because there are generally longer times between commitment and germination with lower rates of germination (20, 29). The fact that ectopic expression of spoVAEa-spoVAF could complement deletions of spoVAF or spoVAEa is consistent with the proteins they encode together playing a role in nutrient germination, although this interpretation is not definitive since (i) we cannot analyze levels of SpoVAF in spores and (ii) expression of spoVAF or spoVAEa alone did not complement the l-valine germination defect of spoVAEa or spoVAF mutant spores. However, the larger effect on l-valine germination of the loss of both SpoVAEa and SpoVAF than that of the loss of SpoVAF alone, the minimal effect of the loss of SpoVAF alone on AGFK and HP germination at 150 MPa, and the very large effect of the loss of both SpoVAEa and SpoVAF on HP germination at 150 MPa all suggest that both of these SpoVA proteins are involved in some fashion in GR-dependent spore germination.
One of the intriguing results of this work is that the main effect of loss of SpoVAEa on spore germination was a much longer slow decrease in the spore's DIC image intensity prior to the end of Tlag. Previous work suggested that this slow DIC image intensity decrease is largely due to slow CaDPA release, and therefore the absence of SpoVAEa greatly prolongs this slow CaDPA release compared to that in wild-type spores. Interestingly, this same extended slow CaDPA release was also observed in the nutrient germination of spores with one of several mutations in the spoVAC gene (29). The major questions that arise from these observations are then (i) how is this slow CaDPA release mediated by GR activation, (ii) how does the presence of SpoVAEa lead to a more rapid transition between slow CaDPA release and the rapid CaDPA release in ΔTrelease, and (iii) mechanistically, what is sensed in the slow CaDPA release period that ultimately triggers very rapid CaDPA release? It is also notable that, given the longer average Tlag in l-valine germination of spoVAEa mutant spores and that commitment of wild-type spores to germination occurs only a few minutes prior to fast CaDPA release (20), there is still much CaDPA release from spoVAEa mutant spores in particular, even before commitment takes place. Consequently, a major period when important events are taking place during spore germination is the period prior to rapid CaDPA release, and understanding what is taking place in spores during this period will be crucial to a more complete understanding of spore germination.
Supplementary Material
ACKNOWLEDGMENT
This report is based upon work supported by a U.S. Department of Defense Multidisciplinary University Research Initiative award through the U.S. Army Research Laboratory and the U.S. Army Research Office under contract W911NF-09-1-0286.
Footnotes
Published ahead of print 28 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01546-14.
REFERENCES
- 1.Setlow P, Johnson EA. 2012. Spores and their significance, p 45–79 In Doyle MP, Buchanan R. (ed), Food microbiology, fundamentals and frontiers, 4th ed. ASM Press, Washington, DC [Google Scholar]
- 2.Errington J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Setlow P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550–556. 10.1016/j.mib.2003.10.001 [DOI] [PubMed] [Google Scholar]
- 4.Vepachedu VR, Setlow P. 2005. Localization of SpoVAD to the inner membrane of spores of Bacillus subtilis. J. Bacteriol. 187:5677–5682. 10.1128/JB.187.16.5677-5682.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vepachedu VR, Setlow P. 2004. Analysis of the germination of spores of Bacillus subtilis with temperature sensitive spo mutations in the spoVA operon. FEMS Microbiol. Lett. 239:71–77. 10.1016/j.femsle.2004.08.022 [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Korza G, Zhang P, Li YQ, Setlow B, Setlow P, Hao B. 2012. Role of a SpoVA protein in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 194:1875–1884. 10.1128/JB.00062-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paredes-Sabja D, Setlow P, Sarker MR. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 19:85–94. 10.1016/j.tim.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 8.Paidhungat M, Setlow B, Driks A, Setlow P. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 182:5505–5512. 10.1128/JB.182.19.5505-5512.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korza G, Setlow P. 2013. Topology and accessibility of germination proteins in the Bacillus subtilis spore inner membrane. J. Bacteriol. 195:1484–1491. 10.1128/JB.02262-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths KK, Zhang J, Cowan AE, Yu J, Setlow P. 2011. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol. Microbiol. 81:1061–1077. 10.1111/j.1365-2958.2011.07753.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fort P, Errington J. 1985. Nucleotide sequence and complementation analysis of a polycistronic sporulation operon, spoVA, in Bacillus subtilis. J. Gen. Microbiol. 131:1091–1105 [DOI] [PubMed] [Google Scholar]
- 12.Velasquez-Guzman JC, Kocer A, Abee T, Poolman B. 2012. SpoVA, putative channels involved in spore germination of Bacillus spores, abstr 010 5th European Spores Conference, 16–19 April 2012, Royal Holloway, University of London, London, United Kingdom [Google Scholar]
- 13.Li Y, Jin K, Setlow B, Setlow P, Hao B. 2012. Crystal structure of the catalytic domain of the Bacillus cereus SleB protein important in cortex degradation during spore germination. J. Bacteriol. 194:4537–4545. 10.1128/JB.00877-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paidhungat M, Setlow P, Driks A, Setlow P. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 182:5505–5512. 10.1128/JB.182.19.5505-5512.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Setlow B, Setlow P. 1996. Role of DNA repair in Bacillus subtilis spore resistance. J. Bacteriol. 178:3486–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tovar-Rojo F, Chander M, Setlow B, Setlow P. 2002. The products of the spoVA operon are involved in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 184:584–587. 10.1128/JB.184.2.584-587.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cutting SM, Vander Horn PB. 1990. Genetic analysis, p 22–74 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom [Google Scholar]
- 18.Steinmetz M, Richter R. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutation in Bacillus subtilis through in vivo recombination. Gene 142:79–83. 10.1016/0378-1119(94)90358-1 [DOI] [PubMed] [Google Scholar]
- 19.Nicholson WL, Sun D, Setlow P. 1990. Sporulation, germination and outgrowth, p 391–450 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom [Google Scholar]
- 20.Yi X, Setlow P. 2010. Studies of the commitment step in the germination of spores of Bacillus species. J. Bacteriol. 192:3424–3433. 10.1128/JB.00326-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butzin XY, Troiana AJ, Coleman WH, Griffiths KK, Doona CJ, Feeherry FE, Wang G, Li YQ, Setlow P. 2012. Analysis of the effects of a gerP mutation on the germination of spores of Bacillus subtilis. J. Bacteriol. 194:5749–5758. 10.1128/JB.01276-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang PF, Kong LB, Wang GW, Setlow P, Li YQ. 2010. Combination of Raman tweezers and quantitative differential interference contrast microscopy for measurement of dynamics and heterogeneity during the germination of individual bacterial spores. J. Biomed. Opt. 15:056010. 10.1117/1.3494567 [DOI] [PubMed] [Google Scholar]
- 23.Wang GW, Zhang PF, Setlow P, Li YQ. 2011. Kinetics of germination of wet heat-treated individual spores of Bacillus species as followed by Raman spectroscopy and differential interference contrast microscopy. Appl. Environ. Microbiol. 77:3368–3379. 10.1128/AEM.00046-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paidhungat M, Setlow P. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 183:3982–3990. 10.1128/JB.183.13.3982-3990.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez-Peralta A, Stewart K-AV, Thomas SK, Setlow B, Chen Z, Li YQ, Setlow P. 2012. Effects of the SpoVT regulatory protein on the germination and germination protein levels of spores of Bacillus subtilis. J. Bacteriol. 194:3417–3425. 10.1128/JB.00504-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart K-AV, Yi X, Ghosh S, Setlow P. 2012. Germination protein levels and rates of germination of spores of Bacillus subtilis with overexpressed or deleted genes encoding germination proteins. J. Bacteriol. 194:3156–3164. 10.1128/JB.00405-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart K-AV, Setlow P. 2013. Numbers of individual nutrient germinant receptors and other germination proteins in spores of Bacillus subtilis. J. Bacteriol. 195:3575–3582. 10.1128/JB.00377-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vellanoweth RL, Rabinowitz JC. 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 6:1105–1114. 10.1111/j.1365-2958.1992.tb01548.x [DOI] [PubMed] [Google Scholar]
- 29.Wang G, Yi X, Setlow P, Li Y-Q. 2011. Germination of individual Bacillus subtilis spores with alterations in the GerD and SpoVA proteins which are important in spore germination. J. Bacteriol. 193:2301–2311. 10.1128/JB.00122-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.