Abstract
The chlamydiae are obligate intracellular parasites that have evolved specific interactions with their various hosts and host cell types to ensure their successful survival and consequential pathogenesis. The species Chlamydia pneumoniae is ubiquitous, with serological studies showing that most humans are infected at some stage in their lifetime. While most human infections are asymptomatic, C. pneumoniae can cause more-severe respiratory disease and pneumonia and has been linked to chronic diseases such as asthma, atherosclerosis, and even Alzheimer's disease. The widely dispersed animal-adapted C. pneumoniae strains cause an equally wide range of diseases in their hosts. It is emerging that the ability of C. pneumoniae to survive inside its target cells, including evasion of the host's immune attack mechanisms, is linked to the acquisition of key metabolites. Tryptophan and arginine are key checkpoint compounds in this host-parasite battle. Interestingly, the animal strains of C. pneumoniae have a slightly larger genome, enabling them to cope better with metabolite restrictions. It therefore appears that as the evolutionarily more ancient animal strains have evolved to infect humans, they have selectively become more “susceptible” to the levels of key metabolites, such as tryptophan. While this might initially appear to be a weakness, it allows these human C. pneumoniae strains to exquisitely sense host immune attack and respond by rapidly reverting to a persistent phase. During persistence, they reduce their metabolic levels, halting progression of their developmental cycle, waiting until the hostile external conditions have passed before they reemerge.
INTRODUCTION
The genus Chlamydia contains a phylogenetically unique group of obligate intracellular parasites that successfully infect humans and a wide range of animals. They are characterized by their unique developmental cycle, which consists of two morphologically distinct forms (reviewed in reference 1). The extracellular, infectious form (elementary body [EB]) is responsible for transmitting infections between cells and between hosts and is generally considered to be metabolically inert (although this has been challenged recently [2]), whereas the intracellular, noninfectious form (reticulate body [RB]) is metabolically active and is responsible for growth and multiplication within the target host cell (Fig. 1). Once an EB attaches to and enters a host cell, it immediately hijacks several host cell pathways (3, 4), preventing phagolysosomal fusion, encasing itself in a unique double-membrane inclusion and rapidly converting to the metabolically active, RB form. The RBs multiply by binary fission around 500-fold during a 24-to-48-h period before converting back to EBs, which subsequently exit the cell to start another round of multiplication. This unique “acute” developmental cycle has both strengths and weaknesses. Once inside a suitable host cell, provided the chlamydiae can subvert several key host cell pathways, they generally have access to a rich pool of nutrients required for growth. While the chlamydiae have retained a diverse set of metabolic pathways, they have also lost many pathways, making them dependent on their host cell for a significant number of key nutrients and intermediates. Of course, eukaryotic cells have evolved a range of strategies to combat pathogens such as Chlamydia (reviewed in reference 5). One main line of defense is that eukaryotic cells sense foreign invaders (via pattern recognition receptor [PRR] signaling), resulting in production of the key cytokine gamma interferon (IFN-γ) (at least in humans), which in turn depletes the host cell tryptophan (TRP) pool and effectively starves the foreign bacteria of this key nutrient.
FIG 1.
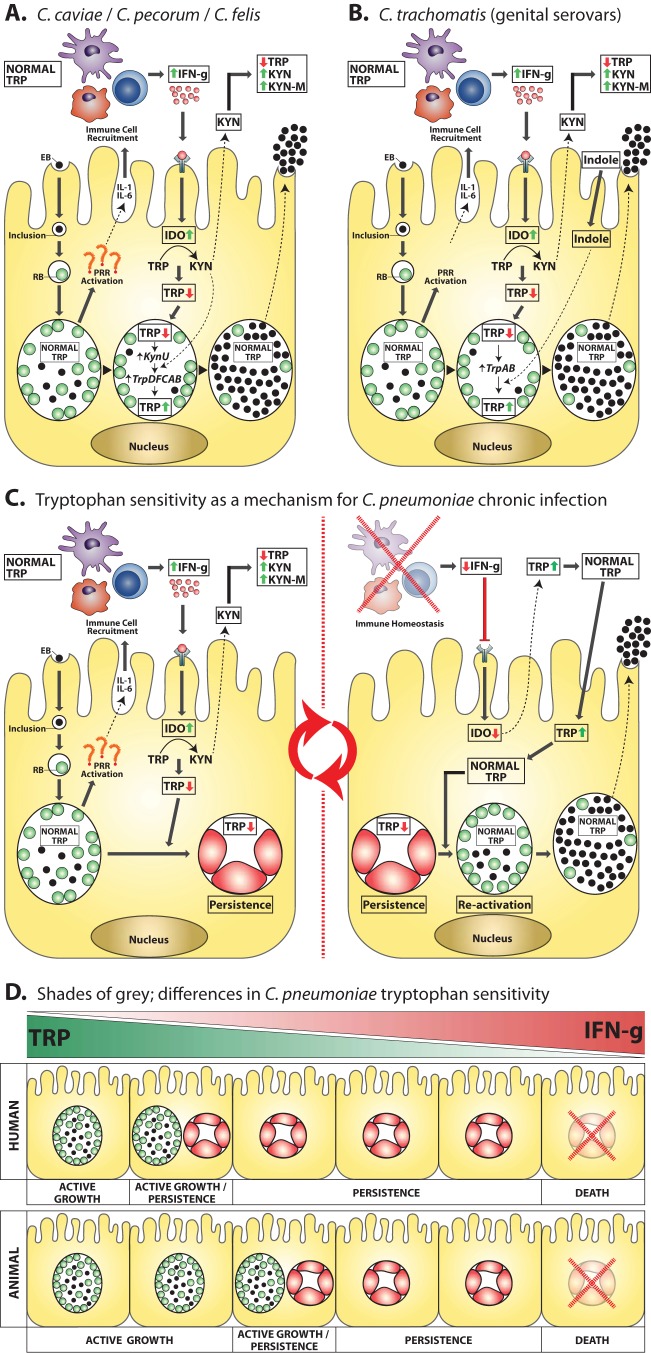
The central role of tryptophan in chlamydial biology. Chlamydial elementary bodies attach to and enter a host epithelial cell, an inclusion is formed, and the elementary body (EB) converts to its replicative form, the reticulate body (RB). RBs undergo replication; however, during infection, PRRs are activated, resulting in secretion of proinflammatory cytokines from the host, which in turn recruits immune cells. Development of an immune response results in host cells being exposed to gamma interferon. IDO is activated, and tryptophan pools are depleted. (A) The species C. caviae, C. pecorum, and C. felis obtain kynurenine (the product of IDO activity) from the host cell and use it to recycle tryptophan via tryptophan biosynthesis genes (kynU and trpD, -F, -C, -A, and -B), meaning that these species are able to actively replicate during IDO activity and host tryptophan depletion. This tryptophan biosynthesis process is regulated by a tryptophan repressor and thus is initiated only when inclusion tryptophan levels become depleted. (B) The urogenital strains of C. trachomatis are believed to respond to IDO-tryptophan depletion by using indole (thought to be supplied by the vaginal microflora) as a precursor for tryptophan synthesis. Tryptophan synthesis is achieved through the tryptophan biosynthesis genes trpA and -B. As with the above-mentioned species, C. trachomatis tryptophan biosynthesis is also regulated via a tryptophan repressor that allows biosynthesis to occur only once inclusion tryptophan levels become depleted. Interestingly, the ocular C. trachomatis strains do not possess the tryptophan biosynthesis genes and are associated with more-chronic disease outcomes. (C) The key role of tryptophan in C. pneumoniae chronic infection. In stark contrast to the above-mentioned species, C. pneumoniae does not contain any tryptophan biosynthesis genes. As a result, tryptophan depletion results in conversion of an active replicating inclusion to a persistent inclusion (a viable but nonreplicating, static phase). In this state, C. pneumoniae becomes refractory to immune insult, allowing the organism to hide in the host cell until the immune response subsides. As the immune response abates, IFN-γ becomes depleted, IDO is no longer induced, local tryptophan pools become recharged, and tryptophan levels eventually return to normal. At this point, C. pneumoniae reverts to an actively replicating inclusion, resulting in the release of infectious progeny ready to reinitiate the chronic infection cycle. (D) Recent studies have found that human and animal (ancestral) strains behave very differently in the face of IFN-γ insult and tryptophan depletion. This finding suggests that the human strains have evolved to become more sensitive to IFN-γ, allowing them to enter a persistent phase at the first signs of an immune response. As a result, they have been able to develop a chronic infection cycle that has enabled them to become, arguably, one of the most successful human pathogens. EB, elementary body; RB, reticulate body; IFN-g, gamma interferon; IL, interleukin; PRR, pattern recognition receptors; IDO, indoleamine 2,3-dioxygenase; TRP, tryptophan; KYN, kynurenine; KYN-M, kynurenine metabolites.
In addition to the acute developmental cycle, chlamydiae have also developed a so-called “persistent phase” (6, 7). While this persistent phase has been best studied in vitro, it can also be observed in vivo (7–12). Chlamydial persistence has been described as “a viable but non-culturable growth stage resulting in a long term relationship with the infected host cell” (13). During persistence, chlamydial metabolism is slowed and RB division, as well as differentiation into EBs, is suspended (6, 14, 15), resulting in the production of so-called aberrant bodies. Overall, this means that while the RBs are metabolically viable, they do not complete the developmental cycle and hence no infectious progeny are produced. This state is reversible. Several different stimuli have been shown to induce persistence, including (i) gamma interferon treatment, (ii) penicillin treatment, (iii) amino acid (e.g., tryptophan), glucose, and iron deprivation, (iv) chlamydiophage infection, and several others (14, 16–26). Persistence therefore appears to be a mechanism that allows the organisms to ride out hostile conditions while maintaining a long-term (chronic) infection within a host cell.
Of the nine species in the genus Chlamydia, C. pneumoniae is arguably the most successful pathogen, infecting humans and an amazingly wide range of animals. In humans, serological studies show that almost all of the world's population is, or had been, infected with C. pneumoniae (27). Respiratory infections are the most common, with seroepidemiological surveys indicating that infection is both endemic and epidemic. Asymptomatic infections are common, usually manifesting as mild upper tract infections that are self-limiting, although they may progress to more-severe upper respiratory tract infections (pharyngitis, sinusitis, and otitis) and lower respiratory tract infections (acute bronchitis, exacerbations of chronic bronchitis and asthma, and community-acquired pneumonia) (28–37). The species was initially “discovered” in Finland in 1985, where it caused epidemics of pneumonia in military barracks (38), and, after its initial worldwide characterization (39–42), was thought to have declined somewhat in prevalence. However, recent reports (both serological studies and studies of outbreaks in confined populations [43, 44]) show that it is still a widespread and successful respiratory pathogen. In addition to respiratory infections, C. pneumoniae has been linked with several other chronic conditions, most notably cardiovascular inflammatory diseases such as atherosclerosis, abdominal aortic aneurisms, and valvular lesions (45–48). While there is a body of published evidence linking C. pneumoniae and cardiovascular disease, a causal link remains to be definitively proven and is considered controversial by some. In addition to the link with cardiovascular diseases, it has also been linked with asthma (49), Alzheimer's disease (50, 51), arthritis (52), lung cancer (53–55), chronic obstructive pulmonary disease (56, 57), and diabetes (58, 59). While there continue to be individual reports of severe respiratory disease associated with C. pneumoniae (60), it appears that the norm is now an association of C. pneumoniae with a range of chronic diseases.
In addition to its widespread prevalence in the human population, C. pneumoniae also causes infection and disease in a wide range of animals. C. pneumoniae infections have been reported in both cold-blooded hosts (frogs, snakes, iguanas, and crocodiles [61–63]) and warm-blooded hosts (horses and marsupials [63–67]). In these animal hosts, it infects a wide range of tissues (lung, liver, heart, eyes, and the female genital tract of koalas) and causes a wide range of disease syndromes (respiratory, vascular, conjunctival, genitourinary, and systemic).
Hypothesis: Chlamydia pneumoniae—a highly successful parasite with “50 shades of gray.”
The hypothesis which we discuss in this review is that the highly successful obligate intracellular pathogen, C. pneumoniae, has actually evolved an increasing susceptibility to tryptophan, the key amino acid. Rather than making this tryptophan auxotroph more vulnerable to host “attack,” it has enabled the evolution of the more ancient, animal infection strains (which are less sensitive to tryptophan availability and cause more acute infections) into the currently successful human strains. The human strains not only are more sensitive to tryptophan availability but primarily cause chronic infections that are exquisitely in balance with their host.
TRYPTOPHAN IS AN ESSENTIAL AMINO ACID AND A CENTRAL REGULATOR FOR CELL BIOLOGY
Humans and animals lack the ability to synthesize the essential amino acid tryptophan. Consequently, tryptophan depletion acts as a major cellular signal. In humans, tryptophan depletion is associated with depression and other neurological consequences (68, 69). At a cellular level, reduction of intracellular tryptophan pools initiates molecular stress response pathways (via GCN2 kinase and mammalian target of rapamycin [mTOR] signaling molecules), resulting in blockage of ribosomal translation and cell cycle arrest. Localized tryptophan deprivation can also initiate these stress response pathways in local T cells, leading to arrest and promotion of regulatory T cell differentiation. Furthermore, bioactive tryptophan metabolites (produced from the kynurenine [KYN] pathway) have been found to have direct immunosuppressive capabilities, facilitating promotion of regulatory T cell differentiation (reviewed in references 70 and 71). In spite of these consequences, tryptophan is also actively depleted by animal cells in a pathogen defense strategy. Most bacteria are able to synthesize tryptophan and can partially overcome the host's efforts to starve it of tryptophan. However, some microbes, such as Chlamydia, are full or partial tryptophan auxotrophs. In fact, Chlamydia have unique host- and tissue niche-specific requirements, intrinsically based on differential abilities to synthesize tryptophan (or not) from metabolites or bioavailable intermediates.
PATHOGEN DEFENCE VIA IDO (INDOLEAMINE 2,3-DIOXYGENASE)-MEDIATED TRYPTOPHAN DEPLETION
Chlamydial infection of mucosal epithelial cells results in activation of innate, pathogen-sensing PRRs. Activation of these receptors initiates an immunological cascade, resulting in expression and secretion of cytokines and chemokines (reviewed in reference 5) and recruitment of leukocytes (NK cells, neutrophils, macrophages, and dendritic cells) to the infected tissue (72). An adaptive immune response is then initiated via antigen-processing cells presenting chlamydial antigens to T cells in local lymph nodes. Animal studies (73–76) and, to a certain degree, human studies (73–78) have found that IFN-γ-producing T cells are essential for resolution of chlamydial infections (72, 79, 80).
The antichlamydial effect of IFN-γ is attributed predominantly to direct inhibition of intracellular growth. Exposure of the host cells to IFN-γ results in induction of a myriad of responses (81). However, it is the induction of IDO (indoleamine 2,3-dioxygenase) (82) and inducible nitric oxide synthase (iNOS) (83) and downregulation of transferrin receptors (84) that is central to intracellular inhibition of chlamydial growth. IDO is a heme-containing, cytosolic, IFN-γ-inducible enzyme that catalyzes the breakdown of the essential amino acid tryptophan to kynurenine via the cleavage of the pyrrole ring. Kynurenine is then released from the cell or further metabolized to the downstream catabolites 3-hydroxykynurenine, 3-hydroxyanthranilic acid, quinolinic acid, and, finally, NAD (i.e., the kynurenine pathway).
More than 2 decades ago, tryptophan depletion, via IFN-γ induction of IDO, was described as a possible mechanism for inhibiting intracellular growth of the parasite Toxoplasma gondii (85). The simplistic paradigm of IDO being a host innate defense mechanism, using essential amino acid deprivation as a means to control tryptophan auxotrophic intracellular pathogen replication, has been superseded recently by a more complex model. Specifically, IDO tryptophan catabolism has been found to have far-ranging effects making it a pivotal component in immune and cellular regulation via the aforementioned signaling mechanisms. Consequently, IDO-tryptophan depletion has been found to have an important role in parasitic (86), viral (87), and bacterial (88, 89) infections. Although most reports document pathogen sensitivity to tryptophan depletion, recent studies are finding a role for IDO activity in chronic or persistent disease. In particular, IDO-tryptophan-directed immunosuppression is thought to play a role in infections by Leishmania major (90), human immunodeficiency virus (reviewed in reference 91), hepatitis C virus (92, 93), human papillomavirus (94), Listeria monocytogenes (95), Mycobacterium tuberculosis (96), Candida albicans (97), and Aspergillus fumigatus (98). Importantly, a recent study investigating the role IDO plays in infections by two protozoal species, T. gondii and L. major, found that IDO facilitates T. gondii clearance and suppresses L. major clearance, highlighting the opposing and pathogen-specific roles that IDO can have in infectious disease (90).
IFN-γ- and IDO-directed tryptophan depletion has had a significant effect on chlamydial evolution. In vitro model studies of epithelial cells, using tryptophan rescue of IFN-γ-treated cultures, indicate that tryptophan depletion is the major antipathogen activity of IFN-γ against Chlamydia (88, 89). The human ocular and urogenital pathogen Chlamydia trachomatis contains tryptophan synthesis (trpBA) genes allowing synthesis of tryptophan from indole (99). However, this adaptation was found to be functional only in the urogenital strains. It was proposed that the urogenital strains gain access to indole via vaginal microflora, thus identifying a tissue tropism mediator (i.e., ocular versus urogenital) (100). Interestingly, the ocular strains (serovars A to C) responsible for the chronic disease trachoma appear to be much more sensitive to the effects of IFN-γ than the urogenital strains (101). Another chlamydial species, Chlamydia caviae (guinea pig), has also evolved an elegant adaptation to tryptophan depletion, utilizing a more complete set of tryptophan synthesis genes (102, 103). These genes allow this species to recycle kynurenine back into tryptophan, ensuring continued propagation in the presence of IFN-γ induced IDO (104). Although not yet experimentally tested, these recycling genes are also conserved on the genomes of Chlamydia pecorum (sheep, cattle, koala), and Chlamydia felis (cats). Even though these tryptophan anabolism capabilities are present across these diverse animal-infecting species of Chlamydia, the situation is different for the murine strain, where IFN-γ inhibition of Chlamydia in murine cells is driven by p47 GTPases and not IDO (105). The murine pathogen (Chlamydia muridarum) produces a species-specific cytotoxin that is thought to degrade the key p47 GTPase, enabling continued propagation in the presence of IFN-γ (105).
In stark contrast, C. pneumoniae, arguably the most successful human pathogen in the Chlamydia genus, does not have any tryptophan biosynthesis genes. As we discuss below, this apparent vulnerability to tryptophan may in fact be a checkpoint system for sensing danger in the form of the presence of some C. pneumoniae strains (particularly the human strains). Nevertheless, C. pneumoniae requires a certain amount of tryptophan for survival and one possibility is that it can perhaps utilize the respiratory microflora to obtain its tryptophan. The microflora of the human respiratory tract is less well characterized than for other body sites, but it is known to be quite complex and contains bacteria that produce tryptophan, which could be transported from the microbiome environment into the Chlamydia-infected cell/inclusion by various C. pneumoniae transporters.
HAVE THE HUMAN STRAINS OF C. PNEUMONIAE ACTUALLY EVOLVED TOWARD AN INCREASED SENSITIVITY TO TRYPTOPHAN AVAILABILITY, ENABLING THEM TO SWITCH FROM ACTIVE GROWTH TO THE LATENT PHASE AND THEREBY BECOME MORE SUCCESSFUL CHRONIC “PATHOGENS”?
The animal strains of C. pneumoniae are evolutionarily older and have a slightly larger genome.
Several strains of C. pneumoniae have now had their full genomes sequenced (106), including four human strains (107–109) and two animal strains (110, 111). While the genomes are remarkably similar (>99% identity in gene content and arrangement), there are some subtle but presumably significant differences between the human and animal strains but also between the various human strains.
One striking feature of all C. pneumoniae strains is that they are complete tryptophan auxotrophs. Perhaps the most interesting outcome of comparisons of the genomes of the human and animal strains of C. pneumoniae, though, is the evidence that the animal strains are evolutionarily more ancient and that the human strains probably evolved from the animal strains, perhaps over a relatively recent and short time period, by reducing their gene content and presumably adapting specifically to the human host (110). The animal C. pneumoniae genome is 1,241,024 bp in size, which is just 10 kb larger than the human C. pneumoniae genome, with just 6,213 single nucleotide polymorphisms (SNPs) separating the strains (110). A potentially important difference, though, is that the animal strains contain a plasmid whereas all human strains do not. While the genes in the C. pneumoniae plasmid have not yet been characterized, they have significant homology to the C. trachomatis plasmid genes. The C. trachomatis plasmid genes are currently the focus of considerable research, and it appears that some of these plasmid genes have roles in regulating chromosomal genes. As chlamydial transformation techniques are refined, it should become possible to test plasmidless animal strains for their response to IFN-γ and tryptophan. The other key difference is that there are several examples of genes that are full length in the animal strain but have truncations or significant disrupting fragmentations in the human strains. These facts have led investigators to conclude that the animal strains are ancestral and that the human strains evolved from them, adapting specifically to the human host as they have evolved (110).
Recent cell biology studies support the suggestion that the human strains of C. pneumoniae are more sensitive to exogenous tryptophan levels than the animal strains.
In 2008, Mitchell et al. analyzed the in vitro growth of the animal strain of C. pneumoniae and compared it to that of the AR39 human strain (112). They observed significant growth differences, with the animal strain growing more aggressively, with a shorter doubling time (3.4 to 4.9 h versus 5.9 to 8.7 h), producing larger inclusions and a higher yield of infectious progeny than the human strain, which generally produced small inclusions with a lower infectious yield (112).
C. pneumoniae human isolates have been reported to be extremely sensitive to tryptophan depletion via IFN-γ-induced IDO (113–115). In vitro studies with epithelial and endothelial cell lines, utilizing IDO inhibition and tryptophan supplementation, have found human C. pneumoniae strains to be highly sensitive to IDO-driven tryptophan depletion (113, 115, 116). A recent study investigated the different effects that amino acid supplementation had on C. trachomatis and C. pneumoniae. Somewhat surprisingly, the addition of excess tryptophan was found to marginally inhibit C. trachomatis growth whereas, in contrast, it significantly enhanced C. pneumoniae growth (117). C. pneumoniae tissue tropism may also be influenced by the ability of the organism to gain access to host tryptophan, as the vascular strains only carry a single copy of the tryptophan-tyrosine permease (versus two copies in the respiratory strain), potentially resulting in reduced amino acid uptake ability in the vascular isolates (118).
Like other chlamydial species, C. pneumoniae can enter a state of persistence (i.e., a noninfectious but viable state) during IFN-γ insult (reviewed in references 13, 119, and 120). This persistent state can be reversed and active replication reinstated through the addition of tryptophan or via the inhibition of IDO (113, 115, 116). Unsurprisingly, due to the lack of tryptophan biosynthesis genes, only very low levels (15 to 25 U/ml) of IFN-γ are required to induce altered inclusion morphology (associated with persistent infection) in in vitro epithelial cell models (20, 113). However, if the organisms are exposed to IFN-γ during infection, a total bactericidal effect requires very high levels (800 U/ml) of IFN-γ (113).
A recent study investigating the cell biology differences between the ancestral animal strains and the more recently evolved human strains of C. pneumoniae has confirmed the sensitivity of the human strains to IFN-γ under controlled in vitro conditions (A. Chacko, C. J. Barker, K. Beagley, P Timms, and W. M. Huston, submitted for publication). The human strains of C. pneumoniae are exquisitely sensitive to the reduction in the tryptophan level via the effects of IFN-γ. The number and size of chlamydial inclusions are decreased (some appear persistent), and their infectious yield is markedly reduced (by greater than 99.99%). By comparison, the ancestral animal strain, under the same conditions, is much less affected by IFN-γ treatment (with approximately 100-fold-greater infectious yield). Interestingly, they also showed that subtle differences in IFN-γ sensitivity exist between the various human strains (comparing a respiratory isolate with a cardiovascular isolate). By using direct depletion of tryptophan from the medium, those researchers demonstrated that the IFN-γ effect was clearly associated with tryptophan depletion (Chacko et al., submitted). Furthermore, adding additional exogenous tryptophan back to the human strains actually increased their infectious yield above that produced by the non-IFN-γ-treated controls (Chacko et al., submitted). The mechanism by which the ancestral animal strains obtain sufficient tryptophan is not yet clear; however, increased host cell autophagy and effective transport from the host cell pool into the chlamydial inclusion is one possibility. This “strategy” enables the animal strains to grow basically as an acute infection, producing highly infectious outcomes, whereas the human strains prefer to sense adverse conditions (such as host immune attack via IFN-γ) by being more sensitive to lowering levels of tryptophan, diverting them from completing their infectious cycle and returning them to their persistent stage, where they wait out the unfavorable conditions.
These recent findings suggest that the ancestral strains are less “dependent” on exogenous tryptophan levels and prefer to grow as acute infections, ensuring their survival by completing their developmental cycle with the production of large numbers of infectious progeny to infect other cells or new hosts. In contrast, the evolution of the human strains has resulted in their being more “sensitive” to decreasing exogenous tryptophan levels, allowing them to sense adverse conditions (such as host immune attack via IFN-γ). These strains use the lowering levels of tryptophan to divert their infectious life cycle to a persistent phase until the adverse conditions subside. Increases in tryptophan levels then trigger reactivation of the developmental cycle, making the human strains more likely to cause chronic infections.
SUBTLE VARIATIONS IN THE TRYPTOPHAN CONTENT OF KEY CHLAMYDIAL PROTEINS COULD BE A FUNDAMENTAL DETERMINANT OF SUSCEPTIBILITY TO LIMITED TRYPTOPHAN AVAILABILITY
Several reviews have emerged recently (121, 122) suggesting that the chlamydial genus as a whole may have evolved a mechanism that mutes or downregulates selected gene products via the tryptophan levels in these key proteins. Those proteins that are required during the acute or active growth phase (i.e., normal development of Chlamydia) have higher tryptophan content, since tryptophan availability is less of an issue in this stage of growth. However, when the host responds to chlamydial infection by producing IFN-γ and tryptophan is depleted, these proteins become a burden to the Chlamydia and this triggers their switch from the acute to the persistent phase. In the persistent phase, a different subset of proteins are required and these have a lower tryptophan content (at least, the high-tryptophan-containing proteins are not highly expressed in this phase), making these proteins less of a metabolic burden to produce during a time when tryptophan levels are limited. While this provocative hypothesis seems to fit the C. trachomatis data best, it may also be partly supported by the limited C. pneumoniae data that are available. Lo et al. (121) reported that MurA is a low-tryptophan-containing enzyme, making it more suited to expression during persistence. Indeed, when Timms et al. (123) evaluated murA transcription in the IFN-γ model of C. pneumoniae, they found that it was upregulated 2-fold under these conditions. While this hypothesis is interesting to consider, at this stage it is built entirely on bioinformatic analysis of chlamydial genomes and is not yet supported by any focused biological data. In addition, while it might be a general strategy used by chlamydiae, a comparison of the human and animal C. pneumoniae genomes does not support an extension of this tryptophan skew in the acute animal strains.
ARGININE AND THE IDO-TRYPTOPHAN NEXUS
C. pneumoniae also appears to have unique arginine-related adaptations. This species has a functional arginine-responsive transcriptional repressor (124) and an arginine decarboxylase and arginine-agmatine antiporter (125). In addition, excess exogenous arginine causes significant growth inhibition in vitro (117). The ability to take up and degrade arginine is most likely a means of protection against the host cell pathogen oxidative attack compound nitric oxide (NO) (125). Immune signaling activates transcription of iNOS, which catabolizes arginine to citrulline, producing NO. Importantly, NO directly inhibits IDO activity via binding to the active-site heme (126). An in vitro study found that exogenous NO is able to inhibit IDO tryptophan catabolism, resulting in nearly normal C. trachomatis L2 growth in epithelial cells (C. trachomatis L2 does not have a functional arginine decarboxylase [127, 128]). Even with these adaptations to prevent NO attack, macrophages from iNOS−/− mice are more susceptible to C. pneumoniae infections in vitro (129, 130). Nonetheless, it is clear that C. pneumoniae can reduce host arginine levels, thereby reducing NO production and, paradoxically, allowing continued IDO activity, perhaps as a direct trigger for chronic or persistent growth. These arginine-based adaptations have not been extensively investigated but seem likely to also function in the shades of gray between acute and chronic disease states and tissue adaptations that exist within the ubiquitous C. pneumoniae strains.
CONCLUSION
Tryptophan availability and the ability to overcome active tryptophan depletion fundamentally define host and tissue tropism and, therefore, pathogenicity for Chlamydia. Here we discuss the hypothesis that sensing tryptophan bioavailabilty to trigger chronic growth or persistence has been selected for as a chronic disease advantage in human C. pneumoniae infections. We already know that ocular C. trachomatis strains are more sensitive to IFN-γ than the closely related genital strains and that they are more chronic in their growth and disease states. At the other end of the spectrum, the ancestral animal C. pneumoniae strains appear to have been selected for a more aggressive, acute disease state. Recent data demonstrate that animal C. pneumoniae strains are less sensitive to tryptophan depletion than the human C. pneumoniae strains, are more aggressive in their growth, and present with more acute disease. The versatile and highly prevalent human C. pneumoniae strains are clearly advantaged by this tryptophan-responsive chronic disease trigger, which may in fact be further adapted with tissue tropism and disease states within these strains. While this model is appealing and is quite well supported by the currently published literature, further studies are needed to confirm it and to better understand the molecular mechanisms involved. As the technique of chlamydial transformation becomes more widely used, evaluating plasmidless or gene knockout strains would be one way of addressing these issues.
ACKNOWLEDGMENT
Our work is supported by funding from the National Health & Medical Research Council (NHMRC).
Footnotes
Published ahead of print 28 March 2014
REFERENCES
- 1.Abdelrahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949–959. 10.1016/j.femsre.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 2.Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. 2012. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc. Natl. Acad. Sci. U. S. A. 109:19781–19785. 10.1073/pnas.1212831109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coombes BK, Mahony JB. 2002. Identification of MEK- and phosphoinositide 3-kinase-dependent signalling as essential events during Chlamydia pneumoniae invasion of HEp2 cells. Cell. Microbiol. 4:447–460. 10.1046/j.1462-5822.2002.00203.x [DOI] [PubMed] [Google Scholar]
- 4.Lane BJ, Mutchler C, Al Khodor S, Grieshaber SS, Carabeo RA. 2008. Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog. 4:e1000014. 10.1371/journal.ppat.1000014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimada K, Crother TR, Arditi M. 2012. Innate immune responses to Chlamydia pneumoniae infection: role of TLRs, NLRs, and the inflammasome. Microbes Infect. 14:1301–1307. 10.1016/j.micinf.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beatty WL, Belanger TA, Desai AA, Morrison RP, Byrne GI. 1994. Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect. Immun. 62:3705–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beatty WL, Byrne GI, Morrison RP. 1993. Morphologic and antigenic characterization of interferon g-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 90:3998–4002. 10.1073/pnas.90.9.3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanada H, Ikeda-Dantsuji Y, Naito M, Nagayama A. 2003. Infection of human fibroblast-like synovial cells with Chlamydia trachomatis results in persistent infection and interleukin-6 production. Microb. Pathog. 34:57–63. 10.1016/S0882-4010(02)00189-4 [DOI] [PubMed] [Google Scholar]
- 9.Nebe CT, Rother M, Brechtel I, Costina V, Neumaier M, Zentgraf H, Bocker U, Meyer TF, Szczepek AJ. 2005. Detection of Chlamydophila pneumoniae in the bone marrow of two patients with unexplained chronic anaemia. Eur. J. Haematol. 74:77–83. 10.1111/j.1600-0609.2004.00353.x [DOI] [PubMed] [Google Scholar]
- 10.Rupp J, Droemann D, Goldmann T, Zabel P, Solbach W, Vollmer E, Branscheid D, Dalhoff K, Maass M. 2004. Alveolar epithelial cells type II are major target cells for C. pneumoniae in chronic but not in acute respiratory infection. FEMS Immunol. Med. Microbiol. 41:197–203. 10.1016/j.femsim.2004.03.004 [DOI] [PubMed] [Google Scholar]
- 11.Sieper J, Braun J. 2002. Diagnosis and antibiotic treatment of reactive arthritis. Dtsch. Med. Wochenschr. 127:1893–1896. 10.1055/s-2002-34068 (In German) [DOI] [PubMed] [Google Scholar]
- 12.Bauriedel G. 2000. Persistence of Chlamydia pneumoniae in coronary plaque tissue. Dtsch. Med. Wochenschr. 125:645 (In German) [PubMed] [Google Scholar]
- 13.Hogan R, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. 2004. Chlamydial persistence: beyond the biphasic paradigm. Infect. Immun. 72:1843–1855. 10.1128/IAI.72.4.1843-1855.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambden PR, Pickett MA, Clarke IN. 2006. The effect of penicillin on Chlamydia trachomatis DNA replication. Microbiology 152:2573–2578. 10.1099/mic.0.29032-0 [DOI] [PubMed] [Google Scholar]
- 15.Byrne GI, Ouellette SP, Wang Z, Rao JP, Lu L, Beatty WL, Hudson AP. 2001. Chlamydia pneumoniae expresses genes required for DNA replication but not cytokinesis during persistent infection of HEp-2 cells. Infect. Immun. 69:5423–5429. 10.1128/IAI.69.9.5423-5429.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones ML, Gaston JSH, Pearce JH. 2001. Induction of abnormal Chlamydia trachomatis by exposure to interferon-gamma or amino acid deprivation and comparative antigenic analysis. Microb. Pathog. 30:299–309. 10.1006/mpat.2001.0433 [DOI] [PubMed] [Google Scholar]
- 17.Wehrl W, Meyer TF, Jungblut PR, Müller EC, Szczepek AJ. 2004. Action and reaction: Chlamydophila pneumoniae proteome alteration in a persistent infection induced by iron deficiency. Proteomics 4:2969–2981. 10.1002/pmic.200400917 [DOI] [PubMed] [Google Scholar]
- 18.Akers JC, Tan M. 2006. Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis. J. Bacteriol. 188:4236–4243. 10.1128/JB.01660-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bragina EY, Gomberg MA, Dmitriev GA. 2001. Electron microscopic evidence of persistent chlamydial infection following treatment. J. Eur. Acad. Dermatol. Venereol. 15:405–409. 10.1046/j.1468-3083.2001.00342.x [DOI] [PubMed] [Google Scholar]
- 20.Mannonen L, Kamping E, Penttila T, Puolakkainen M. 2004. IFN-gamma induced persistent Chlamydia pneumoniae infection in HL and Mono Mac 6 cells: characterization by real-time quantitative PCR and culture. Microb. Pathog. 36:41–50. 10.1016/j.micpath.2003.09.001 [DOI] [PubMed] [Google Scholar]
- 21.Mpiga P, Ravaoarinoro M. 2006. Effects of sustained antibiotic bactericidal treatment on Chlamydia trachomatis-infected epithelial-like cells (HeLa) and monocyte-like cells (THP-1 and U-937). Int. J. Antimicrob. Agents 27:316–324. 10.1016/j.ijantimicag.2005.11.010 [DOI] [PubMed] [Google Scholar]
- 22.Mpiga P, Ravaoarinoro M. 2006. Chlamydia trachomatis persistence: an update. Microbiol. Res. 161:9–19. 10.1016/j.micres.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 23.Moulder JW. 1993. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect. Agents Dis. 2:87–99 [PubMed] [Google Scholar]
- 24.Skilton RJ, Cutcliffen LT, Barlow D, Wang Y, Salim O, Lambden PR, Clarke IN. 2009. Penicillin induced persistence in Chlamydia trachomatis: high quality time lapse video analysis of the developmental cycle. PLoS One 4:e7723. 10.1371/journal.pone.0007723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyrick PB, Knight ST. 2004. Pre-exposure of infected human endometrial epithelial cells to penicillin in vitro renders Chlamydia trachomatis refractory to azithromycin. J. Antimicrob. Chemother. 54:79–85. 10.1093/jac/dkh283 [DOI] [PubMed] [Google Scholar]
- 26.Hoestgaard-Jensen K, Christiansen G, Honore B, Birkelund S. 2011. Influence of the Chlamydia pneumoniae AR39 bacteriophage varphiCPAR39 on chlamydial inclusion morphology. FEMS Immunol. Med. Microbiol. 62:148–156. 10.1111/j.1574-695X.2011.00795.x [DOI] [PubMed] [Google Scholar]
- 27.Villegas E, Sorlozano A, Gutierrez J. 2010. Serological diagnosis of Chlamydia pneumoniae infection: limitations and perspectives. J. Med. Microbiol. 59:1267–1274. 10.1099/jmm.0.020362-0 [DOI] [PubMed] [Google Scholar]
- 28.Beaty CD, Grayston JT, Wang SP, Kuo CC, Reto CS, Martin TR. 1991. Chlamydia pneumoniae, strain TWAR, infection in patients with chronic obstructive pulmonary disease. Am. Rev. Respir. Dis. 144:1408–1410. 10.1164/ajrccm/144.6.1408 [DOI] [PubMed] [Google Scholar]
- 29.Bas S, Cunningham T, Kvien TK, Glennas A, Melby K, Vischer TL. 1996. Synovial fluid and serum antibodies against Chlamydia in different forms of arthritis: intra-articular IgA production in Chlamydia sexually acquired reactive arthritis. Br. J. Rheumatol. 35:548–552. 10.1093/rheumatology/35.6.548 [DOI] [PubMed] [Google Scholar]
- 30.Bauriedel G, Andrie R, Likungu JA, Welz A, Braun P, Welsch U, Luderitz B. 1999. Persistence of Chlamydia pneumoniae in coronary plaque tissue: a contribution to the infection and immune hypothesis concerning unstable angina. Dtsch. Med. Wochenschr. 124:1408–1413 (In German.) 10.1055/s-2007-1024554 [DOI] [PubMed] [Google Scholar]
- 31.Hahn D, Schure A, Patel K, Childs T, Drizik E, Webley W. 2012. Chlamydia pneumoniae-specific IgE is prevalent in asthma and is associated with disease severity. PLoS One 7:e35945. 10.1371/journal.pone.0035945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borel N, Mukhopadhyay S, Kaiser C, Sullivan ED, Miller RD, Timms P, Summersgill JT, Ramirez JA, Pospischil A. 2006. Tissue MicroArray (TMA) analysis of normal and persistent Chlamydophila pneumoniae infection. BMC Infect. Dis. 6:152. 10.1186/1471-2334-6-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clancy R, Ren Z, Pang G, Fletcher P, D'Este C. 2006. Chronic Chlamydia pneumoniae infection may promote coronary artery disease in humans through enhancing secretion of interleukin-4. Clin. Exp. Immunol. 146:197–202. 10.1111/j.1365-2249.2006.03185.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clementsen P, Permin H, Norn S. 2002. Chlamydia pneumoniae infection and its role in asthma and chronic obstructive pulmonary disease. J. Investig. Allergol. Clin. Immunol. 12:73–79 [PubMed] [Google Scholar]
- 35.Cochrane M, Pospischil A, Walker P, Gibbs H, Timms P. 2003. Distribution of Chlamydia pneumoniae DNA in atherosclerotic carotid arteries: significance for sampling procedures. J. Clin. Microbiol. 41:1454–1457. 10.1128/JCM.41.4.1454-1457.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cochrane M, Pospischil A, Walker P, Gibbs H, Timms P. 2005. Discordant detection of Chlamydia pneumoniae in patients with carotid artery disease using polymerase chain reaction, immunofluorescence microscopy and serological methods. Pathology 37:69–75. 10.1080/00313020400011284 [DOI] [PubMed] [Google Scholar]
- 37.Cochrane M, Walker P, Gibbs H, Timms P. 2005. Multiple genotypes of Chlamydia pneumoniae identified in human carotid plaque. Microbiology 151:2285–2290. 10.1099/mic.0.27781-0 [DOI] [PubMed] [Google Scholar]
- 38.Saikku P, Wang SP, Kleemola M, Brander E, Rusanen E, Grayston JT. 1985. An epidemic of mild pneumonia due to an unusual strain of Chlamydia psittaci. J. Infect. Dis. 151:832–839. 10.1093/infdis/151.5.832 [DOI] [PubMed] [Google Scholar]
- 39.Heiskanen-Kosma T, Korppi M, Laurila A, Jokinen C, Kleemola M, Saikku P. 1999. Chlamydia pneumoniae is an important cause of community-acquired pneumonia in school-aged children: serological results of a prospective, population-based study. Scand. J. Infect. Dis. 31:255–259. 10.1080/00365549950163536 [DOI] [PubMed] [Google Scholar]
- 40.Kauppinen M, Saikku P. 1995. Pneumonia due to Chlamydia pneumoniae: prevalence, clinical features, diagnosis, and treatment. Clin. Infect. Dis. 21(Suppl 3):S244–S252. 10.1093/clind/21.Supplement_3.S244 [DOI] [PubMed] [Google Scholar]
- 41.Kauppinen MT, Herva E, Kujala P, Leinonen M, Saikku P, Syrjala H. 1995. The etiology of community-acquired pneumonia among hospitalized patients during a Chlamydia pneumoniae epidemic in Finland. J. Infect. Dis. 172:1330–1335. 10.1093/infdis/172.5.1330 [DOI] [PubMed] [Google Scholar]
- 42.Kauppinen MT, Saikku P, Kujala P, Herva E, Syrjala H. 1996. Clinical picture of community-acquired Chlamydia pneumoniae pneumonia requiring hospital treatment: a comparison between chlamydial and pneumococcal pneumonia. Thorax 51:185–189. 10.1136/thx.51.2.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conklin L, Adjemian J, Loo J, Mandal S, Davis C, Parks S, Parsons T, McDonough B, Partida J, Thurman K, Diaz MH, Benitez A, Pondo T, Whitney CG, Winchell JM, Kendig N, Van Beneden C. 2013. Investigation of a Chlamydia pneumoniae outbreak in a Federal correctional facility in Texas. Clin. Infect. Dis. 57:639–647. 10.1093/cid/cit357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Younes HM. 7 June 2013. High prevalence of Chlamydia pneumoniae infection in an asymptomatic Jordanian population. J. Microbiol. Immunol. Infect. 10.1016/j.jmii.2013.04.004 [DOI] [PubMed] [Google Scholar]
- 45.Haider M, Rizvi M, Malik A, Azam M, Rabbani MU. 2011. Acute and chronic Chlamydia pneumoniae infection and inflammatory markers in coronary artery disease patients. J. Infect. Dev. Ctries. 5:580–586. 10.3855/jidc.1704 [DOI] [PubMed] [Google Scholar]
- 46.Haubitz M, Brunkhorst R. 2001. C-reactive protein and chronic Chlamydia pneumoniae infection–long-term predictors for cardiovascular disease and survival in patients on peritoneal dialysis. Nephrol. Dial. Transplant. 16:809–815. 10.1093/ndt/16.4.809 [DOI] [PubMed] [Google Scholar]
- 47.Hoymans VY, Bosmans JM, Ieven M, Vrints CJ. 2002. Chlamydia pneumoniae and atherosclerosis. Acta Chir. Belg. 102:317–322 [DOI] [PubMed] [Google Scholar]
- 48.Jha HC, Vardhan H, Gupta R, Varma R, Prasad J, Mittal A. 2007. Higher incidence of persistent chronic infection of Chlamydia pneumoniae among coronary artery disease patients in India is a cause of concern. BMC Infect. Dis. 7:48. 10.1186/1471-2334-7-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagy A, Keszei M, Kis Z, Budai I, Tolgyesi G, Ungvari I, Falus A, Szalai C. 2007. Chlamydophila pneumoniae infection status is dependent on the subtypes of asthma and allergy. Allergy Asthma Proc. 28:58–63. 10.2500/aap.2007.28.2957 [DOI] [PubMed] [Google Scholar]
- 50.Balin BJ, Gerard HC, Arking EJ, Appelt DM, Branigan PJ, Abrams JT, Whittum-Hudson JA, Hudson AP. 1998. Identification and localization of Chlamydia pneumoniae in the Alzheimer's brain. Med. Microbiol. Immunol. 187:23–42. 10.1007/s004300050071 [DOI] [PubMed] [Google Scholar]
- 51.Balin BJ, Little CS, Hammond CJ, Appelt DM, Whittum-Hudson JA, Gerard HC, Hudson AP. 2008. Chlamydophila pneumoniae and the etiology of late-onset Alzheimer's disease. J. Alzheimers Dis. 13:371–380 [DOI] [PubMed] [Google Scholar]
- 52.Hannu T, Puolakkainen M, Leirisalo-Repo M. 1999. Chlamydia pneumoniae as a triggering infection in reactive arthritis. Rheumatology (Oxford) 38:411–414. 10.1093/rheumatology/38.5.411 [DOI] [PubMed] [Google Scholar]
- 53.Jackson LA, Wang SP, Nazar-Stewart V, Grayston JT, Vaughan TL. 2000. Association of Chlamydia pneumoniae immunoglobulin A seropositivity and risk of lung cancer. Cancer Epidemiol. Biomarkers Prev. 9:1263–1266 [PubMed] [Google Scholar]
- 54.Kocazeybek B. 2003. Chronic Chlamydophila pneumoniae infection in lung cancer, a risk factor: a case-control study. J. Med. Microbiol. 52:721–726. 10.1099/jmm.0.04845-0 [DOI] [PubMed] [Google Scholar]
- 55.Koyi H, Brandén E, Gnarpe J, Gnarpe H, Steen B. 2001. An association between chronic infection with Chlamydia pneumoniae and lung cancer. A prospective 2-year study. APMIS 109:572–580 [DOI] [PubMed] [Google Scholar]
- 56.Karnak D, Beder S. 2002. Treatment of Chlamydia pneumoniae infection and chronic obstructive pulmonary disease. Exp. Opin. Pharmacother. 3:1461–1470. 10.1517/14656566.3.10.1461 [DOI] [PubMed] [Google Scholar]
- 57.Karnak D, Beng-sun S, Beder S, Kayacan O. 2001. Chlamydia pneumoniae infection and acute exacerbation of chronic obstructive pulmonary disease (COPD). Respir. Med. 95:811–816. 10.1053/rmed.2001.1159 [DOI] [PubMed] [Google Scholar]
- 58.Rizzo A, Paolillo R, Iafusco D, Prisco F, Romano Carratelli C. 2012. Chlamydia pneumoniae infection in adolescents with type 1 diabetes mellitus. J. Med. Microbiol. 61:1584–1590. 10.1099/jmm.0.048512-0 [DOI] [PubMed] [Google Scholar]
- 59.Wang C, Gao D, Kaltenboeck B. 2009. Acute Chlamydia pneumoniae reinfection accelerates the development of insulin resistance and diabetes in obese C57BL/6 mice. J. Infect. Dis. 200:279–287. 10.1086/599796 [DOI] [PubMed] [Google Scholar]
- 60.Chen Z, Ji W, Wang Y, Yan Y, Zhu H, Shao X, Xu J. 2013. Epidemiology and associations with climatic conditions of Mycoplasma pneumoniae and Chlamydophila pneumoniae infections among Chinese children hospitalized with acute respiratory infections. Ital. J. Pediatr. 39:34. 10.1186/1824-7288-39-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berger L, Volp K, Mathews S, Speare R, Timms P. 1999. Chlamydia pneumoniae in a free-ranging giant barred frog (Mixophyes iteratus) from Australia. J. Clin. Microbiol. 37:2378–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hotzel H, Grossmann E, Mutschmann F, Sachse K. 2001. Genetic characterization of a Chlamydophila pneumoniae isolate from an African frog and comparison to currently accepted biovars. Syst. Appl. Microbiol. 24:63–66. 10.1078/0723-2020-00016 [DOI] [PubMed] [Google Scholar]
- 63.Bodetti TJ, Jacobson E, Wan C, Hafner L, Pospischil A, Rose K, Timms P. 2002. Molecular evidence to support the expansion of the hostrange of Chlamydophila pneumoniae to include reptiles as well as humans, horses, koalas and amphibians. Syst. Appl. Microbiol. 25:146–152. 10.1078/0723-2020-00086 [DOI] [PubMed] [Google Scholar]
- 64.Di Francesco A, Donati M, Mattioli L, Naldi M, Salvatore D, Poglayen G, Cevenini R, Baldelli R. 2006. Chlamydophila pneumoniae in horses: a seroepidemiological survey in Italy. New Microbiol. 29:303–305 [PubMed] [Google Scholar]
- 65.Pantchev A, Sting R, Bauerfeind R, Tyczka J, Sachse K. 2010. Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comp. Immunol. Microbiol. Infect. Dis. 33:473–484. 10.1016/j.cimid.2009.08.002 [DOI] [PubMed] [Google Scholar]
- 66.Theegarten D, Sachse K, Mentrup B, Fey K, Hotzel H, Anhenn O. 2008. Chlamydophila spp. infection in horses with recurrent airway obstruction: similarities to human chronic obstructive disease. Respir. Res. 9:14. 10.1186/1465-9921-9-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bodetti TJ, Viggers K, Warren K, Swan R, Conaghty S, Sims C, Timms P. 2003. Wide range of Chlamydiales types detected in native Australian mammals. Vet. Microbiol. 96:177–187. 10.1016/S0378-1135(03)00211-6 [DOI] [PubMed] [Google Scholar]
- 68.Feder A, Skipper J, Blair JR, Buchholz K, Mathew SJ, Schwarz M, Doucette JT, Alonso A, Collins KA, Neumeister A, Charney DS. 2011. Tryptophan depletion and emotional processing in healthy volunteers at high risk for depression. Biol. Psychiatry 69:804–807. 10.1016/j.biopsych.2010.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mace JL, Porter RJ, Dalrymple-Alford JC, Wesnes KA, Anderson TJ. 2011. The effects of acute tryptophan depletion on neuropsychological function, mood and movement in the healthy elderly. J. Psychopharmacol. 25:1337–1343. 10.1177/0269881110389094 [DOI] [PubMed] [Google Scholar]
- 70.Fallarino F, Grohmann U, Puccetti P. 2012. Indoleamine 2,3-dioxygenase: from catalyst to signaling function. Eur. J. Immunol. 42:1932–1937. 10.1002/eji.201242572 [DOI] [PubMed] [Google Scholar]
- 71.Munn DH, Mellor AL. 2013. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 34:137–143. 10.1016/j.it.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roan NR, Starnbach MN. 2008. Immune-mediated control of Chlamydia infection. Cell. Microbiol. 10:9–19. 10.1111/j.1462-5822.2007.01069.x [DOI] [PubMed] [Google Scholar]
- 73.Farris CM, Morrison RP. 2011. Vaccination against Chlamydia genital infection utilizing the murine C. muridarum model. Infect. Immun. 79:986–996. 10.1128/IAI.00881-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rank RG, Whittum-Hudson JA. 2010. Protective immunity to chlamydial genital infection: evidence from animal studies. J. Infect. Dis. 201(Suppl 2):S168–S177. 10.1086/652399 [DOI] [PubMed] [Google Scholar]
- 75.Rothfuchs AG, Kreuger MR, Wigzell H, Rottenberg ME. 2004. Macrophages, CD4+ or CD8+ cells are each sufficient for protection against Chlamydia pneumoniae infection through their ability to secrete IFN-gamma. J. Immunol. 172:2407–2415 [DOI] [PubMed] [Google Scholar]
- 76.Rothfuchs AG, Trumstedt C, Wigzell H, Rottenberg ME. 2004. Intracellular bacterial infection-induced IFN-gamma is critically but not solely dependent on Toll-like receptor 4-myeloid differentiation factor 88-IFN-alpha beta-STAT1 signaling. J. Immunol. 172:6345–6353 [DOI] [PubMed] [Google Scholar]
- 77.Batteiger BE, Tu W, Ofner S, Van Der Pol B, Stothard DR, Orr DP, Katz BP, Fortenberry JD. 2010. Repeated Chlamydia trachomatis genital infections in adolescent women. J. Infect. Dis. 201:42–51. 10.1086/648734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Halme S, Latvala J, Karttunen R, Palatsi I, Saikku P, Surcel HM. 2000. Cell-mediated immune response during primary Chlamydia pneumoniae infection. Infect. Immun. 68:7156–7158. 10.1128/IAI.68.12.7156-7158.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bastidas RJ, Elwell CA, Engel JN, Valdivia RH. 2013. Chlamydial intracellular survival strategies. Cold Spring Harb. Perspect. Med. 3:a010256. 10.1101/cshperspect.a010256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brunham RC, Rey-Ladino J. 2005. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat. Rev. Immunol. 5:149–161. 10.1038/nri1551 [DOI] [PubMed] [Google Scholar]
- 81.Pawliczak R, Logun C, Madara P, Barb J, Suffredini AF, Munson PJ, Danner RL, Shelhamer JH. 2005. Influence of IFN-gamma on gene expression in normal human bronchial epithelial cells: modulation of IFN-gamma effects by dexamethasone. Physiol. Genomics 23:28–45. 10.1152/physiolgenomics.00011.2005 [DOI] [PubMed] [Google Scholar]
- 82.Gupta SL, Carlin JM, Pyati P, Dai W, Pfefferkorn ER, Murphy MJ., Jr 1994. Antiparasitic and antiproliferative effects of indoleamine 2,3-dioxygenase enzyme expression in human fibroblasts. Infect. Immun. 62:2277–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Igietseme JU, Uriri IM, Chow M, Abe E, Rank RG. 1997. Inhibition of intracellular multiplication of human strains of Chlamydia trachomatis by nitric oxide. Biochem. Biophys. Res. Commun. 232:595–601. 10.1006/bbrc.1997.6335 [DOI] [PubMed] [Google Scholar]
- 84.Freidank HM, Billing H, Wiedmann-Al-Ahmad M. 2001. Influence of iron restriction on Chlamydia pneumoniae and C. trachomatis. J. Med. Microbiol. 50:223–227 [DOI] [PubMed] [Google Scholar]
- 85.Pfefferkorn ER. 1984. Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc. Natl. Acad. Sci. U. S. A. 81:908–912. 10.1073/pnas.81.3.908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Murray HW, Szuro-Sudol A, Wellner D, Oca MJ, Granger AM, Libby DM, Rothermel CD, Rubin BY. 1989. Role of tryptophan degradation in respiratory burst-independent antimicrobial activity of gamma interferon-stimulated human macrophages. Infect. Immun. 57:845–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Adams O, Besken K, Oberdorfer C, MacKenzie CR, Takikawa O, Daubener W. 2004. Role of indoleamine-2,3-dioxygenase in alpha/beta and gamma interferon-mediated antiviral effects against herpes simplex virus infections. J. Virol. 78:2632–2636. 10.1128/JVI.78.5.2632-2636.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Byrne GI, Lehmann LK, Kirschbaum JG, Borden EC, Lee CM, Brown RR. 1986. Induction of tryptophan degradation in vitro and in vivo: a gamma-interferon-stimulated activity. J. Interferon Res. 6:389–396. 10.1089/jir.1986.6.389 [DOI] [PubMed] [Google Scholar]
- 89.Byrne GI, Lehmann LK, Landry GJ. 1986. Induction of tryptophan catabolism is the mechanism for gamma-interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect. Immun. 53:347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Divanovic S, Sawtell NM, Trompette A, Warning JI, Dias A, Cooper AM, Yap GS, Arditi M, Shimada K, Duhadaway JB, Prendergast GC, Basaraba RJ, Mellor AL, Munn DH, Aliberti J, Karp CL. 2012. Opposing biological functions of tryptophan catabolizing enzymes during intracellular infection. J. Infect. Dis. 205:152–161. 10.1093/infdis/jir621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boasso A. 2011. Wounding the immune system with its own blade: HIV-induced tryptophan catabolism and pathogenesis. Curr. Med. Chem. 18:2247–2256. 10.2174/092986711795656126 [DOI] [PubMed] [Google Scholar]
- 92.Larrea E, Riezu-Boj JI, Gil-Guerrero L, Casares N, Aldabe R, Sarobe P, Civeira MP, Heeney JL, Rollier C, Verstrepen B, Wakita T, Borras-Cuesta F, Lasarte JJ, Prieto J. 2007. Upregulation of indoleamine 2,3-dioxygenase in hepatitis C virus infection. J. Virol. 81:3662–3666. 10.1128/JVI.02248-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rollier CS, Paranhos-Baccala G, Verschoor EJ, Verstrepen BE, Drexhage JA, Fagrouch Z, Berland JL, Komurian-Pradel F, Duverger B, Himoudi N, Staib C, Meyr M, Whelan M, Whelan JA, Adams VC, Larrea E, Riezu JI, Lasarte JJ, Bartosch B, Cosset FL, Spaan WJ, Diepolder HM, Pape GR, Sutter G, Inchauspe G, Heeney JL. 2007. Vaccine-induced early control of hepatitis C virus infection in chimpanzees fails to impact on hepatic PD-1 and chronicity. Hepatology 45:602–613. 10.1002/hep.21573 [DOI] [PubMed] [Google Scholar]
- 94.Mittal D, Kassianos AJ, Tran LS, Bergot AS, Gosmann C, Hofmann J, Blumenthal A, Leggatt GR, Frazer IH. 2013. Indoleamine 2,3-dioxygenase activity contributes to local immune suppression in the skin expressing human papillomavirus oncoprotein e7. J. Invest. Dermatol. 133:2686–2694. 10.1038/jid.2013.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Popov A, Driesen J, Abdullah Z, Wickenhauser C, Beyer M, Debey-Pascher S, Saric T, Kummer S, Takikawa O, Domann E, Chakraborty T, Kronke M, Utermohlen O, Schultze JL. 2008. Infection of myeloid dendritic cells with Listeria monocytogenes leads to the suppression of T cell function by multiple inhibitory mechanisms. J. Immunol. 181:4976–4988 [DOI] [PubMed] [Google Scholar]
- 96.Desvignes L, Ernst JD. 2009. Interferon-gamma-responsive nonhematopoietic cells regulate the immune response to Mycobacterium tuberculosis. Immunity 31:974–985. 10.1016/j.immuni.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng SC, van de Veerdonk F, Smeekens S, Joosten LA, van der Meer JW, Kullberg BJ, Netea MG. 2010. Candida albicans dampens host defense by downregulating IL-17 production. J. Immunol. 185:2450–2457. 10.4049/jimmunol.1000756 [DOI] [PubMed] [Google Scholar]
- 98.Montagnoli C, Fallarino F, Gaziano R, Bozza S, Bellocchio S, Zelante T, Kurup WP, Pitzurra L, Puccetti P, Romani L. 2006. Immunity and tolerance to Aspergillus involve functionally distinct regulatory T cells and tryptophan catabolism. J. Immunol. 176:1712–1723 [DOI] [PubMed] [Google Scholar]
- 99.Fehlner-Gardiner C, Roshick C, Carlson JH, Hughes S, Belland RJ, Caldwell HD, McClarty G. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893–26903. 10.1074/jbc.M203937200 [DOI] [PubMed] [Google Scholar]
- 100.Caldwell HD, Wood H, Crane C, Bailey R, Jones RB, Mabey D, Maclean I, Mohammed Z, Peeling R, Roshick C, Schachter J, Solomon AW, Stamm WE, Suchland RJ, Taylor L, West SK, Quinn TC, Belland RJ, McClarty G. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Invest. 111:1757–1769. 10.1172/JCI17993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morrison RP. 2000. Differential sensitivities of Chlamydia trachomatis strains to inhibitory effects of gamma interferon. Infect. Immun. 68:6038–6040. 10.1128/IAI.68.10.6038-6040.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Read TD, Myers GS, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, Holtzapple E, Khouri H, Federova NB, Carty HA, Umayam LA, Haft DH, Peterson J, Beanan MJ, White O, Salzberg SL, Hsia RC, McClarty G, Rank RG, Bavoil PM, Fraser CM. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134–2147. 10.1093/nar/gkg321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xie G, Bonner CA, Jensen RA. 2002. Dynamic diversity of the tryptophan pathway in chlamydiae: reductive evolution and a novel operon for tryptophan recapture. Genome Biol. 3:research0051. 10.1186/gb-2002-3-9-research0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wood H, Roshick C, McClarty G. 2004. Tryptophan recycling is responsible for the interferon-gamma resistance of Chlamydia psittaci GPIC in indoleamine dioxygenase-expressing host cells. Mol. Microbiol. 52:903–916. 10.1111/j.1365-2958.2004.04029.x [DOI] [PubMed] [Google Scholar]
- 105.Nelson DE, Virok DP, Wood H, Roshick C, Johnson RM, Whitmire WM, Crane DD, Steele-Mortimer O, Kari L, McClarty G, Caldwell HD. 2005. Chlamydial IFN-gamma immune evasion is linked to host infection tropism. Proc. Natl. Acad. Sci. U. S. A. 102:10658–10663. 10.1073/pnas.0504198102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roulis E, Polkinghorne A, Timms P. 2013. Chlamydia pneumoniae: modern insights into an ancient pathogen. Trends Microbiol. 21:120–128. 10.1016/j.tim.2012.10.009 [DOI] [PubMed] [Google Scholar]
- 107.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397–1406. 10.1093/nar/28.6.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shirai M, Hirakawa H, Kimoto M, Tabuchi M, Kishi F, Ouchi K, Shiba T, Ishii K, Hattori M, Kuhara S, Nakazawa T. 2000. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 28:2311–2314. 10.1093/nar/28.12.2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman RW, Olinger L, Grimwood J, Davis RW, Stephens RS. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385–389. 10.1038/7716 [DOI] [PubMed] [Google Scholar]
- 110.Myers GS, Mathews SA, Eppinger M, Mitchell C, O'Brien KK, White OR, Benahmed F, Brunham RC, Read TD, Ravel J, Bavoil PM, Timms P. 2009. Evidence that human Chlamydia pneumoniae was zoonotically acquired. J. Bacteriol. 191:7225–7233. 10.1128/JB.00746-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Roulis E, Bachmann N, Polkinghorne A, Hammerschlag M, Kohlhoff S, Timms P. 6 February 2014. Draft genome and plasmid sequences of Chlamydia pneumoniae strain B21 from an Australian endangered marsupial, the Western Barred Bandicoot. Genome Announc. 10.1128/genomeA.01223-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mitchell CM, Mathews SA, Theodoropoulos C, Timms P. 2009. In vitro characterisation of koala Chlamydia pneumoniae: morphology, inclusion development and doubling time. Vet. Microbiol. 136:91–99. 10.1016/j.vetmic.2008.10.008 [DOI] [PubMed] [Google Scholar]
- 113.Pantoja LG, Miller RD, Ramirez JA, Molestina RE, Summersgill JT. 2001. Characterization of Chlamydia pneumoniae persistence in HEp-2 cells treated with gamma interferon. Infect. Immun. 69:7927–7932. 10.1128/IAI.69.12.7927-7932.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Summersgill JT, Sahney NN, Gaydos CA, Quinn TC, Ramirez JA. 1995. Inhibition of Chlamydia pneumoniae growth in HEp-2 cells pretreated with gamma interferon and tumor necrosis factor alpha. Infect. Immun. 63:2801–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mehta SJ, Miller RD, Ramirez JA, Summersgill JT. 1998. Inhibition of Chlamydia pneumoniae replication in HEp-2 cells by interferon-gamma: role of tryptophan catabolism. J. Infect. Dis. 177:1326–1331. 10.1086/515287 [DOI] [PubMed] [Google Scholar]
- 116.Pantoja LG, Miller RD, Ramirez JA, Molestina RE, Summersgill JT. 2000. Inhibition of Chlamydia pneumoniae replication in human aortic smooth muscle cells by gamma interferon-induced indoleamine 2, 3-dioxygenase activity. Infect. Immun. 68:6478–6481. 10.1128/IAI.68.11.6478-6481.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Al-Younes HM, Gussmann J, Braun PR, Brinkmann V, Meyer TF. 2006. Naturally occurring amino acids differentially influence the development of Chlamydia trachomatis and Chlamydia (Chlamydophila) pneumoniae. J. Med. Microbiol. 55:879–886. 10.1099/jmm.0.46445-0 [DOI] [PubMed] [Google Scholar]
- 118.Gieffers J, Durling L, Ouellette SP, Rupp J, Maass M, Byrne GI, Caldwell HD, Belland RJ. 2003. Genotypic differences in the Chlamydia pneumoniae tyrP locus related to vascular tropism and pathogenicity. J. Infect. Dis. 188:1085–1093. 10.1086/378692 [DOI] [PubMed] [Google Scholar]
- 119.Schoborg RV. 2011. Chlamydia persistence – a tool to dissect chlamydia-host interactions. Microbes Infect. 13:649–662. 10.1016/j.micinf.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wyrick PB. 2010. Chlamydia trachomatis persistence in vitro: an overview. J. Infect. Dis. 201(Suppl 2):S88–S95. 10.1086/652394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lo CC, Xie G, Bonner CA, Jensen RA. 2012. The alternative translational profile that underlies the immune-evasive state of persistence in Chlamydiaceae exploits differential tryptophan contents of the protein repertoire. Microbiol. Mol. Biol. Rev. 76:405–443. 10.1128/MMBR.05013-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bonner CA, Byrne GI, Jensen RA. 28 February 2014. Chlamydia exploit the mammalian tryptophan-depletion defense strategy as a counter-defensive cue to trigger a survival state of persistence. Front. Cell. Infect. Microbiol. 10.3389/fcimb.2014.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Timms P, Good D, Wan C, Theodoropoulos C, Mukhopadhyay S, Summersgill J, Mathews S. 2009. Differential transcriptional responses between the interferon-gamma-induction and iron-limitation models of persistence for Chlamydia pneumoniae. J. Microbiol. Immunol. Infect. 42:27–37 [PubMed] [Google Scholar]
- 124.Schaumburg CS, Tan M. 2006. Arginine-dependent gene regulation via the ArgR repressor is species specific in Chlamydia. J. Bacteriol. 188:919–927. 10.1128/JB.188.3.919-927.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Giles TN, Graham DE. 2007. Characterization of an acid-dependent arginine decarboxylase enzyme from Chlamydophila pneumoniae. J. Bacteriol. 189:7376–7383. 10.1128/JB.00772-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thomas SR, Terentis AC, Cai H, Takikawa O, Levina A, Lay PA, Freewan M, Stocker R. 2007. Post-translational regulation of human indoleamine 2,3-dioxygenase activity by nitric oxide. J. Biol. Chem. 282:23778–23787. 10.1074/jbc.M700669200 [DOI] [PubMed] [Google Scholar]
- 127.Giles TN, Fisher DJ, Graham DE. 2009. Independent inactivation of arginine decarboxylase genes by nonsense and missense mutations led to pseudogene formation in Chlamydia trachomatis serovar L2 and D strains. BMC Evol. Biol. 9:166. 10.1186/1471-2148-9-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Roshick C, Wood H, Caldwell HD, McClarty G. 2006. Comparison of gamma interferon-mediated antichlamydial defense mechanisms in human and mouse cells. Infect. Immun. 74:225–238. 10.1128/IAI.74.1.225-238.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rothfuchs AG, Gigliotti D, Palmblad K, Andersson U, Wigzell H, Rottenberg ME. 2001. IFN-alpha beta-dependent, IFN-gamma secretion by bone marrow-derived macrophages controls an intracellular bacterial infection. J. Immunol. 167:6453–6461 [DOI] [PubMed] [Google Scholar]
- 130.Rottenberg ME, Gigliotti Rothfuchs A, Gigliotti D, Ceausu M, Une C, Levitsky V, Wigzell H. 2000. Regulation and role of IFN-gamma in the innate resistance to infection with Chlamydia pneumoniae. J. Immunol. 164:4812–4818 [DOI] [PubMed] [Google Scholar]