Abstract
In Sinorhizobium meliloti, catabolite repression is influenced by a noncanonical nitrogen-type phosphotransferase system (PTSNtr). In this PTSNtr, the protein HPr is phosphorylated on histidine-22 by the enzyme EINtr and the flux of phosphate through this residue onto downstream proteins leads to an increase in succinate-mediated catabolite repression (SMCR). In order to explore the molecular determinants of HPr phosphorylation by EINtr, both proteins were purified and the activity of EINtr was measured. Experimentally determined kinetic parameters of EINtr activity were significantly slower than those determined for the carbohydrate-type EI in Escherichia coli. Enzymatic assays showed that glutamine, a signal of nitrogen availability in many Gram-negative bacteria, strongly inhibits EINtr. Binding experiments using the isolated GAF domain of EINtr (EIGAF) showed that it is the domain responsible for detection of glutamine. EINtr activity was not affected by α-ketoglutarate, and no binding between the EIGAF and α-ketoglutarate could be detected. These data suggest that in S. melilloti, EINtr phosphorylation of HPr is regulated by signals from both carbon metabolism (phosphoenolpyruvate) and nitrogen metabolism (glutamine).
INTRODUCTION
The alphaproteobacterium Sinorhizobium meliloti is a free-living soil bacterium capable of forming symbioses with certain leguminous plants, such as Medicago sativa (alfalfa) and Medicago truncatula (barrel medic) (1). S. meliloti is a metabolically diverse organism, capable of catabolizing a wide range of carbon compounds (2, 3). Of these compounds, C4-dicarboxylic acids, such as succinate, play an important role for S. meliloti. During symbiosis, dicarboxylates provided by the plant are required for nitrogen fixation (4). While dicarboxylates are also available in the rhizosphere, they are present in a heterogeneous mixture containing many other carbon sources (5). A likely adaptation to this environment is a form of carbon catabolite repression (CCR) termed succinate-mediated catabolite repression (SMCR), which differs from the well-studied Escherichia coli and Bacillus subtilis CCR. SMCR is a global physiological response to nutrient availability that culminates in the preferential use of succinate over other carbon sources during growth. SMCR can impose inducer exclusion and repression of genes needed for the transport and catabolism of secondary carbons sources, such as lactose and raffinose (6, 7).
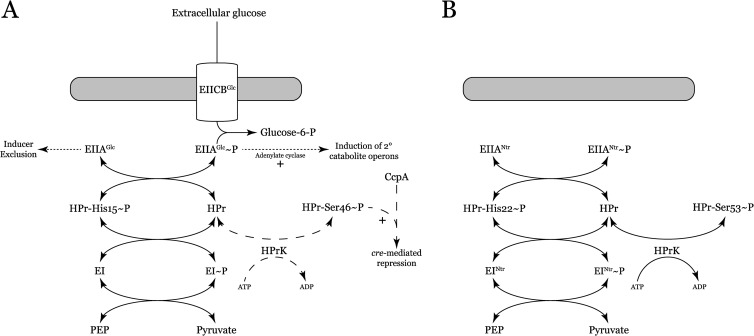
CCR in bacteria is often controlled by a group of proteins collectively termed the phosphotransferase system (PTS). The canonical Gram-negative model of the PTS (based on the PTS of enteric bacteria) includes two general proteins: enzyme I (EI) and HPr, as well as proteins with sugar-specific EIIA, EIIB, and EIIC domains (Fig. 1A). During growth on glucose, EI will autophosphorylate, using phosphoenolpyruvate (PEP) as the phosphate donor (8). EI transfers the phosphate to HPr histidine-15 and then onto EIIAGlc. EIIAGlc will pass the phosphate to the EIICBGlc transport protein, which phosphorylates the incoming sugar (9). When EIIAGlc is unphosphorylated, it is capable of binding secondary solute transporters (e.g., the lactose permease), preventing uptake and ensuring the genes required for secondary catabolism are not induced (inducer exclusion) (10–12). Once the PTS substrate is exhausted, phosphorylated EIIAGlc begins to accumulate. The phosphorylated form of EIIAGlc has a low affinity for secondary substrate transporters and stimulates adenylate cyclase activity, which synthesizes cyclic AMP (cAMP; a global inducer of catabolic operons) (13).
FIG 1.
(A) Model of the canonical PTS. Dotted lines represent pathways specific to E. coli, and dashed lines are those specific to B. subtilis. (B) Proposed model of the S. meliloti PTS. Note the lack of membrane permeases and the presence of an HPrK in a Gram-negative bacterium. Not shown is phosphorylation of EIIAman (ManX) by HPr-His22-P.
The traditional Gram-positive PTS model (based on that of B. subtilis) is similar structurally to the Gram-negative model of PTS, but it differs mechanistically. There is an additional enzyme, HPrK, that phosphorylates HPr at serine-46 by using ATP as a substrate (14). HPr-Ser46-P interacts with the regulatory protein CcpA to repress the genes required for utilization of secondary carbon substrates (15). In certain Gram-positive organisms, HPr-Ser46-P is also responsible for inducer exclusion (16).
Transport operons (largely the ABC type) account for 12% of the S. meliloti genome, allowing the organism to import a wide variety of carbon sources present in soil (17). Despite the abundance of transporters, S. meliloti lacks PTS-type membrane transport proteins. The genome encodes a set of PTS proteins homologous to the E. coli PTSNtr, as well as the enzyme HPrK (initially thought to be present only in Gram-positive organisms) and a mannose-type EIIA (EIIAMan or ManX) (Fig. 1B). N-terminal GAF domains are a hallmark of EINtr, including the enzyme produced by S. meliloti (18). These domains are widely distributed and often responsible for detection of small-molecule signals, such as cGMP and α-ketoglutarate (19, 20). This abbreviated PTS organization that includes a HPrK is found in many of the alphaproteobacteria (21).
Previous work in our lab demonstrated that the PTS proteins expressed by S. meliloti regulate SMCR. HPr of S. meliloti can be phosphorylated on either histidine-22 by EINtr or serine-53 by HPrK (Fig. 1B). An increase in the phosphate flow through HPr histidine-22 results in a concomitant increase in SMCR. In addition to SMCR, we have shown that the PTSNtr regulates symbiosis, succinoglycan production, and trace element requirements (6, 22). To elucidate the mechanism of the PTSNtr, we have characterized the transfer of phosphate from PEP to HPr via EINtr by using biochemical techniques.
MATERIALS AND METHODS
Media, strains, and plasmids.
E. coli XL1-Blue (Stratagene) was used for all cloning steps and was grown in LB supplemented with either ampicillin (100 μg/ml) or kanamycin (25 μg/ml). EINtr was expressed in Tuner (DE3)pLysS (Novagen), and HPr and EIGAF were expressed in BL21(DE3) (Novagen). Media recipes and the nomenclature used for protein expression are those of reference 23, with tryptone used in place of N-Z-Amine, and all cultures contained 100 μg/ml kanamycin.
Construction of expression plasmids.
The genes for HPr and EINtr, hpr (smc02754) and ptsP (smc02437), respectively, were amplified using high-fidelity Phusion polymerase (New England BioLabs) and primers that allowed for cloning into the His-tagging vector pET28a(+) (Novagen). The primers for amplifying hpr were hpr_UP (5′-atgatgGCTAGCcgccccgacacggctct-3′) and hpr_DOWN (5′-atgtttaCTCGAGtcacatctcttcg-3′. The portions of the sequences in capital letters indicate NheI and XhoI sites in the upstream and downstream primers used for cloning the amplified fragment into pET28a(+). The primers for amplifying full-length ptsP were ptsP_UP (5′-cacgcgatgGCTAGcctttccgcaggtccgcg-3′) and ptsP_DOWN (5′-ctcttgGAATTCctaaaccggtatgccgt-3′). The capitalized portions of the sequences indicate NheI and EcoRI sites in the upstream and downstream primers used for cloning the amplified fragment into pET28a(+). The N-terminal GAF domain of EINtr (encoding residues 2 to 162) was amplified using primers ptsP_UP and GAF_DOWN (5′-GGATCCtcaggtcgcgaccatctcgg-3′). The capitalized portion of the sequence indicates the BamHI site in the downstream primer used for cloning the amplified fragment into pET28a(+). Following amplification, the products were A-tailed and cloned into pGEM T-Easy (Promega). Next, the inserts were excised from the plasmid by cutting at the sites indicated in the primers above and cloned into pET28a(+), giving pDG142H, pDG142P, and pRG07, which expressed His6-tagged versions of HPr, EINtr, and EIGAF, respectively. All three plasmids were sequenced to confirm that errors had not been introduced during amplification or cloning.
Expression of recombinant proteins.
All three strains were grown overnight in MDG (medium phosphate plus aspartate and glucose) prior to inoculation of expression media. To express EINtr, Tuner(DE3)pLysS/pDG142P was diluted into ZYM 505 and grown at 37°C for 5 h. The culture was cooled to 18°C and induced overnight with 0.5 mM isopropyl-β-d-galactopyranoside (IPTG). BL21(DE3)/pDG142H was diluted into ZYM 522, grown at 37°C for 2 h, and then incubated for an additional 24 h at 18°C. BL21(DE3)/pRG07 was diluted into ZYM 5052 and grown overnight at 37°C. Following induction, the cultures were harvested by centrifugation and stored as pellets at −20°C.
Purification of recombinant EINtr.
All purification steps were performed at 4°C. Cell pellets were resuspended in 50 mM NaCl, 5 mM MgSO4, 50 mM Tris-Cl (pH 8.0) supplemented with 2.5 U/ml benzonase and 0.1 mg/ml lysozyme and with Halt protease inhibitors (Thermo) and incubated for 2 h. Cells were lysed by sonication for 6 min in 30-s bursts. The extract was clarified by centrifugation at 3,500 × g for 15 min and then 30,000 × g for 30 min. The crude extract was passed through a 0.45-μm syringe filter (Corning), and imidazole was added to 20 mM. The protein was loaded onto a 1-ml HisTrapFF Crude Ni2+ column (GE Healthcare) equilibrated in binding buffer (0.5 M NaCl, 25 mM imidazole, 50 mM Tris-Cl [pH 7.5], 10% glycerol). The column was washed with 20 ml of binding buffer, and then EINtr was eluted in 20 ml of binding buffer containing 0.5 M imidazole. The protein was immediately diluted 2-fold in 50 mM NaCl, 0.1 M imidazole, 1 mM dithiothreitol (DTT), 1 mM EDTA, 5 mM magnesium acetate [Mg(OAc)2], 50 mM Tris-Cl (pH 7.5), and 10% glycerol, and 20 U of bovine thrombin (GE Healthcare) was added to remove the His tag. The protein was dialyzed overnight against the dilution buffer, followed by a 2-hour dialysis against the same buffer with imidazole omitted. The dialyzed protein was centrifuged at 15,000 × g for 30 min, and uncut protein was removed with the HisTrap column after equilibration in dialysis buffer. The untagged protein was loaded onto a 5-ml Q Sepharose FF column (GE Healthcare) equilibrated in Q binding buffer (50 mM NaCl, 1 mM DTT, 50 mM Tris-Cl [pH 7.5], 10% glycerol). The column was washed with 50 ml of Q binding buffer and 5 ml of the same buffer with 150 mM NaCl and then eluted in 20 ml of Q binding buffer with 0.5 M NaCl. The purified EINtr was dialyzed overnight at 4°C against 50 mM NaCl, 5 mM Mg(OAc)2, 1 mM EDTA, 1 mM DTT, 25 mM HEPES (pH 7.5), 10% glycerol, passed through a 0.22-μm syringe filter, and stored in aliquots at −80°C. This resulted in approximately a 10 mg liter−1 culture of >90% pure EINtr, based on Coomassie staining (24). The concentration of EINtr was measured spectrophotometrically at 280 nm by using a molar extinction coefficient (ε280) of 37,360 M−1 cm−1 with a Biotek Synergy HT apparatus and UV-transparent 96-well plates (Falcon).
Purification of recombinant HPr and EIGAF.
For purification of HPr, cells were lysed as described above with DTT omitted from all buffers and with NaHPO4 replacing Tris-Cl at the same concentration and pH. All purification steps were performed at 4°C. After elution from the HisTrap column, HPr was dialyzed overnight against 0.25 M NaCl, 5 mM Na2SO4, 1 mM EDTA, 50 mM NaHPO4 (pH 7.4), 10% glycerol at 4°C. For native electrophoresis assays, the His6 tag was removed during dialysis with 20 U of bovine thrombin, and uncut protein was removed as described above. The protein was passed through a 0.2-μm syringe filter and concentrated to 1 ml with an Amicon 3-kDa centrifugal filter (Millipore), then loaded onto a 65-ml S75 column equilibrated in 150 mM NaOAc, 5 mM Na2SO4, 1 mM EDTA, 25 mM HEPES (pH 7.4), 10% glycerol run at 1 ml min−1. Purified protein was filtered and concentrated as above, then stored in aliquots at −80°C. This resulted in an approximately 5-mg liter−1 culture of homogenous HPr, as determined by Coomassie staining. EIGAF was purified using the same method as for HPr, but the dialysis buffer consisted of 150 mM NaCl, 20 mM NaHPO4 (pH 7.5) and the size exclusion buffer was 150 mM NaOAc, 25 mM HEPES (pH 7.5), 10% glycerol, with a final yield of approximately 25 mg liter−1. The concentrations of HPr and EIGAF were determined by measuring the A205, with extinction coefficients (ε205) of 27 mg−1 cm−1 (HPr) and 27.6 mg−1 cm−1 (EIGAF) (25).
Native PAGE HPr phosphorylation assay.
Reaction mixtures consisting of 0.1 M Tris-OAc (pH 8.0), 10 mM Mg(OAc)2, 3 μg HPr (14 μM final concentration), and the indicated amount of EINtr were set up in 20-μl volumes, which were incubated at room temperature for 15 min to ensure EINtr dimerization (26). PEP (5 mM) was added to initiate the reactions, which were allowed to proceed for 15 min at room temperature and then halted by the addition of ice-cold sample buffer (final concentrations, 180 mM Tris, 120 mM CAPS [N-cyclohexyl-3-aminopropanesulfonic acid], 20 mM EDTA, 12% sucrose). A 12.5-μl aliquot of each reaction mixture was loaded onto a 7.5% acrylamide gel buffered in 180 mM Tris, 120 mM CAPS (pH 9.6) (27). Gels were run for 65 min at 75 V and then Coomassie stained.
EINtr LDH assay.
EINtr lactate dehydrogenase (LDH) activity was assayed as described in reference 28, with modifications. Assays were performed in 96-well UV-transparent microtiter plates (Falcon) at 30°C in a Synergy HT automated plate reader (BioTek). Prior to the assay, 1.6 μM EINtr was preincubated in 0.1 M NaOAc, 10 mM Mg(OAc)2, 2 mM DTT, and 0.1 M HEPES (pH 8.0) at 30°C for 15 min to 1 h (no change in activity was detected during this period). Reactions were performed in the same buffer with 150 μM NADH, 10 U LDH (MP Bio), 10 mM PEP, and the specified amount of HPr in a 200-μl final volume. All solutions were prewarmed to 30°C, and EINtr was added to a final concentration of 40 nM. Phosphorylation was measured by the change in absorbance at 340 nm based on an ε340 for NADH of 6,220 M−1 cm−1 and path length of 0.45 cm for a 200-μl volume. Reaction mixtures were corrected for NADH autooxidation by subtracting the decrease in absorbance of samples lacking EINtr. Reactions were performed in triplicate, and values for kcat and Km were determined by using nonlinear regression curve fitting to the Michaelis-Menten equation using Kaleidagraph 3.1 (Synergy Software). The data were obtained from a single preparation of EINtr and three preparations of HPr. There was no detectable variation between the HPr preparations.
Isothermal titration calorimetry (ITC).
EIGAF was dialyzed overnight at 4°C against 150 mM NaOAc, 50 mM HEPES (pH 7.5), 1% glycerol. After dialysis, the protein was filtered and the final concentration was measured. The ligand for each experiment was dissolved in the dialysis buffer. Binding experiments were performed in a VP-ITC apparatus (MicroCal) at 25°C with 300 rpm stirring. A 2-μl test injection of ligand was performed, followed by 28 injections of 10 μl each. The data were corrected for background by titrating ligand into buffer alone. The data for the test injection were discarded, and the data were analyzed using the Origin software package (MicroCal).
RESULTS
Purification of EINtr, EIGAF, and HPr.
The genes encoding HPr and EINtr were successfully cloned into pET-28a(+), which allowed for overexpression of the His6-tagged proteins. Both EINtr and HPr were largely insoluble when expressed under standard conditions (data not shown), and we subsequently searched for a method that would result in sufficient soluble protein for biochemical characterization. When the induction temperature and IPTG concentration were reduced to 18°C and 0.5 mM, respectively, approximately 10 mg liter−1 of soluble EINtr could be purified from Tuner cells. HPr solubility was not significantly improved by altering the IPTG concentration, by reducing the growth temperature, or by induction in 1 M sorbitol and 2.5 mM betaine (29). Soluble expression of HPr was achieved with an optimized autoinduction protocol (23) that ultimately resulted in approximately 5 mg of purified HPr per liter of culture medium. Autoinduction improved the yield of HPr to the point where we could study its phosphorylation by EINtr. The isolated GAF domain of EINtr could be expressed at levels significantly greater than full-length EINtr, resulting in 25 mg of purified protein per liter of culture medium. The purification procedures described here resulted in highly pure proteins that migrated at their expected molecular masses of 10 kDa, 83 kDa, and 20 kDa for HPr, EINtr, and EIGAF, respectively (Fig. 2). All three proteins remained stable, with no loss of activity, after storage at −80°C for at least 8 months.
FIG 2.
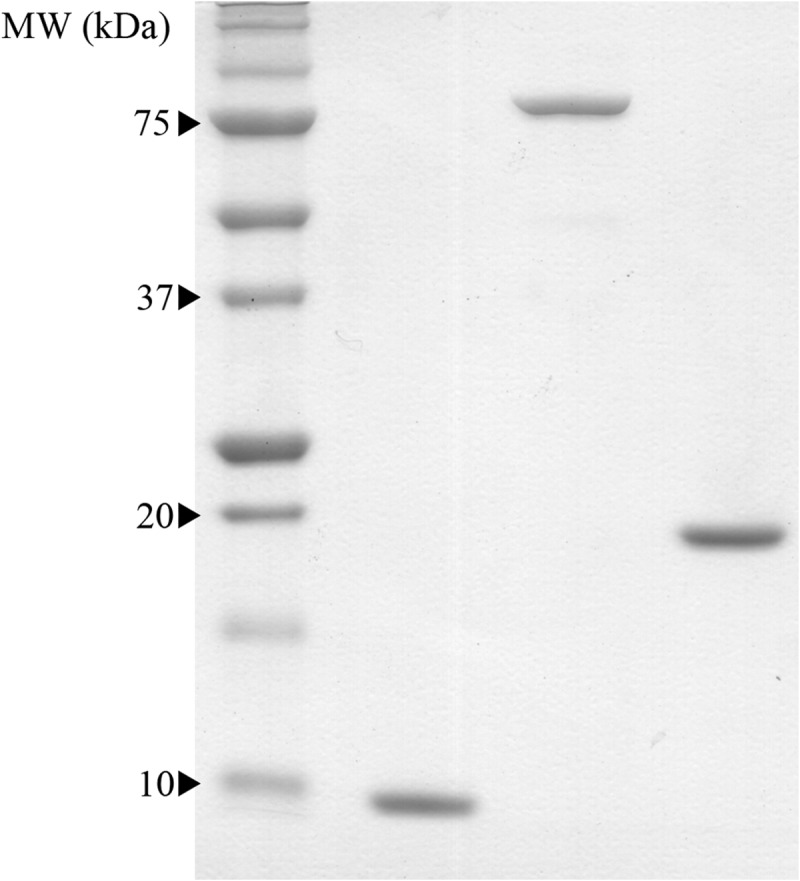
One microgram of purified HPr (left), EINtr (center), and EIGAF (right) were run on a 12% Tris-OAc SDS-polyacrylamide gel (43). All three proteins ran to their expected molecular masses of 10 kDa, 83 kDa, and 20 kDa, respectively.
Phosphorylation of HPr.
Using an in vitro phosphorylation assay that included native electrophoresis, we found that S. meliloti EINtr could use PEP to phosphorylate HPr (Fig. 3). To measure the kinetic parameters of this reaction, an LDH coupled assay was adapted for a microplate reader. The Michaelis-Menten constants of EINtr for HPr were determined: Km of 12.8 ± 1.5 μM, kcat of 1.11 ± 0.4 s−1, and kcat/Km ratio of 8.67 × 104 M−1 s−1. Values are the result of a single fit to all of the data, with the standard error of the fit calculated by the software. The determined kcat/Km ratio is approximately 3 × 103-fold lower than that of the E. coli carbohydrate EI (30), although the two enzymes were purified and assayed under different conditions. The kinetic data were obtained with a single preparation of EINtr. After these experiments were performed, we developed an autoinduction protocol that allowed for soluble expression of EINtr at 37°C. This enzyme was purified using the same protocol and while the yield was improved, its specific activity was unchanged (data not shown).
FIG 3.
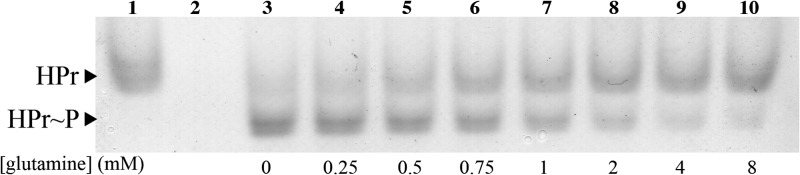
Native gel analysis of HPr phosphorylation by 100 ng of EINtr. Lane 1, reaction mixture contained EINtr and HPr without PEP; lane 2, reaction mixture lacked HPr; lanes 3 to 10, full phosphorylation reaction mixtures containing the indicated concentrations of glutamine.
Inhibition of EINtr by glutamine.
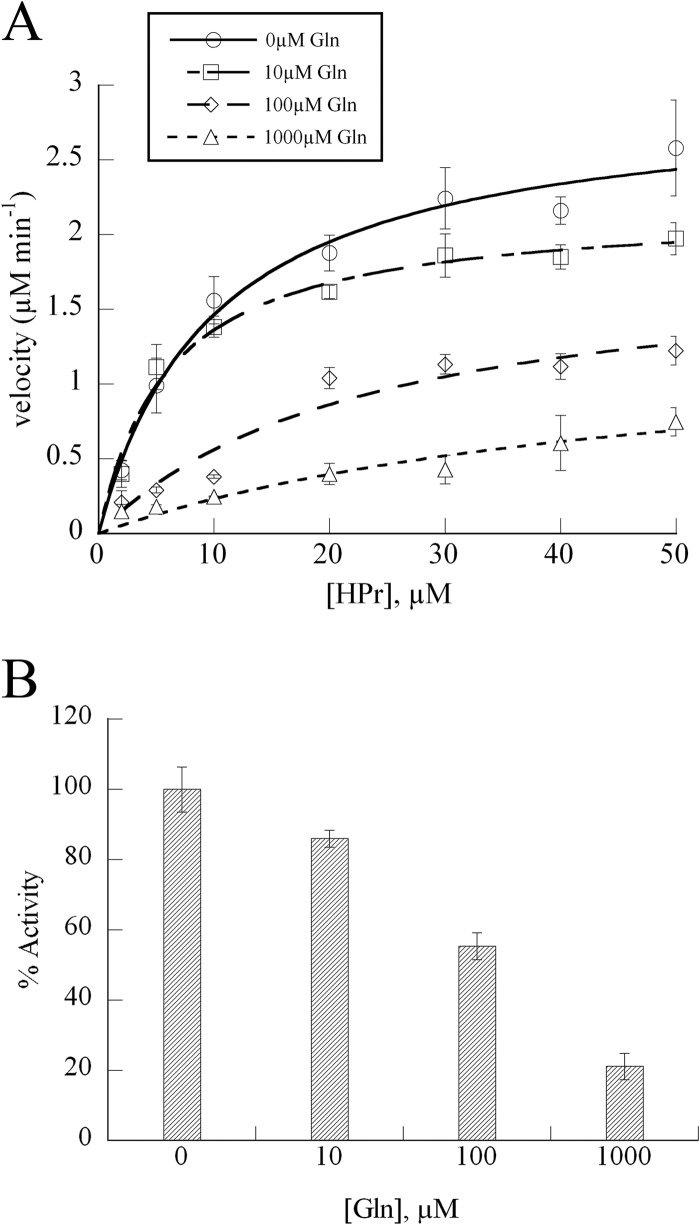
In addition to kinetic characterization, we searched for effector molecules that would alter EINtr activity at physiologically relevant concentrations. Previously, we isolated transposon insertions in gln genes during screens for mutants displaying altered SMCR phenotypes (unpublished results), suggesting that glutamine might modulate SMCR and provide a connection between nitrogen and carbon regulation within the cell. Glutamine was added to phosphorylation reaction mixtures at concentrations similar to those found within E. coli (31), and the reactions were analyzed by native-PAGE. This qualitative assay showed significant inhibition of EINtr when glutamine was present (Fig. 3). For a more precise measure of EINtr inhibition, the enzyme velocity was measured by using the LDH assay across multiple glutamine concentrations. The kinetic assays confirmed the results of the native electrophoresis experiments, showing that EINtr only retained 20% of its activity in 1 mM glutamine (Fig. 4).
FIG 4.
Kinetics of EINtr inhibition by glutamine. (A) Reaction rates were measured with 40 nM EINtr and the indicated HPr concentrations in the coupled LDH assay. Data points represent the means of three independent experiments (bars indicate standard errors) and were fit to the Michaelis-Menten equation. (B) Percent activity of EINtr at 20 μM HPr and the indicated concentration of glutamine. Bars indicate standard errors.
The GAF domain of EINtr binds glutamine.
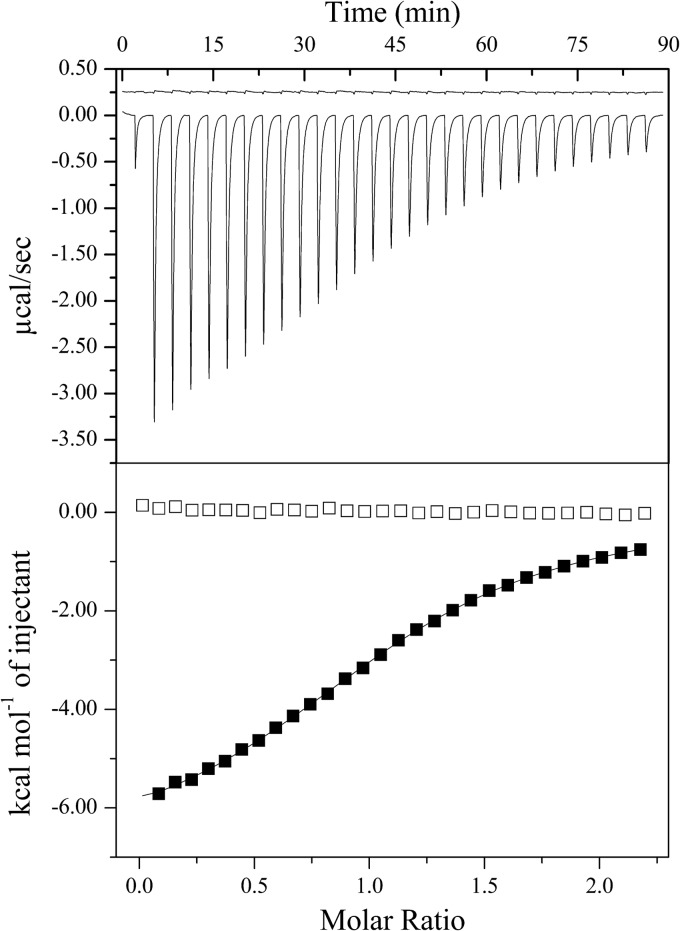
The N-terminal GAF domain is a defining feature of EINtr and was a likely candidate for glutamine binding. In order to detect binding of glutamine by EIGAF and to measure the affinity, we used ITC. Titration of glutamine into isolated EIGAF produced an exothermic binding curve that was fit with a one-site model (Fig. 5). From the fit, we determined that the KD was 35 μM with an n value of 1.1. These values are similar to those reported for the GAF domain of Azotobacter vinelandii NifA binding with α-ketoglutarate (20).
FIG 5.
Titration of 143 μM EIGAF with glutamine and α-ketoglutarate. The upper panel shows the raw heat of titration for α-ketoglutarate (top) and glutamine (bottom). The data for α-ketoglutarate have been shifted upwards by 0.5 μcal/s for clarity. The lower panel shows the binding isotherms for glutamine (filled boxes) and α-ketoglutarate (empty boxes). The isotherm for glutamine was fit with a one-site binding model.
EINtr is insensitive to α-ketoglutarate.
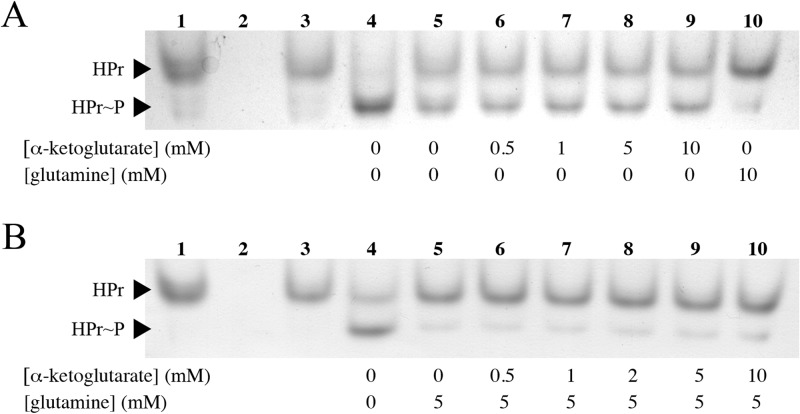
We screened a number of other metabolites for inhibition of EINtr. These included compounds present in the same metabolic pathways as glutamine, such as glutamate, as well as chemically similar amino acids, including lysine, arginine, and asparagine. Other than glutamine, none of the other amino acids caused a measurable decrease in EINtr activity (data not shown). It was recently shown that α-ketoglutarate stimulates the activity of EINtr in E. coli (32). To determine if the S. meliloti enzyme responds the same way to the presence of α-ketoglutarate, we adapted the native electrophoresis assay to detect enzyme activation. In the modified assay, the amount of EINtr was reduced such that it would phosphorylate half of the HPr in 10 min. When the reaction mixtures were incubated in up to 10 mM α-ketoglutarate, the fraction of phosphorylated HPr did not increase (Fig. 6A). To show that all of the HPr could be phosphorylated, a reaction mix was included that contained 10-fold more EINtr. Increasing the enzyme concentration resulted in complete phosphorylation of HPr. Addition of glutamine prevented phosphorylation of HPr, indicating that EINtr was still sensitive to this ligand. Additional experiments were performed to detect an indirect activation of EINtr by α-ketoglutarate via competition with glutamine for the binding site. Inhibition of EINtr by 5 mM glutamine was not relieved by α-ketoglutarate (Fig. 6B). Finally, we could not detect direct binding of α-ketoglutarate by EIGAF based on ITC (Fig. 5).
FIG 6.
Phosphorylation of HPr by 50 ng of EINtr in the presence of α-ketoglutarate. (A) α-Ketoglutarate does not stimulate EINtr activity. Lane 1, no EINtr; lane 2, no HPr; lane 3, no PEP; lane 4, full reaction mixture with 500 ng of EINtr; lanes 5 to 10, reaction mixtures contained the indicated amounts of α-ketoglutarate or glutamine (in mM). (B) α-Ketoglutarate does not compete with glutamine to relieve EINtr inhibition. Lanes 1 to 4, same reaction mixtures as in panel A; lanes 5 to 10, reaction mixtures contained the indicated concentrations of α-ketoglutarate and glutamine (in mM).
DISCUSSION
The nitrogen-type phosphotransferase system is a variant of the carbohydrate PTS found in many Gram-negative bacteria. The systems controlled by the PTSNtr vary greatly among bacteria, from nitrogen fixation and polyhydroxybutyrate accumulation in Azotobacter vinelandii, to oligopeptide transport in Bradyrhizobium japonicum, pathogenesis in Brucella melitensis, biofilm formation in Vibrio cholerae, and potassium homeostasis and tricarboxylic acid (TCA) cycle flux in E. coli (33–39). In S. meliloti, the PTSNtr lacks membrane-bound transporters, making it strictly regulatory, and leaving the uptake of catabolites primarily to ABC and TRAP transporters (17). Understanding how the S. meliloti PTSNtr controls central metabolism and other aspects of its physiology is a crucial step for better understanding how S. meliloti senses and interacts with its environment both in the soil and during symbiosis.
The molecular mechanisms governing the PTSNtr have not been well studied outside of the major model bacteria, and in order to rectify this we have begun biochemical characterization of the phosphotransfer from EINtr to HPr in S. meliloti. In measuring the kinetics of S. meliloti EINtr activity, we found that it phosphorylates HPr at a much lower rate than the carbohydrate EI of E. coli (30). We believe that such a large difference in rates reflects a true difference in enzymatic activities, though we cannot rule out the possibility that the discrepancy was due to purification or assay artifacts. This difference in activities may reflect the function of the PTS in each of these organisms: in a carbohydrate PTS, the activity of the PTS proteins will determine the transport rate of sugar and by extension cell growth, favoring high enzymatic activity; conversely, in the case of a regulatory PTS with no known phosphate sink, a highly active EI would rapidly saturate the system. An oversaturated PTS would subsequently lose sensitivity and become unable to respond to changes in the state of the cell.
In a transposon screen to find mutations that relieved strong SMCR in certain PTS mutants, our lab isolated insertions in the genes glnD and glnE. Since these genes encode proteins that regulate glutamine levels in the cell (40), we assayed glutamine for its ability to alter EINtr activity. These assays showed that glutamine was a potent inhibitor of EINtr, likely coupling the activity of the PTSNtr to a critical signal of nitrogen levels in both free-living and symbiotic S. meliloti (40). Direct binding measurements showed that the GAF domain of EINtr is responsible for binding glutamine. When α-ketoglutarate was tested as an activator of EINtr, we could not find any increase in activity, nor could we detect direct binding of α-ketoglutarate by EINtr.
The data presented here can be used to further refine the model developed for the S. meliloti PTSNtr. Reduction of EINtr velocity relative to carbohydrate EI likely prevents the PTSNtr from becoming saturated with phosphate, which would result in excess SMCR and other regulatory consequences. In Gram-positive bacteria, phosphorylation of HPr by HPrK renders it either a poor substrate for EI or unphosphorylatable (41, 42). If this holds true in S. meliloti, then the presence of HPrK would further reduce the rate of HPr phosphorylation by EINtr in vivo. Currently, our attempts to purify active HPrK have failed, preventing us from exploring this hypothesis.
EINtr senses the cellular carbon/energy state via the [PEP]:[pyruvate] ratio (11). EINtr balances this activation against the glutamine availability in the cell, resulting in an intimate link between the availability of carbon and nitrogen. The GAF domain, which is a defining attribute of EINtr in proteobacteria, is likely responsible for transmitting the signal from glutamine to the catalytic domain in order to coordinate EINtr activity in response to carbon and nitrogen levels in the cell. Unlike the E. coli PTSNtr, S. meliloti does not use α-ketoglutarate to regulate EINtr, possibly because succinate, another TCA cycle intermediate, is a primary carbon source for S. meliloti. Further studies using a combination of genetic and physiological techniques are needed to provide concrete evidence for the role of glutamine inhibition of EINtr in vivo.
ACKNOWLEDGMENTS
We thank Vikki Robinson for the use of her ITC, as well as her advice.
Funding was provided by U.S. Department of Energy contracts DE-FG02-01ER15175 and DE-FG02-06ER15805 to D.J.G.
Footnotes
Published ahead of print 14 March 2014
REFERENCES
- 1.Beringer JE, Brewin N, Johnston AW, Schulman HM, Hopwood DA. 1979. The Rhizobium-legume symbiosis. Proc. R. Soc. Lond. B Biol. Sci. 204:219–233 [DOI] [PubMed] [Google Scholar]
- 2.Mauchline TH, Fowler JE, East AK, Sartor AL, Zaheer R, Hosie AH, Poole PS, Finan TM. 2006. Mapping the Sinorhizobium meliloti 1021 solute-binding protein-dependent transportome. Proc. Natl. Acad. Sci. U. S. A. 103:17933–17938. 10.1073/pnas.0606673103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stowers MD. 1985. Carbon metabolism in Rhizobium species. Annu. Rev. Microbiol. 38:89–108 [DOI] [PubMed] [Google Scholar]
- 4.Finan TM, Wood JM, Jordan DC. 1983. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J. Bacteriol. 154:1403–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinsinger P, Gobran GR, Gregory PJ, Wenzel WW. 2005. Rhizosphere geometry and heterogeneity arising from root-mediated physical and chemical processes. New Phytol. 168:293–303. 10.1111/j.1469-8137.2005.01512.x [DOI] [PubMed] [Google Scholar]
- 6.Pinedo CA, Bringhurst RM, Gage DJ. 2008. Sinorhizobium meliloti mutants lacking phosphotransferase system enzyme HPr or EIIA are altered in diverse processes, including carbon metabolism, cobalt requirements, and succinoglycan production. J. Bacteriol. 190:2947–2956. 10.1128/JB.01917-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bringhurst RM, Gage DJ. 2002. Control of inducer accumulation plays a key role in succinate-mediated catabolite repression in Sinorhizobium meliloti. J. Bacteriol. 184:5385–5392. 10.1128/JB.184.19.5385-5392.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weigel N, Waygood EB, Kukuruzinska MA, Nakazawa A, Roseman S. 1982. Sugar transport by the bacterial phosphotransferase system. Isolation and characterization of enzyme I from Salmonella typhimurium. J. Biol. Chem. 257:14461–14469 [PubMed] [Google Scholar]
- 9.Stolz B, Huber M, Markovic-Housley Z, Erni B. 1993. The mannose transporter of Escherichia coli. Structure and function of the IIABMan subunit. J. Biol. Chem. 268:27094–27099 [PubMed] [Google Scholar]
- 10.Dills SS, Schmidt MR, Saier MH., Jr 1982. Regulation of lactose transport by the phosphoenolpyruvate-sugar phosphotransferase system in membrane vesicles of Escherichia coli. J. Cell. Biochem. 18:239–244 [DOI] [PubMed] [Google Scholar]
- 11.Hogema BM, Arents JC, Bader R, Eijkemans K, Yoshida H, Takahashi H, Aiba H, Postma PW. 1998. Inducer exclusion in Escherichia coli by non-PTS substrates: the role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Mol. Microbiol. 30:487–498 [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Oldham ML, Davidson AL, Chen J. 2013. Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography. Nature 499:364–368. 10.1038/nature12232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saier MH, Jr, Feucht BU, Hofstadter LJ. 1976. Regulation of carbohydrate uptake and adenylate cyclase activity mediated by the enzymes II of the phosphoenolpyruvate:sugar phosphotransferase system in Escherichia coli. J. Biol. Chem. 251:883–892 [PubMed] [Google Scholar]
- 14.Saier MH., Jr 1996. Cyclic AMP-independent catabolite repression in bacteria. FEMS Microbiol. Lett. 138:97–103 [DOI] [PubMed] [Google Scholar]
- 15.Deutscher J, Kuster E, Bergstedt U, Charrier V, Hillen W. 1995. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol. Microbiol. 15:1049–1053 [DOI] [PubMed] [Google Scholar]
- 16.Dossonnet V, Monedero V, Zagorec M, Galinier A, Perez-Martinez G, Deutscher J. 2000. Phosphorylation of HPr by the bifunctional HPr kinase/P-Ser-HPr phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion but not inducer expulsion. J. Bacteriol. 182:2582–2590. 10.1128/JB.182.9.2582-2590.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, Bothe G, Boutry M, Bowser L, Buhrmester J, Cadieu E, Capela D, Chain P, Cowie A, Davis RW, Dreano S, Federspiel NA, Fisher RF, Gloux S, Godrie T, Goffeau A, Golding B, Gouzy J, Gurjal M, Hernandez-Lucas I, Hong A, Huizar L, Hyman RW, Jones T, Kahn D, Kahn ML, Kalman S, Keating DH, Kiss E, Komp C, Lelaure V, Masuy D, Palm C, Peck MC, Pohl TM, Portetelle D, Purnelle B, Ramsperger U, Surzycki R, Thebault P, Vandenbol M, Vorholter FJ, Weidner S, Wells DH, Wong K, Yeh KC, Batut J. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668–672. 10.1126/science.1060966 [DOI] [PubMed] [Google Scholar]
- 18.Hu KY, Saier MH., Jr 2002. Phylogeny of phosphoryl transfer proteins of the phosphoenolpyruvate-dependent sugar-transporting phosphotransferase system. Res. Microbiol. 153:405–415. 10.1016/S0923-2508(02)01339-6 [DOI] [PubMed] [Google Scholar]
- 19.Martinez SE, Beavo JA, Hol WG. 2002. GAF domains: two-billion-year-old molecular switches that bind cyclic nucleotides. Mol. Interv. 2:317–323. 10.1124/mi.2.5.317 [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Argudo I, Little R, Dixon R. 2004. Role of the amino-terminal GAF domain of the NifA activator in controlling the response to the antiactivator protein NifL. Mol. Microbiol. 52:1731–1744. 10.1111/j.1365-2958.2004.04089.x [DOI] [PubMed] [Google Scholar]
- 21.Cases I, Velazquez F, de Lorenzo V. 2007. The ancestral role of the phosphoenolpyruvate-carbohydrate phosphotransferase system (PTS) as exposed by comparative genomics. Res. Microbiol. 158:666–670. 10.1016/j.resmic.2007.08.002 [DOI] [PubMed] [Google Scholar]
- 22.Pinedo CA, Gage DJ. 2009. HPrK regulates succinate-mediated catabolite repression in the Gram negative symbiont Sinorhizobium meliloti. J. Bacteriol. 191:298–309. 10.1128/JB.01115-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234. 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 24.Kang DH, Gho YS, Suh MK, Kang CH. 2002. Highly sensitive and fast protein detection with Coomassie brilliant blue in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Bull. Korean Chem. Soc. 23:1511–1512. 10.5012/bkcs.2002.23.11.1511 [DOI] [Google Scholar]
- 25.Scopes RK. 1974. Measurement of protein by spectrophotometry at 205 nm. Anal. Biochem. 59:277–282 [DOI] [PubMed] [Google Scholar]
- 26.Rabus R, Reizer J, Paulsen I, Saier MH. 1999. Enzyme I-Ntr from Escherichia coli: a novel enzyme of the phosphoenolpyruvate-dependent phosphotransferase system exhibiting strict specificity for its phosphoryl acceptor, NPr. J. Biol. Chem. 274:26185–26191 [DOI] [PubMed] [Google Scholar]
- 27.McLellan T. 1982. Electrophoresis buffers for polyacrylamide gels at various pH. Anal. Biochem. 126:94–99 [DOI] [PubMed] [Google Scholar]
- 28.Waygood EB, Meadow ND, Roseman S. 1979. Modified assay procedures for the phosphotransferase system in enteric bacteria. Anal. Biochem. 95:293–304 [DOI] [PubMed] [Google Scholar]
- 29.Ackerley DF, Caradoc-Davies TT, Lamont IL. 2003. Substrate specificity of the nonribosomal peptide synthetase PvdD from Pseudomonas aeruginosa. J. Bacteriol. 185:2848–2855. 10.1128/JB.185.9.2848-2855.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meadow ND, Mattoo RL, Savtchenko RS, Roseman S. 2005. Transient state kinetics of enzyme I of the phosphoenolpyruvate:glycose phosphotransferase system of Escherichia coli: equilibrium and second-order rate constants for the phosphotransfer reactions with phosphoenolpyruvate and HPr. Biochemistry 44:12790–12796. 10.1021/bi0502846 [DOI] [PubMed] [Google Scholar]
- 31.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5:593–599. 10.1038/nchembio.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CR, Park YH, Kim M, Kim YR, Park S, Peterkofsky A, Seok YJ. 2013. Reciprocal regulation of the autophosphorylation of enzyme INtr by glutamine and alpha-ketoglutarate in Escherichia coli. Mol. Microbiol. 88:473–485. 10.1111/mmi.12196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segura D, Espin G. 1998. Mutational inactivation of a gene homologous to Escherichia coli ptsP affects poly-beta-hydroxybutyrate accumulation and nitrogen fixation in Azotobacter vinelandii. J. Bacteriol. 180:4790–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King ND, O'Brian MR. 2001. Evidence for direct interaction between enzyme I(Ntr) and aspartokinase to regulate bacterial oligopeptide transport. J. Biol. Chem. 276:21311–21316. 10.1074/jbc.M101982200 [DOI] [PubMed] [Google Scholar]
- 35.Lee CR, Cho SH, Yoon MJ, Peterkofsky A, Seok YJ. 2007. Escherichia coli enzyme IIA(Ntr) regulates the K+ transporter TrkA. Proc. Natl. Acad. Sci. U. S. A. 104:4124–4129. 10.1073/pnas.0609897104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houot L, Chang S, Pickering BS, Absalon C, Watnick PI. 2010. The phosphoenolpyruvate phosphotransferase system regulates Vibrio cholerae biofilm formation through multiple independent pathways. J. Bacteriol. 192:3055–3067. 10.1128/JB.00213-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jahn S, Haverkorn van Rijsewijk BR, Sauer U, Bettenbrock K. 2013. A role for EIIA(Ntr) in controlling fluxes in the central metabolism of E. coli K12. Biochim. Biophys. Acta 1833:2879–2889. 10.1016/j.bbamcr.2013.07.011 [DOI] [PubMed] [Google Scholar]
- 38.Luttmann D, Heermann R, Zimmer B, Hillmann A, Rampp IS, Jung K, Gorke B. 2009. Stimulation of the potassium sensor KdpD kinase activity by interaction with the phosphotransferase protein IIA(Ntr) in Escherichia coli. Mol. Microbiol. 72:978–994. 10.1111/j.1365-2958.2009.06704.x [DOI] [PubMed] [Google Scholar]
- 39.Dozot M, Poncet S, Nicolas C, Copin R, Bouraoui H, Maze A, Deutscher J, De Bolle X, Letesson JJ. 2010. Functional characterization of the incomplete phosphotransferase system (PTS) of the intracellular pathogen Brucella melitensis. PLoS One 5(9):e12679. 10.1371/journal.pone.0012679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yurgel SN, Kahn ML. 2008. A mutant GlnD nitrogen sensor protein leads to a nitrogen-fixing but ineffective Sinorhizobium meliloti symbiosis with alfalfa. Proc. Natl. Acad. Sci. U. S. A. 105:18958–18963. 10.1073/pnas.0808048105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casabon I, Couture M, Vaillancourt K, Vadeboncoeur C. 2006. Synthesis of HPr(Ser-P)(His∼P) by enzyme I of the phosphoenolpyruvate:sugar phosphotransferase system of Streptococcus salivarius. Biochemistry 45:6692–6702. 10.1021/bi060278p [DOI] [PubMed] [Google Scholar]
- 42.Reizer J, Sutrina SL, Wu LF, Deutscher J, Reddy P, Saier MH., Jr 1992. Functional interactions between proteins of the phosphoenolpyruvate:sugar phosphotransferase systems of Bacillus subtilis and Escherichia coli. J. Biol. Chem. 267:9158–9169 [PubMed] [Google Scholar]
- 43.Cubillos-Rojas M, Amair-Pinedo F, Tato I, Bartrons R, Ventura F, Rosa JL. 2010. Simultaneous electrophoretic analysis of proteins of very high and low molecular mass using Tris-acetate polyacrylamide gels. Electrophoresis 31:1318–1321. 10.1002/elps.200900657 [DOI] [PubMed] [Google Scholar]