Abstract
Mycoplasma mobile has a unique mechanism that enables it to glide on solid surfaces faster than any other gliding mycoplasma. To elucidate the gliding mechanism, we developed a transformation system for M. mobile based on a transposon derived from Tn4001. Modification of the electroporation conditions, outgrowth time, and colony formation from the standard method for Mycoplasma species enabled successful transformation. A fluorescent-protein tagging technique was developed using the enhanced yellow fluorescent protein (EYFP) and applied to two proteins that have been suggested to be involved in the gliding mechanism: P42 (MMOB1050), which is transcribed as continuous mRNA with other proteins essential for gliding, and a homolog of the F1-ATPase α-subunit (MMOB1660). Analysis of the amino acid sequence of P42 by PSI-BLAST suggested that P42 evolved from a common ancestor with FtsZ, the bacterial tubulin homologue. The roles of P42 and the F1-ATPase subunit homolog are discussed as part of our proposed gliding mechanism.
INTRODUCTION
Mycoplasmas are commensal and occasionally parasitic bacteria that lack peptidoglycan layers and have small genomes (1). Mycoplasma mobile, a fish pathogen, has a membrane protrusion at one pole and exhibits gliding motility in the direction of the protrusion (2–4). The average speed is 2.0 to 4.5 μm/s, or 3 to 7 times the length of the cell per second, with a propulsive force up to 27 pN (5–7). This motility, combined with the ability to adhere to the host cell surface, likely plays a role in infection, as has been suggested for another species, Mycoplasma pneumoniae (2, 3, 8–10). The motor proteins involved in this motility are unlike the motor proteins involved in any other form of bacterial or eukaryotic cell motility (11).
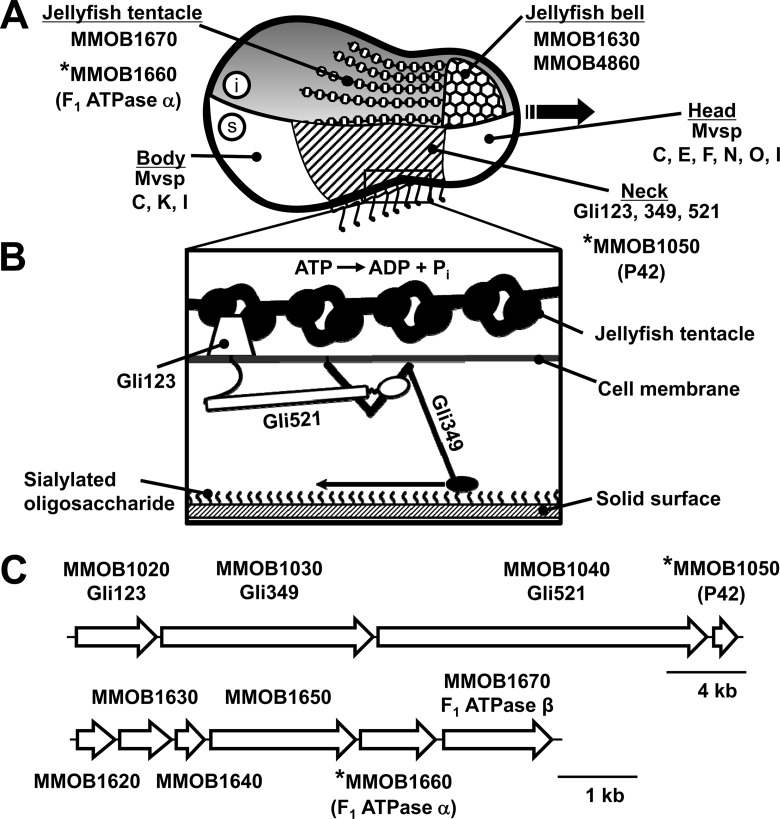
The cell surface can be divided into three parts beginning at the front end, i.e., the head, neck, and body, as shown in Fig. 1A (3, 4, 12–14). Three large proteins, Gli123, Gli349, and Gli521, with respective masses of 123, 349, and 521 kDa, are involved in this gliding mechanism and are localized at the cell neck exclusively, suggesting that this part is specialized for gliding (12, 13, 15–18). Fifty-nanometer legs composed of Gli349 can be seen protruding from the neck surface by electron microscopy (Fig. 1B) (19–21). The surface structure is supported from within the cell by a unique cytoskeleton called the jellyfish structure; its 10 components have been identified by mass spectrometry (Fig. 1C) (22). The energy for motility is supplied by ATP (23, 24), and the direct binding targets for gliding are the sialylated oligosaccharides found on the surface of animal tissue (25–27). On the basis of the above information, we proposed a working model called the “centipede” or “power stroke” model, in which the cells are propelled by “legs” composed of Gli349 that, through repeated cycles driven by the hydrolysis of ATP, catch and release sialylated oligosaccharides (3, 28). However, more information will be needed in order to fully clarify the gliding mechanism.
FIG 1.
Schematic of M. mobile cell architecture (2, 3, 55). The ORFs considered in this study are marked by asterisks. (A) Drawing of the cell showing the inside structure (i, above) and the surface structure (s, below). The cytoskeletal jellyfish structure can be divided into two parts, the “bell” and the “tentacles.” The bell is composed of protein products of the MMOB1630 and MMOB4860 genes, and the tentacles are protein products of the MMOB1670 gene. The cell surface can be divided into three parts, the head, neck, and body, beginning at the front end. The gliding direction is indicated by the black arrow. The neck is covered by the Gli123, Gli349, and Gli521 proteins, all of which are essential for gliding. The head is covered with MvspC, -E, -F, -N, -O, and -I, and the body is covered with MvspC, -K, and -I; these proteins are likely involved in a mechanism for evading the host immune system. (B) Magnified image of the neck surface based on our previous studies, most of which employed electron microscopy. The distal globular part of Gli349 is thought to catch and pull sialylated oligosaccharides fixed on the host surface, a mechanism that hydrolyzes ATP. (C) ORFs for surface and jellyfish structures (above and below, respectively). Ten ORFs are known to encode components of the jellyfish structure, but four are located at other regions of the genome.
Although 10 proteins have been identified as the components of the jellyfish structure, their roles and subcellular localizations are still unclear (22). Interestingly, the amino acid sequences of two of the components, MMOB1660 and MMOB1670, show high similarity to the α- and β-subunits, respectively, of F1-ATPase, the catalytic subunit of proton pumps (22, 29). However, these proteins are unlikely to function in a proton pump because proton pumps require additional subunits—seven in Bacillus subtilis (30)—and the M. mobile genome has another locus containing a complete set of pump subunits. P42 is likely encoded in an operon with Gli123, Gli349, and Gli521 (16). However, the role and subcellular localization of P42 are unknown (31).
Gene manipulation is a powerful tool for clarifying the function of proteins in microbiological studies. In mycoplasma species, methods employing transposons (32), oriC plasmids based on the replication origin of the genome (33), and homologous recombination (34) have been applied. However, this strategy has been hampered by the lack of a genetic system for manipulating M. mobile genes.
In the present study, we developed a transformation system for M. mobile, and we used this method along with fluorescent protein tagging to determine the subcellular localization of two key proteins involved in the gliding mechanism.
MATERIALS AND METHODS
Strains and culture conditions.
The M. mobile 163K (ATCC 43663) and Mycoplasma pulmonis (ATCC 19612) strains were cultured in Aluotto medium at 25°C and 37°C, respectively (35). The sources of plasmids harboring a transposon are listed in Table 1 (36–39). For the selection of transformed M. mobile, tetracycline hydrochloride, gentamicin sulfate, chloramphenicol, or puromycin dihydrochloride was used with a final concentration of 3.0, 30, 18, or 2.5 μg/ml, respectively. The Escherichia coli strain DH5α was used for DNA manipulation.
TABLE 1.
Plasmids used in this study
| Plasmida | Antibiotic resistance | Efficiency (no. of transformants/CFU) | No. of days to colony formation | Reference |
|---|---|---|---|---|
| pKV104 | Chloramphenicol | 2.5E−03 | 9 | 38 |
| pMTnTetM438 | Tetracycline | 1.4E−03 | 9 | 37 |
| pMTnGm | Gentamicin | 3.1E−03 | 9 | 37 |
| Mini-Tn4001PsPuro | Puromycin | 2.3E−03 | 8 | 36 |
| pTK165 | Gentamicin | 4.2E−03 | 7 | 40 |
| pAM120 | Tetracycline | 1.2E−05 | 9 | 39 |
All plasmids contain a transposon and a replication origin of E. coli. pAM120 is based on transposon Tn916, and the others are based on Tn4001.
Plasmid construction.
M. mobile genomic DNA was prepared by the Genomic-tip System (Qiagen, Hilden, Germany). pTK165 was kindly provided by Tsuyoshi Kenri at the National Institute of Infectious Diseases, Tokyo, Japan (40). Plasmid pMobtuf was constructed by replacing the M. pneumoniae tuf promoter sequence (upstream from eyfp, the gene for the enhanced yellow fluorescent protein [EYFP]) by the tuf promoter amplified from the M. mobile genome using BamHI and NcoI sites in the plasmid. The eyfp gene (derived from plasmid pEYFP; Clontech, Palo Alto, CA) of pMobtuf was replaced with a codon-optimized sequence from the pMD19-Myco plasmid using the NcoI and EcoRI sites in the pMobtuf plasmid, resulting in the pMobopt plasmid. The pMD19-Myco plasmid was kindly provided by Itaru Yanagihara at the Osaka Medical Center for Maternal and Child Health, Osaka, Japan. Plasmids containing the p42 (MMOB1050) and MMOB1660 fusion genes were constructed as follows. For the N-terminal fusion of p42, the DNA fragment amplified from the genomic DNA by PCR was inserted into the 5′ end of the eyfp gene of pMobopt by using an In-Fusion EcoDry PCR Cloning Kit (TaKaRa Bio, Shiga, Japan), producing the pMobN-P42 plasmid. For C-terminal fusions, the p42 (MMOB1050) and MMOB1660 genes amplified from the genomic DNA were inserted into the 3′ end of the eyfp gene of plasmid pMobopt using the In-Fusion EcoDry PCR Cloning Kit, producing plasmids pMobC-P42 and pMobC-1660, respectively.
Transformation of M. mobile.
The transformation procedure was modified from the method generally used for mycoplasma (41). The major modifications were the use of stronger conditions for electroporation, a longer incubation period without selection after electroporation in order to permit cell growth, and a lower concentration of serum in the solid medium. M. mobile was grown in liquid medium until the mid-logarithmic phase (∼108 CFU/ml). Cells were collected by centrifugation at 9,000 × g and 4°C for 10 min and then washed three times in an equivalent volume of electroporation buffer (272 mM sucrose, 8 mM HEPES, pH 7.4). The cells were then suspended in 100 μl of electroporation buffer at a concentration of approximately 109 cells per ml, kept on ice with 10 μg of plasmid DNA for 30 min, and transferred to a prechilled 2-mm electroporation cuvette. Electroporation was performed using a Gene Pulser II electroporation system and Pulse Controller II module (Bio-Rad Laboratories, Hercules, CA). Optimized electroporation conditions of 2.5 kV, 800 Ω, and 25 μF were used. Immediately after electroporation, 900 μl of ice-chilled Aluotto medium was added. After incubation for 15 min on ice, the cells were kept at 25°C for 12 h for outgrowth. Aliquots of 100 μl were plated onto solid Aluotto medium containing 6% horse serum and appropriate antibiotics and were cultured at 25°C for 8 to 9 days. The plates were observed by an SZX2-ILLT stereo microscope (Olympus, Tokyo, Japan) and photographed. To isolate the transformants, individual colonies in agar were picked and homogenized by a BioMasher (Iwai Chemicals, Tokyo, Japan).
Colony PCR.
Individual colonies were isolated, homogenized, and used as templates for the colony PCR. The PCR was performed with primers complementary to the gentamicin resistance gene: 5′-ATGAATATAGTTGAAAATGAAATATGTATAAG-3′ and 5′-TCCTTTAATTTCTTTATAACCTAGTATAGAT-3′. The PCR protocol consisted of 5 min at 95°C, followed by 30 cycles of 1 min at 95°C, 30 s at 55°C, and 1 min at 68°C, with a final cycle at 68°C for 5 min. PCR products were checked using a Multina microchip electrophoresis system (Shimadzu, Kyoto, Japan).
Microscopy.
To observe EYFP fluorescence, individual M. mobile transformants were selected and grown in liquid Aluotto medium containing 15 μg/ml gentamicin at 25°C for 3 days. The cells were collected by centrifugation at 12,000 × g for 4 min, suspended in fresh medium, and bound to coverslips washed with saturated ethanolic KOH. The cells were then fixed by 3.0% paraformaldehyde and 0.1% glutaraldehyde for 30 min at room temperature (RT) and observed with a BX51 fluorescence microscope equipped with a YFP filter unit (U-MYFPHQ) and a phase-contrast setup (Olympus, Tokyo, Japan) (12, 40, 42). Immunostaining, staining with 4′,6′-diamidino-2-phenylindole (DAPI) for fluorescence microscopy, and the observation of gliding were performed as described previously (5, 12, 13, 15, 42).
Transposon insertion mapping.
The DNA sequences flanking transposon insertion sites were determined using arbitrary PCR (43, 44). The genomic DNA of the transformant was isolated by the agarose block method, as previously described (45). A DNA fragment covering an end region of a transposon and the flanking region of insertion sites was amplified by a nested PCR. The first-round PCR was performed to isolate genomes using a primer unique to the right end of the Tn4001 element, 5′-CTTTTACACAATTATACGGACTTTATC-3′, and an arbitrary primer, 5′-GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC-3′. The PCR protocol consisted of the following: 10 min at 95°C; six cycles of 30 s at 95°C, 30 s at 30°C, and 1.5 min at 72°C with 5-s increments per cycle; 30 cycles of 30 s at 95°C, 30 s at 45°C, and 2 min at 72°C with 5-s increments per cycle.
The second-round PCR was performed using the primers Tn4001-IR-R-in (5′-GGACTGTTATATGGCCTTTTTACTTTTACACA-3′) and 5′-GGCCACGCGTCGACTAGTAC-3. The second-round PCR protocol consisted of 10 min at 95°C, 35 cycles of 45 s at 95°C, 45 s at 55°C, and 1.5 min at 72°C with 5-s increments per cycle, followed by 10 min at 72°C.
All PCR products were recovered from the agarose gel, and sequencing was carried out using BigDye, version 3.1, with the Tn4001-IR-R-in primer and a 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA). The obtained sequence reads were aligned against the M. mobile whole-genome sequence (11).
The existence of the original p42 gene and MMOB1660 on the genome was confirmed by PCR of regions covering the whole gene and one flanking region. The primer sequences used were 5′-GATGTACCTTCAGGAGCAATTATTG-3′, 5′-TTATCTTAGAAGAATTTCTTCTGTCAAG-3′, 5′-ATGACAAAAAATTGAAATTTCGAAAAT-3′, and 5′-GATAACTATCACTTTTTTTATCTTAGAAG-3′ for p42 and 5′-GTAATTGTACCTATTGCAATTATTTTG-3′, 5′-TTATTTGTAGTCATATGCTATACCTCTTTC-3′, 5′-ATGAAAAATTTAAAAATAACAGCAATTAAAG-3′, and 5′-CTAATTCTGTAAGATTATTTGTAGTCATAT-3′ for MMOB1660.
Measurement of ATP.
Cell suspensions of 50 μl before and after electroporation were centrifuged at 12,000 × g at 4°C for 4 min. The pellets were separated from the supernatant and resuspended in 50 μl of electroporation buffer, and their bioluminescence was determined by using ATP Bioluminescence Assay kit HS II (Roche, Basel, Switzerland) (46, 47). The emission was measured by using a Varioskan Flash reader (Thermo Scientific, Waltham, MA).
Protein analysis.
M. mobile cells were grown in tissue culture flasks to the mid-log phase. The cells were collected by centrifugation at 12,000 × g for 4 min at 4°C and washed three times with phosphate-buffered saline (PBS) consisting of 75 mM Na-phosphate (pH 7.3) and 68 mM NaCl. Cells were lysed by adding sample loading buffer and were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). For immunoblot analysis, the separated proteins were transferred to a nylon membrane (Bio-Rad Laboratories). The EYFP was detected by a monoclonal antibody specific for Aequorea victoria GFP variants (Clontech Laboratories, Mountain View, CA) and a horseradish peroxidase (HRP)-conjugated secondary antibody (40).
Sequence analyses.
Sequence similarities were determined by using a PSI-BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) search against the NCBI nonredundant protein sequences. The sequences of homologs were aligned by T-COFFEE (http://www.igs.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi), and their phylogenetic tree was constructed by ClustalW (http://www.genome.jp/tools/clustalw/).
Nucleotide sequence accession number.
The sequence of the eyfp gene whose codon was optimized for expression in mycoplasma is available from GenBank under accession number AB860250.
RESULTS
Transformation of M. mobile.
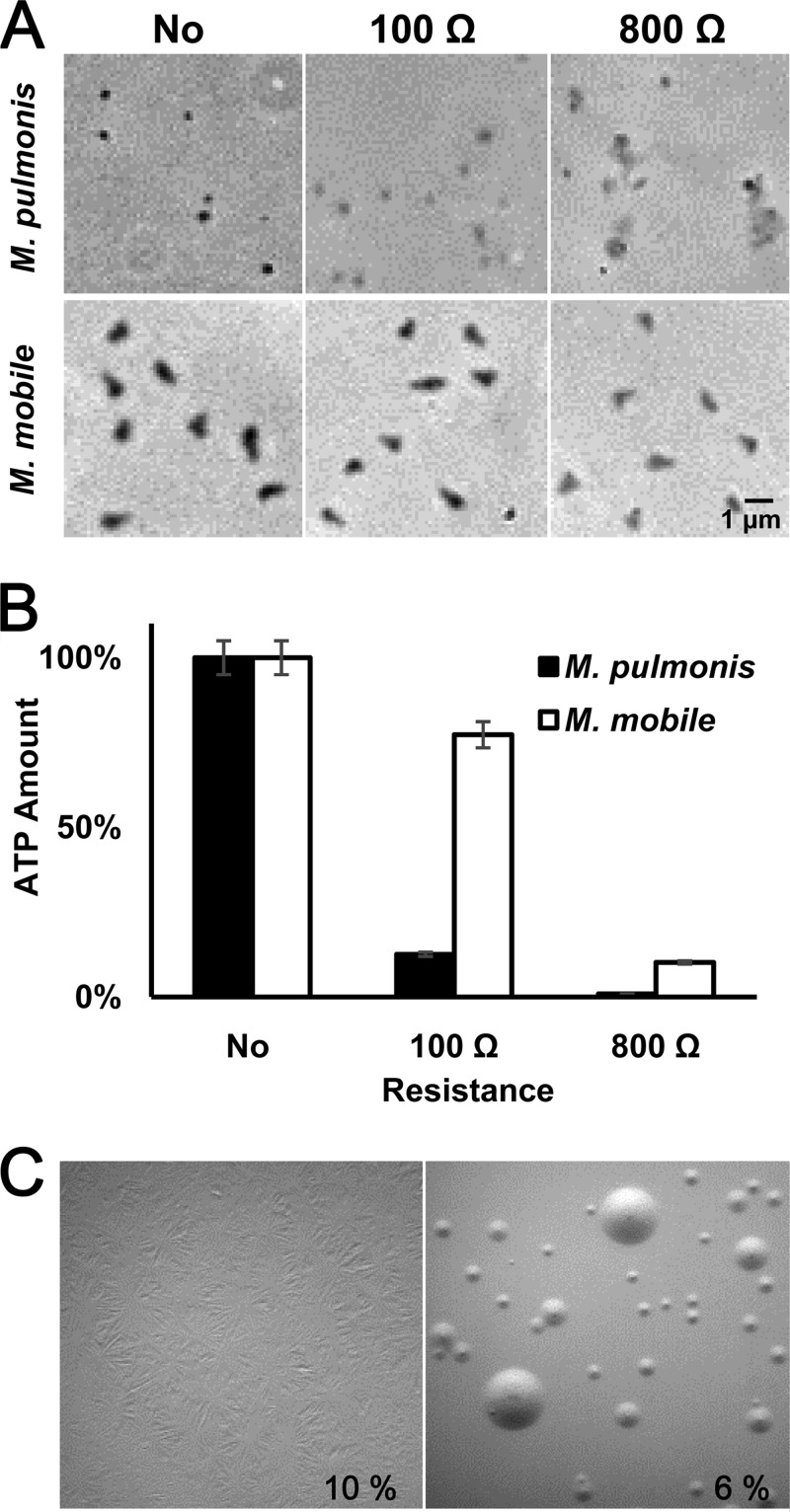
In our initial attempts, we tried unsuccessfully to transform M. mobile by using the transposon vectors used in other mycoplasma species (Table 1) (36, 37, 40, 48) and the electroporation conditions generally used for mycoplasma transformation, i.e., 2.5 kV, 100 Ω, and 25 μF, and a cuvette with a 2-mm gap (41). We then examined and improved each step of the transformation procedure. First, we examined the permeabilization of the cell membrane after the electroporation by microscopy and on the basis of the ATP content remaining in the M. mobile cells. The results were compared with those for M. pulmonis, a relative of M. mobile that can be transformed (Fig. 2A and B) (11, 48–50). Microscopy showed that most of the M. pulmonis cells lost optical density, indicating that the cell membrane was well permeabilized by the electroporation, while M. mobile cells did not show significant damage by microscopy. The amount of ATP in cells was consistent with the results of microscopy. M. pulmonis cells lost 87% of their ATP content, but M. mobile lost only 13%. These results showed that M. mobile cells are more resistant to the applied electric shock than M. pulmonis cells, which was consistent with our observations that the cell shape of M. mobile was more stable than the shapes of other species under a change in osmotic pressure. To permeabilize the M. mobile cell membranes to an extent similar to that in M. pulmonis cells, we varied the resistance for electroporation from 100 to 800 Ω in 100-Ω increments. Using electroporation conditions of 2.5 kV, 800 Ω, and 25 μF resulted in a change in appearance and about an 80% loss of ATP in M. mobile cells, similar to the loss in M. pulmonis (Fig. 2A and B). Then, we modified the conditions of electroporation to transform M. mobile.
FIG 2.
Optimizing the conditions for transformation. (A) Cell images of M. pulmonis and M. mobile after electroporation under different conditions. Cells without electroporation (No) or electroporated using a resistance of 100 Ω or 800 Ω were observed by phase-contrast microscopy. (B) The amounts of ATP in the cells after electroporation under different conditions. The amounts of ATP in the cells without electroporation were normalized to 100%. (C) Formation of film and spots under different concentrations of serum. The serum concentration applied to the solid medium is indicated in each panel. Film and spots formation covers the medium surface only in the left panel. The surface of the solid medium was observed 7 days after inoculation.
During prolonged cultivation on solid growth medium, some mycoplasma species tend to form web-like structures called “film and spots” (Fig. 2C). These structures are thought to be formed by the activity of lipases, which break down phospholipids derived from horse serum contained in the growth medium and form a confluent layer of cells on the surface of the agar (51, 52). Because transformed M. mobile cells grew slowly in the presence of antibiotics, the prolonged cultivation was necessary for colony formation. Then, the serum concentration was reduced to 6% to avoid the formation of film and spots.
Next, we modified the outgrowth time, or the period when the cells are grown in liquid medium without antibiotics to repair the damage caused by the electroporation and to express the resistance gene. We tried different outgrowth durations of 2 to 16 h in 2-h increments and finally succeeded in yielding transformed colonies by using an outgrowth time of 8 h after 7 to 9 days of incubation. The transformation was achieved by all transposon vectors used (Table 1). By varying the outgrowth time after electroporation, we found that 12 h was optimal.
Among the six vectors we tried here, we focused on pTK165 for further studies because it showed the highest transformation efficiency, 4.2 × 10−3 transformants per CFU, and gave rise to colonies in 7 days (Table 1) on medium containing gentamicin. The transformation was confirmed by colony PCR of individual colonies randomly selected from solid medium. These clones yielded the expected 805-bp PCR fragment using Tn4001-specific primers. The PCR detected no bands from the nontransformed M. mobile colonies. The transposon was stable for at least five consecutive inoculations on solid medium with gentamicin.
Fluorescent protein tagging.
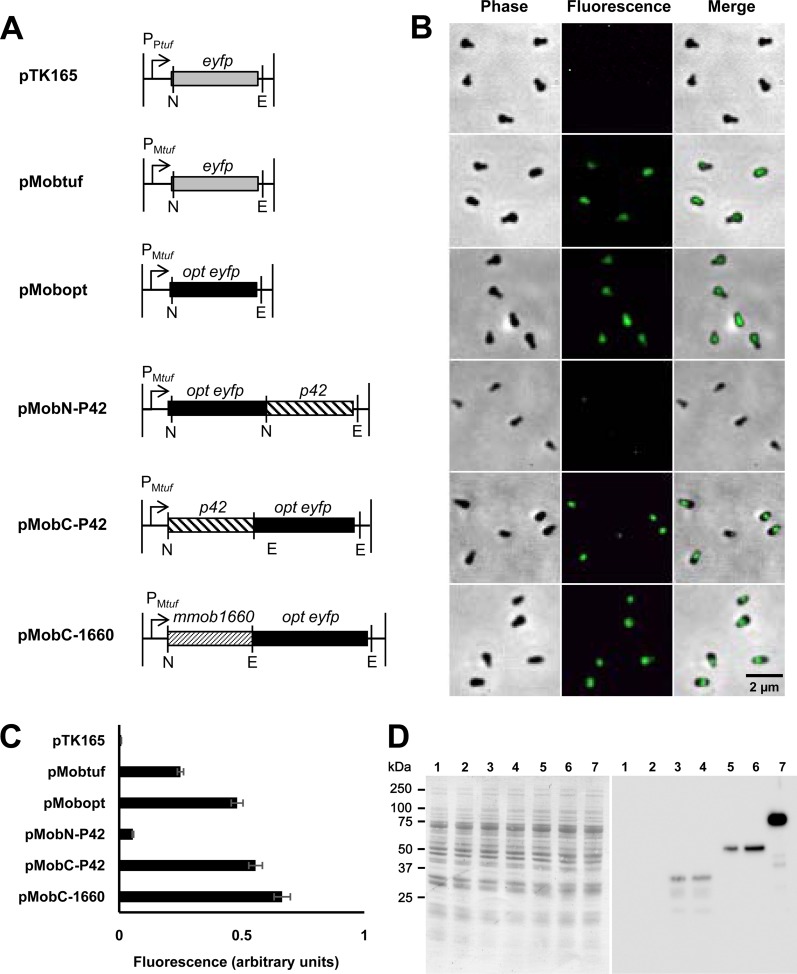
To visualize the subcellular localization of proteins of interest, we developed a fluorescent-protein tagging method. In previous Mycoplasma pneumoniae studies, genes of fluorescent proteins and their fusion proteins were inserted into an arm of a transposon (40, 53). The transposase expressed from the vector recognizes inverted repeats (IRs) and integrates the transposon element between right IR and left IR (R-IR and L-IR, respectively) into the genome (32, 54). Then, the fluorescent proteins are expressed by a strong promoter copied from an M. pneumoniae gene. As the pTK165 plasmid harboring the eyfp gene has been used for this purpose in M. pneumoniae, we examined the fluorescence of M. mobile transformants by fluorescence microscopy in the present study (40). However, the M. mobile transformants did not show fluorescence signals. We then replaced the promoter sequence for the M. pneumoniae gene encoding elongation factor Tu with the sequence of M. mobile to express the eyfp gene, resulting in the plasmid pMobtuf (Fig. 3A) (11). The transformants then showed clear emission of fluorescence (Fig. 3B). To improve the fluorescence intensity, we tried using the promoters for the gene encoding an abundant M. mobile surface protein, MvspI (46, 55, 56), and an abundant Spiroplasma citri surface protein, spiralin (data not shown) (57). However, neither construct showed any fluorescence signal. As mycoplasma genomes are biased toward high-AT contents, the codon usage of mycoplasma genes is different from that of many other organisms. To improve fluorescence intensity, we replaced the eyfp gene with one optimized for codon usage of mycoplasmas, resulting in the pMobopt plasmid, and achieved a 1.9-fold increase in fluorescence intensity (Fig. 3B and C). The fluorescence of cells was stable in medium with gentamicin, but it decreased in cultivation in liquid medium without gentamicin and was totally lost after 3 days, which is a time for about nine generations, suggesting that the transposon could be excised from the chromosome in the absence of gentamicin.
FIG 3.
EYFP and fusion protein expression examined by fluorescence and immunoblotting. (A) Design of constructs. The constructs listed here were inserted into an arm of a transposon in a transposon vector, pTK165 (40). The DNA fragment shown in pTK165 was replaced by that shown for each construct. Plasmid names are on the left. P, promoter of M. pneumoniae tuf (PPtuf) and that of M. mobile (PMtuf). NcoI, BamHI, and EcoRI recognition sites are shown as N, B, and E, respectively. opt eyfp, the codon-optimized eyfp gene. (B) Fluorescence images of cells. Phase-contrast and fluorescence images are merged. The cells were transformed by the plasmid indicated in the corresponding line in panel A. (C) Quantification of the fluorescence intensity of a single cell. The intensity is presented as the average of more than 10 cells with the standard deviation. (D) Immunoblotting analysis of EYFP and fusion proteins. Whole-cell lysates were analyzed by SDS–10% PAGE (left) and blotting (right) for cells transformed by, pTK165 (lane 2), pMobtuf (lane 3), pMobopt (lane 4), pMobN-P42 (lane 5), pMobC-P42 (lane 6), and pMobC-1660 (lane 7). Lane 1, empty vector. Molecular sizes are shown on the left.
Next, we chose the P42 protein as the initial target for fluorescent protein tagging. P42 is encoded in the same operon with three large proteins essential for gliding, suggesting its participation in the gliding mechanism (Fig. 1) (16, 31). However, its subcellular localization is unknown. The p42 gene was fused to the 3′ and 5′ end of the eyfp gene, resulting in p42-eyfp and eyfp-p42 fusion genes, respectively. The level of fluorescence intensity detected from pMobC-P42 was similar to that detected from pMobopt, but we did not detect a signal from pMobN-P42. In all cells transformed by pMobC-P42, the signals were found at the neck part (Fig. 3B and 4).
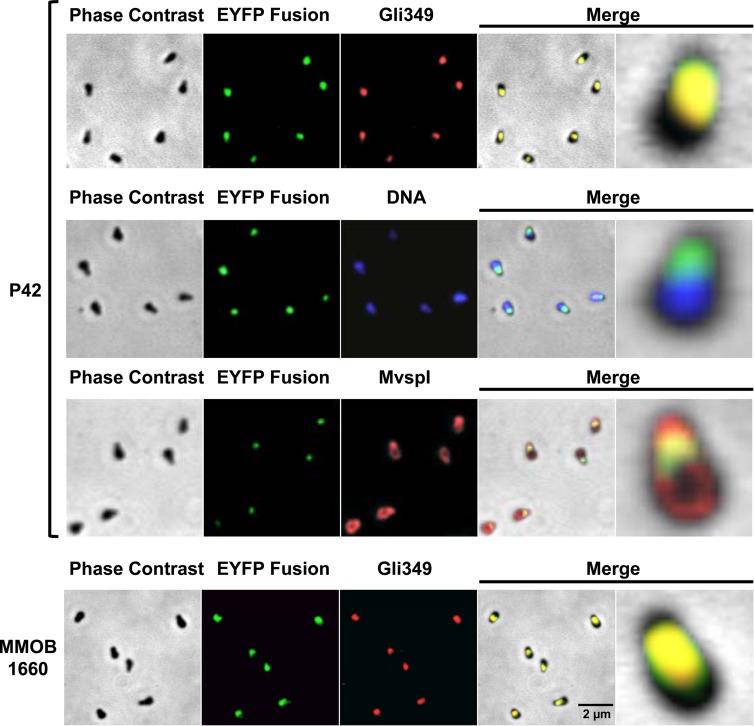
FIG 4.
Subcellular localization of P42 and the F1-ATPase α subunit homolog, MMOB1660. Localizations of P42, MMOB1660, Gli349, DNA, and MvspI were examined by fluorescence microscopy. P42 and MMOB1660 were detected as EYFP fusion proteins. Gli349 and MvspI were detected by immunostaining, and DNA was detected by DAPI staining. Phase-contrast and fluorescence images were merged in the two panels at right. In merged images, a single cell in the left panel is magnified in the right panel. The transposon insertion sites were mapped at nucleotide 374965 on the minus strand and at nucleotide 187879 on the plus strand on the genome, respectively, for P42 and MMOB1660 localization in these images (see Table S1 in the supplemental material).
The next target protein was MMOB1660, which is an F1-ATPase α-subunit homolog and a component of the jellyfish structure (Fig. 1). In the present study, the MMOB1660 gene was fused to the 5′ end of the eyfp gene, resulting in the MMOB1660 gene-eyfp fusion. The signal was found around the neck with a fluorescence intensity slightly higher than that of pMobC-P42 (Fig. 3B and 4).
We next examined the expression levels of EYFP and fusion proteins by immunoblotting (Fig. 3D). EYFP was not detected in the transformants harboring pTK165, but it was detected in all constructs containing the M. mobile tuf promoter. The protein amounts varied significantly although the genes were under the control of the same promoter. The amounts of EYFP and fusion proteins were roughly consistent with the fluorescence intensity.
Mapping of insertion sites on the genome.
The transposon vectors used in the gene manipulation of mycoplasmas harboring pTK165 are unable to replicate in mycoplasma cells because they do not have the replication origin of mycoplasma (40). The genomes were analyzed for 18, 10, and 10 isolates transformed by pTK165, pMobC-P42, and pMobC-1660, respectively (see Fig. S1 in the supplemental material), to determine the insertion sites. Typically, 200- to 300-bp DNA sequences flanking the insertion site were identified by nested arbitrary PCR followed by sequencing. The results showed that the transposon element was integrated into the genome, as expected (32, 54). The insertion sites were all different although four open reading frames (ORFs) were identified as the insertion sites more than once. Obvious localization of insertion sites on the genome was not found, but we could not reach a conclusion about the site distribution because the sample number is not a saturating level. The analyses by PCR of six transformants from each plasmid suggested that the original p42 and MMOB1660 genes remain in the original positions on the genome. The binding and gliding activities of transformed cells did not differ in an obvious way from those of the wild-type strain, suggesting that the fused proteins might function in the same way as the original proteins in these cases.
Subcellular localization of P42 and the F1-ATPase α-subunit homolog.
To determine the subcellular localization of P42, the position of P42 relative to other structures was examined (Fig. 4). Gli349, MvspI, and DNA were localized at the neck, the head and body, and the body, respectively, in good agreement with previous studies (Fig. 1A) (12, 13, 55, 58). P42 was colocalized with Gli349 but not with MvspI or with DNA localized in regions other than the neck, suggesting that P42 is involved in the gliding mechanism. Next, we compared the subcellular localization of MMOB1660, the homolog of the F1-ATPase α-subunit, with that of Gli349 and concluded that the two were localized in the same position, suggesting that MMOB1660 is a component of jellyfish tentacles, part of the intercellular structure of the gliding machinery. We tried several isolates of the same constructs and found no significant differences, suggesting that the fluorescence signal does not depend on the sites of transposon insertion in the genome.
Sequence analysis of P42.
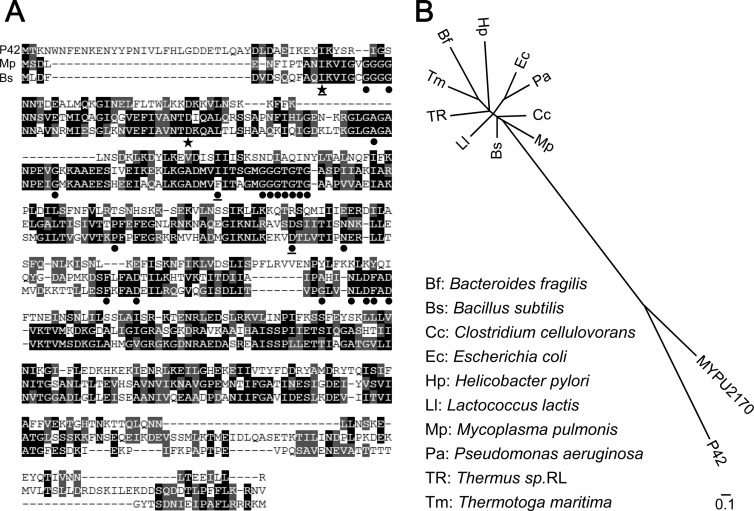
The previous analyses of the amino acid sequence of P42 showed no clear similarity to any proteins other than the plausible ortholog (MYPU2170) of M. pulmonis, a close relative of M. mobile (11, 31). In the present study, we searched for similarities by PSI-BLAST using the amino acid sequences of P42 and MYPU2170. This analysis gave a list of 39 similar proteins, with E values ranging from 0.025 to 1.5; of these proteins, 36 were assigned as FtsZ of Gram-positive bacteria, mainly Clostridium and Bacillus, that are related to Mycoplasma. The homologous region spans at least 228 of the total of 356 amino acids in P42. The sequence alignment of P42 with FtsZ of Mycoplasma pulmonis and Bacillus subtilis is shown in Fig. 5A. Generally, 24 amino acids, most of which were involved in GTPase activity, were well conserved in FtsZ and tubulin (59). Interestingly, however, only two of these amino acids are conserved in P42, suggesting that this protein does not perform the dynamic polymerization and depolymerization observed in FtsZ and tubulin (49). The phylogenetic tree analyzed with 12 related proteins showed that P42 and MYPU2170 form a group apart from the FtsZs and are most closely related to the FtsZ of M. pulmonis (Fig. 5B). These results suggest that P42 originated from the same ancestor as FtsZ but developed to perform different functions.
FIG 5.
Sequence analysis of P42. (A) Similarity between P42 (top rows of sequence) and FtsZs of Mycoplasma pulmonis (Mp; middle rows) and Bacillus subtilis (Bs; bottom rows). Identical and similar amino acids are marked by black and gray boxes, respectively. Twenty-four amino acids that were well conserved in FtsZ and tubulin are marked by filled circles and stars (59); those marked by a solid star were also conserved in P42. Two amino acids were not conserved in FtsZ of Mycoplasma pulmonis. Twenty-one amino acids were involved in GTP binding, and the functions of the other three amino acids (underlined) are unknown. (B) Phylogenetic tree analyzed for P42, the ortholog of M. pulmonis (MYPU2170), and FtsZ proteins from 10 species. The ftsZ gene has not been identified in the genome of M. mobile. The scale bar indicates the number of substitutions by position.
DISCUSSION
Transformation of M. mobile.
Transformants grew more slowly than nontransformants in growth medium without antibiotics; for example, the colony formation of transformants took 7 days with gentamicin but only 3 days without gentamicin. The growth rate did not depend on the insertion site of the transposon in the genome. These facts suggest that the lower growth rate of transformants was not caused by the disruption of important genes but, rather, by the effect of antibiotics although the transformants were transformed by the resistance genes. In other words, the resistance genes were not effective enough to recover the normal growth rate. These features can also be seen in the transformants of other Mycoplasma species. In the case of M. pneumoniae, it takes 4 days to form the colonies of transformants under antibiotics but only 2 days without antibiotics (41). This may be due to the insufficient expression of marker gene products or insufficient protein activity in a mycoplasma cell. The outgrowth of M. mobile took 12 h, much longer than the 2 to 3 h required for other Mycoplasma species. This difference might be related to the lower growth temperature of M. mobile, 25°C, which is distinct from that of other species. The expression of the gentamicin gene in an M. mobile cell may require more time because of the lower growth temperature.
Fluorescence intensity of EYFP.
We replaced the promoter of M. pneumoniae tuf by that of M. mobile and also used the eyfp sequence, which was optimized to that of M. mobile to obtain a higher intensity of fluorescence. These two modifications provided sufficient fluorescence intensity, probably as a result of the higher levels of protein expression, as shown by the immunoblotting analysis (Fig. 3D). The protein amounts depended on the construct although the genes were controlled under the same promoter, showing that protein synthesis and stability depended on the amino acid sequence. Fluorescence intensity also did not precisely parallel protein amount, as shown by the difference between pMobN-P42 and pMobC-P42, suggesting that the environments around the fluorescence moiety also are determinants. The eyfp and its fusion genes were inserted into various sites in the genome. However, we did not detect any significant differences in fluorescence intensity levels among the isolates, suggesting that the high activity of the tuf promoter was not influenced by the genome.
Whole-cell imaging.
To date, the subcellular localizations have been clarified for 13 proteins in M. mobile (Fig. 1), and all proteins localizing at the neck part have been suggested to be involved in the gliding mechanism (3, 22, 55). In the present study, two other proteins were shown to be localized on the gliding machinery (Fig. 4). P42, which appears to have developed from an ancestor shared with FtsZ, has a special role in the gliding mechanism. This protein should be localized inside the cell at some structure other than the jellyfish tentacle or have an occluded position on the surface because a previous study in which surface proteins were biotinylated did not detect exposure of P42 (55), and it was not identified in the fraction of the jellyfish structure (22).
MMOB1660, an F1-ATPase α-subunit homolog and a component of the jellyfish structure, has been shown to localize at a position similar to that of Gli349. As the position of Gli349 corresponds to that of the jellyfish tentacles (5, 22), we concluded that MMOB1660 localizes at the tentacles of the jellyfish structure. Previously, the MMOB1670 protein, an F1-ATPase β-subunit homolog, was shown to localize at the jellyfish tentacles by immunofluorescence microscopy (22). Probably, these two proteins form a complex on the jellyfish tentacles similar to F1-ATPase. On the tentacles of the jellyfish structure, particles about 10 nm wide are aligned periodically with a pitch of about 30 nm. These particles might be composed of MMOB1660 and MMOB1670.
Roles of P42 and the F1-ATPase subunit homolog in the proposed gliding mechanism.
Experimental data have shown that gliding occurs by the repeated catch-pull-release of sialylated oligosaccharides by Gli349, coupled with the movements of Gli521, driven by ATP hydrolysis (Fig. 1) (3, 4, 28). In the present study, we found that a homolog of the F1-ATPase α-subunit, together with a homolog of the F1-ATPase β-subunit, forms the tentacles of the jellyfish structure. This may suggest that the movement for gliding is generated here, inside the cell, by ATP hydrolysis. P42 might not conserve the dynamics of FtsZ because of its lack of amino acids essential for GTPase, but the surfaces needed for protofilament formation might be conserved. This protein could play a role in supporting or bridging parts in the gliding machinery, or in transmitting movements inside the cell to the surface structures, including the legs.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the helpful discussions with Howard C. Berg at Harvard University, Tsuyoshi Kenri at the National Institute of Infectious Diseases, Tokyo, Japan, Itaru Yanagihara and Heng Ning Wu at the Osaka Medical Center for Maternal and Child Health, and Taro Nakamura at Osaka City University.
This work was supported by Grants-in-Aid for Scientific Research in the Priority Area Harmonized Supramolecular Motility Machinery and Its Diversity (24117002 to M.M.), and by Grants-in-Aid for Scientific Research (A, B) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (50209912 and 50209912 to M.M.).
Footnotes
Published ahead of print 7 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01418-13.
REFERENCES
- 1.Razin S, Yogev D, Naot Y. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyata M, Nakane D. 2013. Gliding mechanism of Mycoplasma pneumoniae subgroup implication from Mycoplasma mobile, p 237–252 In Browning G, Citti C. (ed), Molecular and cell biology of Mollicutes. Horizon Press, Norfolk, United Kingdom [Google Scholar]
- 3.Miyata M. 2010. Unique centipede mechanism of Mycoplasma gliding. Annu. Rev. Microbiol. 64:519–537. 10.1146/annurev.micro.112408.134116 [DOI] [PubMed] [Google Scholar]
- 4.Miyata M. 2008. Centipede and inchworm models to explain Mycoplasma gliding. Trends Microbiol. 16:6–12. 10.1016/j.tim.2007.11.002 [DOI] [PubMed] [Google Scholar]
- 5.Nakane D, Miyata M. 2012. Mycoplasma mobile cells elongated by detergent and their pivoting movements in gliding. J. Bacteriol. 194:122–130. 10.1128/JB.05857-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiratsuka Y, Miyata M, Tada T, Uyeda TQP. 2006. A microrotary motor powered by bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:13618–13623. 10.1073/pnas.0604122103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyata M, Ryu WS, Berg HC. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184:1827–1831. 10.1128/JB.184.7.1827-1831.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyata M. 2007. Molecular mechanism of Mycoplasma gliding: a novel cell motility system, p 137–175 In Lenz P. (ed), Cell motility. Springer, New York, NY [Google Scholar]
- 9.Krunkosky TM, Jordan JL, Chambers E, Krause DC. 2007. Mycoplasma pneumoniae host-pathogen studies in an air-liquid culture of differentiated human airway epithelial cells. Microb. Pathog. 42:98–103. 10.1016/j.micpath.2006.11.003 [DOI] [PubMed] [Google Scholar]
- 10.Kirchhoff H. 1992. Motility, p 289–306 In Maniloff J, McElhaney RN, Finch LR, Baseman JB. (ed), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, DC [Google Scholar]
- 11.Jaffe JD, Stange-Thomann N, Smith C, DeCaprio D, Fisher S, Butler J, Calvo S, Elkins T, FitzGerald MG, Hafez N, Kodira CD, Major J, Wang S, Wilkinson J, Nicol R, Nusbaum C, Birren B, Berg HC, Church GM. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447–1461. 10.1101/gr.2674004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uenoyama A, Kusumoto A, Miyata M. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 186:1537–1545. 10.1128/JB.186.5.1537-1545.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kusumoto A, Seto S, Jaffe JD, Miyata M. 2004. Cell surface differentiation of Mycoplasma mobile visualized by surface protein localization. Microbiology 150:4001–4008. 10.1099/mic.0.27436-0 [DOI] [PubMed] [Google Scholar]
- 14.Miyata M, Uenoyama A. 2002. Movement on the cell surface of gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215:285–289. 10.1111/j.1574-6968.2002.tb11404.x [DOI] [PubMed] [Google Scholar]
- 15.Uenoyama A, Seto S, Nakane D, Miyata M. 2009. Regions on Gli349 and Gli521 protein molecules directly involved in movements of Mycoplasma mobile gliding machinery suggested by inhibitory antibodies and mutants. J. Bacteriol. 191:1982–1985. 10.1128/JB.01012-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uenoyama A, Miyata M. 2005. Identification of a 123-kilodalton protein (Gli123) involved in machinery for gliding motility of Mycoplasma mobile. J. Bacteriol. 187:5578–5584. 10.1128/JB.187.16.5578-5584.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seto S, Uenoyama A, Miyata M. 2005. Identification of 521-kilodalton protein (Gli521) involved in force generation or force transmission for Mycoplasma mobile gliding. J. Bacteriol. 187:3502–3510. 10.1128/JB.187.10.3502-3510.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyata M, Yamamoto H, Shimizu T, Uenoyama A, Citti C, Rosengarten R. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311–1320 http://mic.sgmjournals.org/content/146/6/1311.long [DOI] [PubMed] [Google Scholar]
- 19.Nonaka T, Adan-Kubo J, Miyata M. 2010. Triskelion structure of the Gli521 protein, involved in the gliding mechanism of Mycoplasma mobile. J. Bacteriol. 192:636–642. 10.1128/JB.01143-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adan-Kubo J, Uenoyama A, Arata T, Miyata M. 2006. Morphology of isolated Gli349, a leg protein responsible for glass binding of Mycoplasma mobile gliding revealed by rotary-shadowing electron microscopy. J. Bacteriol. 188:2821–2828. 10.1128/JB.188.8.2821-2828.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyata M, Petersen J. 2004. Spike structure at interface between gliding Mycoplasma mobile cell and glass surface visualized by rapid-freeze and fracture electron microscopy. J. Bacteriol. 186:4382–4386. 10.1128/JB.186.13.4382-4386.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakane D, Miyata M. 2007. Cytoskeletal “jellyfish” structure of Mycoplasma mobile. Proc. Natl. Acad. Sci. U. S. A. 104:19518–19523. 10.1073/pnas.0704280104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uenoyama A, Miyata M. 2005. Gliding ghosts of Mycoplasma mobile. Proc. Natl. Acad. Sci. U. S. A. 102:12754–12758. 10.1073/pnas.0506114102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaffe JD, Miyata M, Berg HC. 2004. Energetics of gliding motility in Mycoplasma mobile. J. Bacteriol. 186:4254–4261. 10.1128/JB.186.13.4254-4261.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasai T, Nakane D, Ishida H, Ando H, Kiso M, Miyata M. 2013. Role of binding in Mycoplasma mobile and Mycoplasma pneumoniae gliding analyzed through inhibition by synthesized sialylated compounds. J. Bacteriol. 195:429–435. 10.1128/JB.01141-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lesoil C, Nonaka T, Sekiguchi H, Osada T, Miyata M, Afrin R, Ikai A. 2009. Molecular shape and binding force of Mycoplasma mobile's leg protein Gli349 revealed by an AFM study. Biochem. Biophys. Res. Commun. 391:1312–1317. 10.1016/j.bbrc.2009.12.023 [DOI] [PubMed] [Google Scholar]
- 27.Nagai R, Miyata M. 2006. Gliding motility of Mycoplasma mobile can occur by repeated binding to N-acetylneuraminyllactose (sialyllactose) fixed on solid surfaces. J. Bacteriol. 188:6469–6475. 10.1128/JB.00754-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Neu J, Miyata M, Oster G. 2009. Motor-substrate interactions in Mycoplasma motility explains non-Arrhenius temperature dependence. Biophys. J. 97:2930–2938. 10.1016/j.bpj.2009.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beven L, Charenton C, Dautant A, Bouyssou G, Labroussaa F, Skollermo A, Persson A, Blanchard A, Sirand-Pugnet P. 2012. Specific evolution of F1-like ATPases in mycoplasmas. PLoS One 7:e38793. 10.1371/journal.pone.0038793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santana M, Ionescu MS, Vertes A, Longin R, Kunst F, Danchin A, Glaser P. 1994. Bacillus subtilis F0F1 ATPase: DNA sequence of the atp operon and characterization of atp mutants. J. Bacteriol. 176:6802–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohtani N, Miyata M. 2007. Identification of a novel nucleoside triphosphatase from Mycoplasma mobile: a prime candidate for motor of gliding motility. Biochem. J. 403:71–77. 10.1042/BJ20061439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.French CT, Lao P, Loraine AE, Matthews BT, Yu H, Dybvig K. 2008. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol. Microbiol. 69:67–76. 10.1111/j.1365-2958.2008.06262.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye F, Renaudin J, Bove JM, Laigret F. 1994. Cloning and sequencing of the replication origin (oriC) of the Spiroplasma citri chromosome and construction of autonomously replicating artificial plasmids. Curr. Microbiol. 29:23–29. 10.1007/BF01570187 [DOI] [PubMed] [Google Scholar]
- 34.Dhandayuthapani S, Rasmussen WG, Baseman JB. 1999. Disruption of gene mg218 of Mycoplasma genitalium through homologous recombination leads to an adherence-deficient phenotype. Proc. Natl. Acad. Sci. U. S. A. 96:5227–5232. 10.1073/pnas.96.9.5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aluotto BB, Wittler RG, Williams CO, Faber JE. 1970. Standardized bacteriologic techniques for the characterization of Mycoplasma species. Int. J. Syst. Bacteriol. 20:35–58. 10.1099/00207713-20-1-35 [DOI] [Google Scholar]
- 36.Algire MA, Lartigue C, Thomas DW, Assad-Garcia N, Glass JI, Merryman C. 2009. New selectable marker for manipulating the simple genomes of Mycoplasma species. Antimicrob. Agents Chemother. 53:4429–4432. 10.1128/AAC.00388-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pich OQ, Burgos R, Planell R, Querol E, Pinol J. 2006. Comparative analysis of antibiotic resistance gene markers in Mycoplasma genitalium: application to studies of the minimal gene complement. Microbiology 152:519–527. 10.1099/mic.0.28287-0 [DOI] [PubMed] [Google Scholar]
- 38.Hahn TW, Mothershed EA, Waldo RH, III, Krause DC. 1999. Construction and analysis of a modified Tn4001 conferring chloramphenicol resistance in Mycoplasma pneumoniae. Plasmid 41:120–124. 10.1006/plas.1998.1387 [DOI] [PubMed] [Google Scholar]
- 39.Gawron-Burke C, Clewell DB. 1984. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J. Bacteriol. 159:214–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kenri T, Seto S, Horino A, Sasaki Y, Sasaki T, Miyata M. 2004. Use of fluorescent-protein tagging to determine the subcellular localization of Mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus. J. Bacteriol. 186:6944–6955. 10.1128/JB.186.20.6944-6955.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedreyda CT, Lee KK, Krause DC. 1993. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid 30:170–175. 10.1006/plas.1993.1047 [DOI] [PubMed] [Google Scholar]
- 42.Seto S, Layh-Schmitt G, Kenri T, Miyata M. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621–1630. 10.1128/JB.183.5.1621-1630.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naito M, Sato K, Shoji M, Yukitake H, Ogura Y, Hayashi T, Nakayama K. 2011. Characterization of the Porphyromonas gingivalis conjugative transposon CTnPg1: determination of the integration site and the genes essential for conjugal transfer. Microbiology 157:2022–2032. 10.1099/mic.0.047803-0 [DOI] [PubMed] [Google Scholar]
- 44.Chen T, Dong H, Tang YP, Dallas MM, Malamy MH, Duncan MJ. 2000. Identification and cloning of genes from Porphyromonas gingivalis after mutagenesis with a modified Tn4400 transposon from Bacteroides fragilis. Infect. Immun. 68:420–423. 10.1128/IAI.68.1.420-423.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyata M, Wang L, Fukumura T. 1991. Physical mapping of the Mycoplasma capricolum genome. FEMS Microbiol. Lett. 63:329–333 [DOI] [PubMed] [Google Scholar]
- 46.Wu HN, Kawaguchi C, Nakane D, Miyata M. 2012. “Mycoplasmal antigen modulation,” a novel surface variation suggested for a lipoprotein specifically localized on Mycoplasma mobile. Curr. Microbiol. 64:433–440. 10.1007/s00284-012-0090-y [DOI] [PubMed] [Google Scholar]
- 47.Robertson J, Stemke GW. 1995. Measurement of mollicute growth by ATP-dependent luminometry, p 65–72 In Razin S, Tully JG. (ed), Molecular and diagnostic procedures in mycoplasmology, vol 1 Academic Press, Inc., San Diego, CA [Google Scholar]
- 48.Dybvig K, Gasparich GE, King KW. 1995. Artificial transformation of mollicutes via polyethylene glycol- and electroporation-mediated methods, p 179–184 In Razin S, Tully JG. (ed), Molecular and diagnostic procedures in mycoplasmology, vol 1 Academic Press, San Diego, CA [Google Scholar]
- 49.Chambaud I, Heilig R, Ferris S, Barbe V, Samson D, Galisson F, Moszer I, Dybvig K, Wroblewski H, Viari A, Rocha EP, Blanchard A. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic. Acids Res. 29:2145–2153. 10.1093/nar/29.10.2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahairas GG, Minion FC. 1989. Transformation of Mycoplasma pulmonis: demonstration of homologous recombination, introduction of cloned genes, and preliminary description of an integrating shuttle system. J. Bacteriol. 171:1775–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ichimaru H, Nakamura M. 1979. The components of Mycoplasma salivarium and its growth medium that are responsible for film formation. J. Gen. Microbiol. 112:389–392. 10.1099/00221287-112-2-389 [DOI] [PubMed] [Google Scholar]
- 52.Thorns CJ. 1978. Identification of the products of the “film spots” reaction of Mycoplasma bovis and Mycoplasma bovigenitalium. J. Biol. Stand. 6:127–132. 10.1016/S0092-1157(78)80044-4 [DOI] [PubMed] [Google Scholar]
- 53.Balish MF, Santurri RT, Ricci AM, Lee KK, Krause DC. 2003. Localization of Mycoplasma pneumoniae cytadherence-associated protein HMW2 by fusion with green fluorescent protein: implications for attachment organelle structure. Mol. Microbiol. 47:49–60. 10.1046/j.1365-2958.2003.03282.x [DOI] [PubMed] [Google Scholar]
- 54.Byrne ME, Rouch DA, Skurray RA. 1989. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene 81:361–367. 10.1016/0378-1119(89)90197-2 [DOI] [PubMed] [Google Scholar]
- 55.Wu HN, Miyata M. 2012. Whole surface image of Mycoplasma mobile, suggested by protein identification and immunofluorescence microscopy. J. Bacteriol. 194:5848–5855. 10.1128/JB.00976-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adan-Kubo J, Yoshii SH, Kono H, Miyata M. 2012. Molecular structure of isolated MvspI, a variable surface protein of the fish pathogen Mycoplasma mobile. J. Bacteriol. 194:3050–3057. 10.1128/JB.00208-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chevalier C, Saillard C, Bove JM. 1990. Spiralins of Spiroplasma citri and Spiroplasma melliferum: amino acid sequences and putative organization in the cell membrane. J. Bacteriol. 172:6090–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyata M, Ogaki H. 2006. Cytoskeleton of mollicutes. J. Mol. Microbiol. Biotechnol. 11:256–264. 10.1159/000094059 [DOI] [PubMed] [Google Scholar]
- 59.Erickson HP. 2007. Evolution of the cytoskeleton. Bioessays 29:668–677. 10.1002/bies.20601 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.