Abstract
Evolutionary selection for optimal genome preservation, replication, and expression should yield similar chromosome organizations in any type of cells. And yet, the chromosome organization is surprisingly different between eukaryotes and prokaryotes. The nuclear versus cytoplasmic accommodation of genetic material accounts for the distinct eukaryotic and prokaryotic modes of genome evolution, but it falls short of explaining the differences in the chromosome organization. I propose that the two distinct ways to organize chromosomes are driven by the differences between the global-consecutive chromosome cycle of eukaryotes and the local-concurrent chromosome cycle of prokaryotes. Specifically, progressive chromosome segregation in prokaryotes demands a single duplicon per chromosome, while other “precarious” features of the prokaryotic chromosomes can be viewed as compensations for this severe restriction.
INTRODUCTION
Cells are what their genomes instruct them to be. The observed uniformity, continuity, and robustness of specific life forms reflect how securely their genomes are preserved, how faithfully they are replicated, and how reliably they are expressed to yield specific cellular phenotypes. Formally, the genome is a set of trait-encoding entities (genes) irrespective of how the information is coded, organized, or read. Since the three main functions of genetic information (preservation, replication, and expression) transcend the cell types, one could conservatively expect that, while genome evolution modes might be different between the eukaryotic and prokaryotic cell types (1), the genome organizations would be similar. Indeed, there are basic features of genome organization common to any type of cells: (i) genetic material is always duplex DNA that always replicates semiconservatively; (ii) genetic material is “quantal” in that genes are separate from each other, each gene occupying its own designated stretch of duplex DNA; (iii) besides the gene-encoding DNA, genomes always have noncoding DNA (for example, regulatory regions of genes), as well as selfish elements that use the genome as a habitat in which to multiply; and (iv) major genome changes are either internal rearrangements (usually by DNA repeats) or acquisitions of foreign DNA carrying new genes from the environment (horizontal gene transfer). (Note that, in contrast to cellular life forms, viruses are cell-supported life forms: they are dead outside the cells but can organize their own metabolism and genome replication once inside the host cell. Chemically, viral genomes can be based on RNA or DNA, and either biopolymer can be either single stranded or double stranded. Viral genome organization is diverse and is not covered in this minireview.) There are also differences in genome organization and evolution between eukaryotic cells, which keep their genome in a special compartment called the nucleus, and prokaryotic cells, which keep their genome free-floating in the cytoplasm (1) (with the exception of the membrane-wrapped nucleoid of planctomycetes [2]).
The gene content of the eukaryotic genomes correlates poorly with the genome size (3). There is a lot of noncoding DNA between eukaryotic genes, and the coding sequences of genes themselves are interrupted by introns, both short and long (4). But it is not the random DNA from the environment that inflates the eukaryotic genomes. In fact, eukaryotic genome evolution is not much influenced by horizontal gene transfer, as it is difficult for the unprotected exogenous DNA to reach the nucleus through the cytoplasm, due to the cytoplasmic DNases (5–7) and the cytoplasmic DNA routing that specifically avoids the nucleus (8, 9). The major type of exogenous DNA that has a significant chance of inserting into the eukaryotic genome is the cDNA of retroviruses, single-stranded RNA (ssRNA) viruses that replicate only in the nucleus via the duplex cDNA intermediates integrated into the host genome (10), making retroviral infections a major driver of the eukaryotic genome evolution. The small sizes of the retroviral genomes, the one-enzyme mechanism of retroviral cDNA formation (11), and the rampant recombination during cDNA synthesis (12) breed ever-changing families of simplistic mobile retroelements that infest eukaryotic genomes with thousands of repeats each. These retroelements and the layers of their decaying remnants comprise the bulk of noncoding DNA in the eukaryotic genomes (13–16). The retroelement-derived repeats in eukaryotic genomes facilitate peculiar karyotype fluidity: eukaryotic chromosomes keep exchanging arms with each other, fuse together, or split apart (17, 18). As a result of this constant karyotype reshuffling, even evolutionarily closely related organisms (such as mouse and human) have different numbers of chromosomes and no common genome frame (18, 19). At the same time, “naked” genes from the environment rarely make it into the genomes of higher eukaryotes (20, 21), although horizontal gene transfer does contribute to the genome evolution in unicellular eukaryotes (22).
In contrast, prokaryotic genomes are jam-packed with genes (with minimal intragenic regions and almost no repeats, the genome size becomes an accurate reflection of the gene content) (3), while the very few introns in the prokaryotic genomes are always big, coding for selfish elements (23). In further contrast, prokaryotic genome evolution is dominated by horizontal gene transfer (24, 25), where relatively long uninterrupted chunks of foreign DNA are internalized for food (25) but end up being inserted into the chromosome, becoming part of the genome. Horizontal gene transfer is further enhanced by the “mobilome” (24)—the collection of genes on the extrachromosomal elements (plasmids and phages) staying for a few, or a few thousand, generations within prokaryotic cells. The efficient horizontal gene transfer and the mobilome allow any particular prokaryote to move into any environment compatible with the general metabolism of the newcomer cell: the habitat-specific genes are supplied later by aboriginal neighbors. In yet another stark difference from eukaryotes, prokaryotic genomes have a few active mobile elements (26), and these few are always tightly controlled, as element jumping or repeat-induced recombination in the gene-packed prokaryotic genomes always reduces adaptation and is often lethal. The low activity of mobile elements is a major contributor to the evolutionarily stable common frames in prokaryotic genomes (related prokaryotes show a high degree of syntheny [27]); another major contributor to the genome frame stability has been recently recognized as the spatiotemporal pattern of nucleoid condensation and regulation of gene expression relative to the origin-terminus axis (28, 29). Finally, in yet another contrast to the ever-inflating eukaryotic genomes, prokaryotic genomes strongly prefer to delete rather than insert DNA; this preference, apparently, drives their unrelenting space crunch (30, 31).
DIFFERENCES BETWEEN EUKARYOTIC AND PROKARYOTIC CHROMOSOMES
There are at least four more specific, structural genome organization features common to both eukaryotic and prokaryotic cells: (i) genes are always arranged as unidimensional chains, like beads on a string (genomic DNA is never branched or star-shaped, for example); (ii) these genomic DNA chains, called chromosomes, are always long, comprising hundreds and thousands of genes; (iii) since the length of the chromosomes is always 100 to 1,000× greater than that of the cell or cellular compartment in which these chromosomes are housed (32–35), chromosomes are always highly compacted, in a local fold-back pattern resembling rosettes of radial loops (36, 37); and (iv) while some “genes from the environment” arrive on chromosomal fragments that would be lost unless incorporated into the host chromosomes, some other environmental genes arrive on small autonomously replicating and segregating extrachromosomal elements, called plasmids. Chromosomes, as specific molecular structures performing certain functions and undergoing certain transitions, are practical representations of the cell's vision of how to best organize preservation, replication, and expression of its genetic information. After billions of years of evolution, the specific chemical way to code information (DNA) and the cell's way to organize the genome (chromosomes) must reflect the winning strategy, evolutionarily optimized over an uncountable number of generations. From this perspective, the major details of the chromosome structure or function are also expected to be similar among all cell types. Surprisingly, beyond the four basic structural aspects mentioned above, the chromosome structures and functions are dramatically different between eukaryotes and prokaryotes, the nuclear versus anuclear organization of genetic material having little relevance to this difference. Indeed, both the prokaryotic chromosome organization and the eukaryotic chromosome organization “rules” allow numerous exceptions of the opposite type, suggesting that at the chromosomal level the dichotomy is maintained by a different kind of selection. The structure/function differences between eukaryotes and prokaryotes in the chromosome organization are compared below (for a different view on the dichotomy, see reference 38). As the “opinions” presented at the end of each section argue, this comparison makes it clear that one of the two ways to organize chromosomes is more precarious than the other.
STRUCTURAL DIFFERENCES
The structural differences between eukaryotic and prokaryotic chromosomes are so dramatically obvious that they, together with the presence or absence of the nucleus itself, were offered to secure the concept of the prokaryotic cell some 50 years ago (1).
Eukaryotic cells have multiple chromosomes per karyotype (complete chromosome set), with a typical diploid number of between 10 and 100 (39, 40). The two reported exceptions with a single chromosome per haploid set are the nematode Parascaris equorum univalens (41) and the ant Mirmecia pilosula (42), but they are truly unique, because even their closest relatives are multichromosomal. In contrast, bacteria usually have a single chromosome. A few bacteria, such as Vibrio (43) or Brucella (44) (and a few others [38]), have two chromosomes. All archaea with characterized genomes have a single chromosome (45); Haloarcula marismortui, with two, is the only known exception (46).
Plasmids are extrachromosomal DNA molecules with their own replicons/segregons that carry no sensu stricto essential genes. Plasmids are rare in the eukaryotic genomes (restricted to lower eukaryotes and fungi); many of them are mitochondrial (compartment of the prokaryotic origin), and all of them are small and adaptationally neutral (47, 48). The “instability” of plasmids in the nucleus is likely due to rapid invasion by retroelements, facilitating their terminal integration into one of the chromosomes via repeat-mediated exchanges. In contrast, the unique prokaryotic chromosome is frequently accompanied by one or a few plasmids. Moreover, prokaryotic plasmids tend to carry genes increasing adaptation of their host cells to specific environments, so they are frequently not adaptationally neutral. In fact, a small fraction of bacterial plasmids, carrying niche-specific essential genes and having chromosome-like GC content and codon usage, are now classified as “chromids” (49, 50) (basically, a part of the genome on an auxiliary replicon). Even though they contribute genetically and readily fuse with the chromosome by repeats provided by mobile elements (recall the famous HFR strains of Escherichia coli [51]), for some unclear reason prokaryotic plasmids are not allowed to stay within the chromosome for evolutionarily relevant periods of time, even if their copy number is low.
Eukaryotic chromosomes are always linear. Circular chromosomes can be engineered in eukaryotes but are unstable (52, 53), as there is no mechanism to resolve chromosomal dimers. In contrast, prokaryotic chromosomes are almost always circular (52); at the same time, there are sporadic lineages with linear chromosomes (and/or plasmids) (54). Moreover, circular prokaryotic chromosomes, once made linear using hairpin telomeres, remain stable and fully functional (55). Archaeal chromosomes are always circular (45).
Eukaryotic chromosomes are always equipped with centromeres (either single or multiple ones)—places of attachment of the segregation spindle (56). In contrast, prokaryotic chromosomes are either completely devoid of centromeres or carry the so-called “plasmid centromeres” which are not essential (with a few exceptions, such as Caulobacter) (57–60).
Opinion.
Multiple chromosomes are better than a single chromosome as the gene storage option (to avoid putting all eggs in a single basket), and linear chromosomes are obviously better than the circular ones, because they avoid the potentially lethal problems of chromosome dimerization and catenation. To their credit, prokaryotes have successfully solved both problems (61), but the rationale behind such a precarious chromosomal format as the single circular chromosome without a centromere is unclear. Perhaps the single chromosome in prokaryotes facilitates segregation? The eukaryotic response, having protein-mediated long-lasting sister-chromatid cohesion and allocating at least one centromere per chromosome, guarantees faithful segregation during cell division.
FUNCTIONAL DIFFERENCES
DNA condensation and packing.
Eukaryotic DNA is wrapped around protein nucleosomes and is further organized by histones and other proteins into a toroidal coil of “30-nm fibers” (39, 62–64), bringing the mass ratio of basic proteins to DNA in the eukaryotic chromatin to ∼1 (65). In contrast to this eukaryotic DNA wrapping on spiral rows of histone “bobbins,” prokaryotic DNA appears naked in that the isolated nucleoids look like a collection of wire loops, loosely held together by a proteinaceous core (36, 66, 67). To give these disorganized loops some order, prokaryotes make them braid with the help of unique topoisomerases capable of introducing unconstrained DNA superhelicity. Mesophilic prokaryotes employ DNA gyrase to introduce negative supercoils; thermophilic prokaryotes similarly employ reverse gyrase to introduce positive supercoils (68). No topoisomerase capable of introducing unconstrained supercoiling operates in the eukaryotic nucleus (69), only in prokaryote-descendant mitochondria and chloroplasts (68). Prokaryotic DNA is not “naked” in the strict sense, being complexed by thousands of molecules of the nucleoid-associated proteins and transcription factors, and yet the mass ratio of basic protein to DNA in prokaryotic chromosomes is only ∼0.02, in line with histoneless chromosomes of dinoflagellates (65). Besides this dynoflagellate exception to the eukaryotic histone packaging rule, there is an opposite exception to the prokaryotic “naked DNA” rule: of the two archeael groups, euryarchaea actually use minimalistic histones to pack their DNA (70).
Opinion.
While the eukaryotic DNA looks significantly more secure, the naked prokaryotic DNA is easier to replicate and transcribe.
Replication organization and regulation.
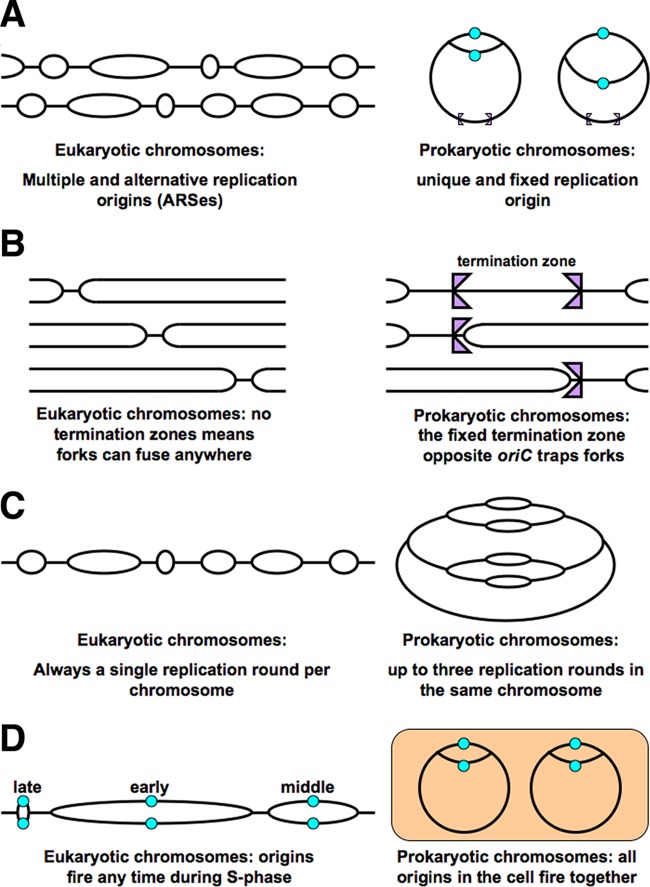
Eukaryotic chromosomes have multiple and alternative replication origins (ARSes), generating up to hundreds of replication bubbles per chromosome (Fig. 1A) (71). There are a few preferred origins that tend to fire every replication round, but most origins fire in only a fraction of replication rounds, and if replication is behind schedule in a particular chromosomal region, “ad hoc” origins fire in the region, accelerating local replication. In contrast, prokaryotic chromosomes typically have a single, unique replication origin that initiates a single replication bubble per chromosome (Fig. 1A) (72). There are examples of archaeal chromosomes with three or even four origins, though (73, 74).
FIG 1.
The differences between eukaryotic and prokaryotic chromosomes in organization and regulation of DNA replication. (A) Replication origins. (B) Replication termini. (C) Replication rounds. (D) Timing of origin firing.
Termination zones where converging replication forks meet are not defined in the eukaryotic chromosomes (Fig. 1B), even though there may be slow-replication zones, revealed in the S-phase checkpoint mutants in yeast (75) and explained by decreased availability of deoxynucleoside triphosphates (dNTPs) (76). The lack of dedicated termination zones is expected, since replication origin usage differs among replication rounds, shifting the location of replication fork fusion. In contrast, prokaryotic chromosomes, with their unique replication origin, have a defined zone, called the terminus, where converging replication forks fuse (Fig. 1B). Unidirectional termination sites bracket this chromosomal zone to form a “replication fork trap” into which replication forks can enter but from which they cannot escape (77, 78).
Notwithstanding the regulation complexity of multiple replication origins, eukaryotic chromosomes always undergo a single replication round at a time, so that during the S phase the ratio of maximally replicated DNA to unreplicated DNA is always 2:1 (Fig. 1C) (79). In contrast, the prokaryotic chromosomes can have several replication rounds in the same chromosome, so that the ratio of maximally replicated DNA to unreplicated DNA (which in prokaryotes can be expressed as the origin/terminus [ori/ter] ratio) can reach 8:1 (Fig. 1C) (80, 81).
Eukaryotic replication origins fire during the whole S phase, so the “early” ARSes fire at the beginning of S, while the “late” ARSes fire toward the end of S (Fig. 1D) (71). In contrast, in prokaryotes, all replication origins in the same cell always fire at once (synchronously) (Fig. 1D) (82, 83).
Opinion.
By all these replication parameters, the eukaryotic organization of replication looks natural, while the prokaryotic way again looks precarious. Why limit the number of replication origins to one? Why insist on a specific termination zone? Why allow the logistical nightmare of several replication rounds in the same cell? And why then demand that they initiate synchronously? With all these arbitrary-looking features, prokaryotes must be experiencing significant stresses in their replication system for an unclear payoff.
SCC.
Sister-chromatid cohesion (SCC) is the postreplication state throughout which separation of the sister chromatids is suppressed, so they appear as a single chromosome (84, 85). SCC guards the critical period of maturation of nascent DNA, during which at least four important tasks must be accomplished: (i) introduction of the regular coiling into the newly synthesized duplexes that emerge from the replisomes essentially paranemic, without coils; (ii) linking of Okazaki fragments together (86); (iii) repairing of persistent single-strand gaps and double-strand breaks (87); and (iv) removing the precatenanes that always accumulate behind replication forks (88, 89). In eukaryotic chromosomes, sister-chromatid cohesion is protein-(cohesin)-mediated and lasts several hours, encompassing the whole S, the whole G2, and part of the M phase until chromatid separation occurs (90). Completely replicated chromosomes do retain a low level of catenation, but it is not responsible for holding sister chromatids together (91). In contrast, in the prokaryotic chromosomes, the duration of sister-chromatid cohesion is short (only 6 min in the rapidly growing E. coli bacterium [92]) and the process is mostly mediated by precatenanes (93, 94) (even though sister chromatids may be held together at late-segregating loci by a special protein [95]).
Opinion.
It is not clear why the prokaryotic cells have to rush through this critical stage of nascent DNA maturation, especially given that prokaryotic DNA has to immediately undergo the stresses of segregation (as described next).
Segregation.
In eukaryotes, chromosomes are segregated once their replication is complete, after additional condensation, by the mitotic spindle, all at once (ensemble segregation), pulled toward the opposite cell poles by microtubules attached to their centromeres (96, 97) (Fig. 2A). In contrast, prokaryotic nucleoids were always known to segregate continuously, as they replicate, and without additional condensation (1, 98–100) (Fig. 2A). These days, we say that prokaryotic chromosomes are segregated locally (maybe even at the level of naked DNA), progressively (via the segregation forks [101] moving along the replicating chromosome), and concurrently with replication, once the short SCC is resolved by decatenation (92, 102–104). Nothing is known about the actual mechanism of prokaryotic chromosome segregation, with several possibilities discussed (61, 67, 96).
FIG 2.
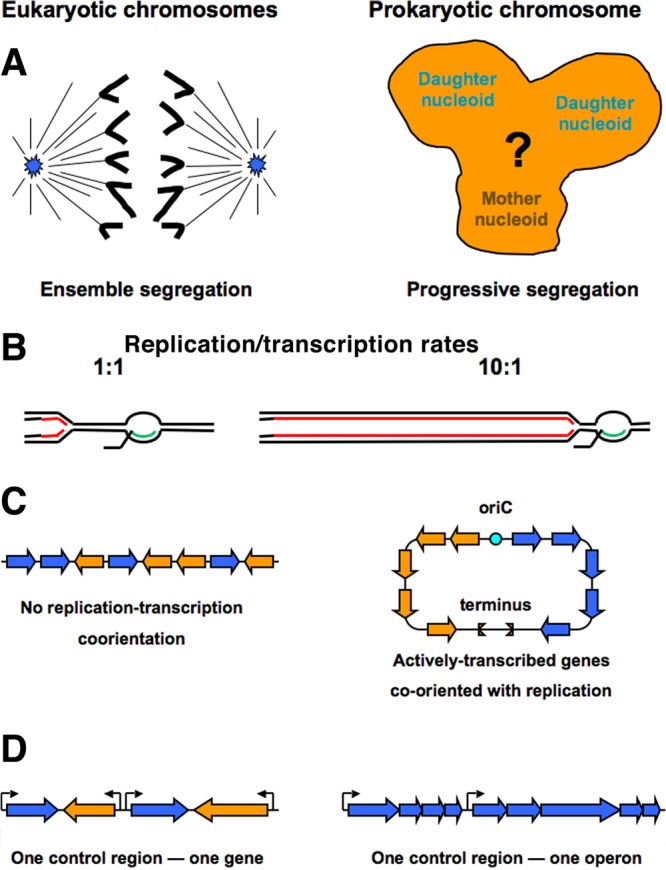
The differences between segregation of eukaryotic and prokaryotic chromosomes and the severity of the replication-transcription conflict. (A) Spindle-driven ensemble segregation in eukaryotes versus the unknown mechanism of progressive segregation in prokaryotes. (B) Schematic differences in rates of replication (red lines) versus transcription (green lines). (C) Gene coorientation with replication through the region. (D) The ratio of control regions to genes.
Opinion.
If confirmed, chromosomal segregation at the DNA level in prokaryotes is a precarious molecular manipulation requiring pulling on the naked DNA with forces strong enough to break it.
Replication-transcription conflict.
In eukaryotes, genome-average DNA replication rates (the size of genome over the replication time × the calculated number of replication forks), at 40 to 100 bp/s in yeast (105) and 10 to 60 bp/s in human cells (106), are similar to the mRNA transcription rates, at 30 to 60 nucleotides (nt)/s (107, 108) (Fig. 2B), so there is no conflict between the two processes except maybe in the rRNA gene arrays, where the rate is regulated by the replication fork barriers (78) and is further reduced due to the extra replication origins. In contrast, in the prokaryotic chromosomes, the genome-average replication rate during fast growth is at least an order of magnitude higher than the transcription rate (in E. coli, 600 to 900 nt/s for DNA synthesis versus 40 to 60 nt/s for transcription [109]) (Fig. 2B), making the conflict between the two processes unavoidable.
The acute reality of this conflict is reflected in the spectacular coorientation of the actively transcribed genes with the direction of replication in prokaryotic chromosomes (110, 111) (Fig. 2C). There are bacterial genomes with more than 80% of all genes cooriented with replication (112). The conflict can be demonstrated experimentally, by inversion of part of the replichore (113–115). In contrast, even though some degree of replication/transcription coorientation was proposed for human chromosomes on the basis of in silico analysis (116), an essentially random orientation of genes was found around experimentally identified replication origins (117) (Fig. 2C). In fact, the bidirectional nature of transcription from the strong eukaryotic promoters (118, 119) makes coorientation of genes with replication in eukaryotic chromosomes irrelevant.
The absence of the replication-transcription conflict in the eukaryotic chromosomes is corroborated by the fact that essentially all eukaryotic genes have their own promoters (“one control region = one gene”) (Fig. 2D) (120). The promoter recognition algorithms predict a promoter consensus in every kbp of DNA in higher eukaryotes, which is too frequent for primary selection of transcription-initiation sites. Instead, transcription-initiation sites are apparently selected because of a particular nucleosome modification; the distance between stretches of such modified nucleosomes corresponds to the observed 30-to-40-kbp distance between experimentally confirmed transcription-initiation sites (121), while the apparently ubiquitous promoters in these nucleosome-depleted DNA regions then initiate transcription (122). Even eukaryotic genes assembled in functional clusters, analogous to prokaryotic operons, still retain their own individual promoters (123). In contrast to this pattern, the number of recognizable promoters in prokaryotic chromosomes is significantly lower than the number of genes (Fig. 2D): the experimentally identified promoter-to-open-reading-frame (ORF) ratio is ∼1:10 for E. coli (124) and ∼1:3 for Bacillus subtilis (111, 125). Chromatin immunoprecipitation with microarray technology (ChIP-chip) analysis of genome sites associated with initiation-poised RNA polymerases in E. coli (the “actual promoters”) brings this ratio in line with the 1:3 ratio of B. subtilis (126). Because of the lower number of available promoters, most prokaryotic genes are assembled into cotranscribed groups called operons, so in prokaryotes, “one control region = one operon.”
The organization of prokaryotic genes into operons is often attributed to frequent horizontal gene transfer, which does play a leading role in prokaryotic genome evolution (see above). Indeed, the several-gene limit of a typical horizontally transferred piece promotes clustering of all the genes required for a particular function: when transferred as a cluster, the new genes instantly provide the recipient cell with a useful function, driving selection for clustering (127). However, horizontal gene transfer explains only the physical proximity of genes (clustering itself) (128) and fails to provide selection for coorientation of the genes in the cluster, let alone for their coregulation via promoter sharing. The few evolutionarily stable “superoperons” in bacteria contain multiple genes involved in the same pathway and may have to be cotranscribed not only because the genes need to be coregulated (as originally proposed [129]) but also because the resulting proteins form a complex and need to be coproduced in a particular order for the complex to have the full activity (130). At the same time, most bacterial operons are evolutionary unstable (131, 132) and the majority of recently formed cotranscribed clusters combine genes coding for proteins of unrelated functions (133–135), suggesting that the main evolutionary drive behind combining genes into operons is to reduce the number of transcription-regulation points. Prokaryotes may need to reduce the number of these points because of their gene regulation logic: whereas the nucleosome packing of eukaryotic DNA automatically maintains a transcriptionally restrictive ground state, the naked DNA in prokaryotes is available for transcription at any time, necessitating multiple repressors to hold in check promoter-bound and initiation-poised RNA polymerases (136), sometimes organized in elaborately looped repressosomes (137, 138). I argue that, having initiation-poised promoter-bound RNA polymerases on their DNA, prokaryotic chromosomes evolved to use fewer transcription-control points to reduce the impediment for replication forks.
Opinion.
Why are prokaryotes forced to race the high-speed trains on the tracks built for horse-drawn carriages?
THE TWO CHROMOSOME CYCLES
In summary, compared to the organization of eukaryotic chromosomes, the organization of prokaryotic chromosomes seems unnecessarily constrained and precarious, raising the questions about its causes and benefits. At the same time, the nuclear organization of the genetic material in eukaryotes, while defining the eukaryotic mode of genome evolution, cannot explain the “safe” state of the eukaryotic chromosomes compared to the precarious state of the prokaryotic chromosomes, as is abundantly demonstrated by various exceptions. Perhaps the variety of differences between the two chromosome organizations hides one primary difference that constrains the system, necessitating compensations that make prokaryotic chromosomes precarious? To discover what is responsible for constraining prokaryotic chromosomes, let us consider the differences between eukaryotes and prokaryotes in their chromosome cycles.
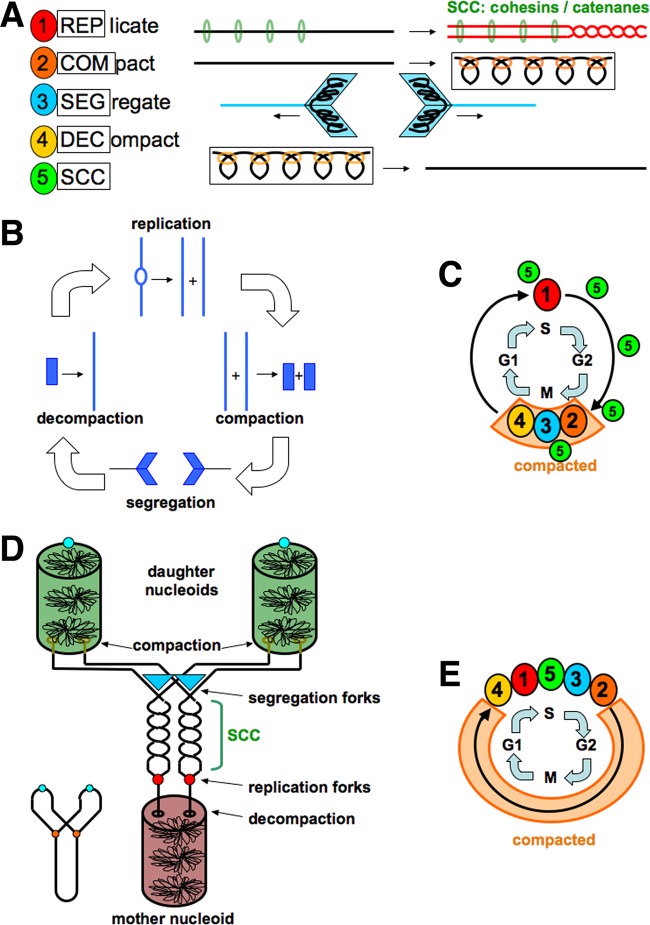
The chromosome cycle is defined as a set of chromosomal transactions following a particular order within the cell cycle (101, 139, 140). The cell cycle with the DNA content per cell as a readout [→G1→S→G2→D(M)→] is invariant across kingdoms, with the two critical stages, S and D(M), corresponding to two critical chromosomal transactions, replication and segregation (141–143). Because of the necessary ∼1,000× degree of intracellular compaction (reviewed in reference 101), chromosomal DNA has to decompact and recompact in order to replicate and segregate. Thus, the full complement of the chromosomal transactions during the cell cycle invariably includes the following steps (Fig. 3A): (i) replication (Rep); (ii) compaction (Com); (iii) segregation (Seg); and (iv) decompaction (Dec) (101, 139, 140). As already mentioned, the fifth transaction, sister-chromatid cohesion (SCC), is a postreplication chromosomal condition that has the DNA component (catenanes) and protein component (cohesins) (Fig. 3A).
FIG 3.
Chromosome transactions and cycles. (A) The five standard chromosome transactions, color coded to correspond to the data in the schemes in panels C and E. (B) The eukaryotic chromosome cycle. (C) Individual transactions of the eukaryotic chromosome cycle over the standard cell cycle grid. (D) The prokaryotic chromosome cycle. (E) Individual transactions of the prokaryotic chromosome cycle over the standard cell cycle grid.
The eukaryotic cell cycle is driven by the cyclin-dependent kinase (CDK) engine (144). The eukaryotic chromosome cycle is →Rep→Com→Seg→Dec→ (Fig. 3B) (139, 140) and is driven by the same CDK engine (145, 146). If laid over the cell cycle grid for reference, Rep corresponds to S, G2 has no chromosomal transactions, and then the Com→Seg→Dec transition happens during M, while G1 is again devoid of chromosomal transactions (Fig. 3C). Sister-chromatid cohesion is a chromosome condition in eukaryotes that starts before S and ends by the end of M, overlapping exactly with the cyclin-regulated part of the eukaryotic cell cycle (Fig. 3C) (145, 146). The eukaryotic chromosome cycle is global and consecutive in that the entire set of chromosomes proceeds through a particular stage or transition together before moving to the next stage or transition (Fig. 3B). Also of notice, relative to the maximal degree of compaction during mitosis, eukaryotic chromosomes stay globally decompacted (still locally compacted) most of the cell cycle (Fig. 3C).
The prokaryotic chromosome cycle is based on the version of the Cairns model of theta replication that emphasizes segregation (147) (Fig. 3D, lower left corner) and features a brief period of sister-chromatid cohesion (Fig. 3D). Its sequence is distinct from the one of the eukaryotic chromosome cycle and goes →Dec→Rep→SCC→Seg→Com→ (101). When laid over the invariant cell cycle grid for reference, a single chromosome cycle transition is revealed that comprises all chromosomal transactions, squeezed together into the same “S phase” of the cell cycle, while no chromosomal transitions happen during the G2, D, and G1 phases of the prokaryotic cell cycle (Fig. 3E). In particular, prokaryotic SCC is a short stage, sandwiched between Rep and Seg (Fig. 3E). In contrast to the global and consecutive chromosome cycle of eukaryotes, the prokaryotic chromosome cycle is local and concurrent in that, at any given time, only a particular and limited part of the chromosome undergoes all the transactions of the chromosome cycle, while all other parts of the chromosome stay compacted (Fig. 3D). The concurrent nature of the prokaryotic chromosome cycle, in which all chromosomal events roll in a single succession once the replication is initiated, is likely why the prokaryotic cell cycle requires no CDK-like engine and is simply driven by replication initiation (148). In contrast to the eukaryotic chromosomes, prokaryotic chromosomes stay maximally compacted for most of the cell cycle (Fig. 3E), but they do not undergo additional condensation.
PROGRESSIVE CHROMOSOME SEGREGATION OBVIATES PRESORTING AND LOGISTIC NEGOTIATION
Comparison of the two chromosome cycles (Fig. 3B and C versus D and E) suggests that the selection for the precarious prokaryotic chromosome organization is driven by the needs of progressive segregation. Although specific segregation mechanisms of the prokaryotic chromosomes are still unknown, the segregation pattern itself is dramatically different from the eukaryotic one (Fig. 2A) and explains the lack of centromeres in prokaryotic chromosomes. I argue that the unique demands of progressive segregation keep prokaryotic chromosomes inadequately protected and hastily replicated, one obvious example being minimizing the duration of the critical period of sister-chromatid cohesion. However, the major and perhaps a related constrain is that progressive segregation strongly favors a single replication bubble (Fig. 1A). The obvious reason is that multiple replication bubbles, under conditions of progressive segregation, necessitate subnucleoid presorting to ensure that all the daughter subnucleoids with the parental “Watson strand” would group into one daughter nucleoid whereas all the daughter subnucleoids with the parental “Crick strand” would group into the other daughter nucleoid (Fig. 4B and C) (147). Such mechanisms of nonrandom segregation of parts of the chromosome are generally unknown, and there is no reason to suspect their existence in prokaryotes. Without subnucleoid presorting, the random assortment of individual subnucleoids forming around corresponding origins should hopelessly entangle sister nucleoids like two strings of beads (Fig. 4B to D and H). The only (theoretical) way to disentangle such fully replicated and intertwined sister nucleoids would be through “logistic negotiation” (Fig. 4D and C), another hypothetical transaction. Thus, progressive segregation should force prokaryotic chromosomes to assume the “single-duplicon” (replicon plus segregon) configuration (Fig. 1A), even discouraging insertion into the chromosome of plasmids with a copy number of 1.
FIG 4.
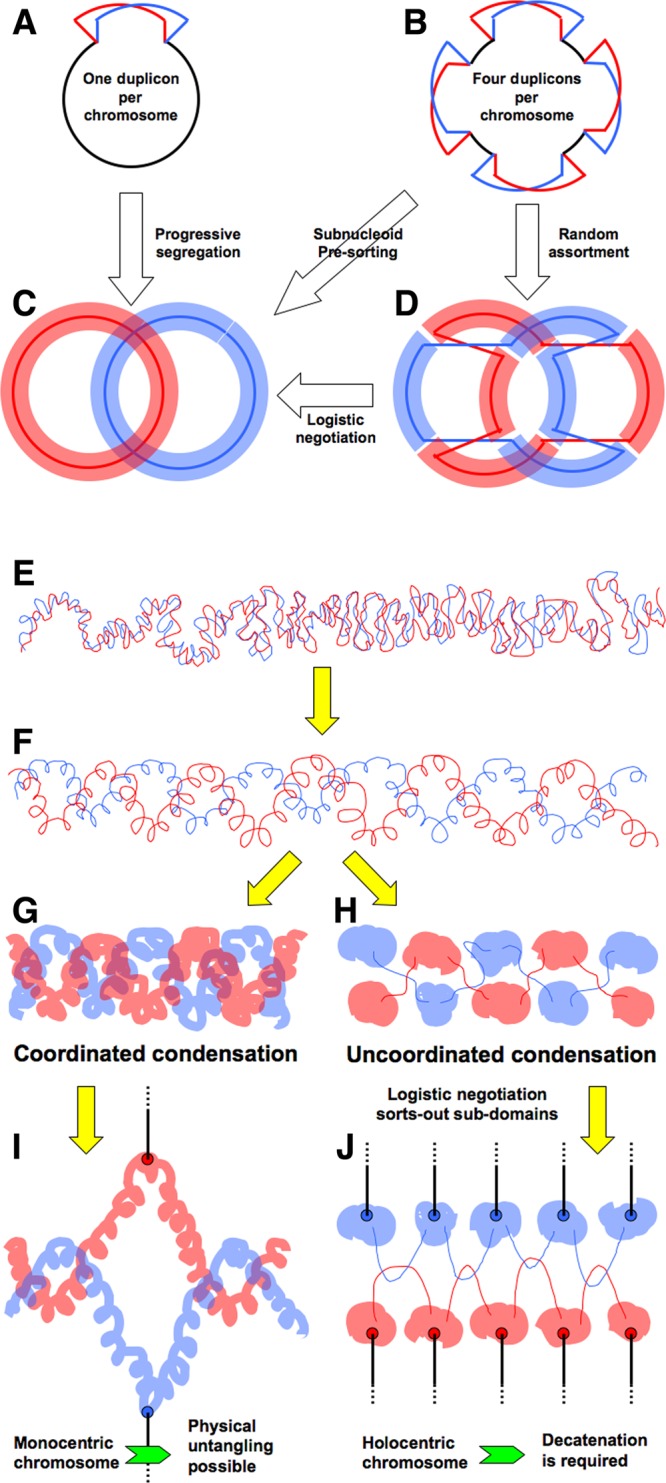
Subdomain presorting and logistic negotiation during chromosome segregation. Red and blue lines designate daughter duplexes containing, correspondingly, “Watson” or “Crick” strands of the parental duplex. (A to D) Prokaryotic chromosome. (A) The theta-replicating chromosome with a single-duplication bubble. (B) A similar replicating chromosome with four duplication bubbles. (C) Progressive segregation from a single-duplication bubble by default yields two completely separate daughter nucleoids. (D) Progressive segregation without subnucleoid presorting yields two daughter nucleoids intertwined due to misclustering of the individual subnucleoids. As a result, the daughter DNA duplex containing, for example, the “Watson” strand of the parental duplex finds itself in both daughter nucleoids. (E to J) Eukaryotic chromosome. (E) Still-to-be-condensed sister chromatids after replication. (F) Gradual condensation sorts sister chromatids out at the level of subdomains. (G) Coordinated condensation results in “single-body” chromosomes ready for segregation. (H) Uncoordinated independent condensation centers produce entangled subdomains. (I) Monocentric chromosomes condensed as “one body” should be able to disentangle during segregation. (J) Holocentric chromosomes likely need logistic negotiation to help sort out all the Watson subdomains (one sister) from all the Crick subdomains (the other sister) before segregation can even take place.
How do eukaryotic cells solve this problem with their multiorigin chromosomes? Presorting may not even be necessary in eukaryotic chromosomes, because they begin condensation in preparation for segregation only after their replication is complete (Fig. 4E). Moreover, with some degree of coordination, some shorter sister chromosomes may be able to condense into continuous bodies (Fig. 4E to G), rather than into a string of several independently condensed domains (Fig. 4H), while unique centromeres on monocentric eukaryotic chromosomes should make it possible to untangle coordinately condensed chromosomes simply by spindle pulling (Fig. 4I). However, the suspected local presegregation in eukaryotic chromosomes (149, 150) and the likely lack of coordination between condensation events in different chromosome subdomains (Fig. 4H), especially in eukaryotes with holocentric chromosomes (Fig. 4J), make entangling of chromosome subdomains a potentially colossal problem for eukaryotic chromosomes. This problem in eukaryotes is likely addressed by the system of logistic negotiation hypothesized above that disentangles condensed sister-chromatid subdomains and groups all subdomains with the “Watson” strand on one side and all those with the “Crick” strand of the other (Fig. 4H to J). I speculate that the extended SCC period in eukaryotes that covers the good half of their cell cycle (Fig. 3C) is required to accomplish this logistic negotiation process. Remarkably, the crenarchaeote Sulfolobus, which has three replication origins in its chromosome (73), does not segregate sister chromatids concurrently with replication, like bacteria (or single-origin archaea [151, 152]), but instead keeps completely replicated daughter chromosomes together during a long G2 phase employing some kind of DNA junctions, rapidly segregating them just before cell division (82, 153, 154). The long G2 phase with no daughter chromosome separation may therefore mark the period of similar logistic negotiation in Sulfolobus.
PROKARYOTIC CHROMOSOME ORGANIZATION COMPENSATES FOR THE SINGLE DUPLICON
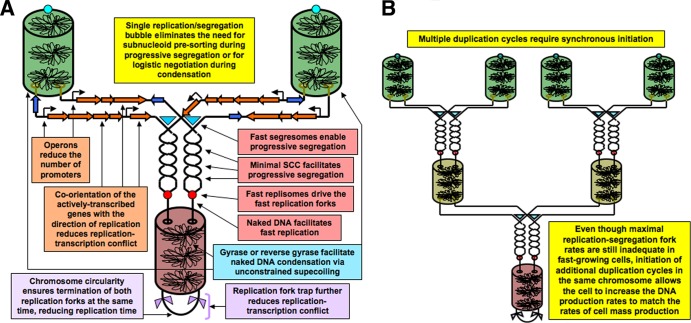
Forcing the entire chromosome to replicate and segregate by a single duplication bubble, while eliminating the need for subnucleoid presorting and logistic negotiation, makes the chromosome duplication round unacceptably long, demanding serious minimization of the limiting stage, which is the chromosome replication time. The minimal replication time in E. coli is an impressive 42 min (109) (translating into the overall DNA synthesis rate of ∼100,000 kbp per min), and yet it is still much longer than E. coli's shortest cell division time of 24 min (109). Minimization of the chromosome replication time in prokaryotes is achieved in multiple ways (Fig. 5A) as follows.
FIG 5.
Prokaryotic chromosome organization compensates for the single duplicon but also creates a strategic opportunity. (A) Various factors minimizing the chromosome duplication time. (B) Multiple relocation cycles in the same chromosome strategically solve the duplication problem.
The enzymatic bacterial replicase rate is at least 10 times higher than that of their eukaryotic counterparts. The in vitro rate of purified main bacterial replicase DNA polymerase (pol) III is ∼500 nt/s (155–157). The directly measured rates of replication fork propagation in vivo, at 620 to 700 nt/s at 30°C (158, 159) and 1,300 nt/s at 42°C (159), are even higher, and there is evidence that the rate is limited by the rate of DNA pol III chain elongation (160). In contrast, the maximal rate of the yeast leading-strand DNA polymerase epsilon in vitro is only 50 nt/s, although under the same conditions the lagging-strand DNA polymerase delta moves faster, at 200 nt/s (157). The directly measured rate of replication fork progression in vivo ranges between 10 and 100 bp/s in yeast (105, 161, 162) and between 5 and 100 bp/s in human cells (106).
Prokaryotes keep their DNA histone free to minimize impeding the progress of replication forks. We know this from E. coli mutants that replicate even faster than wild-type (WT) cells—they make chromosomal DNA even more “naked,” by inactivating nucleoid-associated proteins H-NS and HU (163, 164) or by titrating another DNA-binding protein, DnaA (165).
Prokaryotes coorient reasonably expressed genes with replication, to minimize replication-transcription conflict. The replication fork trap in the terminus region apparently serves the same purpose, by not allowing replication forks to enter chromosomal regions with opposite transcription. The reduction in the number of promoters diminishes the number of idling RNA polymerases on DNA, further reducing the conflict.
The circularity of the prokaryotic chromosomes may have nothing to do with getting rid of telomeres, since linearization of the chromosome does not increase the chromosome replication or cell division time in E. coli (55). However, circularization automatically minimizes the replication time in a chromosome duplicated by a single bubble, by ensuring that the two replication forks always terminate at the same time, so there is no extra wait for one of them. In the circular chromosomes, termination of the two forks is simultaneous by definition, no matter how chromosomal rearrangements displace the origin relative to the terminus.
In contrast to eukaryotic genomes, where deletions and insertions happen at equal rates, deletions outnumber insertions at least 10:1 in the prokaryotic genomes (30, 31), which systematically reduces the amount of DNA to replicate. The specific mechanisms favoring deletions over insertions are not known; the bias might be a mechanistic consequence of the way prokaryotes segregate their DNA (to be discussed elsewhere).
Remarkably, progressive segregation, having brought these serious demands for the fastest possible replication, also created a general solution for the problem of synthesizing enough DNA in case the replication fork rate becomes inadequate for the cell mass growth rate. The elegant solution is to permit multiple replication cycles on the same chromosome (166) (Fig. 5B). Eukaryotes can have only one replication round per chromosome, while prokaryotes have easily three consecutive rounds going on in the same chromosome (81), made possible by concurrent replication-segregation immediately generating daughter nucleoids that, though incomplete, are fully proficient to initiate their own replication round as soon as the eclipse period is over (167). For this system to work smoothly, synchronization of replication initiation would be a necessary feature, as again observed.
WHY DO PROKARYOTES PREFER A SINGLE MASTER CHROMOSOME?
Perhaps the only prevalent prokaryotic chromosomal feature that does not compensate for the burden of progressive segregation, and instead exacerbates it, is the single chromosome in most prokaryotes. In fact, prokaryotic cells could have easily solved their multiple chromosomal problems caused by unique duplicon if, instead of a single big chromosome, they had had multiple smaller (plasmid-like) chromosomes, even if each one of them was still driven by a unique replication/segregation origin. An example of such genome organization could be “multiple minicircles” of the chloroplast genome in dinoflagellates (168). With a genome comprising 10 to 30 small chromosomes, prokaryotic cells would have been able to bring down the rate of replication to match the rate of transcription. Moreover, they would not have had to perform progressive segregation, performing the plasmid “condense-and-polarize” segregation instead (96) and thus being able to extend SCC to guarantee the proper maturation of nascent DNA. The problem of logistic negotiation would have been permanently solved by pairwise and independent segregation of multiple-duplicon chromosomes. Yet, against all these apparent benefits, bacteria still prefer to “put all their eggs in one basket,” evolving a single main chromosome and, sometimes, an additional plasmid-derived chromid, which is always smaller than the master chromosome.
One reason for consolidation of the whole genome into a single chromosome could be that prokaryotic cells have problems handling several independent plasmid segregation systems due to the various incompatibility issues the plasmid systems are known for (169); this explanation is corroborated by the paucity of prokaryotes harboring multiple plasmids. However, the key to the real reason may be the fact that, even with several replicons in the cell, there is always only one replicon driven by the oriC/DnaA pair. The unique oriC/DnaA pair per cell may be behind the preference for a single chromosome in bacteria, for example, due to the fact that it is initiation at oriC by DnaA that is believed to pace the bacterial cell cycle (148). According to this logic, since other replicons in the same cell are driven by their oriP/Rep pairs, they should not influence the cell cycle.
However, this simple idea is inconsistent with the fact that additional oriC/DnaA-driven plasmids are well tolerated in E. coli, at least under laboratory conditions (170). In fact, they initiate replication together with the chromosome (171), at the same time maintaining a higher copy number (170). It could be that, while the oriC/DnaA-specific initiation of the master chromosome starts the cell cycle, replication of the terminus (terC) in the same master chromosome signals its finish. According to this logic, the master duplicon drives the cell cycle by both its initiation and termination events, whereas other duplicons are tolerated as long as they duplicate within the duplication period of the master duplicon—this could be why the chromids are always smaller than the oriC-containing chromosome. If both the initiation and the termination of the master duplicon indeed pace the cell cycle in prokaryotes, this creates selection for the housekeeping genes to relocate from secondary duplicons to the master duplicon as the most stable one. At the same time, multiple oriC-terC duplicons would not be tolerated, because all oirC genes of the cell fire at once (replication synchrony), and if the variously sized terC-containing chromosomes were then to terminate at various times, this could disorient the cell cycle, which is anchored by both the initiation and termination events. Thus, a corollary of the arrangement when the cell cycle is driven by both initiation and termination of the master duplicon is migration of the housekeeping genes from other chromosomes to this particular chromosome, eventually making it a single chromosome in the cell.
CONCLUSION
We have presented an argument that it is the progressive chromosome segregation, possibly operating on naked DNA, that drives the evolution of prokaryotic chromosome organization to be so precarious and so different from the eukaryotic one. Progressive segregation is possible only when the duration of SCC is short and is practical only with a single replication-segregation bubble per chromosome, which, in turn, creates a real chromosome duplication rate crisis. To minimize the chromosome duplication time, prokaryotes employ the fastest known replisomes, keep their DNA naked, coorient most of their transcription with replication, reduce the number of sites where RNA polymerases idle (promoters), and keep the chromosome circular so that the two forks always terminate simultaneously, while the replication fork trap in the terminus prevents replication fork entry into the wrong replichore (Fig. 5A). However, all these features are not enough, and the minimal duplication time of the E. coli chromosome (∼45 min) could be still almost two times longer than the minimal division time under the optimal growth conditions. The major relief comes from the possibility of having multiple duplication rounds in the same chromosome, synchronously initiated from the unique replication origins in the initiation-competent, though incomplete, daughter nucleoids (Fig. 5B).
It should be stressed that, even though the concurrent prokaryotic chromosome cycle was likely developed to minimize the chromosome duplication time and to disengage the chromosome cycle from the cell cycle (101), many bacteria always have a single chromosome cycle per cell, just as in eukaryotes. In fact, among the model bacteria illustrating the prokaryotic chromosome cycle, Caulobacter is incapable of multiple chromosome cycles in the same cell, and yet this does not make its chromosome cycle different (at least in the major aspects) from the one of E. coli. At the same time, the archaeote Sulfolobus, with three replication origins, does have a more eukaryote-like chromosome cycle, in that its segregation is a stage distinct from replication and is separated from it by an extended “postreplicative sister-chromatid synapsis” period. Thus, the chromosome cycle distinction is not between slow-growing versus fast-growing prokaryotes, but it might be between single-origin versus multiple-origin chromosomes.
Challenging the proposed argument with experimental tests should be facilitated by the various exceptions to the eukaryotic versus prokaryotic “chromosome rules.” For example, does the mode of prokaryotic genome evolution apply to planktomycetes that house their nucleoid within the membranous compartment (2)? Dinoflagellates, the eukaryotic protists that, like prokaryotes, maintain condensed chromosomes throughout the interphase and lack histone-based nucleosome packaging of DNA (172), could be predicted to have prokaryote-like fast DNA replication and progressive chromosome segregation (whatever its mechanisms turn out to be). Spectacular pictures of mitosis in dinoflagellates (the so-called “dinomitosis”) are indeed highly suggestive (173). It should be possible, as was demonstrated recently (174), to set up an experimental system to test the central prediction of the “duplicon” argument that the existence of several replication origins in the prokaryotic chromosomes would create a logistical problem with segregation of the resulting subnucleoids (Fig. 4). Even testing the idea that the prokaryotic chromosome evolution is driven by progressive segregation may become possible one day in a fantastic synthetic organism, in which the overall eukaryotic chromosome organization will be asked to evolve under the pressure of the prokaryotic progressive chromosome segregation as the only segregation mechanism available. Without such an experimental test, this otherwise compelling collective argument will retain its mostly philosophical nature.
ACKNOWLEDGMENTS
I thank Francois-Xavier Barre (Centre de Genetique Moleculaire, CNRS) for insightful discussions and encouragement and three anonymous reviewers for clarifying the presentation and strengthening the arguments.
Experimental work in this laboratory is supported by grant GM 073115 from the National Institutes of Health.
I declare that I have no conflicts of interest.
Footnotes
Published ahead of print 14 March 2014
REFERENCES
- 1.Stanier RY, van Niel CB. 1962. The concept of a bacterium. Arch. Mikrobiol. 42:17–35. 10.1007/BF00425185 [DOI] [PubMed] [Google Scholar]
- 2.Fuerst JA, Sagulenko E. 2011. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 9:403–413. 10.1038/nrmicro2578 [DOI] [PubMed] [Google Scholar]
- 3.Koonin EV. 2011. The logic of chance: the nature and origin of biological evolution. FT Press Science, Upper Saddle River, NJ [Google Scholar]
- 4.Cavalier-Smith T. 1985. Eukaryote gene numbers, non-coding DNA and genome size, p 69–103 In Cavalier-Smith T. (ed), The evolution of genome size. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 5.Lechardeur D, Sohn KJ, Haardt M, Joshi PB, Monck M, Graham RW, Beatty B, Squire J, O'Brodovich H, Lukacs GL. 1999. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 6:482–497. 10.1038/sj.gt.3300867 [DOI] [PubMed] [Google Scholar]
- 6.Pollard H, Toumaniantz G, Amos JL, Avet-Loiseau H, Guihard G, Behr JP, Escande D. 2001. Ca2+-sensitive cytosolic nucleases prevent efficient delivery to the nucleus of injected plasmids. J. Gene Med. 3:153–164. 10.1002/jgm.160 [DOI] [PubMed] [Google Scholar]
- 7.Shimizu N, Kamezaki F, Shigematsu S. 2005. Tracking of microinjected DNA in live cells reveals the intracellular behavior and elimination of extrachromosomal genetic material. Nucleic Acid Res. 33:6296–6307. 10.1093/nar/gki946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akita H, Ito R, Khalil IA, Futaki S, Harashima H. 2004. Quantitative three-dimensional analysis of the intracellular trafficking of plasmid DNA transfected by a nonviral gene delivery system using confocal laser scanning microscopy. Mol. Ther. 9:443–451. 10.1016/j.ymthe.2004.01.005 [DOI] [PubMed] [Google Scholar]
- 9.Lechardeur D, Lukacs GL. 2006. Nucleocytoplasmic transport of plasmid DNA: a perilous journey from the cytoplasm to the nucleus. Hum. Gene Ther. 17:882–889. 10.1089/hum.2006.17.882 [DOI] [PubMed] [Google Scholar]
- 10.Goff SP. 1992. Genetics of retroviral integration. Annu. Rev. Genet. 26:527–544. 10.1146/annurev.ge.26.120192.002523 [DOI] [PubMed] [Google Scholar]
- 11.Whitcomb JM, Hughes SH. 1992. Retroviral reverse transcription and integration: progress and problems. Annu. Rev. Cell Biol. 8:275–306. 10.1146/annurev.cb.08.110192.001423 [DOI] [PubMed] [Google Scholar]
- 12.Negroni M, Buc H. 2001. Mechanisms of retroviral recombination. Annu. Rev. Genet. 35:275–302. 10.1146/annurev.genet.35.102401.090551 [DOI] [PubMed] [Google Scholar]
- 13.Boeke JD. 2003. The unusual phylogenetic distribution of retrotransposons: a hypothesis. Genome Res. 13:1975–1983. 10.1101/gr.1392003 [DOI] [PubMed] [Google Scholar]
- 14.Feschotte C, Jiang N, Wessler SR. 2002. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3:329–341. 10.1038/nrg793 [DOI] [PubMed] [Google Scholar]
- 15.Löwer R, Löwer J, Kurth R. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. U. S. A. 93:5177–5184. 10.1073/pnas.93.11.5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smit AF. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9:657–663. 10.1016/S0959-437X(99)00031-3 [DOI] [PubMed] [Google Scholar]
- 17.Eichler EE, Sankoff D. 2003. Structural dynamics of eukaryotic chromosome evolution. Science 301:793–797. 10.1126/science.1086132 [DOI] [PubMed] [Google Scholar]
- 18.Mieczkowski PA, Lemoine FJ, Petes TD. 2006. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair 5:1010–1020. 10.1016/j.dnarep.2006.05.027 [DOI] [PubMed] [Google Scholar]
- 19.Zhao H, Bourque G. 2009. Recovering genome rearrangements in the mammalian phylogeny. Genome Res. 19:934–942. 10.1101/gr.086009.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersson JO. 2005. Lateral gene transfer in eukaryotes. Cell. Mol. Life Sci. 62:1182–1197. 10.1007/s00018-005-4539-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards TA, Soanes DM, Foster PG, Leonard G, Thornton CR, Talbot NJ. 2009. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell 21:1897–1911. 10.1105/tpc.109.065805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersson JO. 2009. Horizontal gene transfer between microbial eukaryotes. Methods Mol. Biol. 532:473–487. 10.1007/978-1-60327-853-9_27 [DOI] [PubMed] [Google Scholar]
- 23.Toro N. 2003. Bacteria and Archaea group II introns: additional mobile genetic elements in the environment. Environ. Microbiol. 5:143–151. 10.1046/j.1462-2920.2003.00398.x [DOI] [PubMed] [Google Scholar]
- 24.Koonin EV, Wolf YI. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 36:6688–6719. 10.1093/nar/gkn668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenz MG, Wackernagel W. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartl DL, Sawyer SA. 1988. Why do unrelated insertion sequences occur together in the genome of Escherichia coli? Genetics 118:537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentley SD, Parkhill J. 2004. Comparative genomic structure of prokaryotes. Annu. Rev. Genet. 38:771–792. 10.1146/annurev.genet.38.072902.094318 [DOI] [PubMed] [Google Scholar]
- 28.Dorman CJ. 2013. Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat. Rev. Microbiol. 11:349–355. 10.1038/nrmicro3007 [DOI] [PubMed] [Google Scholar]
- 29.Sobetzko P, Travers A, Muskhelishvili G. 2012. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl. Acad. Sci. U. S. A. 109:E42–E50. 10.1073/pnas.1108229109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo CH, Ochman H. 2009. Deletional bias across the three domains of life. Genome Biol. Evol. 1:145–152. 10.1093/gbe/evp016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mira A, Ochman H, Moran NA. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589–596. 10.1016/S0168-9525(01)02447-7 [DOI] [PubMed] [Google Scholar]
- 32.Guacci V, Hogan E, Koshland D. 1994. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 125:517–530. 10.1083/jcb.125.3.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes VF, Cozzarelli NR. 2000. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc. Natl. Acad. Sci. U. S. A. 97:1322–1324. 10.1073/pnas.040576797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trask B, Pinkel D, van den Engh G. 1989. The proximity of DNA sequences in interphase cell nuclei is correlated to genomic distance and permits ordering of cosmids spanning 250 kilobase pairs. Genomics 5:710–717. 10.1016/0888-7543(89)90112-2 [DOI] [PubMed] [Google Scholar]
- 35.Trun NJ, Marko JF. 1998. Architecture of a bacterial chromosome. ASM News 64:276–283 http://tigger.uic.edu/~jmarko/Trun.ASMNews.98.pdf. [Google Scholar]
- 36.Kavenoff R, Bowen BC. 1976. Electron microscopy of membrane-free folded chromosomes from Escherichia coli. Chromosoma 59:89–101. 10.1007/BF00328479 [DOI] [PubMed] [Google Scholar]
- 37.Marsden MP, Laemmli UK. 1979. Metaphase chromosome structure: evidence for a radial loop model. Cell 17:849–858. 10.1016/0092-8674(79)90325-8 [DOI] [PubMed] [Google Scholar]
- 38.Bendich AJ, Drlica K. 2000. Prokaryotic and eukaryotic chromosomes: what's the difference? Bioessays 22:481–486. [DOI] [PubMed] [Google Scholar]
- 39.Ris H, Kubai DF. 1970. Chromosome structure. Annu. Rev. Genet. 4:263–294. 10.1146/annurev.ge.04.120170.001403 [DOI] [PubMed] [Google Scholar]
- 40.Wikipedia contributors. 19 September 2013. List of organisms by chromosome count. On Wikipedia, The Free Encyclopedia http://en.wikipedia.org/wiki/List_of_organisms_by_chromosome_count Accessed 17 March 2014 [Google Scholar]
- 41.White MJD. 1973. Animal cytology and evolution, 3rd ed. Cambridge University Press, London, United Kingdom [Google Scholar]
- 42.Crosland MW, Crozier RH. 1986. Myrmecia pilosula, an ant with only one pair of chromosomes. Science 231:1278. 10.1126/science.231.4743.1278 [DOI] [PubMed] [Google Scholar]
- 43.Yamaichi Y, Iida T, Park KS, Yamamoto K, Honda T. 1999. Physical and genetic map of the genome of Vibrio parahaemolyticus: presence of two chromosomes in Vibrio species. Mol. Microbiol. 31:1513–1521. 10.1046/j.1365-2958.1999.01296.x [DOI] [PubMed] [Google Scholar]
- 44.Jumas-Bilak E, Michaux-Charachon S, Bourg G, O'Callaghan D, Ramuz M. 1998. Differences in chromosome number and genome rearrangements in the genus Brucella. Mol. Microbiol. 27:99–106. 10.1046/j.1365-2958.1998.00661.x [DOI] [PubMed] [Google Scholar]
- 45.Mardanov AV, Ravin NV. 2012. The impact of genomics on research in diversity and evolution of archaea. Biochemistry (Mosc) 77:799–812. 10.1134/S0006297912080019 [DOI] [PubMed] [Google Scholar]
- 46.Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW, Shannon P, Chiu Y, Weng RS, Gan RR, Hung P, Date SV, Marcotte E, Hood L, Ng WV. 2004. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 14:2221–2234. 10.1101/gr.2700304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farrar NA, Williams KL. 1988. Nuclear plasmids in the simple eukaryotes Saccharomyces cerevisiae and Dictyostelium discoideum. Trends Genet. 4:343–348. 10.1016/0168-9525(88)90054-6 [DOI] [PubMed] [Google Scholar]
- 48.Griffiths AJ. 1995. Natural plasmids of filamentous fungi. Microbiol. Rev. 59:673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison PW, Lower RP, Kim NK, Young JP. 2010. Introducing the bacterial ‘chromid': not a chromosome, not a plasmid. Trends Microbiol. 18:141–148. 10.1016/j.tim.2009.12.010 [DOI] [PubMed] [Google Scholar]
- 50.Petersen J, Frank O, Göker M, Pradella S. 2013. Extrachromosomal, extraordinary and essential—the plasmids of the Roseobacter clade. Appl. Microbiol. Biotechnol. 97:2805–2815. 10.1007/s00253-013-4746-8 [DOI] [PubMed] [Google Scholar]
- 51.Umeda M, Ohtsubo E. 1989. Mapping of insertion elements IS1, IS2 and IS3 on the Escherichia coli K-12 chromosome. Role of the insertion elements in formation of Hfrs and F' factors and in rearrangement of bacterial chromosomes. J. Mol. Biol. 208:601–614 [DOI] [PubMed] [Google Scholar]
- 52.Ishikawa F, Naito T. 1999. Why do we have linear chromosomes? A matter of Adam and Eve. Mutat. Res. 434:99–107 [DOI] [PubMed] [Google Scholar]
- 53.McClintock B. 1932. A correlation of ring-shaped chromosomes with variegation in Zea mays. Proc. Natl. Acad. Sci. U. S. A. 18:677–681. 10.1073/pnas.18.12.677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hinnebusch J, Tilly K. 1993. Linear plasmids and chromosomes in bacteria. Mol. Microbiol. 10:917–922. 10.1111/j.1365-2958.1993.tb00963.x [DOI] [PubMed] [Google Scholar]
- 55.Cui T, Moro-oka N, Ohsumi K, Kodama K, Ohshima T, Ogasawara N, Mori H, Wanner B, Niki H, Horiuchi T. 2007. Escherichia coli with a linear genome. EMBO Rep. 8:181–187. 10.1038/sj.embor.7400880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schulman I, Bloom KS. 1991. Centromeres: an integrated protein/DNA complex required for chromosome movement. Annu. Rev. Cell Biol. 7:311–336. 10.1146/annurev.cb.07.110191.001523 [DOI] [PubMed] [Google Scholar]
- 57.Draper GC, Gober JW. 2002. Bacterial chromosome segregation. Annu. Rev. Microbiol. 56:567–597. 10.1146/annurev.micro.56.012302.160729 [DOI] [PubMed] [Google Scholar]
- 58.Gordon GS, Wright A. 2000. DNA segregation in bacteria. Annu. Rev. Microbiol. 54:681–708. 10.1146/annurev.micro.54.1.681 [DOI] [PubMed] [Google Scholar]
- 59.Possoz C, Junier I, Espeli O. 2012. Bacterial chromosome segregation. Front. Biosci. (Landmark Ed) 17:1020–1034. 10.2741/3971 [DOI] [PubMed] [Google Scholar]
- 60.Toro E, Shapiro L. 2010. Bacterial chromosome organization and segregation. Cold Spring Harb. Perspect. Biol. 2:a000349. 10.1101/cshperspect.a000349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reyes-Lamothe R, Nicolas E, Sherratt DJ. 2012. Chromosome replication and segregation in bacteria. Annu. Rev. Genet. 46:121–143. 10.1146/annurev-genet-110711-155421 [DOI] [PubMed] [Google Scholar]
- 62.Huberman JA. 1973. Structure of chromosome fibers and chromosomes. Annu. Rev. Biochem. 42:355–378. 10.1146/annurev.bi.42.070173.002035 [DOI] [PubMed] [Google Scholar]
- 63.Sajan SA, Hawkins RD. 2012. Methods for identifying higher-order chromatin structure. Annu. Rev. Genomics Hum. Genet. 13:59–82. 10.1146/annurev-genom-090711-163818 [DOI] [PubMed] [Google Scholar]
- 64.Widom J. 1998. Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 27:285–327. 10.1146/annurev.biophys.27.1.285 [DOI] [PubMed] [Google Scholar]
- 65.Rizzo PJ. 1976. Basic chromosomal proteins in lower eukaryotes: relevance to the evolution and function of histones. J. Mol. Evol. 8:79–94. 10.1007/BF01738884 [DOI] [PubMed] [Google Scholar]
- 66.Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. 2004. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 18:1766–1779. 10.1101/gad.1207504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X, Montero Llopis P, Rudner DZ. 2013. Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 14:191–203. 10.1038/nrg3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Forterre P, Gribaldo S, Gadelle D, Serre MC. 2007. Origin and evolution of DNA topoisomerases. Biochimie 89:427–446. 10.1016/j.biochi.2006.12.009 [DOI] [PubMed] [Google Scholar]
- 69.Bates AD, Maxwell A. 2007. Energy coupling in type II topoisomerases: why do they hydrolyze ATP? Biochemistry 46:7929–7941. 10.1021/bi700789g [DOI] [PubMed] [Google Scholar]
- 70.Luijsterburg MS, White MF, van Driel R, Dame RT. 2008. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 43:393–418. 10.1080/10409230802528488 [DOI] [PubMed] [Google Scholar]
- 71.Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. 2010. Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 79:89–130. 10.1146/annurev.biochem.052308.103205 [DOI] [PubMed] [Google Scholar]
- 72.Sernova NV, Gelfand MS. 2008. Identification of replication origins in prokaryotic genomes. Brief. Bioinform. 9:376–391. 10.1093/bib/bbn031 [DOI] [PubMed] [Google Scholar]
- 73.Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. 2004. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc. Natl. Acad. Sci. U. S. A. 101:7046–7051. 10.1073/pnas.0400656101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pelve EA, Lindås AC, Knöppel A, Mira A, Bernander R. 2012. Four chromosome replication origins in the archaeon Pyrobaculum calidifontis. Mol. Microbiol. 85:986–995. 10.1111/j.1365-2958.2012.08155.x [DOI] [PubMed] [Google Scholar]
- 75.Cha RS, Kleckner N. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297:602–606. 10.1126/science.1071398 [DOI] [PubMed] [Google Scholar]
- 76.Fasullo M, Tsaponina O, Sun M, Chabes A. 2010. Elevated dNTP levels suppress hyper-recombination in Saccharomyces cerevisiae S-phase checkpoint mutants. Nucleic Acids Res. 38:1195–1203. 10.1093/nar/gkp1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duggin IG, Wake RG, Bell SD, Hill TM. 2008. The replication fork trap and termination of chromosome replication. Mol. Microbiol. 70:1323–1333. 10.1111/j.1365-2958.2008.06500.x [DOI] [PubMed] [Google Scholar]
- 78.Mirkin EV, Mirkin SM. 2007. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 71:13–35. 10.1128/MMBR.00030-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Diffley JF. 2011. Quality control in the initiation of eukaryotic DNA replication. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:3545–3553. 10.1098/rstb.2011.0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bird RE, Louarn J, Martuscelli J, Caro L. 1972. Origin and sequence of chromosome replication in Escherichia coli. J. Mol. Biol. 70:549–566. 10.1016/0022-2836(72)90559-1 [DOI] [PubMed] [Google Scholar]
- 81.Martín CM, Guzmán EC. 2011. DNA replication initiation as a key element in thymineless death. DNA Repair 10:94–101. 10.1016/j.dnarep.2010.10.005 [DOI] [PubMed] [Google Scholar]
- 82.Lindås AC, Bernander R. 2013. The cell cycle of archaea. Nat. Rev. Microbiol. 11:627–638. 10.1038/nrmicro3077 [DOI] [PubMed] [Google Scholar]
- 83.Skarstad K, Katayama T. 2013. Regulating DNA replication in bacteria. Cold Spring Harb. Perspect. Biol. 5:a012922. 10.1101/cshperspect.a012922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hirano T. 2000. Chromosome cohesion, condensation, and separation. Annu. Rev. Biochem. 69:115–144. 10.1146/annurev.biochem.69.1.115 [DOI] [PubMed] [Google Scholar]
- 85.Nasmyth K, Peters JM, Uhlmann F. 2000. Splitting the chromosome: cutting the ties that bind sister chromatids. Science 288:1379–1385. 10.1126/science.288.5470.1379 [DOI] [PubMed] [Google Scholar]
- 86.Balakrishnan L, Bambara RA. 2013. Okazaki fragment metabolism. Cold Spring Harb. Perspect. Biol. 5:a010173. 10.1101/cshperspect.a010173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuzminov A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63:751–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. 1998. The structure of supercoiled intermediates in DNA replication. Cell 94:819–827. 10.1016/S0092-8674(00)81740-7 [DOI] [PubMed] [Google Scholar]
- 89.Sundin O, Varshavsky A. 1980. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell 21:103–114. 10.1016/0092-8674(80)90118-X [DOI] [PubMed] [Google Scholar]
- 90.Díaz-Martínez LA, Giménez-Abián JF, Clarke DJ. 2008. Chromosome cohesion - rings, knots, orcs and fellowship. J. Cell Sci. 121:2107–2114. 10.1242/jcs.029132 [DOI] [PubMed] [Google Scholar]
- 91.Farcas AM, Uluocak P, Helmhart W, Nasmyth K. 2011. Cohesin's concatenation of sister DNAs maintains their intertwining. Mol. Cell 44:97–107. 10.1016/j.molcel.2011.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nielsen HJ, Youngren B, Hansen FG, Austin S. 2007. Dynamics of Escherichia coli chromosome segregation during multifork replication. J. Bacteriol. 189:8660–8666. 10.1128/JB.01212-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lesterlin C, Gigant E, Boccard F, Espéli O. 2012. Sister chromatid interactions in bacteria revealed by a site-specific recombination assay. EMBO J. 31:3468–3479. 10.1038/emboj.2012.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang X, Reyes-Lamothe R, Sherratt DJ. 2008. Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 22:2426–2433. 10.1101/gad.487508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Joshi MC, Magnan D, Montminy TP, Lies M, Stepankiw N, Bates D. 2013. Regulation of sister chromosome cohesion by the replication fork tracking protein SeqA. PLoS Genet. 9:e1003673. 10.1371/journal.pgen.1003673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ghosh SK, Hajra S, Paek A, Jayaram M. 2006. Mechanisms for chromosome and plasmid segregation. Annu. Rev. Biochem. 75:211–241. 10.1146/annurev.biochem.75.101304.124037 [DOI] [PubMed] [Google Scholar]
- 97.McIntosh JR, Grishchuk EL, West RR. 2002. Chromosome-microtubule interactions during mitosis. Annu. Rev. Cell Dev. Biol. 18:193–219. 10.1146/annurev.cellbio.18.032002.132412 [DOI] [PubMed] [Google Scholar]
- 98.Løbner-Olesen A, Kuempel PL. 1992. Chromosome partitioning in Escherichia coli. J. Bacteriol. 174:7883–7889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lundgren M, Malandrin L, Eriksson S, Huber H, Bernander R. 2008. Cell cycle characteristics of crenarchaeota: unity among diversity. J. Bacteriol. 190:5362–5367. 10.1128/JB.00330-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Youngren B, Nielsen HJ, Jun S, Austin S. 2014. The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes Dev. 28:71–84. 10.1101/gad.231050.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kuzminov A. 2013. The chromosome cycle of prokaryotes. Mol. Microbiol. 90:214–227. 10.1111/mmi.12372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S. 2006. Progressive segregation of the Escherichia coli chromosome. Mol. Microbiol. 61:383–393. 10.1111/j.1365-2958.2006.05245.x [DOI] [PubMed] [Google Scholar]
- 103.Vallet-Gely I, Boccard F. 2013. Chromosomal organization and segregation in Pseudomonas aeruginosa. PLoS Genet. 9:e1003492. 10.1371/journal.pgen.1003492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. U. S. A. 101:9257–9262. 10.1073/pnas.0402606101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Newlon CS. 1988. Yeast chromosome replication and segregation. Microbiol. Rev. 52:568–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Berezney R, Dubey DD, Huberman JA. 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108:471–484. 10.1007/s004120050399 [DOI] [PubMed] [Google Scholar]
- 107.Ardehali MB, Lis JT. 2009. Tracking rates of transcription and splicing in vivo. Nat. Struct. Mol. Biol. 16:1123–1124. 10.1038/nsmb1109-1123 [DOI] [PubMed] [Google Scholar]
- 108.Marcello A. 2012. RNA polymerase II transcription on the fast lane. Transcription 3:29–34. 10.4161/trns.3.1.19147 [DOI] [PubMed] [Google Scholar]
- 109.Bremer H, Dennis PP. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p 1553–1569 In Neidhardt FC, Curtiss R, III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC [Google Scholar]
- 110.Brewer BJ. 1988. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53:679–686. 10.1016/0092-8674(88)90086-4 [DOI] [PubMed] [Google Scholar]
- 111.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Codani JJ, Connerton IF, Danchin A, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256. 10.1038/36786 [DOI] [PubMed] [Google Scholar]
- 112.Bruggemann H, Baumer S, Fricke WF, Wiezer A, Liesegang H, Decker I, Herzberg C, Martinez-Arias R, Merkl R, Henne A, Gottschalk G. 2003. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl. Acad. Sci. U. S. A. 100:1316–1321. 10.1073/pnas.0335853100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.De Septenville AL, Duigou S, Boubakri H, Michel B. 2012. Replication fork reversal after replication-transcription collision. PLoS Genet. 8:e1002622. 10.1371/journal.pgen.1002622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mirkin EV, Mirkin SM. 2005. Mechanisms of transcription-replication collisions in bacteria. Mol. Cell. Biol. 25:888–895. 10.1128/MCB.25.3.888-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang JD, Berkmen MB, Grossman AD. 2007. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 104:5608–5613. 10.1073/pnas.0608999104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huvet M, Nicolay S, Touchon M, Audit B, d'Aubenton-Carafa Y, Arneodo A, Thermes C. 2007. Human gene organization driven by the coordination of replication and transcription. Genome Res. 17:1278–1285. 10.1101/gr.6533407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Necsulea A, Guillet C, Cadoret JC, Prioleau MN, Duret L. 2009. The relationship between DNA replication and human genome organization. Mol. Biol. Evol. 26:729–741. 10.1093/molbev/msn303 [DOI] [PubMed] [Google Scholar]
- 118.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457:1038–1042. 10.1038/nature07747 [DOI] [PubMed] [Google Scholar]
- 119.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457:1033–1037. 10.1038/nature07728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engström PG, Frith MC, Forrest AR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38:626–635. 10.1038/ng1789 [DOI] [PubMed] [Google Scholar]
- 121.Pedersen AG, Baldi P, Chauvin Y, Brunak S. 1999. The biology of eukaryotic promoter prediction–a review. Comput. Chem. 23:191–207. 10.1016/S0097-8485(99)00015-7 [DOI] [PubMed] [Google Scholar]
- 122.Wei W, Pelechano V, Järvelin AI, Steinmetz LM. 2011. Functional consequences of bidirectional promoters. Trends Genet. 27:267–276. 10.1016/j.tig.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Osbourn AE, Field B. 2009. Operons. Cell. Mol. Life Sci. 66:3755–3775. 10.1007/s00018-009-0114-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hershberg R, Bejerano G, Santos-Zavaleta A, Margalit H. 2001. PromEC: An updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acid Res. 29:277. 10.1093/nar/29.1.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sierro N, Makita Y, de Hoon M, Nakai K. 2008. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 36(Database issue):D93–D96. 10.1093/nar/gkm910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reppas NB, Wade JT, Church GM, Struhl K. 2006. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell 24:747–757. 10.1016/j.molcel.2006.10.030 [DOI] [PubMed] [Google Scholar]
- 127.Lawrence JG, Roth JR. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lawrence JG. 2003. Gene organization: selection, selfishness, and serendipity. Annu. Rev. Microbiol. 57:419–440. 10.1146/annurev.micro.57.030502.090816 [DOI] [PubMed] [Google Scholar]
- 129.Jacob F, Monod J. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3:318–356. 10.1016/S0022-2836(61)80072-7 [DOI] [PubMed] [Google Scholar]
- 130.Dandekar T, Snel B, Huynen M, Bork P. 1998. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 23:324–328. 10.1016/S0968-0004(98)01274-2 [DOI] [PubMed] [Google Scholar]
- 131.Itoh T, Takemoto K, Mori H, Gojobori T. 1999. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol. 16:332–346. 10.1093/oxfordjournals.molbev.a026114 [DOI] [PubMed] [Google Scholar]
- 132.Price MN, Arkin AP, Alm EJ. 2006. The life-cycle of operons. PLoS Genet. 2:e96. 10.1371/journal.pgen.0020096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.de Daruvar A, Collado-Vides J, Valencia A. 2002. Analysis of the cellular functions of Escherichia coli operons and their conservation in Bacillus subtilis. J. Mol. Evol. 55:211–221. 10.1007/s00239-002-2317-1 [DOI] [PubMed] [Google Scholar]
- 134.Price MN, Huang KH, Arkin AP, Alm EJ. 2005. Operon formation is driven by co-regulation and not by horizontal gene transfer. Genome Res. 15:809–819. 10.1101/gr.3368805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22–28. 10.1093/nar/29.1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Struhl K. 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98:1–4. 10.1016/S0092-8674(00)80599-1 [DOI] [PubMed] [Google Scholar]
- 137.Semsey S, Tolstorukov MY, Virnik K, Zhurkin VB, Adhya S. 2004. DNA trajectory in the Gal repressosome. Genes Dev. 18:1898–1907. 10.1101/gad.1209404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Semsey S, Virnik K, Adhya S. 2005. A gamut of loops: meandering DNA. Trends Biochem. Sci. 30:334–341. 10.1016/j.tibs.2005.04.009 [DOI] [PubMed] [Google Scholar]
- 139.Losada A, Hirano T. 2001. Shaping the metaphase chromosome: coordination of cohesion and condensation. Bioessays 23:924–935. 10.1002/bies.1133 [DOI] [PubMed] [Google Scholar]
- 140.Mazia D. 1987. The chromosome cycle and the centrosome cycle in the mitotic cycle. Int. Rev. Cytol. 100:49–92. 10.1016/S0074-7696(08)61698-8 [DOI] [PubMed] [Google Scholar]
- 141.Baserga R, Wiebel F. 1969. The cell cycle of mammalian cells. Int. Rev. Exp. Pathol. 7:1–30 [PubMed] [Google Scholar]
- 142.Bates D, Epstein J, Boye E, Fahrner K, Berg H, Kleckner N. 2005. The Escherichia coli baby cell column: a novel cell synchronization method provides new insight into the bacterial cell cycle. Mol. Microbiol. 57:380–391. 10.1111/j.1365-2958.2005.04693.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bernander R, Poplawski A. 1997. Cell cycle characteristics of thermophilic archaea. J. Bacteriol. 179:4963–4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Murray AW. 2004. Recycling the cell cycle: cyclins revisited. Cell 116:221–234. 10.1016/S0092-8674(03)01080-8 [DOI] [PubMed] [Google Scholar]
- 145.Blow JJ, Tanaka TU. 2005. The chromosome cycle: coordinating replication and segregation. EMBO Rep. 6:1028–1034. 10.1038/sj.embor.7400557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Stillman B. 2005. Origin recognition and the chromosome cycle. FEBS Lett. 579:877–884. 10.1016/j.febslet.2004.12.011 [DOI] [PubMed] [Google Scholar]
- 147.Dingman CW. 1974. Bidirectional chromosome replication: some topological considerations. J. Theor. Biol. 43:187–195. 10.1016/S0022-5193(74)80052-4 [DOI] [PubMed] [Google Scholar]
- 148.Abner K, Aaviksaar T, Adamberg K, Vilu R. 2014. Single-cell model of prokaryotic cell cycle. J. Theor. Biol. 341:78–87 [DOI] [PubMed] [Google Scholar]
- 149.Pflumm MF. 2002. The role of DNA replication in chromosome condensation. Bioessays 24:411–418. 10.1002/bies.10092 [DOI] [PubMed] [Google Scholar]
- 150.Stack SM, Anderson LK. 2001. A model for chromosome structure during the mitotic and meiotic cell cycles. Chromosome Res. 9:175–198. 10.1023/A:1016690802570 [DOI] [PubMed] [Google Scholar]
- 151.Lopez P, Philippe H, Myllykallio H, Forterre P. 1999. Identification of putative chromosomal origins of replication in Archaea. Mol. Microbiol. 32:883–886. 10.1046/j.1365-2958.1999.01370.x [DOI] [PubMed] [Google Scholar]
- 152.Majerník AI, Lundgren M, McDermott P, Bernander R, Chong JP. 2005. DNA content and nucleoid distribution in Methanothermobacter thermautotrophicus. J. Bacteriol. 187:1856–1858. 10.1128/JB.187.5.1856-1858.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Popławski A, Bernander R. 1997. Nucleoid structure and distribution in thermophilic Archaea. J. Bacteriol. 179:7625–7630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Robinson NP, Blood KA, McCallum SA, Edwards PA, Bell SD. 2007. Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO J. 26:816–824. 10.1038/sj.emboj.7601529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.O'Donnell ME, Kornberg A. 1985. Dynamics of DNA polymerase III holoenzyme of Escherichia coli in replication of a multiprimed template. J. Biol. Chem. 260:12875–12883 [PubMed] [Google Scholar]
- 156.Tanner NA, Loparo JJ, Hamdan SM, Jergic S, Dixon NE, van Oijen AM. 2009. Real-time single-molecule observation of rolling-circle DNA replication. Nucleic Acid Res. 37:e27. 10.1093/nar/gkp006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yao NY, Schroeder JW, Yurieva O, Simmons LA, O'Donnell ME. 2013. Cost of rNTP/dNTP pool imbalance at the replication fork. Proc. Natl. Acad. Sci. U. S. A. 110:12942–12947. 10.1073/pnas.1309506110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Breier AM, Weier HU, Cozzarelli NR. 2005. Independence of replisomes in Escherichia coli chromosomal replication. Proc. Natl. Acad. Sci. U. S. A. 102:3942–3947. 10.1073/pnas.0500812102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kouzminova EA, Kuzminov A. 2012. Chromosome demise in the wake of ligase-deficient replication. Mol. Microbiol. 84:1079–1096. 10.1111/j.1365-2958.2012.08076.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pham TM, Tan KW, Sakumura Y, Okumura K, Maki H, Akiyama MT. 18 September 2013. A single-molecule approach to DNA replication in Escherichia coli cells demonstrated that DNA polymerase III is a major determinant of fork speed. Mol. Microbiol. 10.1111/mmi.12386 [DOI] [PubMed] [Google Scholar]
- 161.Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. 2001. Replication dynamics of the yeast genome. Science 294:115–121. 10.1126/science.294.5540.115 [DOI] [PubMed] [Google Scholar]
- 162.Sekedat MD, Fenyö D, Rogers RS, Tackett AJ, Aitchison JD, Chait BT. 2010. GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol. Syst. Biol. 6:353. 10.1038/msb.2010.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Atlung T, Hansen FG. 2002. Effect of different concentrations of H-NS protein on chromosome replication and the cell cycle in Escherichia coli. J. Bacteriol. 184:1843–1850. 10.1128/JB.184.7.1843-1850.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Von Freiesleben U, Rasmussen KV, Atlung T, Hansen FG. 2000. Rifampicin-resistant initiation of chromosome replication from oriC in ihf mutants. Mol. Microbiol. 37:1087–1093. 10.1046/j.1365-2958.2000.02060.x [DOI] [PubMed] [Google Scholar]
- 165.Morigen Løbner-Olesen A, Skarstad K. 2003. Titration of the Escherichia coli DnaA protein to excess datA sites causes destabilization of replication forks, delayed replication initiation and delayed cell division. Mol. Microbiol. 50:349–362. 10.1046/j.1365-2958.2003.03695.x [DOI] [PubMed] [Google Scholar]
- 166.Cooper S, Helmstetter CE. 1968. Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 31:519–540. 10.1016/0022-2836(68)90425-7 [DOI] [PubMed] [Google Scholar]
- 167.von Freiesleben U, Krekling MA, Hansen FG, Løbner-Olesen A. 2000. The eclipse period of Escherichia coli. EMBO J. 19:6240–6248. 10.1093/emboj/19.22.6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Howe CJ, Nisbet RE, Barbrook AC. 2008. The remarkable chloroplast genome of dinoflagellates. J. Exp. Bot. 59:1035–1045. 10.1093/jxb/erm292 [DOI] [PubMed] [Google Scholar]
- 169.Bouet JY, Nordström K, Lane D. 2007. Plasmid partition and incompatibility–the focus shifts. Mol. Microbiol. 65:1405–1414. 10.1111/j.1365-2958.2007.05882.x [DOI] [PubMed] [Google Scholar]
- 170.Jensen MR, Løbner-Olesen A, Rasmussen KV. 1990. Escherichia coli minichromosomes: random segregation and absence of copy number control. J. Mol. Biol. 215:257–265. 10.1016/S0022-2836(05)80344-4 [DOI] [PubMed] [Google Scholar]
- 171.Helmstetter CE, Leonard AC. 1987. Coordinate initiation of chromosome and minichromosome replication in Escherichia coli. J. Bacteriol. 169:3489–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Moreno Díaz de la Espina S, Alverca E, Cuadrado A, Franca S. 2005. Organization of the genome and gene expression in a nuclear environment lacking histones and nucleosomes: the amazing dinoflagellates. Eur. J. Cell Biol. 84:137–149. 10.1016/j.ejcb.2005.01.002 [DOI] [PubMed] [Google Scholar]
- 173.Costas E, Goyanes V. 2005. Architecture and evolution of dinoflagellate chromosomes: an enigmatic origin. Cytogenet. Genome Res. 109:268–275. 10.1159/000082409 [DOI] [PubMed] [Google Scholar]
- 174.Wang X, Lesterlin C, Reyes-Lamothe R, Ball G, Sherratt DJ. 2011. Replication and segregation of an Escherichia coli chromosome with two replication origins. Proc. Natl. Acad. Sci. U. S. A. 108:E243–E250. 10.1073/pnas.1100874108 [DOI] [PMC free article] [PubMed] [Google Scholar]