Abstract
The gut microbiome may modulate intestinal immunity by luminal conversion of dietary amino acids to biologically active signals. The model probiotic organism Lactobacillus reuteri ATCC PTA 6475 is indigenous to the human microbiome, and converts the amino acid L-histidine to the biogenic amine, histamine. Histamine suppresses TNF production by human myeloid cells and is a product of L-histidine decarboxylation, which is a proton-facilitated reaction. A transposon mutagenesis strategy was developed based on a single-plasmid nisin-inducible Himar1 transposase/transposon delivery system for L. reuteri. A highly conserved proton-chloride antiporter gene (eriC), a gene widely present in the gut microbiome was discovered by Himar1 transposon (Tn)-mutagenesis presented in this study. Genetic inactivation of eriC by transposon insertion and genetic recombineering resulted in reduced ability of L. reuteri to inhibit TNF production by activated human myeloid cells, diminished histamine production by the bacteria and downregulated expression of histidine decarboxylase (hdc) cluster genes compared to those of WT 6475. EriC belongs to a large family of ion transporters that includes chloride channels and proton-chloride antiporters and may facilitate the availability of protons for the decarboxylation reaction, resulting in histamine production by L. reuteri. This report leverages the tools of bacterial genetics for probiotic gene discovery. The findings highlight the widely conserved nature of ion transporters in bacteria and how ion transporters are coupled with amino acid decarboxylation and contributed to microbiome-mediated immunomodulation.
Keywords: probiotics, Lactobacillus reuteri, histamine, amino acid decarboxylation, Himar1, transposon mutagenesis
Introduction
Probiotic Lactobacillus reuteri is indigenous to the gastrointestinal tract of humans, pigs, mice, rats, and birds (Walter et al. 2010). Previous studies demonstrated host-specificity within this species, suggesting that physiological and immunological differences between different vertebrates contributed to host-driven evolution of L. reuteri (Frese et al. 2011). L. reuteri is generally regarded as safe and has never been shown to cause diseases in humans (Britton and Versalovic 2008). Human-derived L. reuteri strains hold the potential to be both preventive and therapeutic probiotic treatments of inflammatory diseases. Specifically, L. reuteri ATCC PTA 6475 houses a histidine decarboxylase (hdc) gene cluster important for histamine production, which is characteristic of human-derived L. reuteri strains (Frese et al. 2011). Results from comparative genomics analysis of human-derived L. reuteri strains also demonstrated the presence of hdc gene cluster almost exclusively in strains that are members of multi-locus sequence analysis (MLSA) clade II (Oh et al. 2010) (JK Spinler, unpublished data). L.reuteri 6475-derived histamine activates histamine receptor type 2 (H2) on toll-like receptor 2 (TLR2)-activated human myeloid cells resulting in decreased tumor necrosis factor (TNF) production and inhibition of MEK/ERK MAPK signalling.(Thomas et al. 2012). Supplementation of L-histidine in L. reuteri 6475 growth medium increases the expression of the hdc gene cluster and production of TNF-inhibitory histamine (Thomas et al. 2012). A mutant strain deficient in Lactobacillus reuteri-Specific Immunoregulatory (RsiR) gene did not produce histamine and did not protect mice in a trinitrobenzene sulfonic acid (TNBS)-induced mouse model of acute colitis (Hemarajata et al. 2013). However, little is known about the regulatory mechanism of histidine decarboxylation by L. reuteri and the physiological changes that occur in the bacterial cell during histamine production.
Genetic manipulation of L. reuteri is a challenge and had been limited to a few targeted mutagenesis techniques until now (Walter et al. 2005; van Pijkeren and Britton 2012; Su et al. 2011). Transposon-based mutagenesis strategies can be robust techniques for identifying unknown genes involved in functional areas (Picardeau 2009). The eukaryotic transposable Mariner element, Himar1, from the hornfly Haematobia irritans, has been used successfully in generating mutations in many prokaryotic organisms, such as Enterococcus faecalis (Kristich et al. 2008), Bacillus anthracis (Tam et al. 2006), Bacillus subtilis (Le Breton et al. 2006; Pozsgai et al. 2011) and Clostridium perfringens (Liu et al. 2013). Himar1 transposable elements have also been employed in genetically intractable organisms such as Leptospira interrogans (Bourhy et al. 2005), Francisella tularensis (Maier et al. 2006) and Coxiella burnetii (Beare 2012). Himar1 transposase activity is independent of eukaryotic host cofactors (Lampe et al. 1998), and has little insertion site specificity, requiring only the dinucleotide TA in the target sequence (Akerley et al. 1998), which makes it an attractive candidate for transposon-based mutagenesis in prokaryotic organisms.
There are various approaches for delivering and expressing the Himar1 transposase gene and its transposable elements in prokaryotes. Previous studies have utilized conditionally replicative plasmids as delivery vectors, that can be eliminated after successful transposition (Picardeau 2009). Others have employed Lactococcus lactis-derived nisin-inducible expression systems to control transposase production (Kristich et al. 2008). Most studies approach the Himar1 transposon mutagenesis strategy in dual-plasmid systems (Kristich et al. 2008; Tam et al. 2006; Beare et al. 2009). In this study, we developed a transposon mutagenesis system for L. reuteri with the intent to identify unknown genes important for the anti-inflammatory phenotype of L. reuteri 6475. We constructed a conditionally replicative plasmid to contain both a nisin-inducible Himar1 transposase and transposon and introduced it into L. reuteri. Libraries of transposed mutants were selectively screened for their loss of the ability to inhibit TNF production from human myeloid cells. An insertion in a gene encoding a proton-chloride antiporter (EriC) was identified by whole genome sequencing, and is an important member of the immunomodulatory gene network of L. reuteri.
Materials and methods
Bacterial strains and culture conditions
All bacterial strains used in this study are described in Supplemental Table S1. L. reuteri strains were routinely cultured in deMan, Rogosa, Sharpe media (MRS; Difco, Franklin Lakes, NJ) at 37°C in an anaerobic workstation (MACS MG-500, Microbiology International, Frederick, MD) supplied with a mixture of 10% CO 2, 10% H2, and 80% N2 (Airgas, the Woodlands, TX). Escherichia coli strains were routinely cultured in Luria Broth (LB; Invitrogen, Carlsbad, CA) at 37°C and 220 rpm shaking for 16–18 h. Specific culture conditions for individual experiments are detailed throughout. When appropriate, antibiotics were added in the following concentrations: chloramphenicol (Cm, 10 μg/mL), erythromycin (Erm, 10 μg/mL) for L. reuteri; carbenicillin (Cb, 50 μg/mL), chloramphenicol (30 μg/mL,) erythromycin (400 μg/mL) and kanamycin (Kn, 40 μg/mL) for E. coli.
Construction of a single-plasmid, nisin-inducible Himar1 expression system (pPH-M1)
All plasmids and primers used in this study are detailed in Table S1. A temperature-sensitive, E. coli-L. reuteri shuttle vector containing a nisin-inducible Himar1 transposase and Himar1 transposon (Tn) was constructed in several steps. The nisin-inducible Himar1 expression cassette (NICEHim1) was assembled using the In-Fusion® HD Cloning System (Clontech, Mountain View, CA) in two multi-fragment cloning steps. In the first multi-fragment cloning step, the PnisA promoter and nisRK element were amplified from the chromosome of L. lactis subsp. lactis ATCC 11454 with primers containing an AatII site strategically engineered downstream from the PnisA promoter. Amplicons were cloned into an EcoRI restriction site in pCR®2.1 (Invitrogen, Carlsbad, CA) by standard cloning methods. Next, translational terminator T667 and the Himar1 transposase gene (him1C9) were cloned into this AatII site as follows: T667 was amplified from pTRK847 (a gift from Dr. T. R. Klaenhammer, North Carolina State University, Raleigh, NC), and him1C9 was amplified from pMALC9 (Akerley and Lampe 2002). AatII sites introduced by the primer sets of both T667 and him1C9 enabled the amplicons to be cloned into the AatII site downstream of the PnisA promoter now in pCR®2.1, resulting in the intermediate vector, pCR®2.1-NICEHim1.
The second multi-fragment cloning step involved the assembly of a Himar1 transposon (HimTn) to contain in order: translational stops, a constitutive P23 promoter (Que et al. 2000), and an Erm resistance gene (ermR) from pORI28 (Leenhouts et al. 1996) into a BglII site between the 5′ and 3′ inverted terminal repeats (ITRs) recognized by the Himar1 transposase in pMMOrf (Lampe et al. 1999).
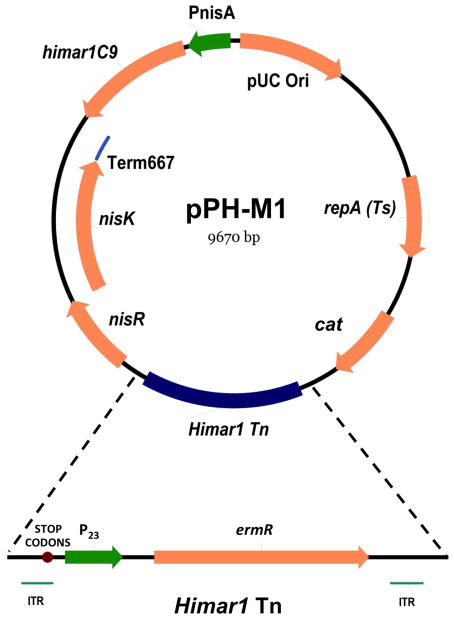
Lastly, elements generated in the above steps along with a pUC origin of replication (ori) were combined into pVE6007 (Law et al. 1995) to create the NICEHim1 system for use in L. reuteri. Plasmid pVE6007 is temperature sensitive and replicates in L. reuteri. The In-Fusion® HD Cloning System was used to transfer the pUC ori from pUC19 into the SacI restriction site in pVE6007 to create a single vector capable of replicating in both L. reuteri and E. coli. Subsequently, the NICEHim1 cassette (from pCR®2.1-NICEHim1) was sub-cloned into the pVE6007 EagI site, followed by HimTn into a BamHI site, resulting in pPH-M1 (Fig. 1).
Fig. 1. The Himar 1 transposase/transposon delivery vector, pPH-M1, generated for Himar1 Tn mutagenesis in L. reuteri.
Genes for the Himar1 transposase, selective antibiotic resistance cassettes, nisin-inducible expression system and elements important for transposition were all combined into a single, conditionally replicative vector for use in L. reuteri. Conditional replication is temperature-dependent, and an increase in temperature to 45°C results in plasmid elimination. cat, chloramphenicol resistance gene; ermR, erythromycin resistance gene; ITR, inverted terminal repeats (ITRs) recognized by the Himar1 transposase; STOP CODONS, translational stop codons; P23, constitutive P23 promoter; nisR and nisK, two-component regulatory elements required for nisin-induced expression system; Term 667, translational stop codons; himar1C9, hyperactive mutant of Himar1 transposase; PnisA, promoter for nisin-induced expression system; pUC Ori, origin of replication for E. coli; repA(Ts), gene encoding temperature sensitive mutant of replication factor RepA, required for replication in L. reuteri
Induction of Himar1 expression and transposition
L. reuteri 6475 pPH-M1 was used to generate a library of Himar1 Tn-mutant clones. L. reuteri 6475 pPH-M1 was generated by electroporation (2 mm cuvettes, 2500 V, 25 μF and 400Ω) and cultured overnight in MRS with 10 μg/ml Cm (Cm 10) and 10 μg/ml Erm (Erm 10) at 30°C anaerobically. The overnight culture was subcultured into fresh MRS Cm 10 Erm 10 at an OD600 = 0.05 and incubated at 30°C anaerobically for 4 hours. Expression of Himar1 transposase was induced by the addition of purified nisin (Sigma-Aldrich, St. Louis, MO) at a final concentration of 50 ng/mL (Wu et al. 2006), followed by anaerobic incubation at 30°C for 16 hours. Elimination of the thermosensitive pPH-M1 was subsequently performed as previously described with some modifications (Walter et al. 2005). Briefly, consecutive subculturing at 45°C (first for 8 hours, then overnight) was carried out in MRS Erm10 using the nisin-induced culture as the initial inoculum. Dilutions of the final subculture were plated on MRS Erm10 agar and incubated anaerobically at 45°C overnight. Isolated colonies were replica-plated onto MRS Erm10 and MRS Cm10 agar, and incubated at 37°C overnight. A diagram of the mutagenesis process is depicted in Supplemental Fig. 1. Erm-resistant, Cm-sensitive isolates were cultured in 200 μL MRS Erm 10 in 96-well plates overnight, mixed with 20% glycerol and stored at −80°C as our Himar1 transposon (Tn)-mutant library.
Southern hybridization
Southern hybridization of digested genomic DNA (gDNA) was performed using DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche, Indianapolis, IN). Briefly, digoxigenin (DIG)-labeled probe was prepared by amplifying ermR from pPH-M1 with primers ErmR-F and ErmR-R according to manufacturer’s instructions. Five micrograms of concentrated gDNA from each mutant and control isolate were digested by HindIII. Digested DNA fragments were separated by 1% agarose gel electrophoresis, transferred to a nylon membrane by capillary transfer, and hybridized to the DIG-labeled probe as per manufacturer’s instructions. The presence of ermR was detected colorimetrically using the anti-digoxigenin-AP conjugate and nitro blue tetrazolium substrate.
High-throughput screening of mutant libraries for reduction in TNF inhibitory activity
High-throughput preliminary screening of Tn-mutants for loss of TNF inhibition was carried out as follows. Using a plate replicator, 96-well plates (200 μL MRS Erm10/well) were inoculated with Tn-mutants from our frozen library and incubated anaerobically for 24 h at 37°C. Overnight cultures (40 μL) were subcultured in 96-well plates of LDMIII (Erm 10, 200 μL/well) and incubated as described. Culture supernatants were filter-sterilized and collected using 0.22 μM PVDF 96-well filter plates (Millipore, Billerica, MA), followed by subsequent size-exclusion using 10 kDa 96-well filter plates (Millipore, Billerica, MA). Supernatants were vacuum dried (Eppendorf, Hamburg, Germany) and resuspended with an equal volume of RPMI prior to use in TNF inhibitory bioassays.
Detection of chromosomal insertion sites using whole genome shotgun (WGS) sequencing
Genomic DNA was isolated from Tn-mutants showing loss of TNF inhibition using the QIAamp DNA Mini Kit with the QIAcube automation system (QIAGEN, Valencia, CA), and was processed and bar-coded using the Illumina TruSeq DNA sample preparation workflow (Illumina Inc, San Diego, CA). Bar-coded samples from all mutants were analyzed in a multiplexed single flow cell on the Illumina MiSeq® platform (Illumina Inc, San Diego, CA) with paired-end 150-bp read lengths. To enhance the efficiency identification of transposon insertion sites and maximize the length of contigs, sequences were assembled de novo using Velvet Assembler (Zerbino and Birney 2008). Assembled genome contigs were aligned to the L. reuteri 6475 draft genome (GenBank accession numbers NZ_ACGX02000001 through NZ_ACGX02000007) and transposon insertion sites were identified by searching for known Himar1 transposon complete sequence (1,365 bp) in genome contigs followed by analysis of the DNA sequences surrounding each insertion.
Construction of L. reuteri eriC recombineered mutant (6475eriC-Stop)
The target gene, eriC, was mutated by recombineering techniques in L. reuteri 6475 as previously described with modifications (van Pijkeren and Britton 2012; van Pijkeren et al. 2012; Hemarajata et al. 2013). Both the control plasmid (pJP577) carrying a chloramphenicol resistance (cat) gene and the eriC mutagenesis recombineering oligo (o384stop) were simultaneously electroporated into L. reuteri 6475 pJP042 (ermR). Plasmid pJP577 was used as a measure of transformation efficiency, and o384stop was designed to introduce translational stops at amino acids 2, 4 and 5 of eriC. Candidate recombineered mutant colonies of L. reuteri 6475 were selected for growth on MRS Cm 10 and successively screened by mismatch amplification mutation assay (MAMA)-PCR using primers homologous to the desired mutation and sequences flanking the mutation site (KJP73, KJP74 and KJP75, respectively). PCR-positive colonies were verified by sequencing. The two recombineering plasmids (pJP042 and pJP577) were cured by successive passaging of overnight cultures in MRS until susceptibility to Cm 10 and Erm 10 was achieved.
TNF inhibition bioassay
TNF inhibition bioassay and TNF ELISA were performed as previously described (Thomas et al. 2012). Briefly, overnight MRS cultures (described above in bacterial strains and culture conditions) were inoculated into 10 mL semi-defined media, LDMIII (OD600 adjusted to 0.1) and grown for 24 hours at 37°C. After centrifugation at 4000 × g for 10 min, bacteria cell-free supernatants were filter-sterilized and size-fractionated to select for factors smaller than 3 kDa in size. Supernatants underwent vacuum drying and normalization by resuspension in RPMI medium to OD600 = 1.5. Normalized supernatants were tested for their ability to modulate TNF production in myeloid cells THP-1 cells. Cells (5×104 cells) were treated with L. reuteri supernatant (5% v/v) and activated by 100 ng/mL Pam3Cys-SKKKK x 3 HCl, a TLR-2 agonist (EMC Microcollections, Tüebingen, Germany) as previously described (Lin et al. 2008). After incubation at 37°C and 5% CO2 for 3.5 hours, quantitative ELISAs were used to determine the concentration of TNF in THP-1 culture supernatants according to the manufacturer’s instructions (R&D Systems, Minneapolis, MN). Data were analyzed using unpaired t-test on GraphPad Prism 5 software (GraphPad Inc, La Jolla, CA).
Histamine ELISA
Quantitative histamine ELISA was performed as previously described (Thomas et al. 2012). Briefly, wild-type (WT) L. reuteri 6475 and 6475eriC-Stop mutant strains were cultured in LDMIII, in the presence or absence of supplemental 4 mg/mL L-histidine. (The addition of 4 mg/mL L-histidine was determined by titration (0.2–4 mg/mL). A plateau in histamine production from WT 6475 was observed when the concentrations of supplemental L-histidine approached 4 mg/mL (data not shown). Culture supernatants were harvested at 24 hours, filter-sterilized and histamine concentrations were determined using the Histamine ELISA kit (Neogen, Lexington, KY) according to the manufacturer’s instructions. Absorbance was measured with a Bio-Rad Spectramax 340PC Spectrophotometer (Bio-Rad, Hercules, CA). Data were normalized to a culture OD of 1.0, background corrected, and analyzed using unpaired t-test and two-way ANOVA on GraphPad Prism 5 software.
Reverse transcription quantitative PCR
WT L. reuteri 6475 and the 6475eriC-Stop mutant strain were grown in LDMIII as described above in the presence or absence of 4 mg/mL supplemental L-histidine. At 16 hours post-inoculation, transcription was stopped using ice-cold methanol and cell pellets were harvested. RNA isolation from collected cell pellets and cDNA synthesis from total RNA were performed as previously described (Thomas et al. 2012). Expression of the hdc cluster genes (hdcA, hdcB, hdcP) and the RNA polymerase β-subunit (rpoB) reference gene was analyzed using reverse transcription quantitative PCR (RT-qPCR) with previously published primers (Table S1). RT-qPCR reactions included 2X FastStart Universal Probe Master (Rox) (Roche Applied Science) and cDNA as template along with corresponding probes and primers at final concentrations of 100 nM and 200 nM, respectively. A standard curve was generated using serially-diluted gDNA from WT L. reuteri 6475. PCR reactions were performed on the ViiA 7 Real-Time PCR System (Life Technologies, Grand Island, NY) with cycling parameters as previously described (Thomas et al. 2012). The relative standard curve method (Larionov et al. 2005) was used to calculate relative changes in gene expression.
Results
Creating a system for probiotic gene discovery by Himar1 transposon mutagenesis
In transposon-mediated mutagenesis, it is essential that the transposing events occur under strict regulation to ensure stability of mutants and prevent multiple insertions. We used a nisin-inducible protein expression system to create such a system for L. reuteri. Nisin is a post-translationally modified antimicrobial peptide produced by some strains of L. lactis (de Vos et al. 1995). Gene expression involved in nisin production by L. lactis is autoregulated by a histidine kinase two-component regulatory system, nisRK (Engelke et al. 1994). Promoter PnisA controls gene expression of the nisin biosynthetic gene cluster by nisin-mediated autoregulation (Kuipers et al. 1995). Nisin-inducible expression systems have been widely used to regulate protein expression in other prokaryotic organisms besides L. lactis (Kleerebezem et al. 1997; Pavan et al. 2000; Wells et al. 2000) including L. reuteri (Wu et al. 2006). For this study, we constructed pPH-M1 (Fig. 1), which is a temperature sensitive, RepA-dependent derivative of pVE6007 (Law et al. 1995). Plasmid pH-M1 contains a PnisA-directed hyperactive variant of Himar1 transposase, him1C9 (Lampe et al. 1999), along with the nisRK elements required for nisin-inducible gene expression. The pH-M1 construct also contains an ErmR Himar1 transposon flanked by the Himar1 cognate inverted terminal repeats (ITRs).
Himar1 transposase expression from pPH-M1 was induced during exponential growth phase of L. reuteri 6475 using purified nisin at a final concentration of 50 ng/mL. After a 16-hour induction, elimination of pPH-M1 from L. reuteri Tn-mutants was carried out by successive overnight culturing at 45°C. L. reuteri::Himar1 Tn-insertion mutants were selected for growth on MRS Erm 10 and a library of 432 Cm-sensitive/Erm-resistant mutants were stored at −80°C. Several L. reuteri::Himar1 mutants were randomly chosen from the library and analyzed for single insertions in different loci per Tn-mutant. PCR on gDNA isolated from these ten L. reuteri::Himar1 mutants using primers specific to the pPH-M1 cat and Himar1 Tn ermR genes confirmed that both pH-M1 elimination and Himar1 Tn-insertion were successful (data not shown). Southern hybridization on restriction enzyme-digested gDNA from a total of 16 L.reuteri::Himar1 Tn-mutants demonstrated single insertions of the Himar1 transposon in different sized DNA fragments between all but one isolate (lane 16, Supplemental Fig. S2). The L. reuteri::Himar1 Tn-mutant represented in lane 16 (Supplemental Fig. S2) clearly demonstrated the presence of the Himar1 transposon in 2 individual chromosomal DNA fragments from the same isolate. This highlights the ability of our Himar1 Tn insert in multiple loci within the L. reuteri 6475 genome and that Himar1 transposase production was reasonably well controlled by our nisin-inducible system.
L. reuteri::Himar1 Tn mutants unable to suppress TNF contained a Himar1 Tn insertion near the promoter region of a proton-chloride antiporter (eriC) gene
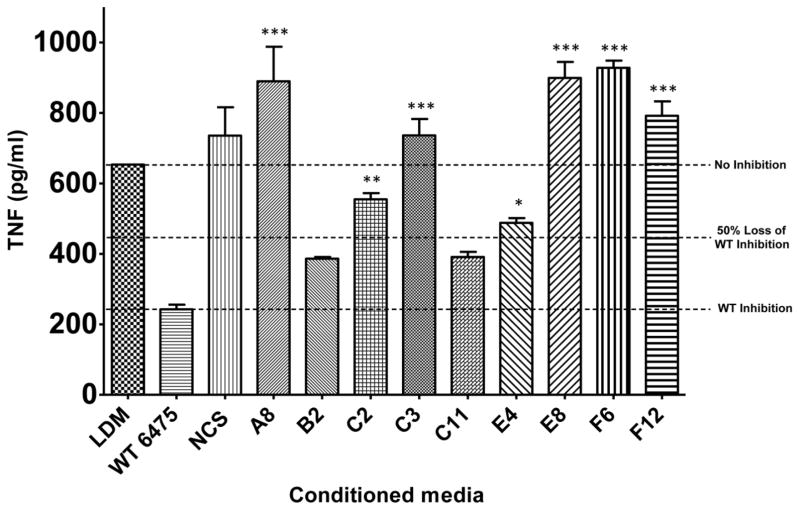
Preliminary high-throughput screening of 216 L. reuteri::Himar1 Tn-mutants for a loss in TNF inhibitory activity revealed 9 isolates that exhibited less than 50% TNF suppression of WT 6475 (see example screening data in Supplemental Fig. S3). These mutants were selected for full-scale preparation of LDMIII culture supernatants and confirmatory testing in a TNF inhibition bioassay (Fig. 2). Six mutants demonstrating complete loss of TNF inhibition (A8, C2, C3, E8, F6 and F12) were selected for whole genome shotgun sequencing to determine transposon insertion sites.
Fig. 2. Himar1 Tn-mutants lacked the ability to suppress TNF production by activated THP-1 cells.
L. reuteri 6475 Tn-mutants that demonstrated loss of TNF inhibition in preliminary high-throughput screening were confirmed for loss of phenotype in a full-scale TNF inhibition assay. Mutants A1, C2, C3, E8, F6 and F12 consistently demonstrated loss of TNF inhibition. Data were analyzed using one-way ANOVA (p<0.0001) followed by Bonferroni’s multiple comparison test against WT 6475 (n=3). LDM, media control; WT 6475, positive control for TNF suppression; NCS, Non-TNF-suppressive L. reuteri strain (6475::rsiR) used as negative control for TNF inhibition, *p < 0.01, **p < 0.001, ***p <0.0001
A total of 14,933,818 150-bp reads from 6 genomes were analyzed, and the primary sequencing data generated 166-fold coverage per genome. This relatively low coverage approach yielded positive results. Analysis of genome contigs from the 6 mutants submitted for WGS sequencing identified transposon insertion sites in mutants A8, C2 and F6. Analysis of sequences from mutants C3, E8 and F12 did not result in significant matches with the reference genome, likely due to gaps in the draft assembly. Transposon insertions in mutants A8, C2 and F6 all occurred in the same position between nucleotides T and A (position 136149 and 136150 in WGS sequence contig 21725, respectively). This insertion is positioned within the intergenic region downstream from the APC family D-serine/D-alanine/glycine transporter gene proY (HMPREF0536_10427) and upstream from the proton-chloride antiporter gene eriC (HMPREF0536_10428) (Fig. 3). The insertion is 7 bp downstream from the stop codon of proY, suggesting that the insertion was unlikely to affect the transcription and function of proY and its gene product. However, this insertion site was located directly upstream from the eriC predicted promoter region (PeriC, 28 bp) (Reese 2001), transcriptional start site (68 bp), and translational start codon (74 bp). The close proximity of the insertion site to PeriC may have disrupted eriC expression and possibly contributed to a diminished effect on TNF suppression by L. reuteri 6475.
Fig. 3. Location of the Himar1 insertion in L. reuteri 6475 chromosome relative to eriC.
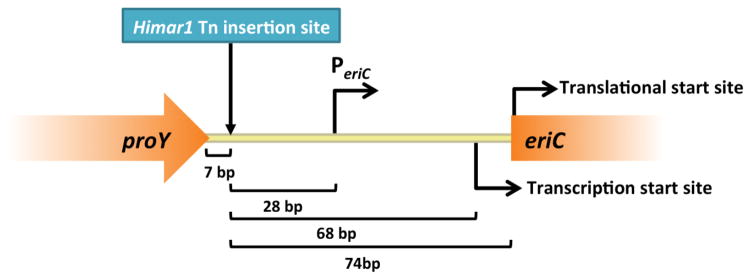
WGS sequencing data of L. reuteri::Himar1 Tn-mutants was analyzed for the presence and location of Himar1 Tn DNA. Himar1 insertion sequence was identified upstream of the eriC predicted promoter region in the L. reuteri 6475 chromosome. proY, APC family D-serine/D-alanine/glycine transporter gene; bp, base pairs; PeriC, predicted eriC promoter; eriC, proton-chloride antiporter gene;
Targeted inactivation of EriC confirmed a role in TNF inhibition and histamine production by L. reuteri
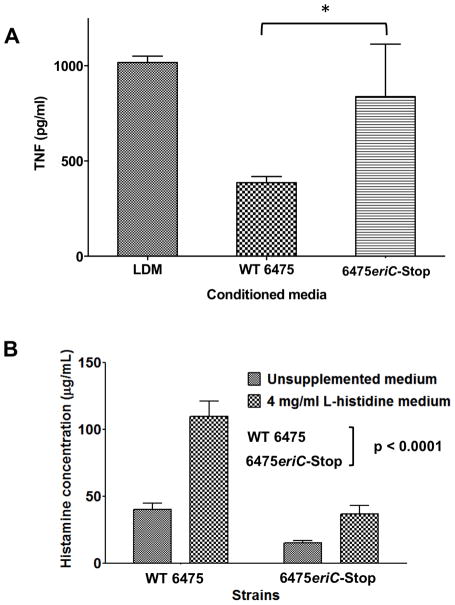
To independently determine whether inactivation of eriC would result in loss of TNF inhibition, we generated stop codons to replace amino acids 2 (Alanine), 4 (Arginine), and 5 (Leucine) in the eriC gene of L. reuteri 6475 by recombineering and tested culture supernatants from the mutant strain (6475eriC-Stop) for its ability to suppress human TNF production by activated human myeloid cells. Culture supernatants from 6475eriC-Stop failed to suppress TNF production from THP-1 cells when compared to WT 6475 (Fig. 4A). This observation confirmed that eriC is important for L. reuteri-mediated immunomodulation, and that the Himar1 Tn-insertion just upstream of eriC resulted in the discovery of a gene important for this probiotic function.
Fig. 4. Measurements of immunomodulatory properties are diminished in 6475eriC-Stop compared to WT 6475.
(A) L. reuteri supernatants were tested for the ability to inhibit TNF by TLR2-activated THP-1 cells. Inhibition of TNF production by recombineered 6475eriC-Stop mutant was significantly reduced when compared to WT 6475. The results are expressed as the mean ± SD, n=3, *p-value < 0.05 compared to WT 6475 using unpaired t-test (B) L. reuteri-derived histamine was quantitated by a histamine-specific ELISA and demonstrated decreased histamine production in 6475eriC-Stop compared to WT 6475 even when grown in histidine-supplemented medium. Data were normalized to final culture OD600 = 1.0 and analyzed by 2-way ANOVA. Results represent the mean ± SD (n = 3), p value < 0.0001 compared to wild-type 6475.
A recent study from our laboratory reported that histamine is an immunomodulatory factor produced by L. reuteri 6475 (Thomas et al. 2012). Because inactivation of eriC resulted in the loss of TNF suppression, we wanted to determine whether histamine production by L. reuteri was affected in the 6475eriC-Stop mutant. Defined media (LDMIII) alone contained approximately 1.5 μg/mL of L-histidine (Nolan 1971) and less than 0.1 μg/mL of histamine, as measured by ELISA (data not shown). Histamine concentrations in culture supernatants from WT 6475 (with an intact hdc gene cluster) were approximately 40 μg/mL, and increased more than 2.5-fold (>100 μg/mL) when growth media was supplemented with 4 mg/mL L-histidine. Histamine production from the 6475eriC-Stop mutant was significantly diminished compared to WT 6475 when grown in the same conditions (62% reduction in unsupplemented media; 67% reduction in L-histidine-supplemented media) (Fig. 4B). These results support a role for eriC in the production of L. reuteri-derived immunomodulatory histamine, and that additional L-histidine was not converted to histamine at the same capacity as wild-type without a functional eriC gene.
Targeted interference with a functional EriC affected histidine decarboxylase (hdc) gene cluster expression
Histamine production by the 6475eriC-Stop mutant was significantly decreased compared to WT 6475. Therefore, we wanted to determine whether preventing EriC translation in 6475eriC-Stop had downstream effects on gene expression within the hdc cluster (hdcA, hdcB and hdcC). Reverse transcription quantitative PCR showed that expression of hdcA and hdcB genes in the 6475eriC-Stop mutant was significantly downregulated (3.14-fold and 1.52-fold downregulation, respectively) compared to that of WT 6475, while the expression of hdcP was not affected (Fig. 5A). L-histidine supplementation elicited significant upregulation of all 3 hdc cluster genes in wild type L. reuteri when compared to no L-histidine supplementation (2.45-fold for hdcA, 3.20-fold for hdcB, and 2.02-fold for hdcP). Nevertheless, changes in the expression of hdcA, hdcB, and hdcP in 6475eriC-Stop were insignificant in the presence of supplemented L-histidine (Fig. 5B). The changes in gene expression of the hdc cluster in either WT 6475 or 6475eriC-Stop was concordant with the measurements of histamine production by ELISA for each strain under the same conditions. These observations showed that inactivation of EriC resulted in the diminished ability of L-histidine to induce hdc genes involved in histamine biosynthesis.
Fig. 5. Expression of the hdc gene cluster was affected by a mutation in eriC.
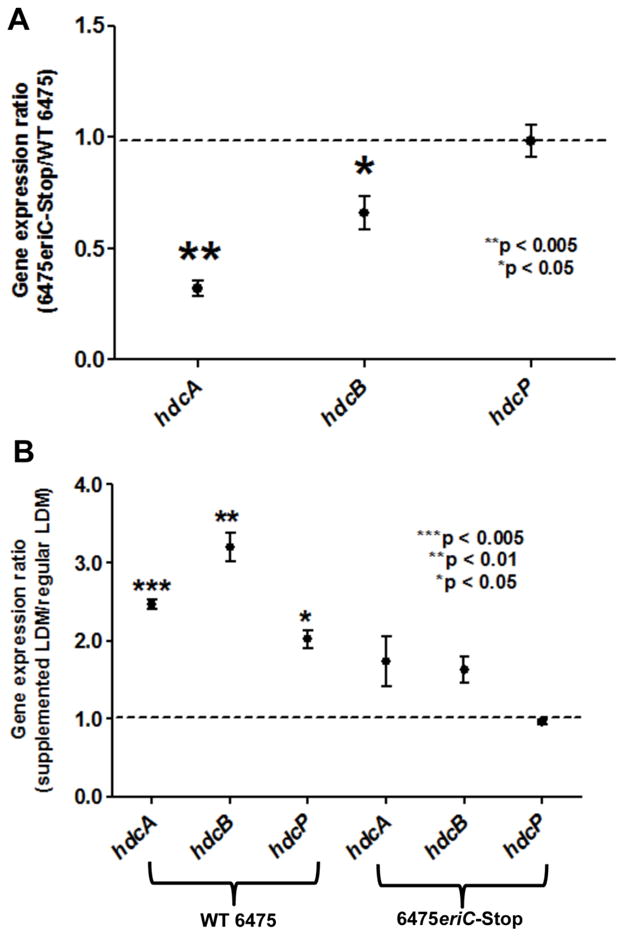
(A) Gene expression of the hdc cluster in 6475eriC-Stop was compared to that of WT 6475. RT-qPCR demonstrated decreased expression of hdcA and hdcB genes in the EriC-deficient mutant compared to WT 6475. Expression ratios of each gene (6475eriC-Stop mutant versus wild-type) were calculated, and results represent the mean ± SD, n=3, *p value < 0.05, **p value < 0.01 using one-sample t-test compared to the theoretical mean of 1.0. (B) Gene expression of the hdc cluster in both 6475eriC-Stop and WT 6475 were compared in the presence and absence of supplemental L-histidine. RT-qPCR demonstrated increased expression of all hdc genes in WT L. reuteri 6475 when grown in the presence or absence of supplemental L-histidine. Changes in expression of the same genes in 6475eriC-Stop were insignificant in the presence of added L-histidine. Expression ratios of each gene (histidine-supplemented versus unsupplemented) were calculated. Results represent the mean ± SD, n=3, ***p value < 0.005, **p value < 0.01, *p value < 0.05 compared to the theoretical mean of 1.0. All RT-qPCR data were normalized to a reference gene rpoB.
Discussion
We identified the proton-chloride antiporter gene (eriC) as a genetic element necessary for L. reuteri-mediated immunomodulation and histamine production. We developed a single-plasmid Tn-mutagenesis system for L. reuteri 6475 using the Himar1 transposon in conjunction with a nisin-inducible Himar1 transposase. The eriC gene was identified through a combination of high-throughput selective screening of randomly selected Himar1 Tn-mutants for a loss of TNF inhibition and WGS sequencing of candidate Tn-mutants. Genetic inactivation of eriC resulted in the loss of the L. reuteri 6475’s ability to suppress TNF production by activated human myeloid cells, a diminished production of histamine, and downregulation of histamine biosynthesis genes as compared to WT 6475.
Transposon mutagenesis systems are powerful genetic tools for gene discovery ranging from those focusing on single organisms to large-scale microbiome studies. Transposon mutagenesis using Himar1 has been used to identify genetic elements in Bacteroides thetaiotaomicron required for stable colonization in mice, in the presence or absence of other members of the intestinal microbiota (Goodman et al. 2009). Few transposon mutagenesis systems have been described for Lactobacillus species, such as the Pjunc-TpaseIS1223 system and the Tn5-based transposon system in L. casei (Ito et al. 2010; Licandro-Seraut et al. 2012). A successful use of a transposon-based mutagenesis for gene discovery in lactobacilli was described in L. plantarum, where the pGh9:ISS1 plasmid system was used to identify genes involved in the regulation of phenolic acid metabolism (Gury et al. 2004). To our knowledge, our single-plasmid Himar1 transposon mutagenesis system was the first successful use of Himar1 in L. reuteri. We have only tested our Himar1 system in the plasmid-free strain, L. reuteri 6475. Other L. reuteri strains containing multiple native plasmids such as L. reuteri ATCC 55730 or DSM 17938 (Rosander et al. 2008) are less amenable to genetic manipulation. However, this Himar1 system was built into a plasmid (pVE6007) known to replicate in multiple L. reuteri strains, and we postulate that it could be used as a gene discovery tool for them as well.
We screened Himar1 Tn-mutants and observed a phenotypic alteration in the loss of TNF suppression. Identification of transposon insertion sites in mutants that failed to inhibit TNF production by THP-1 cells in our preliminary screenings revealed disruptions of the intergenic region near the predicted promoter for proton-chloride antiporter gene (eriC). EriC (also called CIC-ec1) is part of the biologically-ubiquitous chloride ion channel (CIC) superfamily of proteins, which includes chloride channels and proton-chloride antiporters (Chen 2005). Proteins in the CIC superfamily are expressed in species ranging from bacteria to human. However, EriC possesses a unique proton-chloride antiporter activity extensively characterized in E. coli. Despite the differences in the functions between the two types of CIC proteins, all members of this superfamily contain double-barreled architecture and possess voltage-dependent gating mechanism (Maduke et al. 2000). EriC is a homodimeric transmembrane protein and exchanges protons with chlorides in a fixed 2:1 stoichiometry (Kieseritzky and Knapp 2010). Results from protein BLAST with L. reuteri 6475 EriC sequence revealed that proteins with EriC functional domains are ubiquitously present in bacteria including those that are members of the human intestinal microbiome.
As previously discussed in the introduction, we recently revealed histamine as one of the immunomodulatory factors produced by L. reuteri 6475. Production of biogenic amines, including histamine, through microbial decarboxylation have been reported in various types of foods (Naila et al. 2011). Histamine poisoning (or scromboid poisoning), which could result in symptoms that resemble severe allergic reactions, occurs after consumption of foods contaminated with highly elevated quantities of histamine, mostly at concentrations higher than 0.5 mg/mL (Hungerford 2010; Gonzaga et al. 2009). Despite the potential health risk of histamine-contaminated foods whereby histamine is generated in food prior to consumption, histamine may promote healthy development and physiology of the intestinal epithelium and the gut immune system (O’Mahony et al. 2011). Selective blockade of histamine receptors type 1 and 2 resulted in impaired innate immune responses in a mouse model (Metz et al. 2011). Moreover, histamine receptor antagonists have been associated with increased incidence and severity of necrotizing enterocolitis (NEC) in very low birth weight infants (Guillet et al. 2006) and Crohn’s disease (Juillerat et al. 2012). These findings suggest an immunomodulatory role of histamine in the human gastrointestinal tract. L. reuteri 6475 was shown to be able to produce histamine via decarboxylation of L-histidine at low concentrations relative to what is considered toxic when orally consumed (Thomas et al. 2012). Mutants that failed to produce histamine were unable to suppress TNF production by THP-1 cells and yielded diminished protective effects in a TNBS-induced mouse model of acute colitis (Hemarajata et al. 2013; Thomas et al. 2012). Patients suffering from inflammatory bowel disease could benefit from the bacteria-mediated conversion of histidine in the diet to histamine in the lumen of the gastrointestinal tract.
The production of histamine and expression of the hdc gene cluster in an EriC-deficient mutant were diminished compared to WT 6475. Degradative amino acid decarboxylation is a highly efficient bacterial mechanism for withstanding the extreme acidic environment in the gastrointestinal tract and for generating secondary metabolic energy (Gut et al. 2006). Genetic elements involved in amino acid decarboxylation are part of the conserved functional core genes of the healthy human intestinal microbiome, suggesting its essential contribution to the function and maintenance of the gut microbial community as a whole (The Human Microbiome Project Consortium 2012a).
Amino acid decarboxylation results in the production of biogenic amines and helps regulate bacterial intracellular pH in the presence of acid stress. A single proton is consumed during the decarboxylation of each amino acid molecule, resulting in alkalinization of the cytoplasm (Romano et al. 2012). Many gut microbes, including lactic acid bacteria, are able to convert glutamate to γ-aminobutyrate (Cotter and Hill 2003), arginine to agmatine (Iyer et al. 2003), lysine to cadaverine (Lin et al. 1995), ornithine to putrescine (Romano et al. 2012; Azcarate-Peril et al. 2004) and histidine to histamine (Romano et al. 2012). An intestinal bacterium, E. coli, was shown to have at least 4 different mechanisms for acid resistance and maintenance of transmembrane potential, and 3 of those mechanisms specifically rely on the decarboxylation of glutamate, arginine, and lysine and involve EriC (Richard and Foster 2004). L. reuteri 100-23, a murine-derived L. reuteri strain, contains a glutamate decarboxylase system involved in acid resistance during sourdough fermentations (Su et al. 2011). The human microbiome resident, L. reuteri 6475, also possesses genetic elements required for decarboxylation of glutamate and histidine but the role of these processes in regulation of membrane potential and response to acid stress has not been characterized. Decarboxylation of L-histidine and the subsequent histamine production by lactobacilli was first identified and characterized in a horse isolate of Lactobacillus saerimneri ATCC 33222 (formerly known as Lactobacillus sp. strain 30a). (Rodwell 1953). A functional hdc gene cluster, with the histidine decarboxylase pyruvoyl type (hdcA), a putative helper protein (hdcB) and a histidine/histamine antiporter (hdcP), is found in histaminogenic strains of Lactobacillus (Rossi et al. 2011). Metabolic factors such as pH, the concentration of free L-histidine and the presence of other carbon sources could affect the expression of genes in the hdc gene cluster (Pessione et al. 2005; Landete et al. 2006, 2008). Genetic elements such as rsiR may regulate the expression of these hdc genes as well (Hemarajata et al. 2013).
Amino acid decarboxylation is an electrogenic process where an accumulation of intracellular negative transmembrane potential occurs as protons are virtually driven out of the bacterial cell. The increasing negative transmembrane potential may antagonize amino acid decarboxylation and ultimately lead to the decreased production of biogenic amines (Iyer et al. 2002). EriC in E. coli functions as an electrical shunt, facilitating proton entry with the simultaneous export of chloride, relieving the accumulated negative intracellular charge and allowing amino acid decarboxylation to continue (Supplement Fig. S4). We propose that EriC may perform the same function in L. reuteri. EriC-deficient L. reuteri may be unable to alleviate increasing intracellular negative charges by failing to replenish the intracellular protons needed for histidine decarboxylation. In turn, this may result in the inhibition of histidine decarboxylation, diminished histamine production and loss of TNF suppression in human immune cells. Inactivation of eriC also affected hdc cluster gene expression. Expression of genes involved in glutamate decarboxylation and the acid resistance mechanism in L. lactis (Sanders et al. 1998) and E. coli (De Biase and Pennacchietti 2012) were influenced by the amounts of chloride in growth media and intracellular pH. In the EriC-deficient mutant, regulation of intracellular anions and pH may also be altered and result in an indirect effect upon hdc gene cluster expression. The role of L. reuteri EriC in transmembrane potential maintenance could be further characterized by comparing intracellular pH (using 2,7-bis-(2-carboxyethyl)-5(6)-carboxyfluorescein (Molenaar et al. 1991)) and membrane potential (with 3,3′-dipropylthiodicarbocyanine (Sip et al. 1990)) between those of WT 6475 and the EriC-deficient mutant in the presence or absence of the recently discovered CIC-specific inhibitor, 4,4′-octanamidostilbene-2,2′-disulfonate (OADS) (Howery et al. 2012).
The successful implementation of our novel Himar1 Tn-mutagenesis system in immunomodulatory gene discovery is a significant advance in the field of L. reuteri functional genomics. This system enables the identification of genetic elements from large Tn-mutant libraries that can be screened in high-throughput methods for any loss-of-function phenotype. Further screening of random mutants generated from this system could help identify additional genes that may be involved in L. reuteri-mediated immunomodulation. We identified EriC as one of the major players in a putative regulatory mechanism for histidine decarboxylation in L. reuteri. The importance of EriC in histidine metabolism highlights the significance of EriC as a member in the immunomodulatory gene network of L. reuteri 6475, and the potential importance of ion transporters in amino acid metabolism and immunomodulation by the gut microbiome. Future manipulation of immunomodulatory gene networks important for L-histidine decarboxylation and histamine biosynthesis could facilitate the development of next generation probiotics with enhanced immunoregulatory functions. More broadly, dietary modifications can be coupled with specific probiotic strains to maximize immunomodulatory functions of gut-derived microbes in future microbiome-based therapeutics for disorders of intestinal inflammation including inflammatory bowel disease.
Supplementary Material
L. reuteri 6475 was electroporated with pPH-M1 (2 mm cuvettes at 2500 V, 25 μF and 400 Ω) and transformants were cultured in MRS with Cm 10 and Erm 10 in the presence of nisin and overnight at 30°C to induce for Himar1-transposase expression and transposition. Consecutive subculturing at 45°C in the absence of Cm and presence of Erm was carried out to cure Himar1 Tn-insertion mutants of pPH-M1. Finally, L. reuteri::Himar1 Tn-mutants were selected for growth on Erm 10 and corresponding susceptibility to Cm 10. L. reuteri::Himar1 Tn-mutants meeting this susceptibility pattern were included in our Tn-mutant library and stored in 96-well plates at −80°C.
HindIII digestions of gDNA from 16 randomly chosen Tn-mutants (lanes 1–16) were subjected to Southern hybridization with a DIG-labeled probe specific to ermR in the Himar1 Tn. Bands in each lane represent the presence of the Himar1 Tn in DNA fragments. DIG-labeled molecular weight marker VII (Roche, Indianapolis, IN) is shown on the left and are indicated in bp. +, positive control/HindIII-digested pPH-M1; −, negative control/HindIII-digested WT 6475 gDNA;
Conditioned media from Himar1 Tn-mutants were tested for the ability to inhibit TNF by TLR2-activated THP-1 cells in a 96-well format. L.reuteri::Himar1 Tn-mutants with ≥50% reduction in TNF suppression as compared to wild type 6475 were selected for phenotype confirmation in the full-scale TNF inhibition assay. The results are expressed as the mean ± SD, n=3, *p-value < 0.01, **p-value < 0.005 compared to WT 6475 using unpaired t-test. Data from 72 of the 216 screened L. reuteri::Himar1 mutants are shown here. Data from the remaining 144 L. reuteri::Himar1 Tn mutants are not shown.
We propose that EriC simultaneously coordinates the import of protons with the export of electrons to maintain a neutral membrane potential, and is constantly relieving the inside-negative membrane potential that accumulates during histidine decarboxylation. This process would also replenish intracellular proton concentrations required to continue producing histamine. HdcB, a putative helper protein proposed to convert HdcA into a mature enzyme, is not depicted. HdcA, histidine decarboxylase enzyme; HdcP, histidine-histamine antiporter
Acknowledgments
This work was supported in part by research support from National Institutes of Health (R01 AT004326, R01 DK065075, UH3 DK083990). We also acknowledge NIH support (DK56338) for the Texas Medical Center Digestive Diseases Center.
We thank Eamonn Connolly (BioGaia AB, Stockholm) for providing the L. reuteri strains, David Lampe (Duquesne University, Pittsburgh, PA) and Todd Klaenhammer (North Carolina State University, Raleigh, NC) for providing plasmids used in this study, Kathryn Pflughoeft for designing recombineering oligonucleotides and scientific expertise, and Michael White for his assistance in generating Himar1 Tn-mutants.
Footnotes
Conflict of interest
J. Versalovic receives unrestricted research support from Biogaia AB (Stockholm, Sweden).
References
- Akerley BJ, Lampe DJ. Analysis of gene function in bacterial pathogens by GAMBIT. Methods Enzymol. 2002;358:100–108. doi: 10.1016/s0076-6879(02)58082-4. [DOI] [PubMed] [Google Scholar]
- Akerley BJ, Rubin EJ, Camilli A, Lampe DJ, Robertson HM, Mekalanos JJ. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc Natl Acad Sci U S A. 1998;95 (15):8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azcarate-Peril MA, Altermann E, Hoover-Fitzula RL, Cano RJ, Klaenhammer TR. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Applied and environmental microbiology. 2004;70(9):5315–5322. doi: 10.1128/AEM.70.9.5315-5322.2004. 70/9/5315 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare PA. Genetic manipulation of Coxiella burnetii. Adv Exp Med Biol. 2012;984:249–271. doi: 10.1007/978-94-007-4315-1_13. [DOI] [PubMed] [Google Scholar]
- Beare PA, Howe D, Cockrell DC, Omsland A, Hansen B, Heinzen RA. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J Bacteriol. 2009;191 (5):1369–1381. doi: 10.1128/JB.01580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy P, Louvel H, Saint Girons I, Picardeau M. Random insertional mutagenesis of Leptospira interrogans, the agent of leptospirosis, using a mariner transposon. J Bacteriol. 2005;187 (9):3255–3258. doi: 10.1128/JB.187.9.3255-3258.2005. 187/9/3255 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Versalovic J. Probiotics and gastrointestinal infections. Interdiscip Perspect Infect Dis. 2008;2008:290769. doi: 10.1155/2008/290769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TY. Structure and function of clc channels. Annual review of physiology. 2005;67:809–839. doi: 10.1146/annurev.physiol.67.032003.153012. [DOI] [PubMed] [Google Scholar]
- Cotter PD, Hill C. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiology and molecular biology reviews : MMBR. 2003;67 (3):429–453. doi: 10.1128/MMBR.67.3.429-453.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biase D, Pennacchietti E. Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol Microbiol. 2012 doi: 10.1111/mmi.12020. [DOI] [PubMed] [Google Scholar]
- de Vos WM, Kuipers OP, van der Meer JR, Siezen RJ. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by gram-positive bacteria. Mol Microbiol. 1995;17 (3):427–437. doi: 10.1111/j.1365-2958.1995.mmi_17030427.x. [DOI] [PubMed] [Google Scholar]
- Engelke G, Gutowski-Eckel Z, Kiesau P, Siegers K, Hammelmann M, Entian KD. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Applied and environmental microbiology. 1994;60 (3):814–825. doi: 10.1128/aem.60.3.814-825.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese SA, Benson AK, Tannock GW, Loach DM, Kim J, Zhang M, Oh PL, Heng NC, Patil PB, Juge N, Mackenzie DA, Pearson BM, Lapidus A, Dalin E, Tice H, Goltsman E, Land M, Hauser L, Ivanova N, Kyrpides NC, Walter J. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 2011;7 (2):e1001314. doi: 10.1371/journal.pgen.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzaga VE, Lescano AG, Huaman AA, Salmon-Mulanovich G, Blazes DL. Histamine levels in fish from markets in Lima, Peru. J Food Prot. 2009;72 (5):1112–1115. doi: 10.4315/0362-028x-72.5.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6 (3):279–289. doi: 10.1016/j.chom.2009.08.003. S1931-3128(09)00281-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet R, Stoll BJ, Cotten CM, Gantz M, McDonald S, Poole WK, Phelps DL. Association of H2-blocker therapy and higher incidence of necrotizing enterocolitis in very low birth weight infants. Pediatrics. 2006;117 (2):e137–142. doi: 10.1542/peds.2005-1543. peds.2005-1543 [pii] [DOI] [PubMed] [Google Scholar]
- Gury J, Barthelmebs L, Cavin JF. Random transposon mutagenesis of Lactobacillus plantarum by using the pGh9:IS S1 vector to clone genes involved in the regulation of phenolic acid metabolism. Arch Microbiol. 2004;182 (5):337–345. doi: 10.1007/s00203-004-0705-1. [DOI] [PubMed] [Google Scholar]
- Gut H, Pennacchietti E, John RA, Bossa F, Capitani G, De Biase D, Grutter MG. Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB. The EMBO journal. 2006;25 (11):2643–2651. doi: 10.1038/sj.emboj.7601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemarajata P, Gao C, Pflughoeft KJ, Thomas CM, Saulnier DM, Spinler JK, Versalovic J. Lactobacillus reuteri-Specific Immunoregulatory Gene rsiR Modulates Histamine Production and Immunomodulation by Lactobacillus reuteri. J Bacteriol. 2013;195 (24):5567–5576. doi: 10.1128/JB.00261-13. JB.00261-13 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howery AE, Elvington S, Abraham SJ, Choi KH, Dworschak-Simpson S, Phillips S, Ryan CM, Sanford RL, Almqvist J, Tran K, Chew TA, Zachariae U, Andersen OS, Whitelegge J, Matulef K, Du Bois J, Maduke MC. A designed inhibitor of a CLC antiporter blocks function through a unique binding mode. Chemistry & biology. 2012;19 (11):1460–1470. doi: 10.1016/j.chembiol.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hungerford JM. Scombroid poisoning: a review. Toxicon. 2010;56 (2):231–243. doi: 10.1016/j.toxicon.2010.02.006. S0041-0101(10)00045-0 [pii] [DOI] [PubMed] [Google Scholar]
- Ito M, Kim YG, Tsuji H, Kiwaki M, Nomoto K, Tanaka R, Okada N, Danbara H. A practical random mutagenesis system for probiotic Lactobacillus casei using Tn5 transposition complexes. Journal of applied microbiology. 2010;109 (2):657–666. doi: 10.1111/j.1365-2672.2010.04690.x. [DOI] [PubMed] [Google Scholar]
- Iyer R, Iverson TM, Accardi A, Miller C. A biological role for prokaryotic ClC chloride channels. Nature. 2002;419 (6908):715–718. doi: 10.1038/nature01000. [DOI] [PubMed] [Google Scholar]
- Iyer R, Williams C, Miller C. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J Bacteriol. 2003;185 (22):6556–6561. doi: 10.1128/JB.185.22.6556-6561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juillerat P, Schneeweiss S, Cook EF, Ananthakrishnan AN, Mogun H, Korzenik JR. Drugs that inhibit gastric acid secretion may alter the course of inflammatory bowel disease. Aliment Pharmacol Ther. 2012;36 (3):239–247. doi: 10.1111/j.1365-2036.2012.05173.x. [DOI] [PubMed] [Google Scholar]
- Kieseritzky G, Knapp EW. Charge transport in the ClC-type chloride-proton anti-porter from Escherichia coli. J Biol Chem. 2010;286 (4):2976–2986. doi: 10.1074/jbc.M110.163246. M110.163246 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M, Beerthuyzen MM, Vaughan EE, de Vos WM, Kuipers OP. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Applied and environmental microbiology. 1997;63 (11):4581–4584. doi: 10.1128/aem.63.11.4581-4584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristich CJ, Nguyen VT, Le T, Barnes AM, Grindle S, Dunny GM. Development and use of an efficient system for random mariner transposon mutagenesis to identify novel genetic determinants of biofilm formation in the core Enterococcus faecalis genome. Applied and environmental microbiology. 2008;74 (11):3377–3386. doi: 10.1128/AEM.02665-07. AEM.02665-07 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers OP, Beerthuyzen MM, de Ruyter PG, Luesink EJ, de Vos WM. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270 (45):27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci U S A. 1999;96 (20):11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe DJ, Grant TE, Robertson HM. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 1998;149 (1):179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landete JM, Pardo I, Ferrer S. Histamine, histidine, and growth-phase mediated regulation of the histidine decarboxylase gene in lactic acid bacteria isolated from wine. FEMS Microbiol Lett. 2006;260 (1):84–90. doi: 10.1111/j.1574-6968.2006.00294.x. [DOI] [PubMed] [Google Scholar]
- Landete JM, Pardo I, Ferrer S. Regulation of hdc expression and HDC activity by enological factors in lactic acid bacteria. Journal of applied microbiology. 2008;105 (5):1544–1551. doi: 10.1111/j.1365-2672.2008.03865.x. [DOI] [PubMed] [Google Scholar]
- Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol. 1995;177 (24):7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton Y, Mohapatra NP, Haldenwang WG. In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Applied and environmental microbiology. 2006;72 (1):327–333. doi: 10.1128/AEM.72.1.327-333.2006. 72/1/327 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, Mierau I, Dabrowska M, Venema G, Kok J. A general system for generating unlabelled gene replacements in bacterial chromosomes. Molecular & general genetics : MGG. 1996;253 (1–2):217–224. doi: 10.1007/s004380050315. [DOI] [PubMed] [Google Scholar]
- Licandro-Seraut H, Brinster S, van de Guchte M, Scornec H, Maguin E, Sansonetti P, Cavin JF, Serror P. Development of an efficient in vivo system (Pjunc-TpaseIS1223) for random transposon mutagenesis of Lactobacillus casei. Applied and environmental microbiology. 2012;78 (15):5417–5423. doi: 10.1128/AEM.00531-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol. 1995;177 (14):4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Thibodeaux CH, Pena JA, Ferry GD, Versalovic J. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflammatory bowel diseases. 2008;14 (8):1068–1083. doi: 10.1002/ibd.20448. [DOI] [PubMed] [Google Scholar]
- Liu H, Bouillaut L, Sonenshein AL, Melville SB. Use of a Mariner-Based Transposon Mutagenesis System To Isolate Clostridium perfringens Mutants Deficient in Gliding Motility. J Bacteriol. 2013;195 (3):629–636. doi: 10.1128/JB.01288-12. JB.01288-12 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduke M, Miller C, Mindell JA. A decade of CLC chloride channels: structure, mechanism, and many unsettled questions. Annual review of biophysics and biomolecular structure. 2000;29:411–438. doi: 10.1146/annurev.biophys.29.1.411. [DOI] [PubMed] [Google Scholar]
- Maier TM, Pechous R, Casey M, Zahrt TC, Frank DW. In vivo Himar1-based transposon mutagenesis of Francisella tularensis. Applied and environmental microbiology. 2006;72 (3):1878–1885. doi: 10.1128/AEM.72.3.1878-1885.2006. 72/3/1878 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz M, Doyle E, Bindslev-Jensen C, Watanabe T, Zuberbier T, Maurer M. Effects of antihistamines on innate immune responses to severe bacterial infection in mice. Int Arch Allergy Immunol. 2011;155 (4):355–360. doi: 10.1159/000321614. 000321614 [pii] [DOI] [PubMed] [Google Scholar]
- Molenaar D, Abee T, Konings WN. Continuous measurement of the cytoplasmic pH in Lactococcus lactis with a fluorescent pH indicator. Biochimica et biophysica acta. 1991;1115 (1):75–83. doi: 10.1016/0304-4165(91)90014-8. [DOI] [PubMed] [Google Scholar]
- Naila A, Flint S, Fletcher G, Bremer P, Meerdink G. Control of biogenic amines in food--existing and emerging approaches. J Food Sci. 2011;75 (7):R139–150. doi: 10.1111/j.1750-3841.2010.01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan RA. Amino acids and growth factors in vitamin-free casamino acids. Mycologia. 1971;63 (6):1231–1234. [PubMed] [Google Scholar]
- O’Mahony L, Akdis M, Akdis CA. Regulation of the immune response and inflammation by histamine and histamine receptors. J Allergy Clin Immunol. 2011;128 (6):1153–1162. doi: 10.1016/j.jaci.2011.06.051. S0091-6749(11)01082-7 [pii] [DOI] [PubMed] [Google Scholar]
- Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, Walter J. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. The ISME journal. 2010;4 (3):377–387. doi: 10.1038/ismej.2009.123. [DOI] [PubMed] [Google Scholar]
- Pavan S, Hols P, Delcour J, Geoffroy MC, Grangette C, Kleerebezem M, Mercenier A. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Applied and environmental microbiology. 2000;66 (10):4427–4432. doi: 10.1128/aem.66.10.4427-4432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessione E, Mazzoli R, Giuffrida MG, Lamberti C, Garcia-Moruno E, Barello C, Conti A, Giunta C. A proteomic approach to studying biogenic amine producing lactic acid bacteria. Proteomics. 2005;5 (3):687–698. doi: 10.1002/pmic.200401116. [DOI] [PubMed] [Google Scholar]
- Picardeau M. Transposition of fly mariner elements into bacteria as a genetic tool for mutagenesis. Genetica. 2009 doi: 10.1007/s10709-009-9408-5. [DOI] [PubMed] [Google Scholar]
- Pozsgai ER, Blair KM, Kearns DB. Modified mariner transposons for random inducible-expression insertions and transcriptional reporter fusion insertions in Bacillus subtilis. Applied and environmental microbiology. 2011;78 (3):778–785. doi: 10.1128/AEM.07098-11. AEM.07098-11 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que YA, Haefliger JA, Francioli P, Moreillon P. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infection and immunity. 2000;68 (6):3516–3522. doi: 10.1128/iai.68.6.3516-3522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem. 2001;26(1):51–56. doi: 10.1016/s0097-8485(01)00099-7. S0097-8485(01)00099-7 [pii] [DOI] [PubMed] [Google Scholar]
- Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186(18):6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. 186/18/6032 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodwell AW. The histidine decarboxylase of a species of Lactobacillus; apparent dispensability of pyridoxal phosphate as coenzyme. Journal of general microbiology. 1953;8 (2):233–237. doi: 10.1099/00221287-8-2-233. [DOI] [PubMed] [Google Scholar]
- Romano A, Trip H, Lonvaud-Funel A, Lolkema JS, Lucas PM. Evidence of two functionally distinct ornithine decarboxylation systems in lactic acid bacteria. Applied and environmental microbiology. 2012;78 (6):1953–1961. doi: 10.1128/AEM.07161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosander A, Connolly E, Roos S. Removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730 and characterization of the resulting daughter strain, L. reuteri DSM 17938. Applied and environmental microbiology. 2008;74 (19):6032–6040. doi: 10.1128/AEM.00991-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F, Gardini F, Rizzotti L, La Gioia F, Tabanelli G, Torriani S. Quantitative analysis of histidine decarboxylase gene (hdcA) transcription and histamine production by Streptococcus thermophilus PRI60 under conditions relevant to cheese making. Appl Environ Microbiol. 2011;77 (8):2817–2822. doi: 10.1128/AEM.02531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JW, Leenhouts K, Burghoorn J, Brands JR, Venema G, Kok J. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol. 1998;27 (2):299–310. doi: 10.1046/j.1365-2958.1998.00676.x. [DOI] [PubMed] [Google Scholar]
- Sip M, Herman P, Plasek J, Hrouda V. Transmembrane potential measurement with carbocyanine dye diS-C3-(5): fast fluorescence decay studies. Journal of photochemistry and photobiology B, Biology. 1990;4 (3):321–328. doi: 10.1016/1011-1344(90)85037-w. [DOI] [PubMed] [Google Scholar]
- Su MS, Schlicht S, Ganzle MG. Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb Cell Fact 10 Suppl. 2011;1:S8. doi: 10.1186/1475-2859-10-S1-S8. 1475-2859-10-S1-S8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam C, Glass EM, Anderson DM, Missiakas D. Transposon mutagenesis of Bacillus anthracis strain Sterne using Bursa aurealis. Plasmid. 2006;56 (1):74–77. doi: 10.1016/j.plasmid.2006.01.002. S0147-619X(06)00013-8 [pii] [DOI] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012a;486 (7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CM, Hong T, van Pijkeren JP, Hemarajata P, Trinh DV, Hu W, Britton RA, Kalkum M, Versalovic J. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PloS one. 2012;7 (2):e31951. doi: 10.1371/journal.pone.0031951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pijkeren JP, Britton RA. High efficiency recombineering in lactic acid bacteria. Nucleic acids research. 2012;40 (10):e76. doi: 10.1093/nar/gks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pijkeren JP, Neoh KM, Sirias D, Findley AS, Britton RA. Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri. Bioengineered. 2012;3 (4):209–217. doi: 10.4161/bioe.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Britton RA, Roos S. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci U S A. 2010;108(Suppl 1):4645–4652. doi: 10.1073/pnas.1000099107. 1000099107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Chagnaud P, Tannock GW, Loach DM, Dal Bello F, Jenkinson HF, Hammes WP, Hertel C. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Applied and environmental microbiology. 2005;71 (2):979–986. doi: 10.1128/AEM.71.2.979-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CL, Moore EA, Hoag JA, Hirt H, Dunny GM, Erlandsen SL. Inducible expression of Enterococcus faecalis aggregation substance surface protein facilitates bacterial internalization by cultured enterocytes. Infection and immunity. 2000;68 (12):7190–7194. doi: 10.1128/iai.68.12.7190-7194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CM, Lin CF, Chang YC, Chung TC. Construction and characterization of nisin-controlled expression vectors for use in Lactobacillus reuteri. Biosci Biotechnol Biochem. 2006;70(4):757–767. doi: 10.1271/bbb.70.757. JST.JSTAGE/bbb/70.757 [pii] [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18 (5):821–829. doi: 10.1101/gr.074492.107. gr.074492.107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
L. reuteri 6475 was electroporated with pPH-M1 (2 mm cuvettes at 2500 V, 25 μF and 400 Ω) and transformants were cultured in MRS with Cm 10 and Erm 10 in the presence of nisin and overnight at 30°C to induce for Himar1-transposase expression and transposition. Consecutive subculturing at 45°C in the absence of Cm and presence of Erm was carried out to cure Himar1 Tn-insertion mutants of pPH-M1. Finally, L. reuteri::Himar1 Tn-mutants were selected for growth on Erm 10 and corresponding susceptibility to Cm 10. L. reuteri::Himar1 Tn-mutants meeting this susceptibility pattern were included in our Tn-mutant library and stored in 96-well plates at −80°C.
HindIII digestions of gDNA from 16 randomly chosen Tn-mutants (lanes 1–16) were subjected to Southern hybridization with a DIG-labeled probe specific to ermR in the Himar1 Tn. Bands in each lane represent the presence of the Himar1 Tn in DNA fragments. DIG-labeled molecular weight marker VII (Roche, Indianapolis, IN) is shown on the left and are indicated in bp. +, positive control/HindIII-digested pPH-M1; −, negative control/HindIII-digested WT 6475 gDNA;
Conditioned media from Himar1 Tn-mutants were tested for the ability to inhibit TNF by TLR2-activated THP-1 cells in a 96-well format. L.reuteri::Himar1 Tn-mutants with ≥50% reduction in TNF suppression as compared to wild type 6475 were selected for phenotype confirmation in the full-scale TNF inhibition assay. The results are expressed as the mean ± SD, n=3, *p-value < 0.01, **p-value < 0.005 compared to WT 6475 using unpaired t-test. Data from 72 of the 216 screened L. reuteri::Himar1 mutants are shown here. Data from the remaining 144 L. reuteri::Himar1 Tn mutants are not shown.
We propose that EriC simultaneously coordinates the import of protons with the export of electrons to maintain a neutral membrane potential, and is constantly relieving the inside-negative membrane potential that accumulates during histidine decarboxylation. This process would also replenish intracellular proton concentrations required to continue producing histamine. HdcB, a putative helper protein proposed to convert HdcA into a mature enzyme, is not depicted. HdcA, histidine decarboxylase enzyme; HdcP, histidine-histamine antiporter