SUMMARY
Stimulator of interferon genes (STING, also named MITA, MYPS or ERIS) is an intracellular DNA sensor that induces type I interferon through its interaction with TANK-binding kinase 1 (TBK1). Here we found that the nucleotide-binding, leucine-rich repeat containing protein, NLRC3, reduced STING-dependent innate immune activation in response to cytosolic DNA, cyclic di-GMP (c-di-GMP) and DNA viruses. NLRC3 associated with both STING and TBK1, and impeded STING-TBK1 interaction and downstream type I interferon production. Using purified recombinant proteins NLRC3 was found to interact directly with STING. Furthermore, NLRC3 prevented proper trafficking of STING to perinuclear and punctated region, known to be important for its activation. In animals, herpes simplex virus 1 (HSV-1)-infected Nlrc3−/− mice exhibited enhanced innate immunity, reduced morbidity and viral load. This demonstrates the intersection of two key pathways of innate immune regulation, NLR and STING, to fine tune host response to intracellular DNA, DNA virus and c-di-GMP
INTRODUCTION
Nucleic acid and small nucleic acid-like molecules can act as pathogen associated molecular patterns (PAMPs) to cause type I interferon (IFN-I) and other cytokine induction (Takeuchi and Akira, 2007). Among DNA sensors, STING (also referred to as mediator of IRF3 activation (MITA), plasma membrane tetraspanner (MPYS) or endoplasmic reticulum IFN stimulator (ERIS) has emerged as central for DNA-induced IFN-I activation (Ishikawa et al., 2009; Jin et al., 2008; Sun et al., 2009; Zhong et al., 2008). Other DNA sensors such as gamma-interferon-inducible protein 16 (IFI16) and DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (DDX41) are known to activate IFN-I in a STING-dependent manner (Unterholzner et al., 2010; Zhang et al., 2011).
The nucleotide binding domain and leucine-rich repeat-containing (NLR) protein family are intracellular sensors that regulate inflammatory responses (Eisenbarth and Flavell, 2009; Shaw et al., 2010). Most NLRs positively influence inflammatory responses, particularly the inflammasome NLRs. However emerging studies of gene-deficient mice have revealed that several NLRs negatively affect innate immune responses (Allen et al., 2011; Allen et al., 2012; Anand et al., 2012; Cui et al., 2010; Schneider et al., 2012; Xia et al., 2011; Zaki et al., 2011). Notably, we have previously shown that NLRC3 reduces LPS-induced nuclear factor kappa B (NF-κB) activation through inhibiting the adaptor protein TNF receptor associated factor 6 (TRAF6)(Schneider et al., 2012). However, the intersection of NLRs with DNA-sensing molecules has not been described. In this report, we find that NLRC3 deficiency also leads to increased innate immune response to intracellular DNA and c-di-GMP in both hematopoietic and non-hematopoietic cells. NLRC3 interacts with STING and the protein kinase TBK1, leading to reduced STING-TBK1 association, improper STING trafficking and decreased activation of innate immune cytokines. However this interference is separate from the previously described function of NLRC3 in impeding TRAF6 activation during LPS response. This work reveals the intersection of NLR with STING-mediated DNA sensing and unveils the multi-facet function of NLR family.
RESULTS
NLRC3 deficiency leads to elevated of DNA- and HSV-1-induced IFN-I and cytokine production
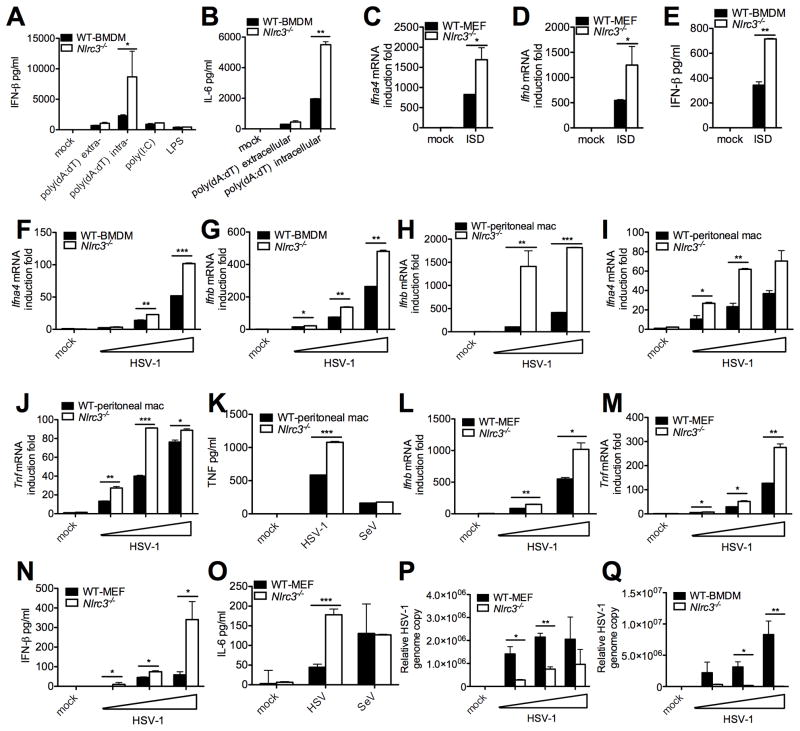
During our screening of NLR-deficient cells for new functions, we observed that IFN-I protein (Figure 1A) was higher in Nlrc3−/− bone marrow-derived macrophages (BMDM) than wildtype (WT) cells. This enhancement was observed in response to transfected poly(dA:dT) but not to extracellular poly(dA:dT), poly(I:C) or LPS (Figure 1A). Interleukin-6 (IL-6) protein was also higher in Nlrc3−/− BMDM in the presence of intracellular poly(dA:dT) but not extracellular poly(dA:dT) (Figure 1B). In addition, the effect of NLRC3 was extended to the interferon stimulatory DNA (ISD), which has been used to more specifically demonstrate cytoplasmic DNA sensing (Chiu et al., 2009; Stetson and Medzhitov, 2006). NLRC3 also negatively regulates IFN-I (Figure 1C–E) and IL-6 (Figure S1A) responses to ISD in mouse embryonic fibroblasts (MEFs). These results suggest that NLRC3 functions as a negative regulator of cytoplasmic DNA sensing. To determine its role in a more physiologic setting, Ifna4 and Ifnb response to a DNA virus, Herpes simplex virus 1 (HSV-1) was tested and found to be higher in Nlrc3−/− BMDMs (Figure 1F–G) and peritoneal macrophages (Figure 1H–I). The impact of NLRC3 is not limited to type I IFN because tumor necrosis factor (TNF) protein and transcript were similarly increased (Figure 1J–K). However, NLRC3 did not affect various responses to the Sendai RNA virus (SeV) (Figure 1K).
Figure 1. NLRC3 attenuates DNA- and HSV-1-induced cytokines.
(A–B) WT and Nlrc3−/− BMDMs were transfected (intracellular) or incubated (extracellular) with poly(dA:dT), transfected with poly(I:C) or treated with LPS. IFN-β and IL-6 were measured 16 hours after treatment. (C–D) Nlrc3+/+ and Nlrc3−/− MEFs were transfected with ISD and Ifna4 and Ifnb transcripts or (E) IFN-β measured 6 hours post-transfection. HSV-1 was used at MOI 0.05, 0.1 and 1 for panels F-J unless specified. (F–G) BMDMs were infected with HSV-1 and Ifna4 and Ifnb1 transcripts were measured. (H–J) Peritoneal macrophages were infected with HSV-1 and Ifna4, Ifnb1 and Tnf transcripts were measured. (K) Peritoneal macrophages were infected with HSV-1 (MOI 1) or SeV (80 HA unit/ml) and TNF assayed. (L–M) Primary MEFs were infected with HSV-1, and Ifnb1 and Tnf transcripts were measured. (N–O) Primary MEFs were infected with HSV-1 and IFNβ and IL-6 proteins were measured. (P) Primary MEFs or (Q) BMDMs were infected with HSV-1, genomic DNA was extracted and HSV relative genome copy number was determined by real-time PCR. All cytokines were determined by ELISA and transcripts determined by real-time PCR. All data are representative of at least three independent experiments.
See also Figure S1–S3
To assess if the suppressive role of NLRC3 on DNA-induced IFN-I and cytokine response is exhibited in non-immune cells, pairs of Nlrc3+/+ and Nlrc3−/− MEFs were isolated from siblings from heterozygous matings. Ifnb and Tnf transcripts were significantly increased in Nlrc3−/− MEFs in response to HSV-1 (Figure 1L–M), as were IFN-β and IL-6 proteins (Figure 1N–O). However, Nlrc3−/− MEFs responded normally to SeV (Figure 1O). The lack of an effect of NLRC3 on poly(I:C) or RNA virus-induced cytokine responses was more extensively analyzed. Wildtype and Nlrc3−/− cells responded similarly to Sendai virus, intracellular or extracellular poly(I:C), and vesicular stomatitis virus (VSV) under a variety of test conditions (Figure S2).
Due to concerns about variations in MEFs, we isolated a second pair of sibling-matched MEFs, and identical effects of Nlrc3 deletion on Ifna4 and Ifnb transcripts was observed, indicating that the suppressive effect of NLRC3 was not due to artificial differences in one specific pair of gene-sufficient and deficient MEFs (Figure S1B–D). Similar results were observed when IFNβ protein was measured. Consistent with increased cytokines which would be expected to reduce viral load, HSV-1 genomic DNA copy number was significantly reduced in Nlrc3−/− MEFs (Figure 1P) and BMDMs (Figure 1Q). However HSV-1-mediated cell death was not altered in Nlrc3−/− MEFs, indicating that the observed differences were not due to different cell viability (Figure S3). These data demonstrate that NLRC3 attenuates cytokine response to intracellular DNA without affecting cell viability.
NLRC3 deficiency causes increased IFN-β and IL-6 production in response to c-di-GMP and c-di-GMP
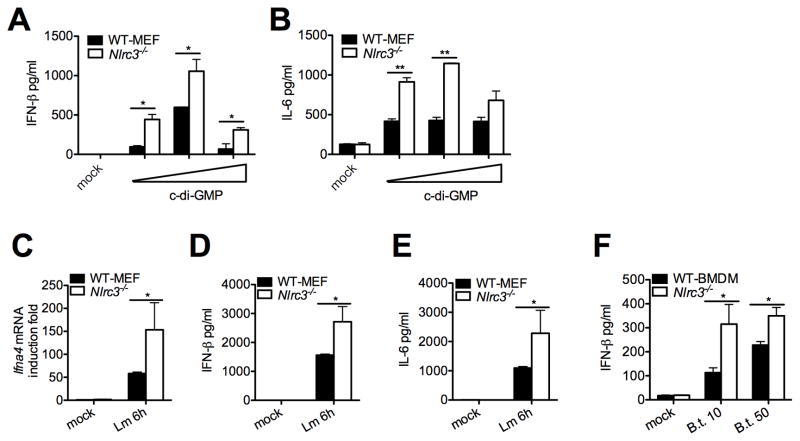
C-di-GMP, a small di-nucleotide monophosphate, is a second messenger of bacteria such as Listeria monocytogenes and Burkholderia thaildensis, and activates the IFN-I response via interaction with STING (Burdette et al., 2011; Jin et al., 2011; Sauer et al., 2011). Nlrc3−/− MEFs produced more IFN-β and IL-6 proteins in response to transfected c-di-GMP (Figure 2A–B). Additionally, Nlrc3−/− MEFs produced increased IFN-I and IL-6 in response to infection with c-di-GMP producing L. monocytogenes (Figure 2C–E). Increased IFNβ was also observed in Nlrc3−/− cells infected with another c-di-GMP producing bacteria, B. thaildensis (Figure 2F). Thus Nlrc3-deficiency leads to increased innate immune response to cytoplasmic DNA, c-di-GMP, and bacteria that produce c-di-GMP.
Figure 2. NLRC3 deficiency causes increased IFN-β and IL-6 production in response to c-di-GMP and c-di-GMP.
(A–B) C-di-GMP was transfected into primary MEFs, and IFNβ and IL-6 were measured 24 and 48 hours post-transfection respectively by ELISA. Primary MEFs were infected with L. monocytogenes at MOI of 10, and (C) Ifna4 transcripts or (D–E) IFNβ and IL-6 proteins were measured 6 hours post-infection. (F) BMDMs were infected with a second bacterial strain, B. thaildensis, and IFNβ was measured 6 hours post-infection. All data are representative of at least three independent experiments.
NLRC3 inhibits the STING-dependent pathway
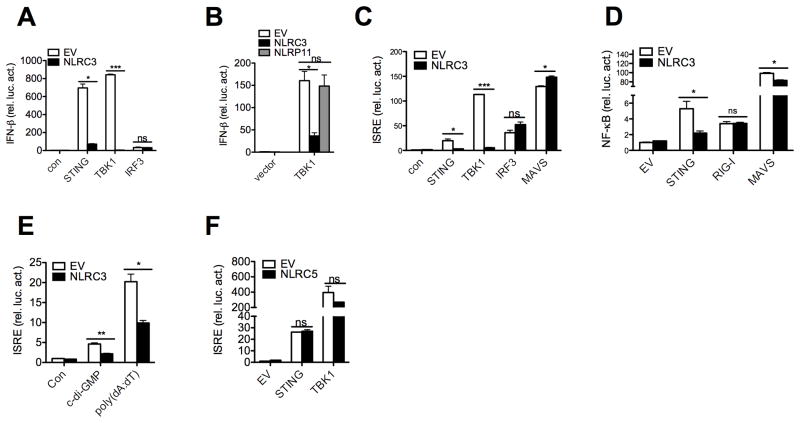
Cytoplasmic DNA and c-di-GMP induce IFN-I through the STING molecule, which led us to examine both functional and molecular interactions between NLRC3 and STING (Burdette et al., 2011; Huang et al., 2012; Ouyang et al., 2012; Shang et al., 2012; Shu et al., 2012). To investigate if NLRC3 affects the STING pathway, we examined the impact of NLRC3 on the activation of IFN-β promoter-luciferase by STING. This reporter assay was internally controlled by the co-transfection of a Renilla luciferase construct. NLRC3 inhibited IFN-β promoter activation by STING by 9.72 fold. STING operates by interaction and activation of its downstream kinase, TBK1 (Tanaka and Chen, 2012). NLRC3 dramatically reduced IFN-β promoter activation by TBK1. However NLRC3 had no direct effect on the downstream interferon regulatory transcription factor 3 (IRF3), indicating that NLRC3 likely functions at the upstream STING-TBK level (Figure 3A). As a specificity control, another NLR, NLRP11, did not reduce IFN-β promoter activation by TBK1 (Figure 3B). NLRC3 also inhibited a second promoter driven by the canonical interferon-stimulated responsive element (ISRE), which is known to be activated by STING and TBK1 (Ishikawa and Barber, 2008; Zhong et al., 2008) (Figure 3C). However NLRC3 had no effect on the activation of the ISRE promoter by mitochondrial antiviral signaling protein (MAVS) (also referred to as interferon-beta promoter stimulator 1 (IPS-1), virus-induced signaling adapter (VISA) and CARD adaptor inducing IFN-β (CARDIF)), which is important for RNA sensing, nor did it affect promoter activation by the downstream IRF3 (Figure 3C). In addition, NLRC3 inhibited NF-κB promoter activated by STING, and reduced MAVS activation slightly but did not affect retinoic acid-inducible gene 1 (RIG-I)(Figure 3D). We also observed that NLRC3 inhibited c-di-GMP and poly(dA:dT)-induced ISRE activation (Figure 3E). These experiments indicate that the predominant effect of NLRC3 is on the STING pathway. As an additional specificity control for NLR proteins, overexpression of NLRC5, which has been reported to inhibit various innate immune pathways when tested in an overexpression system (Cui et al., 2010) did not inhibit STING or TBK1-induced ISRE activation (Figure 3F). These experiments suggest that NLRC3 down-regulates innate immunity caused by STING and TBK1.
Figure 3. NLRC3 suppresses STING-TBK1-mediated IFN-β, ISRE and NF-κB reporter activation.
(A–D) HEK293T cells were transfected with the IFN-β, ISRE or NF-κB promoter reporter with the internal control Renilla luciferase reporter pLR-TK and empty vector or NLRC3 plasmid as indicated. Luciferase assays were performed 24 hours post transfection. (E) HEK293T cells were transfected with the ISRE promoter reporter with the internal control Renilla luciferase reporter pLR-TK. Stimulatory ligands were transfected 24 hours post plasmids transfection. Luciferase assays were performed 16 hours post ligand stimulation. (F) Same as A–D, except cells were transfected with empty vector or NLRC5. Data are representative of at least two independent experiments.
NLRC3 associates with STING and TBK1 and alters the STING-TBK1 interaction after stimulation
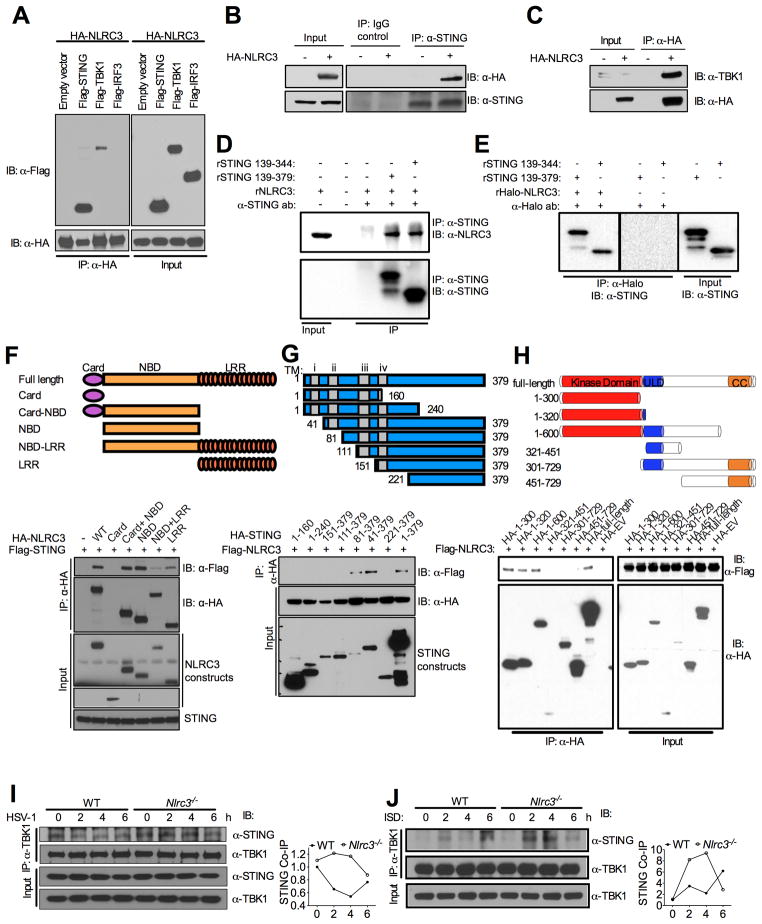
To explore the mechanism by which NLRC3 interferes with STING and TBK1 function, we tested if NLRC3 interacts with STING and/or TBK1. Transient transfection and co-immuno-precipitation followed by immunoblot showed that HA-NLRC3 strongly associated with Flag-STING and more modestly with Flag-TBK1, but not with Flag-IRF3 (Figure 4A), suggesting it interacts with the upstream STING-TBK complex but not with the downstream IRF3. This agrees with earlier data indicating that NLRC3 affected STING and TBK1 function but not IRF3 function (Figure 3A). Immunoblot of the input protein indicates that all of the proteins are expressed in readily detectable amounts (Figure 4A, right panel). In a more physiologic approach, HA-NLRC3 also associated with endogenous STING (Figure 4B, top lane) and TBK1 (Figure 4C) in a hemi-endogenous system, but not with IRF3 (data not shown). These experiments indicate that NLRC3 can associate with STING and TBK1.
Figure 4. NLRC3 associates with STING and TBK1 to interfere with their interaction.
(A) HEK293T cells were transfected with indicated plasmids and co-immunoprecipitation assay was performed 24 hours post-transfection. (B–C) HEK293T cells were transfected with HA-tagged NLRC3 or empty vector. Immunoprecipitation and immunoblots in A–C were performed with indicated antibodies. (D–E) Recombinant NLRC3 and STING in vitro binding assay was performed. Western blots were performed using the antibodies indicated. (F–H) HEK293T cells were transfected with indicated plasmids, constructs are shown at the top, the corresponding blots are shown below. The smaller peptides in (G) produced by 1–379 might be degradative products. (I–J) WT and Nlrc3−/− BMDMs were infected with HSV-1 (I) or stimulated with ISD (J), immunoprecipitation and western blot were performed with the indicated antibodies at the indicated time-points. Densitometric measurements of STING-TBK1 co-precipitation are shown to the right of each immunoblot. Data are representative of at least three independent experiments.
See also Figure S4–S5
To further investigate whether the association between NLRC3 and STING is direct, we prepared purified, recombinant full length NLRC3 and truncated STING protein (amino acid 139–379 and 139–344) and performed a protein pull-down assay. The results show NLRC3 and STING directly bind to each other in a reciprocal pull-down assay (Figure 4D–E).
Next, a domain mapping experiment was conducted with NLRC3 deletion constructs (Figure 4F). Full-length NLRC3, caspase activation and recruitment domain (CARD)-nucleotide binding domain (NBD) and NBD strongly associated with STING, while the CARD or leucine-rich repeats (LRR) domain alone either did not associate, or did not associate strongly, with STING (Figure 4F). The CARD domain alone did not express in high amounts, however a prolonged exposure did not reveal any interaction (not shown). The presence of LRR reduced the association of NBD with STING suggesting that the LRR is an inhibitory domain. These data indicate that the primary interaction domain in NLRC3 is the region that includes the NBD domain. A reciprocal experiment was performed to map the interaction domain in STING (Figure 4G). The first 240 residues of the N-terminus or the C-terminal 111–379 residues did not interact with NLRC3, while the C-terminal residues 81–379 interacted with NLRC3. This indicates that the STING c-terminus soluble tail and residues 81–111 are required for interaction with NLRC3. The C terminal residues 139–344 was shown to directly bind NLRC3 as demonstrated in Figure 4D–E, thus this region contains residues necessary and sufficient for association with NLRC3. However, a confounding issue with STING is that it is membrane bound and the transmembrane domain is required for STING localization to the ER. To examine this with the truncation mutants, we performed sub-cellular fractionation assay and showed that truncations 41–379 and 81–379 are membrane associated while 111–379 and 221–379 lose their membrane localization, indicating that residues 81–111 contained a sequence important for membrane-localization (Figure S4A). These results indicate that only the membrane-associated form of STING interacted with NLRC3. The interaction of STING with TBK1 produced the same results in that STING truncation mutant 81–379 but not 111–379 interacted with TBK1 (Figure S4B), which is also consistent with previous findings (Zhong et al., 2008). We also mapped the domains on TBK1 that bind to NLRC3. The result shows that N-terminus of TBK-1, which contained the kinase domain, is required for NLRC3 association (Figure 4H).
Upon DNA stimulation, the association of STING with TBK1 is essential to activate downstream signals (Ishikawa and Barber, 2008; Sun et al., 2009; Tanaka and Chen, 2012; Zhong et al., 2008). Thus we tested if the presence of NLRC3 interfered with the association of STING and TBK1. To pursue this in a physiologic system that did not involve overexpressed proteins, the association of STING and TBK1 was tested in Nlrc3−/− and control BMDMs in response to HSV-1 infection. The avoidance of over-expressed protein for this analysis is because overexpressed NLRs are prone to artifacts. The results show stronger STING-TBK1 association in Nlrc3−/− cells than WT controls 2–4 hours post-infection (Figure 4I, top lane; quantitation to the right). However, the association of STING-TBK1 was not enhanced by HSV-1. Because HSV-1 encodes a complex array of immune evasion and regulatory proteins that might obscure the outcome, we resort to ISD as a simplified system to examine responses to DNA without the confounding regulatory functions associated with HSV-1. The result shows enhanced STING-TBK1 association in WT cells after ISD stimulation, which was further potentiated in Nlrc3−/− cells 2–4 hours post-stimulation (Figure 4J, top lane; quantitation to the right). However at the six hour timepoint, STING-TBK1 interaction was more pronounced in WT cells. These results indicate that NLRC3 interfered with STING-TBK1 association at the 2–4 hr timepoint.
NLRC3 blocks STING trafficking
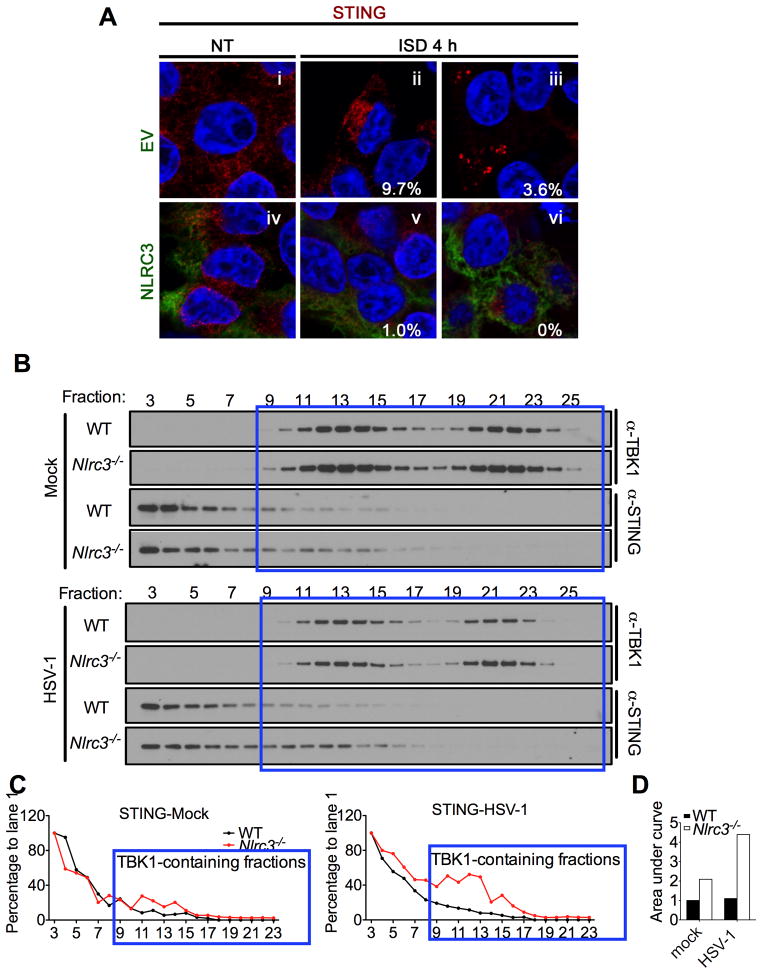
STING has been shown to traffic from the ER to a perinuclear/golgi location and to endoplasmic-associated puncta after DNA stimulation (Ishikawa et al., 2009; Saitoh et al., 2009). STING is reported to colocalize with TBK1 at these puncta, which represent the proposed platform for TBK1-mediated IRF3 activation. In cells transfected with an empty vector (EV), ISD caused STING to present in a perinuclear pattern (Figure 5A panel ii) followed by a punctated appearance (Figure 5A panel iii). However the presence of NLRC3 dramatically reduced the trafficking of STING to the perinuclear region (12-fold) (Figure 5A panel v) and completely prevented STING’s movement to puncta (Figure 5A panel vi). Thus NLRC3 reduced STING trafficking after ISD stimulation.
Figure 5. NLRC3 blocks STING trafficking.
(A) HEK293T cells were transfected with STING with an empty vector (EV) or with NLRC3 followed by stimulation with ISD. The percentages indicate the number of cells showing STING localized to the perinuclear region (ii and v) or forming puncta (iii and vi). A minimum of 100 cells were counted per group. (B) WT and Nlrc3−/− MEFs were mock-treated or infected with HSV-1 (MOI=1) for 4 hours. Cell lysate were subjected to FPLC, and fractions were collected and immunoblotted for STING and TBK1. The blue boxes indicate fractions that contained both TBK1 and STING. (C) Densitometric tracing of STING in WT (black line) and Nlrc3−/− cells (red) was performed using ImageJ. (D) The areas depicted in Fig. 5C were calculated using area under curve in Prism. The areas where STING-TBK1 were localized in the same fractions are depicted for uninfected WT and Nlrc3−/− MEFs, and HSV-1 infected WT and Nlrc3−/− MEFs. In each case, the value for WT cells was set as 1.0. Data are representative of two independent experiments.
To further pursue this finding using a biochemical approach, we examined if the absence of NLRC3 affected STING and TBK1 co-localization by fast protein liquid chromatography (FPLC). This was performed using cell lysates prepared from HSV-1 infected and un-infected WT and Nlrc3−/− primary MEFs. Similar to Figure 4I–J, this strategy did not involve any over-expressed proteins, thus providing a physiologically relevant condition to test the impact of NLRC3 on STING and TBK1. Whole cell lysates were fractionated by FPLC followed by immunoblotting of the fractions for STING and TBK1. In mock, uninfected wildtype controls (Figure 5B, top four rows, densitometry results in Figure 5C left panel, quantitation in Figure 5D), a majority of TBK1 and STING resided in different fractions and only a small portion of STING and TBK1 was detected in the same fractions. In uninfected Nlrc3−/− cells, 2.09-fold more STING and TBK1 were found in the same fractions compared to wildtype controls. Upon HSV-1 stimulation, 4.41-fold more STING and TBK1 were detected in the same fractions in Nlrc3−/− cells than controls (Figure 5B, bottom four rows, densitometry results in Figure 5C right panel, quantitation in Figure 5D). The cumulative data in this Figure are consistent with a model where NLRC3 interacts with STING and TBK1 to impede the interaction, since removal of NLRC3 by gene deletion led to more association of these two proteins. The inhibitory effect of NLRC3 on STING-TBK1 association was observed at the uninfected state, and became more pronounce upon HSV-1 infection.
Nlrc3−/− cells exhibit elevated signal transduction after HSV-1 infection
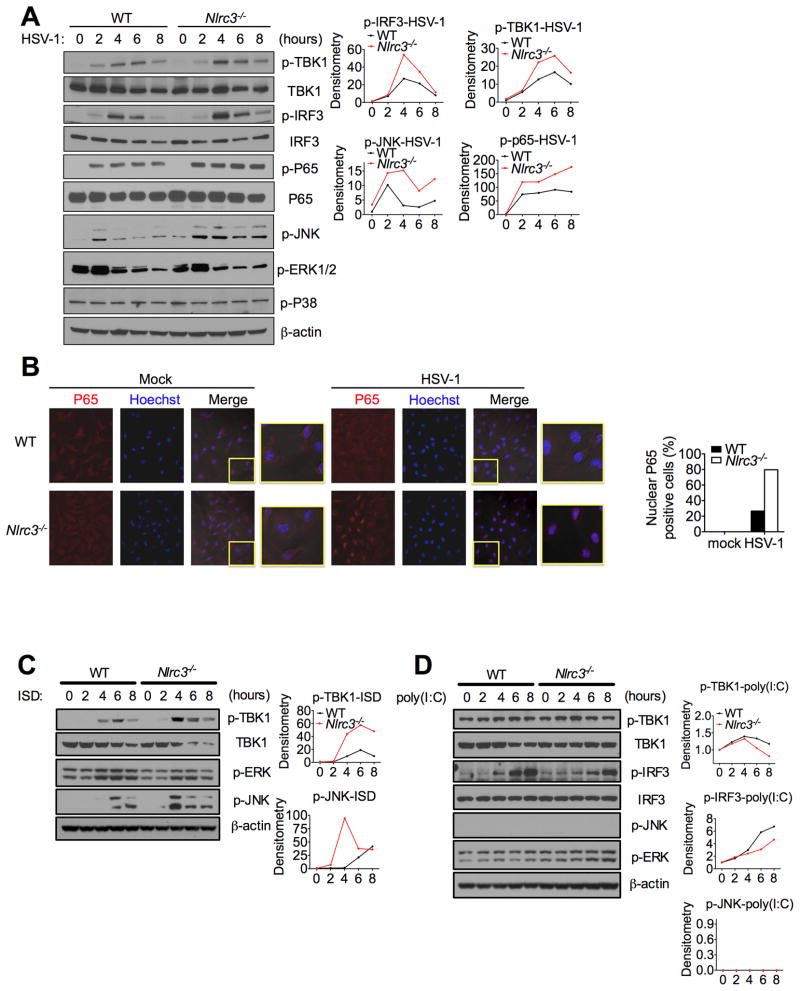
To examine for changes in downstream signals that are known to be activated by STING and TBK1, we examined for changes in protein phosphorylation that lie downstream of STING activation post-HSV-1 infection. Phosphorylation of TBK1, IRF3, p65 and JNK were induced 4–6 hours post-infection in wildtype controls (Figure 6A). The amount of phospho-TBK1 and phospho-IRF3 4–6 hours post-infection were higher in Nlrc3−/− than control MEFs, while the phosphorylation of JNK was enhanced throughout all of the timepoints measured in Nlrc3−/− cells. HSV-1 infection did not increase phosphorylation of ERK or p38, and NLRC3 did not alter these signals. HSV-1 infection induced p65 nuclear translocation was also visualized by confocal microscopy and was found to be significantly augmented in Nlrc3−/− cells (Figure 6B). Our earlier data indicate that NLRC3 affected the sensing of intracellular DNA. To study if downstream signals induced by DNA are affected by NLRC3, we assessed phosphorylation induced by ISD transfected into MEFs. Intracellular ISD caused increased phosphorylation of TBK1 and p-JNK in wildtype controls, and these responses, but not p-ERK, were further augmented in Nlrc3−/− cells, supporting the model that NLRC3 regulates signaling responses caused by intracellular DNA (Figure 6C). As a specificity control, intracellular poly(I:C) was transfected into cells, and it did not cause increases in the phosphorylation of multiple key pathways in Nlrc3−/− cells relative to controls (Figure 6D). These data suggest that NLRC3 is a negative regulator of innate immune signals generated upon HSV-1 infection and ISD stimulation. However, this function of NLRC3 is distinct from its regulation of NF-κB signaling induced by TRAF6 during an LPS response (Schneider et al., 2012), as TRAF6 was not required for HSV-1-induced IFN-I activation (Figure S5A–B). TRAF6 also did not associate with STING in co-IP assays (Figure S5C).
Figure 6. NLRC3 deficiency enhances immune signaling.
(A) Immunoblot of phosphorylated (p-) TBK1, IRF3, p65, JNK, ERK and p38 in lysates of WT and Nlrc3−/− MEFs infected with HSV-1 (MOI 1) for indicated time points. Densitometric measurements are depicted to the right. (B) BMDMs isolated from WT or Nlrc3−/− mice were infected with HSV-1 (MOI 1) for 2.5 hours. Cells were fixed and stained for endogenous p65 (red) or hoechst which stains the nucleus (blue). The merged purple color is indicative of nuclear p65. (C) Similar to (A), except cells were transfected with ISD. (D) Similar to (A) except cells were transfected with poly(I:C). Data are representative of at least two independent experiments.
NLRC3 deficiency augments host response to HSV-1 in vivo
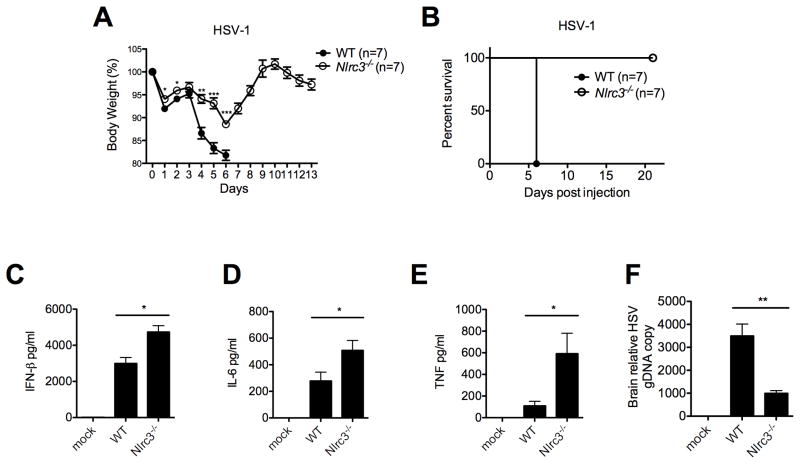
Next, to examine the in vivo importance of NLRC3, Nlrc3−/− and control mice were infected intravenously (i.v.) with HSV-1, and survival, weight change and morbidity were monitored (Figure 7A–B). Infected control mice exhibited significant lethargy and lack of movement (Movie S1), while infected Nlrc3−/− mice were active and mobile (Movie S2). Many control mice had to be euthanized 6–8 days post-infection when their body temperature was <32 °C, whereas 100% of similarly infected Nlrc3−/− mice showed a more modest temperature drop ranging from 34.2°C to 35.9 °C. Control mice also exhibited rapid weight loss after HSV-1 infection and had to be sacrificed due to a >20% weight loss. In contrast, Nlrc3−/− mice maximally lost up to 11% of body weight and recovered 100% of body weight by day 9. Sera from HSV-1-infected Nlrc3−/− mice showed increased IFNβ, TNF and IL-6 six hours post-infection when compared to controls (Figure 7C–E). HSV-1 genomic DNA copy number was significantly reduced in Nlrc3−/− mice (Figure 7F). In contrast, weight loss or serum IFNβ level in Nlrc3−/− mice was not significantly different from WT mice after infection with VSV (Figure S6). Thus NLRC3 attenuates physiologic host response to HSV-1, a DNA virus, but not VSV, a RNA virus.
Figure 7. Nlrc3−/− mice are more resistant to HSV-1 infection.
WT and Nlrc3−/− mice were infected i.v. with HSV-1 (2×107 pfu) and tested for (A) body weight, (B) survival rate, (C–E) serum cytokines (6 hours post-infection), and (F) HSV-1 genomic DNA (harvested 5 days post-infection and measured using real-time PCR analysis). Data are representative of at least three independent experiments.
DISCUSSION
This study identifies NLRC3 as a negative regulator of type I IFN and proinflammatory cytokine production triggered by cytoplasmic DNA and HSV-1. It also reduced the response caused by c-di-GMP, which provided us with the clue that linked NLRC3 to the STING pathway. Mechanistically, NLRC3 inhibits type I IFN promoter activation by STING and TBK, but not by the RIGI-MAV pathway. NLRC3 can directly interact with STING to reduce STING-TBK1 association, which is normally required for interferon induction. Furthermore, NLRC3 blocks ISD-induced STING trafficking to perinuclear and punctated regions, which is important for signal transduction downstream of STING (Ishikawa et al., 2009; Saitoh et al., 2009). Ablation of the Nlrc3 gene led to enhanced anti-viral cytokine production and viral clearance in culture. Most important, HSV-1-infected Nlrc3−/− mice exhibited greatly reduced morbidity, enhanced interferon and cytokine production and reduced viral load. This work demonstrates that NLR is a negative regulator of innate immunity triggered by the STING pathway.
There are multiple papers by several group that identify the negative regulatory functions of NLRs. Studies of gene deletion strains show that NLRX1 inhibits RNA virus and LPS induced cytokines in a cell-specific fashion (Allen et al., 2011; Xia et al., 2011), NLRP12 reduces canonical and non-canonical NF-κB (Allen et al., 2012; Zaki et al., 2011), NLRP6 impedes MAPK and NF-κB activation (Anand et al., 2012), and NLRC5 inhibits NF-κB and MAPK activation in some, but not all, gene deletion strains (Cui et al., 2010; Kumar et al., 2011). In addition, an in vitro study shows that NLRP4 reduces IFN production induced by nucleic acids (Cui et al., 2012). These findings indicate a broad function for NLRs in attenuating innate immune responses. However, none of the previously studied NLRs have been linked to the STING-mediated DNA-sensing pathway.
While our previous work showed a function of NLRC3 in reducing the activation of TRAF6 in response to LPS (Schneider et al., 2012), this report shows that intracellular DNA sensing during HSV-1 infection is independent of TRAF6. Furthermore, the present report also shows that NLRC3 does not affect IFN-I induction by LPS. Thus the impact of NLRC3 on LPS-induced cytokines such as TNF and IL-6 shown in our previous work (Schneider et al., 2012) likely occurs via a different path from IFN-I production caused by intracellular DNA. However, a recent paper indicates that TRAF6 is involved in cellular response to DNA and RNA (Konno et al., 2009). This may likely explain the more robust impact of NLRC3 in some experiments that used ISD instead of HSV-1. Further investigation is needed to fully assess the contribution of each pathway in response to nucleic acids in a NLRC3-dependent fashion.
The involvement of NLRC3 in two different responses (LPS-induced proinflammatory cytokines and intracellular DNA induced IFN-I response) is in line with other NLRs, which serve multiple functions. For example, NLRP3 and NLRP1 are involved in inflammasome function, but also in pyroptosis (Eisenbarth and Flavell, 2009; Kovarova et al., 2012; Masters et al., 2012). NOD2 activates NF-κB, MAV-induced type I IFN and autophagy (Cooney et al., 2010; Homer et al., 2010; Sabbah et al., 2009; Travassos et al., 2010). NLRP6 mediates inflammasome activation (Elinav et al., 2011), inhibits NF-κB activation (Anand et al., 2012) and promotes epithelium repair and renewal (Chen et al., 2011; Normand et al., 2011).
It is well-accepted that cytosolic DNA is immune stimulatory, and STING is the central adaptor protein for multiple intracellular DNA-sensing pathways (Ishikawa and Barber, 2008; Ishikawa et al., 2009; Jin et al., 2008; Sun et al., 2009; Zhong et al., 2008). Additionally, STING also mediates responses to RNA (Ishikawa et al., 2009; Sun et al., 2009; Zhong et al., 2008), cyclic dinucleotides (Jin et al., 2011; Sauer et al., 2011), cyclic GMP-AMP (Wu et al., 2013), bacterial (Gratz et al., 2011; Ishikawa and Barber, 2008; Ishikawa et al., 2009; Jin et al., 2011; Manzanillo et al., 2012; Watson et al., 2012), viral (Holm et al., 2012; Ishikawa and Barber, 2008; Ishikawa et al., 2009; Sun et al., 2009; Zhong et al., 2008), eukaryotic pathogen-derived (Sharma et al., 2011) and self DNA (Gall et al., 2012). It also intersects with other DNA sensors such as IFI16 and DDX41 (Unterholzner et al., 2010; Zhang et al., 2011). Thus it is significant that NLRC3 impacts this central DNA sensing molecule. In contrast to its intersection with STING-TBK1, we have not found a direct effect of NLRC3 on IFI16 or DXD41 (not shown). We also have not found a consistent function for NLRC3 in altering host response to intracellular poly(I:C) or the RNA viruses tested. While previous work has shown a consistent role for STING in host response to DNA virus, the results are less consistent for RNA virus. For example, IFNβ production and IRF3 nuclear translocation status are comparable between VSV-infected WT and Sting−/− MEFs and BMDMs, while Sting−/− dendritic cells produced less IFNα after VSV infection (Ishikawa et al., 2009). It is possible that an investigation of IFNα in dendritic cells might reveal a function for NLRC3 in response to VSV. It is also possible that NLRC3 inhibits RNA virus in a time- and dose-dependent fashion which was missed. Finally, NLRC3 only partially shuts off STING function, hence residual function might promote anti-RNA viral response.
The main finding of this work is that NLRC3 interacts with STING biochemically and functionally. It would follow that NLRC3 should reduce signals that lie downstream of STING activation. This is supported by the observation that Nlrc3−/− cells showed increased p-IRF3 (Figure 6A) and NF-κB phosphorylation/translocation (Figures 6A–B) after HSV-1 infection. The luciferase data showed that NLRC3 did not affect IRF3 activation of an ISRE promoter, hence the impact of NLRC3 is not directly on IRF3. We further showed that NLRC3 affected NF-κB activation by STING but not RIG-I or MAVS (Figure 3D), hence NLRC3 did not indiscriminately inhibit NF-κB activation. Instead it only inhibited NF-κB activation downstream of STING activation. Together, these data lead to the conclusion that NLRC3 negatively impacts STING, which then affects downstream events such as IRF3 and NF-κB activation.
In addition to pathogen-driven responses, DNA-dependent immune response triggered by self-DNA is associated with several diseases. As an example, DNase II deficient mice were unable to digest self-DNA from apoptotic cells and mice lacking DNase II died during embryonic development partly due to anemia (Kawane et al., 2001), which was rescued when STING was also removed (Ahn et al., 2012). This suggests that the cytosolic DNA-sensing pathway is involved in the pathology evoked by DNA sensing by STING.
In summary, our findings show the attenuation of DNA and c-di-GMP sensing by NLRC3 and reveal the intersection two pivotal pathways, NLR and STING in the control of innate immune responses. This work expands the function of NLRs to the important task of regulating host response elicited by intracellular DNA and c-di-GMP.
EXPERIMENTAL PROCEDURES
Cell culture
HEK293T cells were purchased from ATCC and maintained in DMEM (Gibco) supplemented with 10% fetal bovine serum, 1% penicillin and 100μg/ml streptomycin. Nlrc3+/+ and Nlrc3−/− MEFs were generated from 13.5-day embryos and maintained in the complete DMEM medium described above with 1 mM sodium pyruvate, 4 mM L-glutamine and non-essential amino acid. BMDMs were generated in the presence of L-929 conditional medium as previously described. All cells were grown in a 37°C incubator supplied with 5% CO2.
Reagents and antibodies
Poly (dA:dT) was purchased from InvivoGen, c-di-GMP from KeraFast, cytotoxicity detection kit from Roche, HSV-1 (KOS strain) from ATCC and propagated in Vero cells, Sendai virus (Cantell strain) from Charles River and VSV (Indiana strain) from ATCC and propagated in Vero cells. TNF and IL-6 ELISA kits were from BD Biosciences, mouse anti-IFN-β antibody was from Cosmo Bio, anti-IFN-β was from R&D system, anti-phospho-TBK1, TBK1, phospho-IRF3, IRF3, phospho-p65, p65, phospho-JNK, phospho-ERK, phospho-p38 were from Cell Signaling Technology, and anti-β-actin antibody was from Santa Cruz.
Experimental animals and and in vivo virus infection
The C57BL/6 Nlrc3−/− mice have been described (Schneider et al., 2012). C57BL/6 littermates were produced and used in selected experiments. Mice were treated in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill. Eight-10 weeks old mice were infected with HSV (2×107 pfu of viruses per mouse) or VSV (5×107 pfu of viruses per mouse) by intravenous injection. The temperature and weight of the mice were monitored accordingly. For cytokine studies, mice were sacrificed 6 hours post-infection and sera were collected through cardiac puncture. For HSV-1 genome copy number measurement, brains were harvested 5 days post-infection. For all experiments, wild-type and Nlrc3−/− mice were matched for age and sex.
Plasmids and molecular cloning
FLAG-tagged STING (originally cloned and named as MITA) full-length and domain truncation expression plasmids have been described (Zhong et al., 2008). HA-tagged NLRC3 full-length and domain truncation expression plasmids were cloned into pcDNA3.1.
Real time RT-PCR
Total RNA was extracted and assayed by Real-time PCR as described using SYBR green master mix or Tagman assay (Schneider et al., 2012). Primers used: mIFNβ: 5′-ATGAGTGGTGGTTGCAGGC-3′, 5′-ATGAGTGGTGGTTGCAGGC-3′; mIFN-α4: 5′-CCTGTGTGATGCAGGAACC-3′, 5′-TCACCTCCCAGGCACAGA-3′; actin: 5′-AGGGCTATGCTCTCCCTCAC-3′, 5′-CTCTCAGCTGTGGTGGTGAA-3′; mTNF: Mm00443258_m1, mIL-6: Mm00446190_m1.)
HSV-1 genomic DNA copy number measurement
Genomic DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen) according to manufacturer’s protocol. HSV-1 genomic DNA copy numbers were then determined by real-time PCR using HSV-1 specific primer: 5′-TGGGACACATGCCTTCTTGG-3′, 5′-ACCCTTAGTCAGACTCTGTTACTTACCC-3′.
Virus, bacteria infection and poly(dA:dT) stimulation
BMDMs or MEFs were plated in 24-well plates and grown to 80% confluence overnight. The cells were washed with PBS and infected with HSV-1 (KOS strain), SeV (Cantell strain) or VSV (Indiana strain) at the indicated multiplicity of infection (MOI) at 37 °C in serum-free DMEM for 1h. The cells were then washed with warm PBS and cultured in complete DMEM.
L. monocytogenes (43251) or B. thaildensis were grown to log phase and added to the cell cultures at a MOI of 10. After 30 min, gentamicin (50 μg/ml; Life Technologies) was added to the medium. The medium was changed after 1 hour of infection. Poly(dA:dT), poly(I:C) or ISD were added or transfected using lipofectamine 2000 (Invitrogen) at 4 μg/ml to BMDMs or MEFs for the indicated time. LPS were added into cell culture at 100 ng/ml. c-di-GMP were transfected at 1 μg/ml, 2 μg/ml or 4 μg/ml.
Co-immunoprecipitation
Procedures were done following the previous publication. (Schneider et al., 2012)
Transfection and Luciferase reporter analysis
HEK293T cells were seeded in 24-well plates at the density of 1.0×105 per well and transfected the following day by lipofectamine 2000 following the manufacturer’s instruction. 10 ng of pRT-TK Renilla luciferase reporter plasmid and 100 ng of firefly luciferase reporter plasmids were transfected together with indicated expression plasmids. Luciferase activity was measured 24 hours post-transfection using the Dual-Glo® Luciferase Assay System.
Recombinant protein purification and in vitro pulldown
Recombinant baculovirus expressing NLRC3 carrying an N-terminal Halo-Tag™ (HALO) and C-terminal hexahistidine tag (6XHIS) was generated as described by Mo et al. (Mo et al., 2012). For assays using immobilized Halo ligand capture (HaloLink™, Promega) for protein capture, recombinant HALO-NLRC3-6XHIS was partially purified using immobilized Ni2+ chromatography (PrepEase ® Ni-TED, USB). HaloLink™ resin or HaloLink™ resin incubated with 1 μg of HALO-NLRC3-6XHIS was added to IP buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM β-mercaptoethanol and 0.1% CHAPS and 1μg of recombinant STING protein (Ouyang et al., 2012). After 4 hours rotating at 4°C, the HALO-resin associated proteins were isolated by centrifugation for 30s at 13k × g, the resins were washed 3 times with IP buffer supplemented with 300 mM NaCl. The resin associated STING proteins were liberated with 1x SDS-PAGE loading buffer and analyzed using immunoblot analysis. For assays using anti-STING antibody co-immunoprecipitation tag-free NLRC3 was purified to near homogeneity using tandem immobilized Ni2+ chromatography and HALO-ligand affinity chromatography followed by TEV–protease treatment to remove both the HALO and 6XHIS as previously described. Purified NLRC3 (1 μg) and purified STING proteins (1 μg) were combined in IP buffer and incubated with anti-STING antibody while rotating at 4° C overnight. The STING protein and associated NLRC3 were captured using protein A paramagnetic beads (Miltenyi) according to the manufacturers protocols and assayed by immunoblot for STING (Cell Signaling) and NLRC3 (Sigma) as described.
Fast protein liquid chromatography (FPLC)
Nlrc3+/+ or Nlrc3−/− MEFs were harvested after indicated treatment and lysed in hypotonic buffer (25 mM Tris-HCL, pH 7.5, 2 mM DTT) with a dounce homogenizer on ice. The lysate was then centrifuged at 16,000g for 10 minutes. The precipitate was lysed in CHAPS buffer (50 mM Tris-HCL, pH 7.5, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, 5% glycerol and 0.1% CHAPS) with a dounce homogenizer on ice and then centrifuged at 16,000 g for 10 minutes. The protein concentrations of the supernatant were determined by Bio-Rad Protein Assay. Equal amount of protein from Nlrc3+/+ or Nlrc3−/− cells were subjected to size exclusion chromatography (Superose 6). The indicated fractions were subjected to western blot analysis with the indicated antibodies. Densitometry for western blot was performed using ImageJ.
Confocal microscopy
Nlrc3+/+ or Nlrc3−/− BMDMs (4 × 105) were seeded onto coverslips in 24-well dishes and were grown overnight before inoculation with HSV-1 at the MOI of 1 for 3 hours. Cells were fixed with 4% paraformaldehyde (PFA), followed by ice-cold methanol. Cells were stained with anti-p65 (D14E12; Cell Signaling) and Alexa Fluor 546-conjugated anti-rabbit antibody (A-11035; Invitrogen) and then were counterstained for nucleic acids with Hoechst 33342. In experiments using overexpressed protein, HEK293T cells (2.5 × 105) were reverse transfected using Lipofectamine 2000 with STING-HA (100 μg) and NLRC3-FLAG (375 μg) directly onto poly-L-lysine coated coverslips. After 24 h, cells were transfected with ISD (4μg/ml) for 4 h, followed by PFA fixation. Cells were stained with anti-HA (3724S; Cell Signaling) and anti-FLAG (F1804, Sigma) followed with AF546-conjugated anti-rabbit antibody and AF488-conjugated anti-mouse IgG1 antibody (A-11035 and A11029; Invitrogen), and then counterstained for nucleic acids with Hoechst 33342. Cells were analyzed with a Zeiss LSM 710 laser-scanning confocal microscope.
Statistical Analysis
Statistical analysis was carried out with Prism 5.0 for Macintosh. All data are shown as mean ± s.d. The mean values for biochemical data from each group were compared by Student’s t-test. Comparisons between multiple time points were analyzed by repeated-measurements analysis of variance with Bonferroni post-tests. In all tests, P-values of less than 0.05 were considered statistically significant. *P< 0.05, **P<0.01, ***P<0.001.
Supplementary Material
HIGHLIGHTS.
NLRC3 reduced IFN-I induced by intracellular DNA, HSV-1 and c-di-GMP
NLRC3 directly associates with STING and can block STING trafficking to puncta.
NLRC3 also associates with TBK1 and interferes with STING-TBK1 interaction.
Nlrc3−/− mice are more resistant to HSV-1 infection and exhibit elevated cytokines.
Acknowledgments
Supported by NIH grants CA156330, U54 AI057157, R37-AI029564 and P01DK094779 (J.P.-Y.T); AI088255 (J.A.D) and DE-018281 (B.D. and J.P-Y.T); Burroughs Wellcome Fund Career Award for Medical Scientists (J.A.D); MOST grants 2014CB910400, 2013CB911103 and NSFC grants 31200559, 31330019 (S. O. and Z-J. L.). We thank Dr. Tak W. Mak for sharing Traf6+/+, Traf6+/− and Traf6−/− cells, Drs. Albert Baldwin and Lishan Su for materials, Dr. Edward Miao for Burkholderia thaildensis, Dr. Rui Chen for support and discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci U S A. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, Gris D, Roney KE, Zimmermann AG, Bowzard JB, et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-κB signaling pathways. Immunity. 2011;34:854–865. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, Wilson JE, Schneider M, Lich JD, Roberts RA, Arthur JC, Woodford RM, Davis BK, Uronis JM, Herfarth HH, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity. 2012;36:742–754. doi: 10.1016/j.immuni.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand PK, Malireddi RKS, Lukens JR, Vogel P, Bertin J, Lamkanfi M, Kanneganti TD. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012;488:389–393. doi: 10.1038/nature11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Liu M, Wang F, Bertin J, Nunez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol. 2011;186:7187–7194. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nature Medicine. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- Cui J, Li Y, Zhu L, Liu D, Songyang Z, Wang HY, Wang RF. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol. 2012;13:387–395. doi: 10.1038/ni.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, Shen P, Zheng S, Chen ZJ, Wang RF. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth SC, Flavell RA. Innate instruction of adaptive immunity revisited: the inflammasome. EMBO Mol Med. 2009;1:92–98. doi: 10.1002/emmm.200900014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A, Treuting P, Elkon KB, Loo YM, Gale M, Jr, Barber GN, Stetson DB. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz N, Hartweger H, Matt U, Kratochvill F, Janos M, Sigel S, Drobits B, Li XD, Knapp S, Kovarik P. Type I interferon production induced by Streptococcus pyogenes-derived nucleic acids is required for host protection. PLoS pathogens. 2011;7:e1001345. doi: 10.1371/journal.ppat.1001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moeller HB, Gonzalez-Dosal R, Rasmussen SB, Christensen MH, Yarovinsky TO, et al. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol. 2012;13:737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. 2010;139:1630–1641. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol. 2011;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawane K, Fukuyama H, Kondoh G, Takeda J, Ohsawa Y, Uchiyama Y, Nagata S. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science. 2001;292:1546–1549. doi: 10.1126/science.292.5521.1546. [DOI] [PubMed] [Google Scholar]
- Konno H, Yamamoto T, Yamazaki K, Gohda J, Akiyama T, Semba K, Goto H, Kato A, Yujiri T, Imai T, et al. TRAF6 establishes innate immune responses by activating NF-kappaB and IRF7 upon sensing cytosolic viral RNA and DNA. PLoS One. 2009;4:e5674. doi: 10.1371/journal.pone.0005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarova M, Hesker PR, Jania L, Nguyen M, Snouwaert JN, Xiang Z, Lommatzsch SE, Huang MT, Ting JP, Koller BH. NLRP1-dependent pyroptosis leads to acute lung injury and morbidity in mice. J Immunol. 2012;189:2006–2016. doi: 10.4049/jimmunol.1201065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Pandey S, Zou J, Kumagai Y, Takahashi K, Akira S, Kawai T. NLRC5 deficiency does not influence cytokine induction by virus and bacteria infections. J Immunol. 2011;186:994–1000. doi: 10.4049/jimmunol.1002094. [DOI] [PubMed] [Google Scholar]
- Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters SL, Gerlic M, Metcalf D, Preston S, Pellegrini M, O’Donnell JA, McArthur K, Baldwin TM, Chevrier S, Nowell CJ, et al. NLRP1 inflammasome activation induces pyroptosis of hematopoietic progenitor cells. Immunity. 2012;37:1009–1023. doi: 10.1016/j.immuni.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J, Boyle JP, Howard CB, Monie TP, Davis BK, Duncan JA. Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J Biol Chem. 2012;287:23057–23067. doi: 10.1074/jbc.M112.344283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand S, Delanoye-Crespin A, Bressenot A, Huot L, Grandjean T, Peyrin-Biroulet L, Lemoine Y, Hot D, Chamaillard M. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc Natl Acad Sci U S A. 2011;108:9601–9606. doi: 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. Structural Analysis of the STING Adaptor Protein Reveals a Hydrophobic Dimer Interface and Mode of Cyclic di-GMP Binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Zimmermann AG, Roberts RA, Zhang L, Swanson KV, Wen H, Davis BK, Allen IC, Holl EK, Ye Z, et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-kappaB. Nat Immunol. 2012;13:823–831. doi: 10.1038/ni.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, Gu L. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- Sharma S, DeOliveira RB, Kalantari P, Parroche P, Goutagny N, Jiang Z, Chan J, Bartholomeu DC, Lauw F, Hall JP, et al. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 2011;35:194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Lamkanfi M, Kanneganti TD. NOD-like receptor (NLR) signaling beyond the inflammasome. Eur J Immunol. 2010;40:624–627. doi: 10.1002/eji.200940211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol Rev. 2007;220:214–224. doi: 10.1111/j.1600-065X.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Science Signaling. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Cui J, Wang HY, Zhu L, Matsueda S, Wang Q, Yang X, Hong J, Songyang Z, Chen ZJ, Wang RF. NLRX1 negatively regulates TLR-induced NF-kappaB signaling by targeting TRAF6 and IKK. Immunity. 2011;34:843–853. doi: 10.1016/j.immuni.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Vogel P, Malireddi RK, Body-Malapel M, Anand PK, Bertin J, Green DR, Lamkanfi M, Kanneganti TD. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 2011;20:649–660. doi: 10.1016/j.ccr.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.