Abstract
BACKGROUND
Microarray technology is becoming a powerful tool for diagnostic, therapeutic, and prognostic applications. There is at present no consensus regarding the optimal technique to isolate nucleic acids from blood leukocyte populations for subsequent expression analyses. Current collection and processing techniques pose significant challenges in the clinical setting. Here, we report the clinical validation of a novel microfluidic leukocyte nucleic acid isolation technique for gene expression analysis from critically ill, hospitalized patients that can be readily used on small volumes of blood.
METHODS
We processed whole blood from hospitalized patients after burn injury and severe blunt trauma according to the microfluidic and standard macroscale leukocyte isolation protocol. Side-by-side comparison of RNA quantity, quality, and genome-wide expression patterns was used to clinically validate the microfluidic technique.
RESULTS
When the microfluidic protocol was used for processing, sufficient amounts of total RNA were obtained for genome-wide expression analysis from 0.5 mL whole blood. We found that the leukocyte expression patterns from samples processed using the 2 protocols were concordant, and there was less variability introduced as a result of harvesting method than there existed between individuals.
CONCLUSIONS
The novel microfluidic approach achieves leukocyte isolation in <25 min, and the quality of nucleic acids and genome expression analysis is equivalent to or surpasses that obtained from macroscale approaches. Microfluidics can significantly improve the isolation of blood leukocytes for genomic analyses in the clinical setting.
As genome-wide expression analyses are becoming increasingly common in clinical medicine for diagnostic, therapeutic, and prognostic application in patients with trauma, cancer, and other diseases, assuring adequate data quality is taking on progressively more importance (1-4). Translating the sensitive and complex gene expression technologies from research laboratory to the clinical arena requires particular attention to the protocols and methodologies for all individual steps between blood sample collection, RNA processing, and nucleic acid hybridization. Although considerable attention has been devoted to developing standardized analytical protocols for nucleic acid labeling and microarray hybridization (5, 6), much less attention has been devoted to processing of the blood sample and separation of cells of interest. Currently, most blood sample processing in clinical settings is done manually, requires skilled personnel, and is prone to variability due to complexity and time constraints in the hospital environment.
Improved technologies are required that are fast, reproducible, and do not activate or perturb the cell populations. Among emerging technologies, microfluidics and “lab-on-a-chip” devices have the potential to spur the development of protocols and affordable instruments for performing specific blood analyses with minimal perturbation of individual cell populations (7). Previously, we reported a microfluidic system for the enrichment and isolation of leukocytes from whole blood for genomic analysis (8, 9). The device, consisting of a serpentine channel, enables rapid depletion of erythrocytes from whole blood via continuous deionized water lysis while enriched leukocytes are readily recovered for downstream genomic analyses. We tested this device with whole blood obtained from healthy volunteers or normal whole blood treated ex vivo with a bacterial toxin (9). However, the critical clinical validation of the microfluidic technology in patient samples collected at the point-of-care has not been addressed to date.
In this study, we tested the hypothesis that the newly developed microfluidic technology, when applied for cell separation by skilled medical personnel with no previous experience with microfluidic devices, can generate samples of equal quality to those of traditional cell separation technologies. We provided 4 h of training on the use of the microfluidic device to research nurses and other personnel with previous experience in macroscale leukocyte isolation techniques. The nurses, following both the microfluidic and the macroscale protocols, processed whole blood samples from critically ill patients with severe burn or trauma injuries. The extracted RNA from these samples was processed further in parallel for microarray analysis. We show that the expression patterns from samples processed using the microfluidic and the macroscale buffy-coat procedures are similar, and there is less variability between the 2 methods than there is between individuals. To our knowledge, this is the first successful application of microfluidic techniques for processing clinical samples obtained from critically ill, hospitalized patients, and the first validation of a microfluidic technique in a multiple hospital setting.
Materials and Methods
STUDY DESIGN
We performed side-by-side comparisons of RNA quantity, quality, and genome-wide expression patterns obtained from whole blood leukocytes isolated by either the microfluidic or standard macroscale leukocyte isolation protocol (Fig. 1C) on 4 groups of participants: healthy controls (n = 4); healthy controls receiving an intravenous administration of reference E. coli endotoxin (2 ng/kg lipopolysaccharide) (n = 3); hospitalized patients with burn injury in excess of 30% total body surface area (n = 5); and hospitalized patients with severe blunt trauma (n = 5).
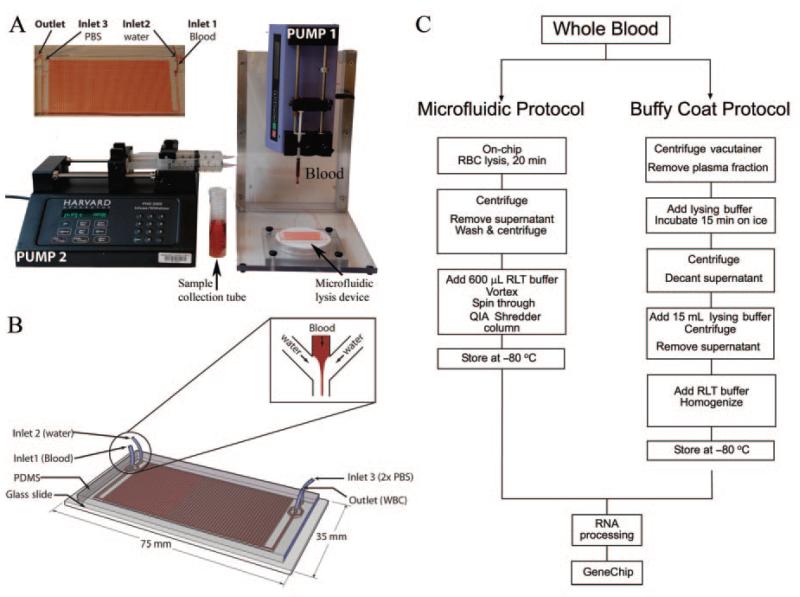
Fig. 1. Microfluidic cassette and setup for blood processing and overview of the experimental protocol.
The setup consists of 2 syringe pumps connected to the microfluidic device (A). The upper left corner shows an enlarged picture of the device during experiment. One pump drives the blood at 20 μL/min through inlet 1 and a second pump deionized water (lysing buffer) and 2× PBS (restoring buffer) at 600 μL/min through inlets 2 and 3, respectively. The enriched leukocyte population and debris of lysed RBCs is collected at the outlet. The process is automated and takes about 25 min to process 500 μL whole blood. Schematic device design is shown in B. The channel dimension is 500 by 200 μm with a total internal volume of 70 μL. The enlarged box shows details at the inlet: deionized water branches into 2 streams, 1 on either side of the entry stream of whole blood in the channel. This focuses the whole blood into a narrow stream flanked on both sides by deionized water. Overview of the major experimental steps is illustrated in C. The microfluidic isolation protocol is fairly automated and takes approximately 35 min, whereas standard buffy-coat isolation can take up to 2 h.
The single-use microfluidic chips were manufactured at the BioMEMS resource center (Boston, MA) as described (9). These were assembled and shipped to 4 clinical sites (Gainesville, FL; New Brunswick, NJ; Galveston, TX; Seattle WA). Blood samples were processed at each clinical site and isolated cells were lysed in RNA isolation buffer (RLT buffer™; Qiagen) and frozen at −80 °C. The frozen RNA samples were shipped to the sample coordination site at the University of Florida. Depending on the experiment, samples were either processed locally or sent to Stanford University for processing of RNA and hybridization to HU133 plus v2 GeneChip™.
MICROFLUIDIC DEVICE FABRICATION
The device (Fig. 1) and the experimental setup have been described (9). Briefly, the microfluidic device was fabricated by casting polydimethylsiloxane (PDMS8; Dow Corning) polymer on a resist-structured silicon wafer according to standard soft lithographic techniques. Structures in SU-8 resist (MicroChem) were produced according to the supplier’s recipe using standard MEMS technology. The PDMS was mixed (10:1, wt/wt) with a cross linker, poured on top of the silicon wafer, degassed, and cured at 65 °C for 6 h. The PDMS with the replicated channels was peeled off from the master, and channel access holes were punched with a 22-gauge needle. The PDMS replica was bonded to a glass slide via oxygen plasma. Access tubing (Tygon; Small Parts) of slightly larger diameter was press-fitted into the holes.
PARTICIPANTS
We obtained permission to obtain venous blood from healthy volunteers and patients from the institutional review boards of the participating institutions. Informed consent was obtained from 4 healthy individuals at University of Florida, 5 severely injured trauma patients at Washington University, 5 severely injured burn patients at University of Texas, and 3 healthy volunteers to be administered endotoxin at UMDNJ-Robert Wood Johnson Medical School.
BLOOD PROCESSING
Venous blood (7 mL) was collected into Vacutainer™ tubes containing EDTA (Becton Dickinson), and 0.5 mL blood was collected directly from the Vacutainer into a 1-mL syringe (Becton Dickinson) and processed according to the microfluidic protocol, whereas the remaining 6.5 mL blood was processed in parallel according to the macroscale protocol. Three healthy volunteers selected for the endotoxin study were intravenously administered NIH Clinical Center Reference Endotoxin (CC-RE-Lot 2) at a dose of 2 ng/kg body weight over a 5-min period. Blood samples were collected before endotoxin infusion (0 h) and at 3, 6, and 24 h after infusion and processed according to the microfluidic and standard bulk method in parallel. The patients’ clinical characteristics are shown in Table S1 in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol54/issue5.
MICROFLUIDIC BLOOD PROCESSING
The experimental setup for the microfluidic technique consists of the microfluidic device and 2 syringe pumps (Fig. 1). The device is first primed by filling with 1× PBS and connected to respective syringes without trapping air bubbles. One syringe pump (Harvard Pump 11 Plus; Harvard Apparatus) drives the whole blood at 20 μL/min through 1 inlet, and the second pump (Harvard PHD 22/2000 Syringe Pump; Harvard Apparatus) drives deionized water and 2× PBS (double the regular PBS concentration) at 600 μL/min through the other 2 inlets. The deionized water branches into 2 streams, 1 on either side of the entry stream of whole blood to focus it into a narrow stream. We optimized the flow rates such that each cell in the blood was exposed to the lysing solution for approximately 10 s before restoring the osmolarity of blood using equal volumes of 2× PBS. The enriched leukocyte population and debris of lysed erythrocytes were collected at the outlet in 50-mL Eppendorf tubes, and cell debris were removed in the supernatant by low-speed centrifugation. The cell pellets were washed in 1 mL PBS, immediately lysed in 600 μL RLT buffer (Qiagen) with 0.1% vol/vol β-mercaptoethanol, and spun through a QIAShreder column (Qiagen). The samples were immediately stored at −80 °C until shipped to the sample collection and coordination site in Florida.
CONVENTIONAL BUFFY-COAT BLOOD PROCESSING
The standard buffy-coat leukocyte isolation protocol has been described (1, 10, 11). Briefly, we centrifuged the blood sample at 400g for 10 min at room temperature and transferred the buffy-coat layer to 50-mL conical tubes (Corning Labs). We lysed residual erythrocytes with 45 mL lysing buffer (EL Buffer; Qiagen) for 15 min at 4 °C and collected the leukocyte-rich fraction by centrifugation at 400g for 10 min at 4 °C. The leukocyte pellets were washed in ice-cold PBS, immediately lysed with 2 mL RLT Buffer (RNeasy® Midi Kit; Qiagen), and homogenized by aspirating and forcing the sample repeatedly through a 10-mL syringe (Monoject) with an 18-gauge needle (Becton Dickinson). The sample was immediately stored at −80 °C until shipped to the sample collection and coordination site in Florida.
LEUKOCYTE PHENOTYPING
We took 50 μL of the cell suspension for flow cytometry analysis before the cell-lysis step. Different subpopulations were assessed by staining with fluorescein isothiocyanate–anti-CD66b, phycoerythrin–anti-CD2, and PerCP-CY5.5–anti-CD33 antibodies (BD Biosciences). Stained cells were washed in PBS without calcium or magnesium, plus 1% BSA and 0.1% sodium azide, then pelleted, resuspended in PBS, and analyzed on a FACSCalibur™ flow cytometer (BD Biosciences), using CellQuest™ software (BD Pharmingen). We used 3-color analyses to identify granulocytes as side-scatter high, CD66b high; monocytes as side-scatter intermediate, CD33 high; and T lymphocytes as side-scatter low, CD2 high.
RNA ISOLATION, cRNA SYNTHESIS, AND MICROARRAY HYBRIDIZATION
We extracted total cellular RNA using a commercial RNA purification kit (RNeasy; Qiagen) and amplified biotinylated cRNA (12) from 1 μg total RNA. cRNA was transcribed in vitro incorporating biotinylated nucleotides using the Affymetrix High Yield RNA Transcript T7 Kit (Affymetrix), and the product was hybridized onto an Affymetrix HU133 plus v.2 GeneChip™ for 16 h at 45 °C in an Affymetrix hybridization oven. Arrays were stained and washed using an Affymetrix fluidics station and EukGEWSv4 Affymetrix protocol and analyzed on an Affymetrix scanner. We determined quality and purity of total RNA and biotinylated-synthesized cRNA by UV spectroscopy and capillary electrophoresis using the Bioanalyzer 2001 system (Agilent). An absorbance ratio (A260/A280) of 1.8 to 2.1 is indicative of highly purified RNA.
MICROARRAY DATA ANALYSIS
We performed low- and high-level statistics using dChip. Expression was modeled using Perfect Match Only dChip algorithms. Affymetrix Microarray Suite GeneChip Operating Software identified probe sets whose signal intensities were at or below background as “absent.” Absent probe sets were discarded from the high-level analysis on all arrays.
We applied an unsupervised analysis to assess similarities and gene expression profile differences. We performed unsupervised analyses on the expression matrix by selecting probe sets whose hybridization signal varied across the data set with a CV (SD/mean ratio) >1.0. We then subjected the mean-centered and variance-normalized levels to hierarchical cluster analyses using the dChip algorithm. We calculated Pearson’s correlation coefficient to assess similarity of gene expression among probe sets, with the results displayed as a dendrogram on top of the cluster image. Significance analysis of microarrays identified probe sets whose intensities differed between the 2 groups at a false discovery rate of 1% (B = 0.01).
Results
We compared microfluidic and conventional macroscale isolation techniques by evaluating the correlation between global gene expression patterns in total leukocyte samples. First, we tested and characterized the overall experimental design using blood from healthy individuals. To confirm that results from healthy individuals could be extrapolated to patients, we evaluated the 2 methods using a human in vivo endotoxemia model. Finally, we isolated leukocyte RNA from critically ill burn and trauma patients at 2 clinical institutions. We used clustering analysis to describe the overall changes in gene expression between individuals using the 2 leukocyte isolation protocols.
LEUKOCYTE COMPOSITION ISOLATED FROM WHOLE BLOOD
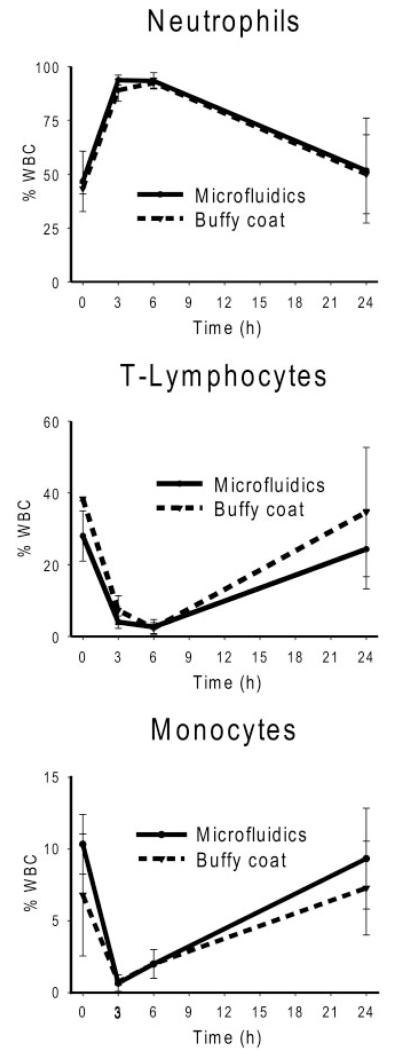
Complete lysis of erythrocytes was achieved within 10 s, and close to 100% of the leukocytes were recovered in the microfluidic device. We have earlier demonstrated that the microfluidic system is better than macroscale protocols for total and differential leukocyte recovery, owing to more precise control of time of exposure to the lysing conditions (9). In this study, we further examined the differential leukocyte composition by flow cytometry to evaluate the ability of the microfluidic isolation protocol to identify phenotypic changes in blood leukocyte populations secondary to in vivo endotoxin administration. Whole blood leukocytes were isolated according to the 2 protocols from 3 volunteers immediately before and at 3, 6, and 24 h after intravenous administration of bacterial endotoxin. Fig. 2 shows differential results of the leukocyte subpopulations during the 24-h time course. Comparison of the differential profiles of the subpopulations reveals that the 2 protocols generated similar results. As reported (13), endotoxin administration produces a rapid and transient leukopenia, followed by a release of mature and immature neutrophils from the bone marrow at 3 h and a sustained T lymphopenia and monocytopenia by 6 h. After 24 h, the differential subpopulation cells return to initial values, indicating that the host response to the bacterial toxin has restored homeostasis.
Fig. 2. Differential leukocyte recovery: side-by-side comparison of the microfluidic and macroscale protocols in whole blood obtained from human volunteers administered endotoxin.
Three healthy volunteers were intravenously administered endotoxin (2 ng/kg), and samples were drawn at baseline (0 h) and 3, 6, and 24 h. The cell counts generated from the 2 protocols were similar, and the responses to the endotoxin occurred in a consistent temporal sequence, although the magnitude varied among individuals.
RNA YIELD AND QUALITY
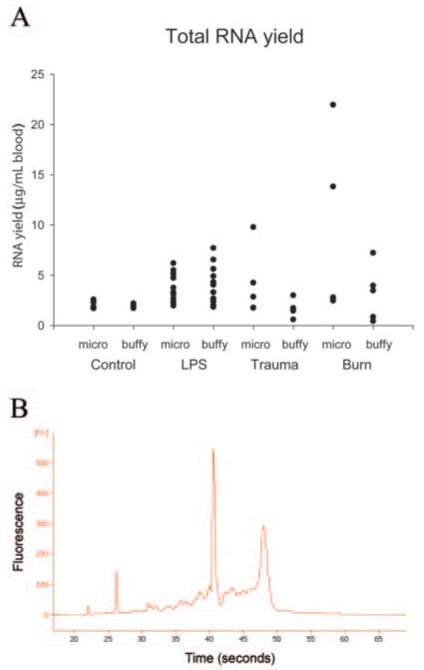
We used RNeasy Qiagen RNA purification kits to extract total RNA from both the microfluidics cassettes and the macroscale isolation procedure. We generated 26 RNA samples from each of the 2 protocols: healthy controls (n = 4) and healthy controls who received the in vivo administration of endotoxin (n = 3) at 4 time points. Results of RNA yield and quality are shown in Fig. 3. Although there were some variations in the amount of RNA recovered among the protocols and individuals, all samples provided sufficient RNA for subsequent microarray analysis. The quality of isolated RNA was acceptable for all samples generated by both protocols, as confirmed by capillary electrophoresis using the Agilent 2100 Bioanalyzer system. Spectrometric analysis (A260/A280 ratio) of the 12 samples from the healthy volunteers administered endotoxin averaged 1.8 (SD 0.1) for both methods, indicating that the samples were free of contaminating protein.
Fig. 3. RNA yield and quality.
The samples were analyzed on an Agilent Bioanalyzer 2100. (A), Total RNA yield for all participants from the 2 protocols. Individual samples are shown with solid symbols. (B), Capillary electrophoresis of an RNA sample purified from whole blood using the microfluidic protocol. The 18S and 28S peaks are visible at 42 and 47 s, respectively.
GENE EXPRESSION ANALYSIS
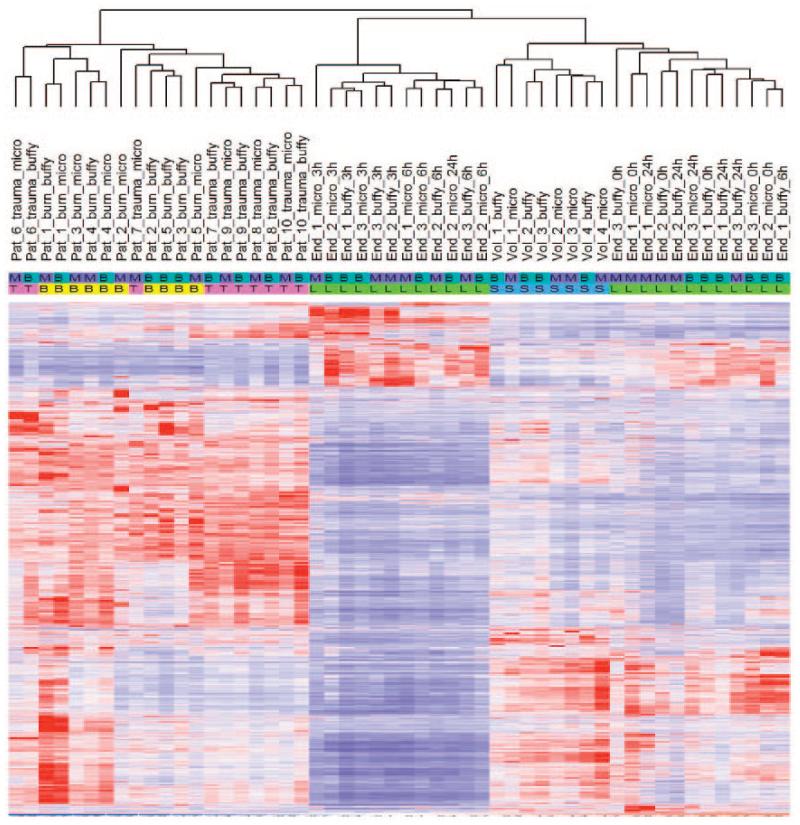
We generated 52 arrays (26 for each protocol) and normalized the readings from the GeneChips by applying model-based algorithms from dChip 2006 to create an expression matrix. We identified 8766 probe sets with CVs of >1.0 across the data set. Their expression values were mean-centered and variance-normalized before hierarchical clustering. We analyzed hybridization signals from the different samples to determine the extent of gene expression differences between the 2 methods. The unsupervised hierarchical clustering with 8766 probe sets revealed grouping according to the clinical source of samples rather than according to differences in the leukocyte isolation methods (Fig. 4). This is noticeable from the heat map of the burn and trauma patient samples, where the pattern of the individual probes and individual samples were properly ordered by hierarchical clustering. Fig. 5 shows the dendrogram from cluster analysis of the patients, reflecting the degree of similarity between the connected samples. For the samples obtained from volunteers administered endotoxin, the unsupervised analysis showed that the major node of separation tends to separate the 3- and 6-h time periods from the 0- and 24-h time points, and that within a time point there was a tendency for these individuals to subcluster over protocols (Fig. 4). As expected, gene expression patterns from the healthy controls were subclustered with the samples from the endotoxin volunteers obtained at the 0- and 24-h time points. Pearson correlation coefficients of the gene expression profiles, estimating the degree of correlation between the isolation methods and intramethod between persons, are summarized in Table 1. For a given patient, the correlation between the protocols was calculated and then averaged over the various individuals. For a given protocol (microfluidics or buffy coat), the correlation coefficients of the gene expression profiles between the individuals were calculated in a pairwise fashion and then averaged. The correlation coefficients for the 2 isolation protocols from the healthy controls and patients [>0.98 (0.01)] were consistently higher than the correlation between individuals for the same protocol. When the buffy coat and microfluidics protocols were compared, supervised analyses for samples obtained from volunteers administered endotoxin or from critically ill burn and trauma patients identified 76, 33, and 43 probe sets, respectively, significant at P < 0.001, which is below the number expected to exceed the significance threshold by chance. Therefore, the supervised analysis comparing the 2 protocols detected no apparent differences in gene expression profiles.
Fig. 4. Summary of the unsupervised hierarchical cluster analysis of leukocyte genes differentially expressed using the 2 protocols in the 4 different experimental systems (healthy volunteer controls; endotoxin time-course 0, 3, 6, 24 h; burn injury patients; and trauma injury patients).
The analysis was performed on the expression matrix by selecting probe sets whose hybridization signal varied across the data set with a CV >1.0. The heat map depicts the hybridization values of the individual samples, with rows representing individual probes and columns representing individual samples ordered by hierarchical clustering. There were 8766 probe sets identified with a CV of >1.0 across the data set. The expression values were then mean-centered and variance-normalized before hierarchical clustering. Based on the cluster analysis, the patient-to-patient differences are greater than the differences between the protocols. Comparing the patterns between the patients and healthy controls reveals distinct differences in gene expression. Protocols: M, microfluidic; B, buffy coat; participants: B, burn; T, trauma; L, endotoxin; S, volunteers.
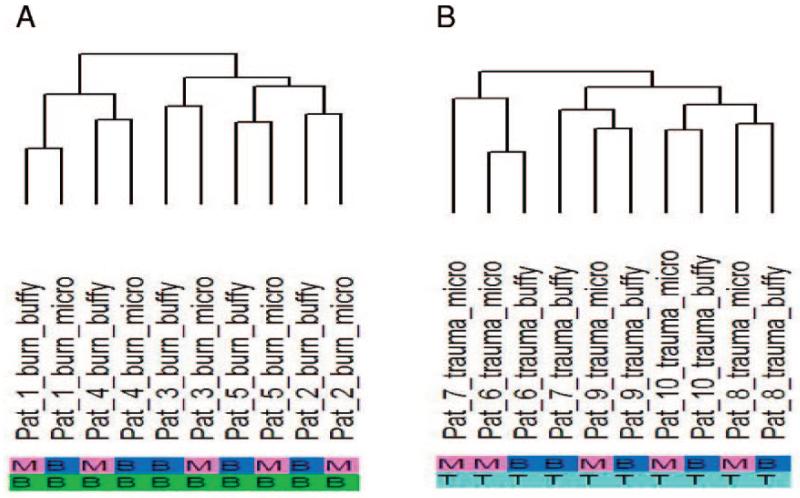
Fig. 5. Dendrogram from cluster analysis performed on the expression matrix from the burn (A) and trauma (B) patient samples.
Inspection of the dendrogram shows that the differences between individuals are greater than the difference in the protocols. Protocols: M, microfluidic; B, buffy coat; participants: B, burn; T, trauma.
Table 1. Summary of the average correlation coefficients (SD) of gene expression profiles obtained between the 2 isolation protocols and among different participants.
| Participants | Samples, n |
Between methods |
Between participants | |
|---|---|---|---|---|
| Microfluidic | Buffy coat | |||
| Healthy controls | 4 | 0.989 (0.003) | 0.987 (0.005) | 0.980 (0.009) |
| Endotoxin-administered controls |
||||
| 0 h | 3 | 0.978 (0.005) | 0.978 (0.005) | 0.967 (0.015) |
| 3 h | 3 | 0.971 (0.017) | 0.976 (0.008) | 0.977 (0.004) |
| 6 h | 3 | 0.919(0.062) | 0.939 (0.064) | 0.973 (0.006) |
| 24 h | 3 | 0.944(0.041) | 0.938 (0.037) | 0.967 (0.002) |
| Burn patients | 5 | 0.983 (0.007) | 0.954(0.019) | 0.955 (0.017) |
| Trauma patients | 5 | 0.980(0.021) | 0.972 (0.017) | 0.972 (0.015) |
In the case of correlation coefficients between protocols, we calculated the correlation between the arrays that were prepared via microfluidics or buffy coat for a given patient and then averaged the correlations over the various individuals. In cases of correlating patient gene expression profiles within a given protocol (between participants), we calculated the correlation coefficients of the gene expression profiles between the individuals in a pairwise fashion and then averaged the correlations.
Discussion
Blood continues to be the most informative and readily available source of tissue from hospitalized patients. Recently, genome-wide expression patterns obtained from whole blood leukocytes have been shown to be a valuable diagnostic and prognostic tool in critically ill patients (1, 4, 14-16). However, extending these observations from research laboratories to the clinical setting requires reliable methods for isolating leukocytes from blood and isolating and processing RNA.
Current macroscale techniques for isolating nucleic acids from whole blood and blood leukocytes use either commercial preparations such as PAXGene™, which introduce considerable variance in the gene expression profiles (10), or complex lysis and density centrifugation protocols that require substantial technical skill. In addition, with macroscale isolation procedures, the cells are often exposed for extended periods to known stressors including altered temperature, tonicity, and G-forces (17-21), all of which can activate leukocytes. There is, therefore, a compelling need for standardized, fast, and robust methods to isolate leukocytes for subsequent proteomic and genomic analyses in clinical settings.
An entirely new class of analysis systems for chemical and biological applications is emerging, enabled by microfluidic technologies (22-24), with increasing impact on the sorting, handling, and analysis of mammalian cells (24-26). The miniaturization of analytical equipment may allow several shortcomings associated with bulky and expensive instrumentation to be overcome. The new microscale devices would enable reduction in sample and reagent volumes and provide results faster, at the bedside. The technologies, derived from the electronics industry, would allow batch-fabrication, reducing costs, and resulting in disposable instruments that are used once and thrown away to prevent sample contamination. Finally, apart from having high accuracy, microfluidic-based methods should minimize the number of manual steps and provide rapid and reproducible results to compete with present technologies and be fully implemented in the clinical setting. Consequently, clinical diagnostics is one of the fastest developing areas for microfluidic applications (27). Despite the fact that microfluidics approaches hold great potential for clinical application, their validation and actual application in clinical settings are infrequent.
In this report, we critically evaluated the ability of our newly developed microfluidic technique (9) to isolate leukocytes for proteomic and genomic analyses in the clinical setting. We provided brief training at 4 clinical sites to research nurses with no prior knowledge in microfluidic techniques and compared the quality of the samples generated by the new and traditional cell separation methods. The microfluidic devices require minimal operator intervention and enable separation of leukocytes from whole blood using protocols that are faster and more efficient than standard techniques. Nurses and laboratory personnel were able to generate reliable clinical data during the brief training session using the microfluidic protocol. This rapid and single-step method does not require prior manipulation of whole blood and has paved the way for the translation of an assay from the research laboratory into the clinical setting.
The quality of RNA from all samples, after storage at −80 °C for several months, and microarray analyses was determined to be excellent. Although we observed variability in RNA yield, the amount of RNA available was always larger than the amount required for analysis. Only a fraction of the total amount of RNA isolated (1 μg total RNA) was used to prepare the microarrays. Overall, 500 μL whole blood was shown to produce sufficient quantities of RNA for genome-wide expression analysis. Hence, this microfluidic approach dramatically reduces the blood requirements compared with macroscale protocols. In the future, it may be possible to reduce this amount of blood even further, which would open the possibility for blood collection from a simple fingerprick, avoiding the need for phlebotomy and its risk of complications. These new capabilities could make microfluidic technologies extremely attractive for pediatric and neonatal patients and allow more frequent sampling from adult patients with conditions that require frequent monitoring.
Side-by-side comparison of the standard bulk and new microscale methods produced highly concordant results in healthy controls and in response to endotoxin administration. As confirmed by gene expression analysis, both isolation techniques were capable of differentiating the changes in response to endotoxin administration at 3- and 6-h time points from the 0- and 24-h time points. As expected, traumatic and burn injuries in patients induced dramatic changes in apparent gene expression. In addition, a correlation coefficient >0.98 (Table 1) in the gene expression results for the patients by each protocol strongly suggested that there were no substantive differences between the protocols. Hence, the microfluidic technique should be readily applicable for detecting changes in gene expression that occur in response to burn and trauma injury. Because leukocytes are capable of mounting rapid responses and altering their gene expression accordingly, the ability to process blood samples quickly after collection from patients free of potential operator handling errors is extremely important. Microfluidic techniques that could be implemented as portable systems operated at the bedside would offer advantages in this area.
In summary, the 4-clinical-site comparison reveals essentially no differences between the microfluidic technique and the standard leukocyte isolation method with respect to RNA yield and quality and genome-wide expression analysis. The microfluidic technique makes the routine processing of whole blood more efficient with an automated and easy-to-use system, ideal for genome-wide expression analysis studies involving patients.
Supplementary Material
Acknowledgments
We thank Octavio Hurtado for support with microfabrication. We acknowledge Megan Knoll, Keir, Deb Benjamin, Robert Granier, and Liz Taylor for their help processing blood samples. Additional participating investigators in the Large Scale Collaborative Research Program, “Inflammation and the Host Response to Injury,” are as follows: Paul E. Bankey, Timothy R. Billiar, Bernard H. Brownstein, David G. Camp II, George Casella, Irshad H. Chaudry, Mashkoor Choudhry, J. Perren Cobb, Asit De, Constance Elson, Bradley Freeman, Richard L. Gamelli, Nicole S. Gibran, Douglas L. Hayden, Brian G. Harbrecht, Jureta W. Horton, William Hubbard, Jeffrey Johnson, Matthew B. Klein, James A. Lederer, Tanya Logvinenko, John A. Mannick, Philip H. Mason, Grace P. McDonald-Smith, Bruce A. McKinley, Carol Miller-Graziano, Joseph P. Minei, Ernest E. Moore, Frederick A. Moore, Avery B. Nathens, Grant E. O’Keefe, Laurence G. Rahme, Daniel G. Remick, David A. Schoenfeld, Michael B. Shapiro, Martin Schwacha, Geoffrey M. Silver, Richard D. Smith, John Storey, H. Shaw Warren, Michael A. West, and Wenzhong Xiao.
Grant/Funding Support: These studies were supported by the National Institutes of Health (Inflammation and the Host Response to Injury Large Scale Collaborative Project, U54 GM-062119) and the National Institutes of Health BioMEMS Resource Center (P41 grant, P41 EB-002503).
Footnotes
Nonstandard abbreviations: PDMS, polydimethylsiloxane.
Financial Disclosures: None declared.
References
- 1.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, et al. A network-based analysis of systemic inflammation in humans. Nature (Lond) 2005;437:1032–7. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 2.Cobb JP, Mindrinos MN, Miller-Graziano C, Calvano SE, Baker HV, Xiao W, et al. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A. 2005;102:4801–6. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cobleigh MA, Tabesh B, Bitterman P, Baker J, Cronin M, Liu ML, et al. Tumor gene expression and prognosis in breast cancer patients with 10 or more positive lymph nodes. Clin Cancer Res. 2005;11:8623–31. doi: 10.1158/1078-0432.CCR-05-0735. [DOI] [PubMed] [Google Scholar]
- 4.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 5.Irizarry RA, Warren D, Spencer F, Kim IF, Biswal S, Frank BC, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–50. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- 6.Larkin JE, Frank BC, Gavras H, Sultana R, Quackenbush J. Independence and reproducibility across microarray platforms. Nat Methods. 2005;2:337–44. doi: 10.1038/nmeth757. [DOI] [PubMed] [Google Scholar]
- 7.Toner M, Irimia D. Blood-on-a-chip. Annu Rev Biomed Eng. 2005;7:77–103. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sethu P, Anahtar M, Moldawer LL, Tompkins RG, Toner M. Continuous flow microfluidic device for rapid erythrocyte lysis. Anal Chem. 2004;76:6247–53. doi: 10.1021/ac049429p. [DOI] [PubMed] [Google Scholar]
- 9.Sethu P, Moldawer LL, Mindrinos MN, Scumpia PO, Tannahill CL, Wilhelmy J, et al. Microfluidic isolation of leukocytes from whole blood for phenotype and gene expression analysis. Anal Chem. 2006;78:5453–61. doi: 10.1021/ac060140c. [DOI] [PubMed] [Google Scholar]
- 10.Feezor RJ, Baker HV, Mindrinos MN, Hayden D, Tannahill CL, Brownstein BH, et al. Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genomics. 2004;19:247–54. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- 11.Laudanski K, Miller-Graziano C, Xiao W, Mindrinos MN, Richards DR, De A, et al. Cell-specific expression and pathway analyses reveal alterations in trauma-related human T cell and monocyte pathways. Proc Natl Acad Sci U S A. 2006;103:15564–9. doi: 10.1073/pnas.0607028103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelder RNV, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A. 1990;87:1663–7. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson RP, Rhyne CD, Fong Y, Hesse DG, Tracey KJ, Marano MA, et al. Peripheral blood leukocyte kinetics following in vivo lipopolysaccharide (LPS) administration to normal human subjects: influence of elicited hormones and cytokines. Ann Surg. 1989;210:239–45. doi: 10.1097/00000658-198908000-00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Achiron A, Gurevich M, Friedman N, Kaminski N, Mandel M. Blood transcriptional signatures of multiple sclerosis: unique gene expression of disease activity. Ann Neurol. 2004;55:410–7. doi: 10.1002/ana.20008. [DOI] [PubMed] [Google Scholar]
- 15.Batliwalla FM, Li W, Ritchlin CT, Xiao X, Brenner M, Laragione T, et al. Microarray analyses of peripheral blood cells identifies unique gene expression signature in psoriatic arthritis. Mol Med. 2005;11:21–9. doi: 10.2119/2006-00003.Gulko. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–47. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 17.Fearon DT, Collins LA. Increased expression of C3b receptors on polymorphonuclear leukocytes induced by chemotactic factors and by purification procedures. J Immunol. 1983;130:370–5. [PubMed] [Google Scholar]
- 18.Glasser L, Fiederlein RL. The effect of various cell separation procedures on assays of neutrophil function: a critical appraisal. Am J Clin Pathol. 1990;93:662–9. doi: 10.1093/ajcp/93.5.662. [DOI] [PubMed] [Google Scholar]
- 19.Kouoh F, Levert H, Gressier B, Luyckx M, Brunet C, Dine T, et al. Reduced ammonium chloride haemolysis time enhances the number of isolated functional rabbit polymorphonuclear neutrophils. Apmis. 2000;108:417–21. doi: 10.1034/j.1600-0463.2000.d01-77.x. [DOI] [PubMed] [Google Scholar]
- 20.Lundahl J, Hallden G, Hallgren M, Skold CM, Hed J. Altered expression of CD11b/CD18 and CD62L on human monocytes after cell preparation procedures. J Immunol Methods. 1995;180:93–100. doi: 10.1016/0022-1759(94)00303-e. [DOI] [PubMed] [Google Scholar]
- 21.Macey MG, McCarthy DA, Vordermeier S, Newland AC, Brown KA. Effects of cell purification methods on CD11b and L-selectin expression as well as the adherence and activation of leucocytes. J Immunol Methods. 1995;181:211–9. doi: 10.1016/0022-1759(95)00003-s. [DOI] [PubMed] [Google Scholar]
- 22.Reyes DR, Iossifidis D, Auroux PA, Manz A. Micro total analysis systems. 1. Introduction, theory, and technology. Anal Chem. 2002;74:2623–36. doi: 10.1021/ac0202435. [DOI] [PubMed] [Google Scholar]
- 23.Dittrich PS, Manz A. Lab-on-a-chip: microfluidics in drug discovery. Nat Rev Drug Discov. 2006;5:210–8. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- 24.Dittrich PS, Tachikawa K, Manz A. Micro total analysis systems. Latest advancements and trends. Anal Chem. 2006;78:3887–908. doi: 10.1021/ac0605602. [DOI] [PubMed] [Google Scholar]
- 25.Andersson H, van den Berg A. Microfluidic devices for cellomics, a review. Sensors Actuators. 2003;92:315–25. [Google Scholar]
- 26.Beebe D, Folch A. The science and applications of cell biology in microsystems. LabChip. 2005;5:10–1. [Google Scholar]
- 27.Clayton J. Go with the flow. Nature Methods. 2005;2:622–5. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.