Abstract
The apoptotic actions of p53 require its phosphorylation by a family of phosphoinositide-3-kinase-related-kinases (PIKKs), which include DNA-PKcs and ATM. These kinases are stabilized by the TTT (Tel2, Tti1, Tti2) co-chaperone family, whose actions are mediated by CK2 phosphorylation. The inositol pyrophosphates, such as 5-diphosphoinositol pentakisphosphate (IP7), are generated by a family of inositol hexakisphosphate kinases (IP6Ks) of which IP6K2 has been implicated in p53-associated cell death. In the present study we report a novel apoptotic signaling cascade linking CK2, TTT, the PIKKs, and p53. We demonstrate that IP7, formed by IP6K2, binds CK2 to enhance its phosphorylation of the TTT complex thereby stabilizing DNA-PKcs and ATM. This process stimulates p53 phosphorylation at serine-15 to activate the cell death program in human cancer cells and in murine B cells.
INTRODUCTION
Following genotoxic stress, cell arrest or apoptosis is typically mediated by the tumor suppressor transcription factor p53 (Vousden and Prives, 2009). p53 enhances the transcription of genes, such as p21, which arrest the cell cycle and facilitate cell repair. In instances of prolonged insult or irreparable cell damage, p53 activates genes such as Noxa, PUMA, and Bax, which initiate pro-death programs that proceed through mitochondrial signaling leading to caspase-3 activation. The regulation of p53 dynamics is complex, involving multiple stimuli at the level of transcription, translation and post-translational modifications (Kruse and Gu, 2009; Vousden and Prives, 2009). Nonetheless, in response to DNA damage, the initial activation trigger is phosphorylation of p53 at serine-15 (S15) (Zhang et al., 2011) by three kinases in the family of phosphoinositide-3-kinase-related protein kinases (PIKKs): ATM (ataxia-telangiectasia mutated), ATR (ATM- and Rad3-related), and DNA-PKcs (DNA-dependent protein kinase catalytic subunit) (Banin et al., 1998; Canman et al., 1998; Shieh et al., 1997; Tibbetts et al., 1999). S15-phosphorylation of p53, especially when mediated by DNA-PKcs, facilitates transcription of apoptotic genes such as PUMA, while down-regulating p21 levels (Hill et al., 2008; Hill et al., 2011; Sluss et al., 2004; Wang et al., 2000).
PIKKs are large proteins (270–470 kDa) whose C-terminal portion contains catalytic domains resembling PI3 kinase (Abraham, 2004), and whose N-terminal area includes 40–50 HEAT repeats of >2,000 amino acids (Perry and Kleckner, 2003). In mammals, there are six PIKKs: ATM, ATR, DNA-PKcs, mTOR, TRRAP, and SMG1. PIKKs typically function in the context of multi-protein complexes (Lovejoy and Cortez, 2009). Maintaining physiologic conformations of such large, repeat-rich proteins and assembling their complexes requires the actions of chaperone proteins. Recently, the TTT complex, comprising Tel2, Tti1 and Tti2 (Hurov et al., 2010), has been demonstrated to stabilize the PIKK enzymes (Takai et al., 2007) in conjunction with chaperone systems (Horejsi et al., 2010; Izumi et al., 2010; Takai et al., 2010), with Tti1 (Kaizuka et al., 2010) and Tel2 (Takai et al., 2007) binding to newly synthesized PIKKs (Takai et al., 2010).
Mechanisms regulating functions of the TTT complex have not been well characterized. Recently, it was reported that Tti1 and Tel2 are physiologically phosphorylated by casein kinase-2 (CK2), thereby influencing the stability and function of PIKKs (Fernandez-Saiz et al., 2013; Horejsi et al., 2010). Phosphorylation of Tti1 at Ser828 and Tel2 at Ser485 facilitates their ubiquitination and degradation when in complex with mTORC1 (Fernandez-Saiz et al., 2013; Horejsi et al., 2010), whereas phosphorylation of Tel2 at Ser487/491 augments interaction with the RT2P/prefoldin-like chaperone complex (Horejsi et al., 2010).
CK2 has been regarded as a constitutive kinase not subject to regulation (Pinna and Allende, 2009). Tobin and associates identified inositol polyphosphates as potential regulators of CK2 (Solyakov et al., 2004). They reported that exogenous inositol hexakisphosphate (IP6) stimulates CK2 activity in liver preparations by competing with an endogenous tissue constituent, which was later identified as hNopp140 (Kim et al., 2006), a protein with very high affinity and binding capacity for CK2 (Lee et al., 2008). Although the stimulation by IP6 suggests that CK2 normally exists in an inhibited state with low constitutive activity, it remains unclear whether IP6 physiologically regulates CK2.
Inositol pyrophosphates are inositol polyphosphate derivatives containing highly energetic diphosphate bonds that turn over rapidly in cells (Chakraborty et al., 2011a; Menniti et al., 1993; Stephens et al., 1993). The most extensively characterized member of this class is diphosphoinositol pentakisphosphate in which five of the inositol hydroxyls are monophosphates, while a sixth, at the 5-position, contains a pyrophosphate moiety so that the molecule is typically referred to as 5-diphosphoinositol pentakisphosphate (5-IP7, henceforth simply designated IP7) (Albert et al., 1997). In mammals, IP7 is generated from IP6 by a family of IP6 kinases (IP6K1-3) (Saiardi et al., 1999; Saiardi et al., 2001). Another isomer of IP7, 1-IP7, can also be formed by a more recently identified enzyme termed VIP, though VIP appears to be predominantly associated physiologically with the formation of IP8 (Choi et al., 2007). IP6Ks display substantial sequence homology but different physiologic functions (Barker et al., 2009; Chakraborty et al., 2011a). IP6K2, in particular, physiologically mediates apoptotic cell death. Thus, Lindner and associates (Morrison et al., 2002; Morrison et al., 2001) and ourselves (Koldobskiy et al., 2010; Nagata et al., 2005) demonstrated that overexpression of IP6K2 sensitizes cells to apoptotic stressors, while deletion of IP6K2, but not IP6K1 or IP6K3, confers resistance. Mice with targeted deletion of IP6K2 display a predisposition to the formation of certain tumors and are resistant to cell death elicited by ionizing radiation (Morrison et al., 2009). In human cancers there are also reports of IP6K2 mutation (Jones et al., 2012), deletion (Kay et al., 2010), and allelic-specific expression (Zhao et al., 2010). Recently, we found that IP6K2 is required for p53-mediated apoptosis (Koldobskiy et al., 2010). Thus, gene disruption of IP6K2 selectively impairs p53-mediated apoptosis, instead favoring cell-cycle arrest. However, molecular mechanisms whereby IP6K2 controls p53-mediated cell fate determination remain unclear.
Earlier, York et al (York et al., 2005) and we (Saiardi et al., 2005) reported that yeast deficient in Kcs1, the only yeast IP6 kinase, display altered telomere length and sensitivity to wortmannin and caffeine, both mediated by the yeast ATM homolog Tel1, indicating a functional linkage between IP6Ks and PIKKs. In the present study we employed tandem affinity purification method to screen for IP6K interaction partners and found that human IP6K2 directly binds to Tti1. We delineate a signaling system whereby IP7, generated by IP6K2, interacts selectively with CK2 to stimulate phosphorylation of the Tel2/Tti1/Tti2 complex thereby enhancing the ability of DNA-PKcs/ATM to phosphorylate and activate p53. We further demonstrate that this cascade accounts for the pro-apoptotic influences of IP6K2, establishing inositol pyrophosphates as physiologic mediators of the DNA-PK/ATM-p53 cell death axis.
RESULTS
IP6K2 binds to the Tti1/Tti2/Tel2 complex through direct interactions with Tti1
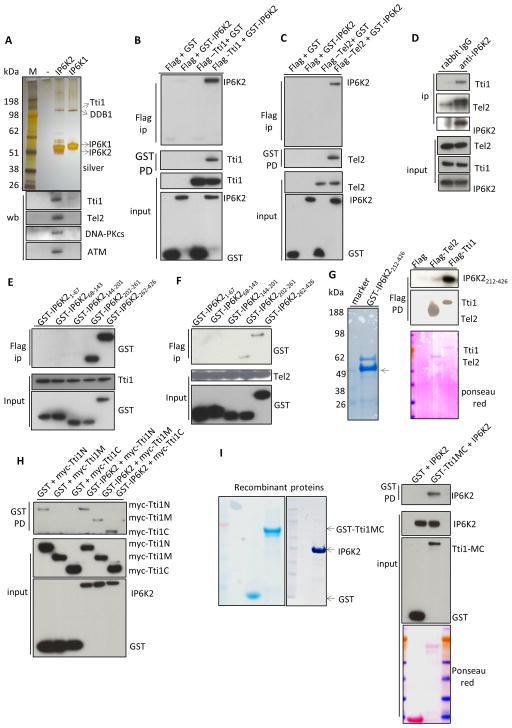
Earlier we provided evidence that apoptotic actions of IP6K2 involve p53, but underlying molecular mechanisms were not characterized (Koldobskiy et al., 2010). To clarify this signaling system, we sought to identify binding partners for IP6K2 (Figure 1). Tandem affinity purification followed by mass spectrometry reveals selective binding of IP6K2 to Tti1 (Figure 1A, Figure S1A). By contrast, IP6K1 but not IP6K2 binds to DDB1, part of an E3 ubiquitin ligase system (Iovine et al., 2011). Since yeast IP6K regulates PIKKs (Saiardi et al., 2005; York et al., 2005), and Tti1 is known to bind and stabilize PIKKs as part of a Tti1/Tti2/Tel2 complex (Hurov et al., 2010; Kaizuka et al., 2010), we chose to investigate the functional relevance of the Tti1-IP6K2 interaction. Western blot analysis reveals co-precipitation of IP6K2 with Tti1, Tel2, DNA-PKcs and ATM (Figure 1A), but not ATR, mTOR, SMG1 or TRRAP (data not shown), suggesting that IP6K2 exists in a ternary complex with DNA-PKcs/ATM and the TTT proteins. Since DNA-PKcs and ATM phosphorylate p53 to activate its apoptotic program (Callen et al., 2009), these binding interactions might underlie the apoptotic influences of IP6K2.
Figure 1. IP6K2 binds to the TTT complex.
(A) SDS-PAGE of tandem-affinity purified IP6Ks, followed by silver-staining. Lower panel, western-blot of tandem-affinity purified IP6Ks identifies binding partner. (B) Co-IP between GST-IP6K2 and Flag-Tti1 expressed in HEK293 cells. (C) Co-IP between GST-IP6K2 and Flag-Tel2 expressed in HEK293 cells. (D) Endogenous Co-IP between IP6K2 and Tel2/Tti1 in HCT116 cells. (E) Co-IP between GST-IP6K2 fragments and Flag-Tti1. (F) Co-IP between GST-IP6K2 fragments and Flag-Tel2. (G) Direct binding between recombinant GST-IP6K2212-426 and purified Flag-Tti1 in vitro. (H) Co-IP between myc-Tti1 N (aa1-460), M (aa 461-825), and C (aa 826-1089) fragments and GST-IP6K2. (I) Direct binding between purified recombinant full-length IP6K2 and recombinant GST-Tti1MC (aa 461-1089) in vitro. see also Figure S1.
We verified the interaction between IP6K2 and the TTT complex by co-immunoprecipitation (Co-IP). Transiently transfected GST-IP6K2 binds to Flag-Tti1, Flag-Tel2 (Figure 1B, 1C), and myc-Tti2 (Figure S1B). We also detect binding between myc-IP6K2 and Flag-Tti1 (Figure S1C), and between GST-IP6K2 and myc-Tel2 (Figure S1D). The physiologic relevance of these interactions is supported by the binding of endogenous Tti1 and Tel2 to IP6K2 (Figure 1D).
Mapping studies reveal that the C-terminus of IP6K2 binds Tti1 (Figure 1E) and Tel2 (Figure 1F). These binding interactions appear to be direct, as we observe in vitro binding of recombinant IP6K2’s C-terminus to purified Tti1 (Figure 1G). We do not detect IP6K2 binding to purified Tel2, suggesting that Tel2 may interact with IP6K2 indirectly, via Tti1.
We also mapped areas of Tti1 that interact with IP6K2 (Figure 1H), revealing that the middle (Tti1M) and C-terminal (Tti1C) portions of Tti1 differentially bind GST-IP6K2 but not GST controls. Direct binding of recombinant IP6K2 and recombinant Tti1MC is established by in vitro experiments (Figure 1I).
IP7 enhances phosphorylation of Tti1/Tel2 by influences upon CK2
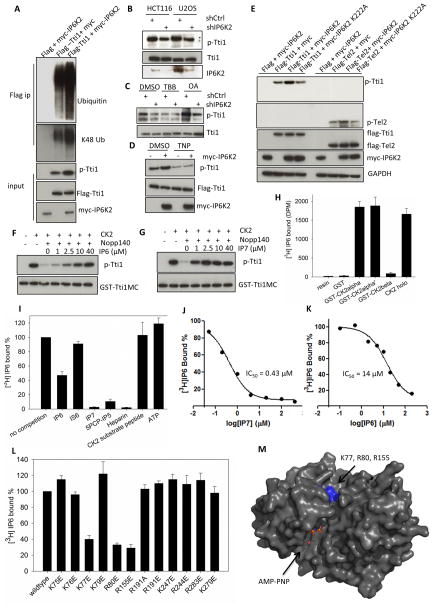
In the Co-IP experiments, we noticed that overexpression of IP6K2 consistently decreases levels of Tti1 (Figure 1B, S1C), but not Tti2 or Tel2. Recently, Bassermann and colleagues (Fernandez-Saiz et al., 2013) described S828-phosphorylation of Tti1 by CK2, leading to its ubiquitination and degradation with specific influences on mTORC1 signaling. We find that IP6K2 co-expression enhances phosphorylation and K48-linked ubiquitination of Tti1 (Figure 2A, S2A). Conversely, in both HCT116 and U2OS cells, depletion of IP6K2 by RNA interference diminishes endogenous Tti1 phosphorylation (Figure 2B) without affecting total Tti1 levels. Inhibition of protein phosphatase activity by okadaic acid increases levels of phospho-Tti1, while 4,5,6,7-tetrabromo-1H-benzotriazole (TBB), a selective CK2 inhibitor, decreases phospho-Tti1 levels (Figure 2C). TBB, but not okadaic acid, nullifies the effect of IP6K2 depletion, suggesting that IP6K2 impacts Tti1 phosphorylation by enhancing CK2 activity.
Figure 2. IP7 binds to CK2 and enhances CK2-mediated phosphorylation of Tti1/Tel2.
(A) Coexpression of IP6K2 increases the phosphorylation (S828) and ubiqutination of Tti1. Cells were harvested 28 h after transfection. The amount of Tti1 plasmids used for transfection was adjusted to achieve equal expression levels. (B) Lentiviral shRNA knockdown of IP6K2 diminishes endogenous Tti1 phosphorylation. (C) Effect of CK2 inhibitor TBB (20 μM, 4 h) and protein phosphatase inhibitor okadaic acid (OA) (50 nM, 1 h) on Tti1 phosphorylation. U2OS cells were transfected with the indicated lentiviral shRNA constructs and selected with puromycin (1 μg/ml) prior to drug treatment. (D) Effect of TNP treatment (10 μM, 1.5 h) on the phosphorylation of Tti1, with/without IP6K2 co-expression. (E) Co-expression of IP6K2 wildtype, but not the K222A mutant, increases phospho-Tti1 (S828) and phospho-Tel2 (S487/S491). (F–G) Concentration-dependent effects of IP6 (F) and IP7 (G) on reversing hNopp140 (500 nM) inhibition of GST-Tti1MC phosphorylation by CK2. Reaction conditions were: 20 mM MgCl2 (pH 7.5), 50 mM KCl, 10 mM MgCl2, 200 μM ATP, 30 °C, 15 min. (H) [3H]IP6 binding to various CK2 preparations. Twenty μg purified recombinant CK2 proteins on glutathione beads were incubated with 10 μl [3H]IP6 (65 μCi/ml) overnight at 4 °C. Controls included buffer or GST alone. (I–J) Concentration-dependent competition of [3H]IP6 binding to CK2α by unlabeled IP6 (I) and IP7 (J). (K) [3H]IP6 binding to CK2α in the presence of various competitors. The concentrations of small molecules used were: ATP (100 μM), the rest (25 μM). (L) [3H]IP6 binding to CK2α mutants. (M) Location of K77, R80 and R155 in the 3D structure of CK2α displayed in surface mode (PDB id: 2PVR). The ATP analog AMPPNP is located in the catalytic active site. Residues K77, R80 and R155 are highlighted in blue. The figure was generated by Pymol. see also Figure S2.
To determine whether IP6K2’s catalytic activity is required for Tti1 phosphorylation, we employed N2-(m-(trifluoromethy)lbenzyl)N6-(p-nitrobenzyl)purine (TNP), a specific inhibitor of IP6 kinase activity (Padmanabhan et al., 2009), and the IP6K2 K222A kinase-dead mutant (Saiardi et al., 2001). TNP markedly reduces Tti1 phosphorylation (Figure 2D). Moreover, kinase-dead IP6K2 does not enhance Tti1 phosphorylation (Figure 2E). IP6K2 also increases Tel2 phosphorylation at S487/491 in a catalytic activity-dependent manner (Figure 2E).
The requirement of IP6K2’s catalytic activity for enhancing Tti1 and Tel2 phosphorylation suggests that inositol pyrophosphates activate this process. However, in vitro experiments fail to reveal augmentation by IP6 or IP7 of CK2 phosphorylation of Tti1, Tel2, or the classical CK2 substrate peptide (RRRADDSDDDDD) (Figure S2 B–F). These findings are consistent with observations of Tobin and colleagues (Solyakov et al., 2004) that IP6 stimulates CK2 activity in partially purified cell lysates but not with pure CK2. They noted that IP6 acts by reversing inhibition of CK2 by an endogenous factor, which appears to be hNopp140 (Kim et al., 2006). We find that recombinant hNopp140 inhibits CK2 phosphorylation of Tti1 in a concentration-dependent manner (Figure S2G). Importantly, while IP6 reverses this inhibition with a 50% effect at about 10 μM, IP7 appears to be approximately 10 times more potent, with an EC50 of about 1 μM (Figure 2F, 2G). Thus, IP7 selectively enhances CK2 activity by counteracting the inhibitory influences of hNopp140.
We speculate that IP6/7 directly binds CK2, an acidophilic kinase with abundant positively charged surface residues (Niefind et al., 2001). To evaluate such molecular interactions, we monitored binding of [3H]IP6 to HEK293-purified CK2 linked to myc beads. [3H]IP6 binds to myc-CK2α, the catalytic subunit, but not the β-subunit or myc-Tel2 (Figure S2H). To exclude the possibility that [3H]IP6 binds to a CK2α-associated factor and to decrease background binding to beads, we assessed binding of [3H]IP6 to purified recombinant GST-CK2 attached to glutathione beads. [3H]IP6 binds robustly to holo-CK2 and CK2α, but not CK2β (Figure 2H). At 25 μM concentration IP7 abolishes [3H]IP6 binding to CK2, whereas IP6 reduces binding only about 55% and inositol hexasulfate is inactive (Figure 2I). 5PCP-IP5, a non-hydrolyzable derivative of IP7 (Wu et al., 2013), is as active as IP7 itself. Using this binding paradigm we conducted detailed concentration-response analysis to ascertain the relative affinities of IP6 and IP7 for CK2 (Figure 2J, 2K). IP7 displays an IC50 of about 0.4 μM, 30-times more potent than IP6 whose IC50 value is 14 μM.
IP7 appears to impact CK2 at a site distinct from the substrate binding site (Figure 2I). Thus, ATP and a CK2 substrate peptide fail to compete with [3H]IP6 binding, which might explain the lack of influence of IP6/IP7 upon pure CK2 activity. On the other hand, heparin, which binds CK2 over an extended surface, abolishes [3H]IP6 binding. To ascertain regions of CK2 required for [3H]IP6 binding, we made a variety of CK2 mutations of which three, mutations of Lys77, Arg80, and Arg 151 to glutamate, reduce binding (Figure 2L). When mapped to the crystal structure of CK2 (Niefind et al., 2001), these three residues lie close to each other and form a positively charged pocket adjacent to the active site (Figure 2M). While this work was under revision, Lee et al reported the crystal structure of the CK2-IP6 complex (Lee et al., 2013). Consistent with our mutagenesis data, residues Lys77, Arg80, and Arg 151 are located in proximity to the phosphate groups of IP6 (Figure S2I)
IP6K2 stabilizes DNA-PKcs/ATM by enhancing phosphorylation-dependent binding of Tti1/Tel2 to DNA-PKcs/ATM
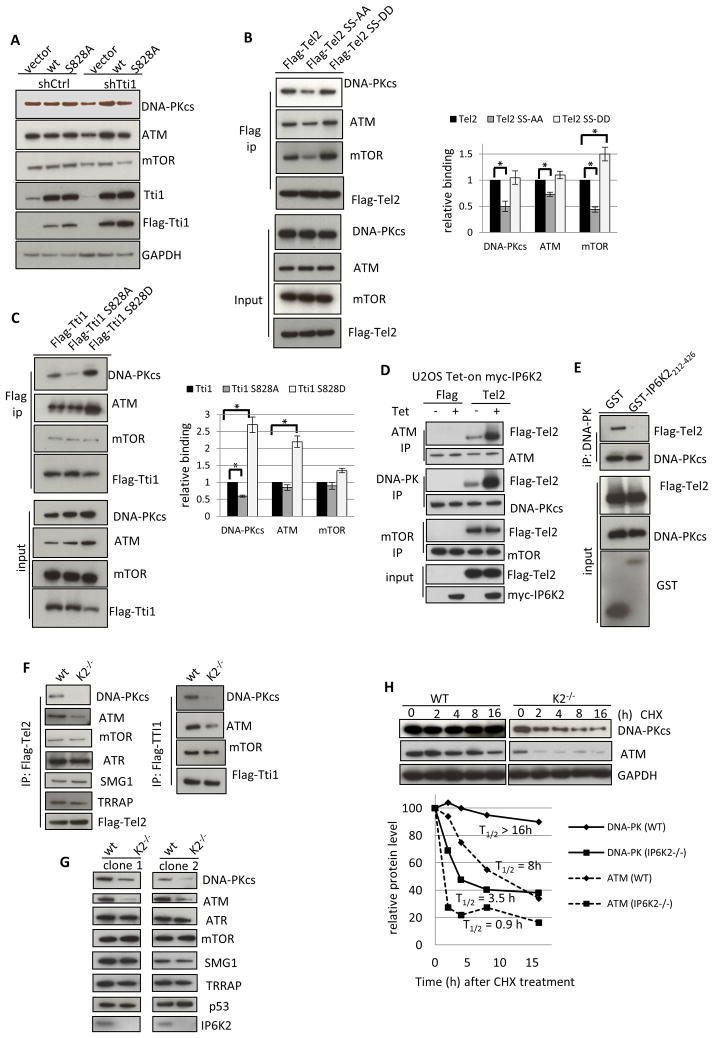
What is the physiological implication of the finding that IP7 enhances CK2 phosphorylation of Tti1/Tel2? Tti1/Tel2 binds and stabilizes PIKKs (Hurov et al., 2010). CK2 phosphorylation of Tel2 is required for its binding to chaperones and for PIKK stabilization (Horejsi et al., 2010). Whether CK2 phosphorylation of Tti1 also mediates the stability of PIKKs is unknown. We therefore measured the levels of PIKKs in Tti1 knockdown cells that have been complemented with RNAi-resistant wildtype Tti1 or Tti1S828A. While wildtype Tti1 rescues the loss of DNA-PKcs, ATM and mTOR resulted from Tti1 depletion, the S828A mutant does so to a much lesser extent (Figure 3A). Thus, CK2 phosphorylation of both Tel2 and Tti1 is required for them to stabilize PIKKs.
Figure 3.
IP6K2 is required for the binding of the TTT complex to DNA-PKcs/ATM and for the stability of DNA-Pkcs/ATM. (A) Protein levels of DNA-PKcs, ATM and mTOR in HCT116 cells that were transfected with shTti1 for 3 d, and with siRNA-resistant flag-Tti1 or flag-Tti1-S828A for 2 d. (B) Immunoprecipitation of Tel2 wildtype, S478A/491A, and S478D/S491D mutant proteins, followed by western-blot analysis. The bar graph represents average data from three different experiments. Relative binding was determined as the amount of co-immunoprecipitated PIKKs divided by the levels of Flag-Tti1 variants. (C) Immunoprecipitation of Tel2 wildtype, S828A, and S828D mutant proteins, followed by western-blot analysis. (D) Overexpression of myc-IP6K2 in U2OS cells increases the co-IP between Flag-Tel2 and ATM/DNA-PKcs, but not that between Flag-Tel2 and mTOR. Four h after cells were transfected with Flag-Tel2 (lipofectamine), tetracycline (1 μM) was added for another 20 h. (E) Overexpression of GST-IP6K2212-426 abolishes the co-IP between Flag-Tel2 and DNA-PKcs. (F) Flag-Tel2 and Flag-Tti1 immunoprecipitation from wildtype and isogenic IP6K2−/− HCT116 cells, followed by western-blot analysis. (G). Western-blot analysis of the expression levels of PIKKs in two clones of IP6K2 wildtype and null cell lines. (H) Cycloheximide treatment (50 μM) decreases DNA-PKcs and ATM protein levels more rapidly in IP6K2−/− than wild-type cells. see also Figure S3.
To understand the role of Tti1/Tel2 phosphorylation, we examined whether phospho-acceptor site mutants of Tti1 and Tel2 display altered binding properties to DNA-PKcs, ATM and mTOR. Mutation to alanine of Tel2’s two serine residues that are CK2 phosphorylation sites decreases binding of Tel2 to DNA-PKcs, ATM, and mTOR, an effect not elicited by the phospho-mimicking DD mutation (Figure 3B). The parallel S828A mutation of Tti1 also diminishes Tti1 binding to DNA-PKcs, whereas the phospho-mimic S828D mutation stimulates binding to both DNA-PKcs and ATM (Figure 3C). Thus, both for Tti1 and Tel2, phosphorylation at the CK2-sensitive sites augments binding of these proteins to DNA-PKcs/ATM, which is associated with their stabilization.
The presence of DNA-PKcs and ATM in our IP6K2-TAP pulldown (Figure 1A) suggests that IP6K2 participates in stabilizing DNA-PKcs and ATM. We first examined whether IP6K2 regulates the interaction between the TTT complex and DNA-PKcs/ATM. Tetracycline-induced IP6K2 overexpression in U2OS cells markedly increases binding of Tel2 to ATM/DNA-PKcs but not mTOR (Figure 3D). Conversely, the dominant-negative IP6K2 (aa 212-426), designed from our mapping studies (Figure 1E, 1F), substantially reduces binding of Tel2 to DNA-PKcs (Figure 3E). Thus, IP6K2 augments binding of Tel2 to DNA-PKcs/ATM.
To validate that Tel2/Tti1 binding to DNA-PKcs/ATM depends on IP6K2, we employed the IP6K2 null HCT116 cell line generated by homologous recombination (Koldobskiy et al., 2010). IP6K2 deletion greatly diminishes binding between Tel2 and DNA-PKcs/ATM, with no apparent effect on other PIKK members such as mTOR, ATR, SMG1, and TRRAP (Figure 3F). Similar selectivity is evident for IP6K2 deletion-associated diminution of Tti1 binding to DNA-PKcs/ATM (Figure 3F). Notably, protein levels of DNA-PKcs/ATM, but not the other PIKKs, are markedly reduced in IP6K2 knockouts (Figure 3G), suggesting that decreased binding of Tel2/Tti1 to DNA-PKcs/ATM leads to their destabilization. Consistent with this notion, IP6K2 deletion does not alter mRNA levels of DNA-PKcs/ATM (Figure S3B). Rather, the half-life of DNA-PKcs/ATM, assessed by rates of protein loss with cycloheximide treatment, is greatly accelerated with IP6K2 deletion (Figure 3G). Similarly, the half-life of DNA-PKcs/ATM is decreased in Tti1 knock-down cells rescued with Tti1S828A compared with cells complemented with wildtype Tti1 (Figure S3C).
We explored protein degradative mechanisms that might account for IP6K2-mediated stabilization of DNA-PKcs/ATM (Figure S3D). Depletion of DNA-PKcs/ATM associated with IP6K2 deletion is not altered by the proteasomal inhibitor MG132 or the caspase inhibitor z-VAD-fmk, similar to findings in Tel2 knockout cells (Takai et al., 2007). By contrast, leupeptin, a lysosomal protease inhibitor, does partially prevent DNA-PKcs/ATM degradation.
IP6K2 is required for the DNA-PKcs/ATM-p53 signaling axis
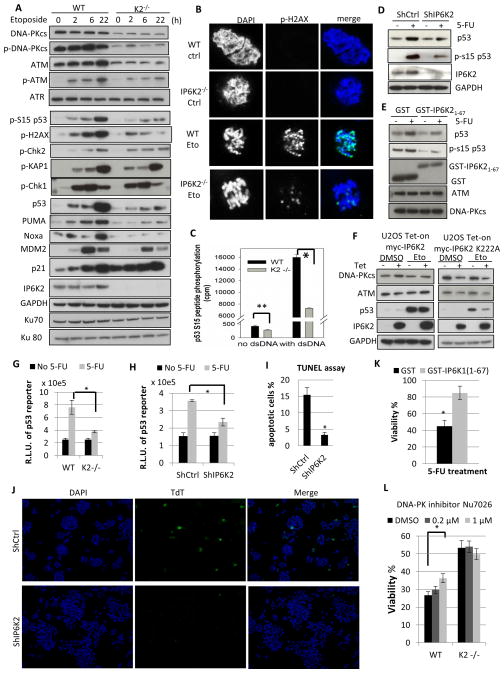
Thus far we have identified a signaling system whereby IP7, generated by IP6K2, enhances CK2 phosphorylation of Tti1 and Tel2 to facilitate their stabilization of DNA-PKcs/ATM. To understand the physiological consequences of these signaling events, we monitored the phosphorylation of DNA-PK/ATM downstream substrates in response to DNA-damage (Figure 4A). At multiple times following etoposide treatment IP6K2 deletion leads to marked depletion of total and phosphorylated levels of DNA-PKcs/ATM as well as total and phospho-S15 p53, consistent with a role for DNA-PKcs/ATM in phosphorylating and stabilizing p53. Diminished p53 stabilization following IP6K2 deletion is also observed with another DNA damage inducer, 5-fluorouracil (5-FU) (Figure S4A). Downstream targets of p53 associated with cell death, such as PUMA and NOXA, are depleted by IP6K2 deletion, while p21 levels, associated with cell arrest, are increased, which might be due to the specific repressive role for DNA-PKcs in p21 transcription (Hill et al., 2011). Consistent with this notion, the DNA-PKcs inhibitor Nu7026 increases p21 levels in wild type but not IP6K2 knockout cells (Figure S4B). Phosphorylation of the histone variant H2AX, an early marker of DNA damage loci and another common substrate of DNA-PKcs and ATM, is also attenuated in IP6K2 knockouts (Figure 4A, 4B). By contrast, IP6K2 has negligible effect on phosphorylation of the ATM-specific substrates checkpoint kinase 2 (Chk2) and KAP1, or signaling through the ATR-Chk1 axis, suggesting that IP6K2’s actions are rather selective.
Figure 4.
IP6K2 influence the p53 cell-death pathway by promoting p53 S15-phosphorylation by DNA-PKcs/ATM. (A) Western blots of DNA-PKcs, ATM, their active phosphorylated forms, and their downstream targets after treatment with etoposide (20 μM) for 2, 6, and 22 h, respectively. (B) Immunofluorescence staining for p-H2AX in WT and IP6K2−/− HCT116 cells with or without etoposide treatment. (C) [32P]ATP-dependent phosphorylation of a p53 S15-peptide by wild-type and IP6K2−/− cell lysate in vitro, in the presence or absence of dsDNA. For experimental details see Materials and Methods. (D) Knockdown of IP6K2 in HCT116 cells by shRNA (lipofectamine) abolishes p53 induction upon DNA damage. (E) Effect of GST-IP6K21-67 expression on induction of p53 and its S15 phosphorylation upon 5-FU treatment. (F) Kinase dead IP6K2 destabilizes DNA-PKcs/ATM and diminishes p53 induction. IP6K2 wildtype and K222A mutant were overexpressed (16 h) in a Tet-on U2OS cell system, followed by etoposide treatment (20 μM, 16 h). (G) Luciferase activity from the p53 consensus promoter driven reporter PG13-luc. R.L.U.: relative luciferase unit. (H) Knockdown of IP6K2 attenuates p53 transcriptional activation of the PG13-luc luciferase reporter. (I–J) Knockdown of IP6K2 increases cell viability as measured by TUNEL assay. Forty-eight h after transfection (lipofectamine) of shRNA constructs, cells were treated with etoposide (40 μM, 24 h). Cells were stained using the Terminal deoxynucleotidyl Transferase (TdT)-mediated dUTP-biotin Nick End Labeling (TUNEL) assay system. Fragmented apoptotic cell nuclei were visualized by TUNEL (TdT, green), and the nucleus was stained with DAPI (blue). (K). Viability of HCT116 cells with or without overexpression of IP6K21-67 followed by 5-FU treatment. (L) Viability of HCT116 wildtype and IP6K2 null cells upon treatment with cytotoxic concentrations of 5-FU (400 μM) with or without pre-treatment with the DNA-PK inhibitor Nu7026 for 1 h. see also Figure S4.
The above findings are consistent with our previous report of a role for IP6K2 in p53 downstream target expression (Koldobskiy et al., 2010), and also imply that IP6K2 regulates p53 at the level of its S15-phosphorylation. To explore this possibility, we first verified the role of IP6K2 in phospho-S15-p53 generation in vitro using cell lysates. Both basal and double-stranded DNA-stimulated phosphorylation of a p53-derived peptide containing S15 is markedly reduced upon IP6K2 deletion (Figure 4C). We then determined whether IP6K2 regulates p53 phosphorylation only at Ser15. Multiple phosphorylation sites on p53 influence its apoptotic actions (Vousden and Prives, 2009). IP6K2 deletion virtually abolishes 5-FU-induced phospho-S15-p53 but has negligible effect on phosphorylation of p53 at S20, S37 and S392 (Figure S4C). The effects of IP6K2 genetic deletion are substantiated by experiments depleting IP6K2 transiently with RNA interference in HCT116 and U2OS cells (Figure 4D, S4D). This means of reducing IP6K2 levels also leads to substantial decreases in both p53 protein and phospho-S15-p53. IP6K2 directly binds to p53 (Koldobskiy et al., 2010). To assess whether this binding is required for p53 S15-phosphorylation, we employed IP6K21-67, which disrupts the IP6K2-p53 association (Koldobskiy et al., 2010). Expression of IP6K21-67 blocks both basal and 5-FU induced p53 S15-phosphorylation and stabilization without affecting the stability of ATM or DNA-PKcs (Figure 4E). Together, data employing IP6K2 deletion, depletion and dominant-negative fragment competition suggest that IP6K2 is required for DNA-PKcs/ATM-mediated p53 S15 phosphorylation at two levels: stabilizing DNA-PKcs and ATM, and also keeping the kinase complexes in proximity to p53.
We then examined whether IP6K2 overexpression potentiates the DNA-PKcs/ATM-p53 pathway (Figure 4F). While overexpressing wild-type IP6K2 does not alter levels of DNA-PKcs/ATM or p53 induction, overexpressed IP6K2 K222A mutant acts as a dominant-negative to diminish DNA-PKcs/ATM levels as well as p53 induction, reinforcing the importance of IP6K2’s catalytic activity in this process. Consistent with this requirement for IP7, in Tti1 depleted cells, expression of wildtype Tti1 results in more extensive p53 induction and S15-phosphorylation than the expression of Tti1S828A (Figure S4E).
IP6K2 appears to regulate apoptotic cell death via the DNA-PKcs/ATM-p53 signaling cascade we have characterized. Thus, activation by 5-FU of p53 transcriptional reporter activity is virtually abolished in HCT116 cells with IP6K2 deletion (Figure 4G) or depletion by shRNA (Figure 4H). Consequently, cell viability is increased in IP6K2 deficient cells treated with 5-FU or etoposide (Figure S4F–G). Apoptotic cell death monitored by TUNEL assay is reduced by 80% in cells depleted of IP6K2 by shRNA (Figure 4J, 4K). IP6K2 knockdown fails to improve the viability of p53−/− HCT116 cells (Figure S4H), consistent with the notion that apoptotic action of IP6K2 requires the DNA-PKcs/ATM-p53 pathway. Expression of IP6K21-67 also enhances cell viability (Figure 4K), reinforcing the importance of IP6K2-mediated p53 S15-phosphorylation in cell death.
If IP6K2 deletion influences cell viability via the DNA-PKcs/ATM signaling cascade, then drugs that inhibit DNA-PKcs or ATM should not exert additive effects upon cell viability. We monitored viability of HCT116 cells with IP6K2 deletion as well as treatment with the DNA-PKcs inhibitor Nu7026 or the ATM inhibitor Ku55933 (Figure 4L, S4I). Nu7026 increases viability in wild type cells but not in IP6K2 knockouts; whereas Ku55933 negatively influences cell viability in both wildtype and knockouts (Figure S4I). Together, these findings suggest that IP6K2 influences cell viability by signaling through DNA-PKcs and ATM, with a specific pro-death contribution from DNA-PKcs, consistent with the emerging apoptotic role of DNA-PKcs (Callen et al., 2009; Cooper et al., 2013; Hill and Lee, 2010).
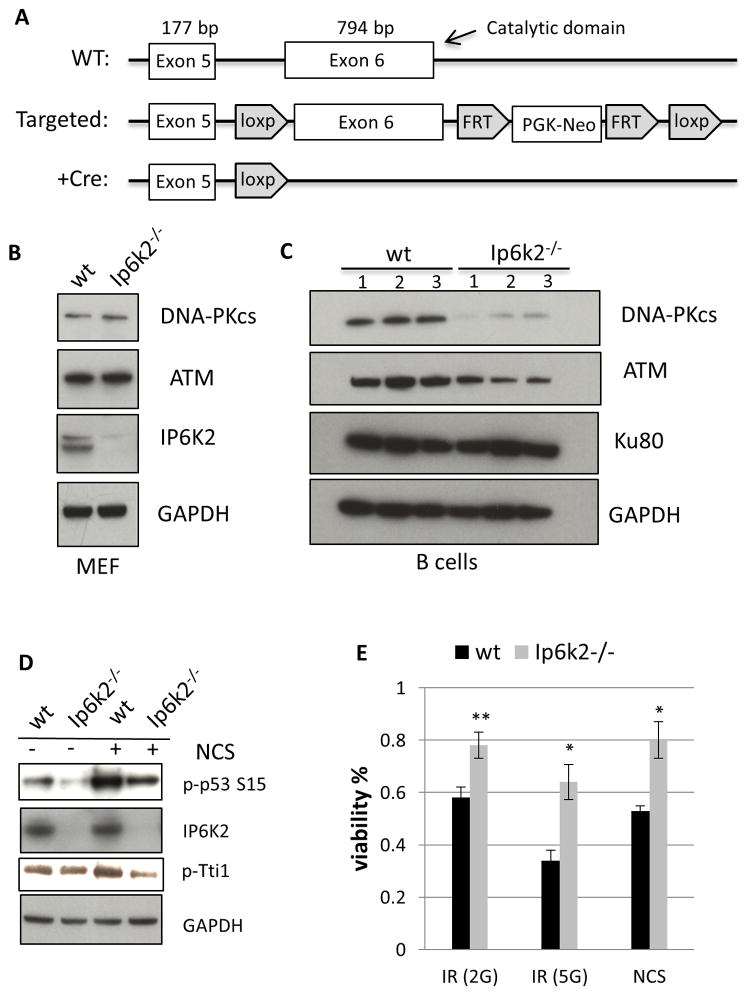
We extended our analysis of this signaling pathway to intact animals by generating mice with targeted deletion of IP6K2, using a Cre-loxp methodology (Figure 5A). The mutant mice are viable, comparable in size to wild-type mice and are born at normal Mendelian ratios. Both male and female mutants are fertile. This phenotype is similar to IP6K2 knockout mice generated by Lindner and associates (Morrison et al., 2009). Mouse embryonic fibroblasts (MEFs) from the mutant mice display about a 20% decrease in generation of IP7 based on [3H]inositol profiling (Figure S5A), confirming findings of the Lindner group (Morrison et al., 2009). This finding is consistent with the observation that in these cells most IP7 is generated by IP6K1 (Chakraborty et al., 2010).
Figure 5.
IP6K2 mediates the DNA-PKcs/ATM-p53 pathway in murine B cells. (A) Schematic depiction of the targeting strategy used to generate Ip6k2−/− mice. Exon 6, containing the majority of the catalytic domain and the 3′ UTR, was flanked by loxP sites to generate the Flox/Flox mice. Breading with a Cre driver mice leads to targeted deletion of exon 6 and loss of IP6K2 expression. (B) Western-blot analysis of primary MEFs prepared from littermate wildtype and Ip6k2−/− mice. (C) Western-blot analysis of resting B cells prepared from littermate wildtype and Ip6k2−/− mice. (D) Western-blot analysis of resting B cells treated with or without NCS (20 μM, 1 h). (E) Viability of B cells after treatment with ionizing radiation (2G, 5 G) or NCS (40 μM). see also Figure S5.
In the IP6K2 deleted MEFs, we fail to detect any alteration in levels of DNA-PKcs or ATM (Figure 5B), which might be due to the minimal contribution of IP6K2 to IP7 formation in MEFs. Whereas murine cells generally display very low levels of DNA-PKcs, human cells possess high concentrations (Figure S5B), consistent with the known difference in DNA-PK activity between human and rodents (Achari and Lees-Miller, 2000). Nussenzweig and associates (Callen et al., 2009) detected substantially higher DNA-PKcs expression in murine B cells than MEFs. Accordingly, we examined DNA-PKcs and ATM in B cell preparations from mice (Figure 5C). Expression of DNA-PKcs and ATM is markedly diminished in IP6K2 deleted B cells, whereas levels of Ku80, a subunit of the DNA-PK holo-complex, are unaffected.
We examined the influence of IP6K2 deletion on the disposition of p53 and cell viability in B cells (Figure 5D, 5E). Neocarzinostatin (NCS), which elicits double-stranded DNA breaks that mimic effects of ionizing radiation, markedly enhances S15-phosphorylation of p53 and phosphorylation of Tti1 with both effects substantially diminished in IP6K2 knockout cells. IP6K2 deletion also significantly increases viability of the B cells subjected to ionizing radiation or NCS treatment. Thus, in cells of intact mice, as in continuous cell lines, IP6K2 regulates p53 phosphorylation and cell viability via influences upon DNA-PKcs/ATM.
DISCUSSION
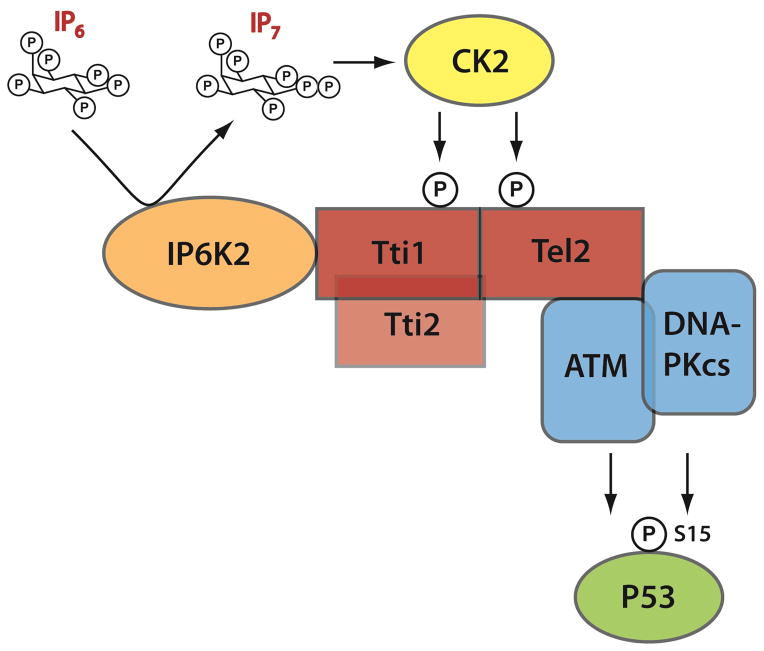
In the present study we elucidate a signaling pathway whereby IP6K2 mediates p53-associated cell death (Figure 6). In this cascade, IP6K2 directly binds Tti1 via its C-terminus, and the IP7 generated by IP6K2 binds with high affinity and selectivity to CK2 enhancing its ability to phosphorylate Tti1/Tel2. This phosphorylation enables the TTT complex to bind and stabilize DNA-PKcs and ATM, which in turn S15-phosphorylates and activates p53. It is now well established that the Tti1/Tti2/Tel2 complex binds to DNA-PKcs/ATM providing stabilization (Takai et al., 2007), and that these latter two proteins stabilize and activate p53 (Vousden and Prives, 2009). Coordination of the pathway had not been previously elucidated. The interface between CK2 and IP6K2/IP7 facilitates this signaling cascade.
Figure 6.
Schematic depiction of IP6K2’s apoptotic actions involving the DNA-PKcs/ATM-p53 pathway. IP7 generated by IP6K2 enhances CK2 phosphorylation of Tti1 and Tel2, leading to their binding of DNA-PKcs/ATM. This binding augments the stability and catalytic activities of DNA-PKcs and ATM, leading to the activating S-15 phosphorylation of p53. S-15 phosphorylation stabilizes p53 and promotes the transcription of apoptotic effectors.
Our earlier study showed that IP6K2 binds directly to p53 via its N-terminus and decreases expression of the pro-arrest gene target p21 (Koldobskiy et al., 2010). Here we show that this interaction is required for p53 to be phosphorylated by DNA-PKcs/ATM, which might contribute to the selective effect of IP6K2 on p53. However, IP6K2 also mediates phosphorylation of H2AX, but not the other substrates examined. The detailed mechanism of substrate specificity remains unclear, but may reflect the fact that H2AX and p53 are common substrates of both DNA-PK and ATM. The depletion of p21 elicited by IP6K2 presumably relates to the known down-regulation of p21 by DNA-PKcs (Hill and Lee, 2010; Hill et al., 2008; Hill et al., 2011; Sluss et al., 2004; Wang et al., 2000), and so constitutes a component of the present signaling cascade.
Several aspects of this cascade are notably selective. It is already established that IP6K2 is unique in its pro-apoptotic actions. Thus, while overexpression of all IP6Ks can augment apoptosis, deletion of IP6K2 but not IP6K1 or IP6K3 augments cell viability (Nagata et al., 2005). IP6K2, but not IP6K1, binds Tti1, generating IP7 in proximity to Tti1 to enhance the CK2-mediated phosphorylation of Tti1. The influence of IP7 upon CK2 phosphorylation is potent and highly selective, with about 10-fold greater potency than IP6. This specific interaction between IP7 and CK2 provides a physiologic means for CK2 regulation. Although CK2 is considered a constitutive enzyme (Pinna and Allende, 2009), its activity can be modulated by IP6 in vitro (Kim et al., 2006; Solyakov et al., 2004). Our in vitro and in vivo studies demonstrate that IP7 and IP6K2 are physiological regulators of CK2. IP7 does not directly interfere with CK2 binding of substrates (peptide or ATP). Rather, IP7 binds to an allosteric site, which also binds heparin, to modulate CK2 interaction with the regulatory protein hNopp140. Two other proteins, nucleolin (Li et al., 1996) and HSP90 (Miyata and Yahara, 1995), also tightly associate with CK2 in a heparin-sensitive, substrate-insensitive manner. Whether they coordinate with IP7 in regulating CK2 activity toward certain substrates is unclear. The highly energetic nature of IP7 and its rapid turnover (Menniti et al., 1993; Stephens et al., 1993) imply that such targets, like Tti1, should reside in the vicinity of IP6Ks. Mice with targeted deletion of IP6K1 or CK2α′ both have defects in male spermatogenesis and fertility (Bhandari et al., 2008; Xu et al., 1999), which might reflect a loss of IP7/CK2 interactions.
There are a few other instances in which IP7 is comparably potent and selective over IP6. In one case, IP7 inhibits Akt phosphorylation by PDK1 with great potency and selectivity over IP6 (Chakraborty et al., 2010). In Dictyostelium, IP7 is about 50–100 times more potent than IP6 in inhibiting membrane translocation of PH domains (Luo et al., 2003). In yeast, a different isomer of IP7 selectively binds and inhibits a cyclin-CDK-CDK-inhibitor complex, with no appreciable effect of IP6 (Lee et al., 2007).
Selectivity is also evident in the specific association of IP6K2 with DNA-PKcs/ATM, but not the other PIKKs. While the mechanism is not entirely clear, it may be relevant that ATR, mTOR, SMG1, and TRRAP form highly stable complexes even under basal conditions (Cortez et al., 2001; Laplante and Sabatini, 2012; Murr et al., 2007; Yamashita et al., 2009), whereas the binding of ATM to its cognate activator complex Mre11-Rad50-Nbs1, and that of DNA-PKcs to Ku70/80, occur after DNA damage and at the damage site (Dvir et al., 1992; Gottlieb and Jackson, 1993; Lee and Paull, 2005). Perhaps, following exit from the ribosomes, mTOR, ATR, SMG-1, and TRRAP dissociate from the TTT chaperone complex and enter into the various complexes. By contrast, DNA-PKcs and ATM appear to remain associated with the TTT proteins and IP6K2. IP6K2 deletion only decreases Tti1 phosphorylation by half but virtually abolishes binding of DNA-PKcs and ATM to Tel2/Tti1, consistent with the notion that IP6K2 selectively impacts a subpopulation of the TTT complex.
Functional interactions between IP6Ks and ATM are also evident in yeast (Saiardi et al., 2005; York et al., 2005). Thus, maintenance of telomere length and cell death induced by wortmannin or caffeine, both mediated by Tel1, the yeast homolog of ATM, are altered in mutants deficient in Kcs1 (yeast IP6K), suggesting that Kcs1 regulates Tel1 activity. Unlike ATM or the other PIKKs, DNA-PKcs is phylogenetically recent, occurring in zebrafish, Xenopus, mouse and human, but not in yeast, C. elegans or Drosophila (Figure S5A). IP6K2 appears to have evolved at the same time as DNA-PKcs and occurs only in species that manifest DNA-PKcs (Figure S5A, S5B). This co-occurrence supports a close functional relationship between the two proteins.
In summary, we define a novel role for IP6K2 in regulating DNA-PKcs and ATM, two PIKKs that are at the center of genome integrity maintenance and apoptosis induction. IP6K2 is therefore a potential cancer chemotherapeutic target. In support of this possibility are genetic alterations in IP6K2 that are associated with cancer (Jones et al., 2012; Kay et al., 2010; Zhao et al., 2010). Accordingly, strategies to activate IP6K2 might afford therapeutic benefit.
EXPERIMENTAL PROCEDURES
Reagents
Chemicals are from Sigma (5-FU, etoposide, cycloheximide, TNP, Mg132, leupeptin, Nu7026, IP6, IS6, Neocarzinostatin), Tocris Biosciences (Ku55933, TBB), and Promega (z-VAD-fmk). IP7 and PCP-IP5 were synthesized as previously described (Wu et al., 2013; Zhang et al., 2009). IP6K2 knockdown shRNA constructs were from Sigma (NM_016291.2).
Antibodies and plasmids
The primary antibodies used were: DNA-PKcs (Abcam, Cell Signaling), ATM (ECM Biosciences), IP6K2, p21, ATR, p-DNA-Pkcs (T2609) (Santa Cruz), p-ATM (S1981) (Epitomics), PUMA, MDM2 and GAPDH (Calbiochem), p53, phospho-p53 (S15, S20, S37, S392), Ku70, Ku80, mTOR, SMG1, TRRAP, p-Chk2, p-Chk1, p-H2AX, p-KAP1 (Cell Signaling), Noxa (Imgenex), Tel2 (Proteintech Group), Tti1 (Bethyl labs), β-tubulin (Hybridoma bank), Flag and GST (sigma), Myc (Roche). Antibody used for IP6K2 immunoprecipitation was generated in our lab.
Plasmids encoding genes studied were obtained from Addgene (Tel2, Tti1, CK2α′, CK2β, pMD2.G, pAX2, and PG13-luc), Source Bioscience (Tti2), and DF/HCC DNA resource (pDNR-dual-His6-hNopp140). Cloning and subcloning of IP6K2, Tti1, and CK2 subunits employed standard procedures. The shRNA construct targeting Tti1 was as described (Hurov et al., 2010), and the siRNA-resistant Flag-Tti1 construct was constructed after 7 silent mutations in the targeting region.
Tandem affinity purification
IP6Ks were cloned into the pcDNA3.1-STAP vector containing streptavidin-binding peptide and Protein A at the C-terminus. After plasmid transfection, cells were lysed, incubated with IgG beads, washed, and then incubated with TEV protease. The supernatants were incubated with streptavidin bead, and the purified complexes were eluted using D-biotin. The eluted samples were fractionated by SDS-PAGE and silver-stained. Bands were sent for LC-MS/MS analysis at the Hopkins Proteomics Core.
Immunoblotting and Immunoprecipitation
Standard methods for cell lysis, immune-precipitation, SDS-PAGE, and Western Blot were as described (Koldobskiy et al., 2010). Blots were quantified using ImageJ.
Cell culture and transfection conditions
HEK23, U2OS and HCT116 cells were cultured and transfected as described (Koldobskiy et al., 2010). Cell viability was assessed in six-well plates by MTT assay, as described previously (Chakraborty et al., 2011b; Koldobskiy et al., 2010). TUNEL assay employed the In Situ Cell Death Detection Kit, Fluorescein (Roche).
Luciferase reporter assay
Cells transfected with PG13-luc were treated with 5-FU (400 μM) for 16 h and lysed for luciferase assay using the Luciferase Assay System (Promega).
In vitro protein kinase assay
To determine dsDNA dependent kinase activity for p53 S15-phosphorylation, whole cell lysates were prepared as described (Achari and Lees-Miller, 2000). Kinase activity was measured using the SignaTECT DNA-dependent Protein Kinase Assay System (Promega).
Expression and purification of recombinant protein from E. coli
The purification of GST- and His6-tagged recombinant proteins were as previously described (Rao et al., 2010). Holo-CK2 was prepared as described (Turowec et al., 2010).
[3H] IP6 binding
Recombinant GST-tagged CK2 proteins on glutathione beads were incubated with [3H] IP6 (Perkin Elmer, specific activity: 10–20 Ci/mmol) overnight in binding buffer containing 40 mM Tris (pH 8.0), 150 mM NaCl, 1mM MgCl2, 4% glycerol and 0.1% Triton. Beads were washed 5 times, and analyzed by scintillation counting.
Reverse Transcript ion PCR
RNA was extracted using the RNeasy Plus Mini Kit (Qiagen), reverse transcribed using the iScript Reverse Transcription Supermix (Bio-Rad). PCR employed 2XPCR master polymerase (Fermentas) for 30–33 cycles.
Generation of IP6K2 Knockout Mice, MEFs and B cell prepration
IP6K2−/− mice were generated and housed similar to IP6K1−/− mice (Chakraborty et al., 2010). Experimental protocols were approved by the Johns Hopkins University Animal Care and Use Committee. Primary MEFs were prepared as previously described (Xu et al., 2013). Radiolabeling with [3H]inostiol and inositol phosphate detection were done as previously described (Chakraborty et al., 2010). B cells were selected from spleens of 8-week old mice, using anti-CD43 Microbeads (Milteny Biotech). Cells were irradiated (2 and 5 Gray) using 137Cs as radiation source. Cell viability was measured by the trypan blue exclusion method (Vandiver et al., 2013).
Statistical analysis
All results are presented as the mean and standard error of at least three independent experiments. Statistical significance was calculated by Student’s t-test (**p < 0.05, *p < 0.01). For detailed experimental procedures, please see supplementary information.
Supplementary Material
Highlights.
IP7 binds with high affinity to CK2.
IP7 binding enhances CK2-mediated phosphorylation of Tel2/Tti1.
IP6K2 binds Tti1 to facilitate stabilization of DNA-PKcs/ATM.
The IP7/CK2/TTT cascade activates the DNA-PKcs/ATM-p53 apoptotic program.
Acknowledgments
We thank Drs S. J. Boulton, F. Basserman, and S. J. Elledge for kindly providing the phospho-Tel2 (S487/491) antibody, phospho-Tti1 (S828) antibody, and shTti1 construct, respectively. We also acknowledge Drs J. Barbi, F. Pan, Y. Dang and J. Liu for kindly providing experimental tools, and members of the Snyder laboratory for reagents and discussions. This work was supported by US Public Health Service Grant DA-000266 (to S.H.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham RT. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Achari Y, Lees-Miller SP. Detection of DNA-dependent protein kinase in extracts from human and rodent cells. Methods Mol Biol. 2000;99:85–97. doi: 10.1385/1-59259-054-3:85. [DOI] [PubMed] [Google Scholar]
- Albert C, Safrany ST, Bembenek ME, Reddy KM, Reddy K, Falck J, Brocker M, Shears SB, Mayr GW. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem J. 1997;327(Pt 2):553–560. doi: 10.1042/bj3270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Barker CJ, Illies C, Gaboardi GC, Berggren PO. Inositol pyrophosphates: structure, enzymology and function. Cell Mol Life Sci. 2009;66:3851–3871. doi: 10.1007/s00018-009-0115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari R, Juluri KR, Resnick AC, Snyder SH. Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc Natl Acad Sci U S A. 2008;105:2349–2353. doi: 10.1073/pnas.0712227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Jankovic M, Wong N, Zha S, Chen HT, Difilippantonio S, Di Virgilio M, Heidkamp G, Alt FW, Nussenzweig A, et al. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell. 2009;34:285–297. doi: 10.1016/j.molcel.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Kim S, Snyder SH. Inositol pyrophosphates as mammalian cell signals. Sci Signal. 2011a;4:re1. doi: 10.1126/scisignal.2001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Koldobskiy MA, Bello NT, Maxwell M, Potter JJ, Juluri KR, Maag D, Kim S, Huang AS, Dailey MJ, et al. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Werner JK, Jr, Koldobskiy MA, Mustafa AK, Juluri KR, Pietropaoli J, Snowman AM, Snyder SH. Casein kinase-2 mediates cell survival through phosphorylation and degradation of inositol hexakisphosphate kinase-2. Proc Natl Acad Sci U S A. 2011b;108:2205–2209. doi: 10.1073/pnas.1019381108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Williams J, Cho J, Falck JR, Shears SB. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J Biol Chem. 2007;282:30763–30775. doi: 10.1074/jbc.M704655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Garcia M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013 doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci U S A. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Fernandez-Saiz V, Targosz BS, Lemeer S, Eichner R, Langer C, Bullinger L, Reiter C, Slotta-Huspenina J, Schroeder S, Knorn AM, et al. SCFFbxo9 and CK2 direct the cellular response to growth factor withdrawal via Tel2/Tti1 degradation and promote survival in multiple myeloma. Nat Cell Biol. 2013;15:72–81. doi: 10.1038/ncb2651. [DOI] [PubMed] [Google Scholar]
- Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- Hill R, Lee PW. The DNA-dependent protein kinase (DNA-PK): More than just a case of making ends meet? Cell Cycle. 2010;9:3460–3469. doi: 10.4161/cc.9.17.13043. [DOI] [PubMed] [Google Scholar]
- Hill R, Leidal AM, Madureira PA, Gillis LD, Waisman DM, Chiu A, Lee PW. Chromium-mediated apoptosis: involvement of DNA-dependent protein kinase (DNA-PK) and differential induction of p53 target genes. DNA Repair (Amst) 2008;7:1484–1499. doi: 10.1016/j.dnarep.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Hill R, Madureira PA, Waisman DM, Lee PW. DNA-PKCS binding to p53 on the p21WAF1/CIP1 promoter blocks transcription resulting in cell death. Oncotarget. 2011;2:1094–1108. doi: 10.18632/oncotarget.378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Horejsi Z, Takai H, Adelman CA, Collis SJ, Flynn H, Maslen S, Skehel JM, de Lange T, Boulton SJ. CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol Cell. 2010;39:839–850. doi: 10.1016/j.molcel.2010.08.037. [DOI] [PubMed] [Google Scholar]
- Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24:1939–1950. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine B, Iannella ML, Bevilacqua MA. Damage-specific DNA binding protein 1 (DDB1): a protein with a wide range of functions. Int J Biochem Cell Biol. 2011;43:1664–1667. doi: 10.1016/j.biocel.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci Signal. 2010;3:ra27. doi: 10.1126/scisignal.2000468. [DOI] [PubMed] [Google Scholar]
- Jones S, Wang TL, Kurman RJ, Nakayama K, Velculescu VE, Vogelstein B, Kinzler KW, Papadopoulos N, Shih Ie M. Low-grade serous carcinomas of the ovary contain very few point mutations. J Pathol. 2012;226:413–420. doi: 10.1002/path.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizuka T, Hara T, Oshiro N, Kikkawa U, Yonezawa K, Takehana K, Iemura S, Natsume T, Mizushima N. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem. 2010;285:20109–20116. doi: 10.1074/jbc.M110.121699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay NE, Eckel-Passow JE, Braggio E, Vanwier S, Shanafelt TD, Van Dyke DL, Jelinek DF, Tschumper RC, Kipps T, Byrd JC, et al. Progressive but previously untreated CLL patients with greater array CGH complexity exhibit a less durable response to chemoimmunotherapy. Cancer Genet Cytogenet. 2010;203:161–168. doi: 10.1016/j.cancergencyto.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Lee KJ, Jeon H, Yu YG. Protein kinase CK2 is inhibited by human nucleolar phosphoprotein p140 in an inositol hexakisphosphate-dependent manner. J Biol Chem. 2006;281:36752–36757. doi: 10.1074/jbc.M604785200. [DOI] [PubMed] [Google Scholar]
- Koldobskiy MA, Chakraborty A, Werner JK, Jr, Snowman AM, Juluri KR, Vandiver MS, Kim S, Heletz S, Snyder SH. p53-mediated apoptosis requires inositol hexakisphosphate kinase-2. Proc Natl Acad Sci U S A. 2010;107:20947–20951. doi: 10.1073/pnas.1015671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee WK, Lee SY, Kim WI, Rho YH, Bae YS, Lee C, Kim IY, Yu YG. Characterization of the InsP6-dependent interaction between CK2 and Nopp140. Biochem Biophys Res Commun. 2008;376:439–444. doi: 10.1016/j.bbrc.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Lee WK, Son SH, Jin BS, Na JH, Kim SY, Kim KH, Kim EE, Yu YG, Lee HH. Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140. Proc Natl Acad Sci U S A. 2013;110:19360–19365. doi: 10.1073/pnas.1304670110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Dobrowolska G, Krebs EG. The physical association of casein kinase 2 with nucleolin. J Biol Chem. 1996;271:15662–15668. doi: 10.1074/jbc.271.26.15662. [DOI] [PubMed] [Google Scholar]
- Lovejoy CA, Cortez D. Common mechanisms of PIKK regulation. DNA Repair (Amst) 2009;8:1004–1008. doi: 10.1016/j.dnarep.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, Huang Y, Nagata E, Devreotes P, Snyder SH. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–572. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- Menniti FS, Miller RN, Putney JW, Jr, Shears SB. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J Biol Chem. 1993;268:3850–3856. [PubMed] [Google Scholar]
- Miyata Y, Yahara I. Interaction between casein kinase II and the 90-kDa stress protein, HSP90. Biochemistry. 1995;34:8123–8129. doi: 10.1021/bi00025a019. [DOI] [PubMed] [Google Scholar]
- Morrison BH, Bauer JA, Hu J, Grane RW, Ozdemir AM, Chawla-Sarkar M, Gong B, Almasan A, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 sensitizes ovarian carcinoma cells to multiple cancer therapeutics. Oncogene. 2002;21:1882–1889. doi: 10.1038/sj/onc/1205265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-beta in ovarian carcinoma cells. J Biol Chem. 2001;276:24965–24970. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison BH, Haney R, Lamarre E, Drazba J, Prestwich GD, Lindner DJ. Gene deletion of inositol hexakisphosphate kinase 2 predisposes to aerodigestive tract carcinoma. Oncogene. 2009;28:2383–2392. doi: 10.1038/onc.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Vaissiere T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene. 2007;26:5358–5372. doi: 10.1038/sj.onc.1210605. [DOI] [PubMed] [Google Scholar]
- Nagata E, Luo HR, Saiardi A, Bae BI, Suzuki N, Snyder SH. Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J Biol Chem. 2005;280:1634–1640. doi: 10.1074/jbc.M409416200. [DOI] [PubMed] [Google Scholar]
- Niefind K, Guerra B, Ermakowa I, Issinger OG. Crystal structure of human protein kinase CK2: insights into basic properties of the CK2 holoenzyme. EMBO J. 2001;20:5320–5331. doi: 10.1093/emboj/20.19.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan U, Dollins DE, Fridy PC, York JD, Downes CP. Characterization of a selective inhibitor of inositol hexakisphosphate kinases: use in defining biological roles and metabolic relationships of inositol pyrophosphates. J Biol Chem. 2009;284:10571–10582. doi: 10.1074/jbc.M900752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Kleckner N. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell. 2003;112:151–155. doi: 10.1016/s0092-8674(03)00033-3. [DOI] [PubMed] [Google Scholar]
- Pinna LA, Allende JE. Protein kinase CK2 in health and disease: Protein kinase CK2: an ugly duckling in the kinome pond. Cell Mol Life Sci. 2009;66:1795–1799. doi: 10.1007/s00018-009-9148-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, Qi Y, Murugan E, Pasunooti S, Ji Q. 2′,3′-cAMP hydrolysis by metal-dependent phosphodiesterases containing DHH, EAL, and HD domains is non-specific: Implications for PDE screening. Biochem Biophys Res Commun. 2010;398:500–505. doi: 10.1016/j.bbrc.2010.06.107. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Nagata E, Luo HR, Snowman AM, Snyder SH. Identification and characterization of a novel inositol hexakisphosphate kinase. J Biol Chem. 2001;276:39179–39185. doi: 10.1074/jbc.M106842200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc Natl Acad Sci U S A. 2005;102:1911–1914. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Sluss HK, Armata H, Gallant J, Jones SN. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol Cell Biol. 2004;24:976–984. doi: 10.1128/MCB.24.3.976-984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solyakov L, Cain K, Tracey BM, Jukes R, Riley AM, Potter BV, Tobin AB. Regulation of casein kinase-2 (CK2) activity by inositol phosphates. J Biol Chem. 2004;279:43403–43410. doi: 10.1074/jbc.M403239200. [DOI] [PubMed] [Google Scholar]
- Stephens L, Radenberg T, Thiel U, Vogel G, Khoo KH, Dell A, Jackson TR, Hawkins PT, Mayr GW. The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s) J Biol Chem. 1993;268:4009–4015. [PubMed] [Google Scholar]
- Takai H, Wang RC, Takai KK, Yang H, de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131:1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- Takai H, Xie Y, de Lange T, Pavletich NP. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev. 2010;24:2019–2030. doi: 10.1101/gad.1956410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowec JP, Duncan JS, French AC, Gyenis L, St Denis NA, Vilk G, Litchfield DW. Protein kinase CK2 is a constitutively active enzyme that promotes cell survival: strategies to identify CK2 substrates and manipulate its activity in mammalian cells. Methods Enzymol. 2010;484:471–493. doi: 10.1016/B978-0-12-381298-8.00023-X. [DOI] [PubMed] [Google Scholar]
- Vandiver MS, Paul BD, Xu R, Karuppagounder S, Rao F, Snowman AM, Ko HS, Lee YI, Dawson VL, Dawson TM, et al. Sulfhydration mediates neuroprotective actions of parkin. Nat Commun. 2013;4:1626. doi: 10.1038/ncomms2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wang S, Guo M, Ouyang H, Li X, Cordon-Cardo C, Kurimasa A, Chen DJ, Fuks Z, Ling CC, Li GC. The catalytic subunit of DNA-dependent protein kinase selectively regulates p53-dependent apoptosis but not cell-cycle arrest. Proc Natl Acad Sci U S A. 2000;97:1584–1588. doi: 10.1073/pnas.97.4.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Dul BE, Trevisan AJ, Fiedler D. Synthesis and characterization of non-hydrolysable diphosphoinositol polyphosphate second messengers. Chem Sci. 2013;4:405–410. doi: 10.1039/C2SC21553E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Sen N, Paul BD, Snowman AM, Rao F, Vandiver MS, Xu J, Snyder SH. Inositol polyphosphate multikinase is a coactivator of p53-mediated transcription and cell death. Sci Signal. 2013;6:ra22. doi: 10.1126/scisignal.2003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Toselli PA, Russell LD, Seldin DC. Globozoospermia in mice lacking the casein kinase II alpha′ catalytic subunit. Nat Genet. 1999;23:118–121. doi: 10.1038/12729. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280:4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Thompson J, Prestwich GD. A scalable synthesis of the IP7 isomer, 5-PP-Ins(1,2,3,4,6)P5. Org Lett. 2009;11:1551–1554. doi: 10.1021/ol900149x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XP, Liu F, Wang W. Two-phase dynamics of p53 in the DNA damage response. Proc Natl Acad Sci U S A. 2011;108:8990–8995. doi: 10.1073/pnas.1100600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Kirkness EF, Caballero OL, Galante PA, Parmigiani RB, Edsall L, Kuan S, Ye Z, Levy S, Vasconcelos AT, et al. Systematic detection of putative tumor suppressor genes through the combined use of exome and transcriptome sequencing. Genome Biol. 2010;11:R114. doi: 10.1186/gb-2010-11-11-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.