Abstract
Tendon formation and repair rely on specific combinations of transcription factors, growth factors, and mechanical parameters that regulate the production and spatial organization of type I collagen. Here, we investigated the function of the zinc finger transcription factor EGR1 in tendon formation, healing, and repair using rodent animal models and mesenchymal stem cells (MSCs). Adult tendons of Egr1–/– mice displayed a deficiency in the expression of tendon genes, including Scx, Col1a1, and Col1a2, and were mechanically weaker compared with their WT littermates. EGR1 was recruited to the Col1a1 and Col2a1 promoters in postnatal mouse tendons in vivo. Egr1 was required for the normal gene response following tendon injury in a mouse model of Achilles tendon healing. Forced Egr1 expression programmed MSCs toward the tendon lineage and promoted the formation of in vitro–engineered tendons from MSCs. The application of EGR1-producing MSCs increased the formation of tendon-like tissues in a rat model of Achilles tendon injury. We provide evidence that the ability of EGR1 to promote tendon differentiation is partially mediated by TGF-β2. This study demonstrates EGR1 involvement in adult tendon formation, healing, and repair and identifies Egr1 as a putative target in tendon repair strategies.
Introduction
Repair of tendon and ligament injuries represents a major clinical challenge for orthopedic medicine (1). A better understanding of the molecular aspects of tendon cell differentiation during tendon development, postnatal formation, healing, and repair is expected to lead to better clinical management of tendon and ligament injuries.
Tendons in adults are composed mostly of a dense extracellular matrix of collagen fibrils that are arranged parallel to the bone-muscle axis (2). The collagen fibrils are hierarchically organized to withstand tensile forces transmitted from muscle to bone (3). The fibrils contain mostly type I collagen, which is a triple helical molecule having 2 α1(I) and 1 α2(I) polypeptide chains that are encoded by the genes Col1a1 and Col1a2, respectively. One of the difficulties in studying tendon development is that tendon specificity is determined not by the expression of Col1a1 or Col1a2 genes, but rather by the specific parallel organization of type I collagen fibrils. Other tendon components important for tendon collagen fibrillogenesis, such as type III, V, VI, XII, and XIV collagens (referred to herein as tendon-associated collagens) and other tendon-associated molecules (proteoglycans, tenascin, elastin), are also present in a wide range of connective tissues. The absence of specific tendon markers explains why our knowledge of tendon development lags behind that of other mesoderm-derived tissues, such as muscle, cartilage, and bone. The discovery of the basic helix-loop-helix (bHLH) transcription factor scleraxis (Scx) as being a specific tendon and ligament marker revolutionized the field of tendon development (4, 5). Scx labels tendon progenitors and differentiated cells in embryos and is also expressed in adult tendons (6, 7). The type II transmembrane glycoprotein, tenomodulin (TNMD), is also a marker of differentiated tenocytes downstream of Scx (5, 8, 9). Two main signaling pathways, TGF-β and FGF, have been identified as being involved in vertebrate tendon development (reviewed in refs. 10, 11). Two DNA-binding molecules, the zinc finger protein EGR1 (early growth response-1) and the homeodomain protein Mohawk, have recently been shown to be involved in tendon development (12–14). However, the molecular interactions between the signaling pathways and transcription factors underlying tendon development have not been fully elucidated.
In addition to the use of animal models to study tendon development, mesenchymal stem cells (MSCs) have been used to study tendon cell differentiation in vitro (15–18). MSCs are multipotent progenitor cells that can be induced to differentiate into various tissues of mesodermal origin (19). MSCs have been extensively used for cartilage and bone repair, since they readily differentiate into chondrocytes and osteocytes under appropriate conditions (19). Consistent with TGF-β and FGF involvement during tendon development, components of the TGF-β and FGF signaling pathways have been shown to increase the expression of Scx or tendon-associated markers in mouse and rat MSCs (15–18). Recently, lentiviral overexpression of Scx in human MSCs has been shown to activate the tendon differentiation gene, Tnmd (17). In addition to being a powerful in vitro system to study the differentiation processes in the tendon lineage, MSCs are also attractive tools for cell therapy in tendon injury (20–23).
In this manuscript, we describe the involvement of Egr1 in adult mouse tendon formation, healing, and repair. EGR1 is a multifunctional transcription factor involved in developmental processes, including cell proliferation, migration, and differentiation, as well as in pathological processes such as fibrosis (24). It is activated by external stimuli such as mechanical forces, stress situations, growth factors, or hormones (25). We have recently shown the remarkable property of Egr1 to be sufficient for tendon gene expression, including Scx and Col1a1, in ectopic contexts in chick embryos (14). Based on this ability to induce tendon gene expression ectopically in the embryo, we evaluated Egr1 as a therapeutic target for tendon repair.
Results
Egr1 is involved in type I collagen production in postnatal tendons by directly regulating Col1a1 and Col1a2 gene transcription.
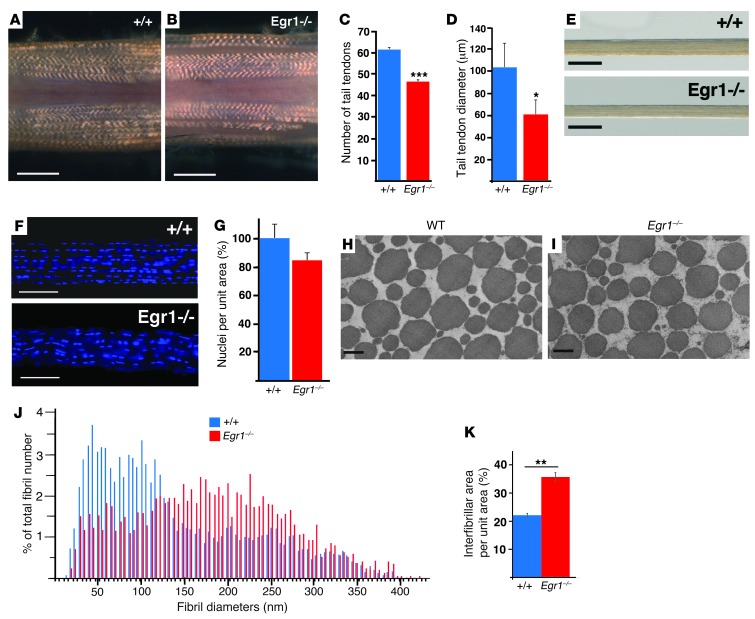
Egr1 is expressed in adult mouse tendons (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI67521DS1). Macroscopic observation of Egr1–/– tails did not show any obvious modification in gross tendon hierarchy, including tendon crimp (Figure 1, A and B). However, we observed a significantly lower number of individual tail tendons in Egr1–/– mice compared with control mice (Figure 1C). There was also a significant diminution in the diameters of tail tendons (Figure 1, D and E), but no change in the number of nuclei per unit area (Figure 1, F and G). However, since the tail tendon diameters of the Egr1–/– mice were smaller than those of the control tendons, we concluded that the absolute number of nuclei was smaller in the Egr1-deficient tendons compared with the controls. Electron microscopic analyses of tail and Achilles tendons of adult Egr1–/– mutant mice showed differences in the repartition of collagen fibril diameters (Figure 1, H–J, for tail tendons, Supplemental Figure 2 for Achilles tendons). In addition, the total interfibrillar area percentage was higher in the Egr1-deficient tendons compared with the control tendons (Figure 1K).
Figure 1. Multiscale analysis of tendons from adult Egr1–/– mice.
(A and B) Macroscopic views showing tail tendons of mouse tails close to the body from 2-month-old WT (A) and Egr1–/– (B) mice. (C) Number of tail tendons counted at the same level (close to the body) from 2- to 3-month-old WT and Egr1–/– mutant mice. (D) Tail tendon diameters from 2- to 3-month-old WT and Egr1–/– mice. (E) Examples of isolated tail tendons from WT and Egr1–/– mice. (F) Hoechst staining of individual tail tendons from WT and Egr1–/– mice. (G) Number of nuclei per unit area in tail tendon sections from 2- to 3-month-old age-matched WT and Egr1–/– mice. Numbers of nuclei were normalized to those in WT mice. (H and I) Electron microscopic analysis of tail tendons from 2-month-old WT and Egr1–/– mice. (J) Histograms showing the frequencies of fibril diameters from WT versus Egr1–/– mice. For WT mice, the mean of the diameters = 143.89 nm; SD = 87.63. For Egr1–/–, the mean of the diameters = 183.73 nm; SD = 84.88. (K) Interfibrillar area (per unit area) of tail tendons from WT and Egr1–/– mice. The error bars represent the SEM. *P < 0.05; **P < 0.01; ***P < 0.001. Scale bars: 1 mm (A and B); 100 μm (E and F); 200 nm (H and I).
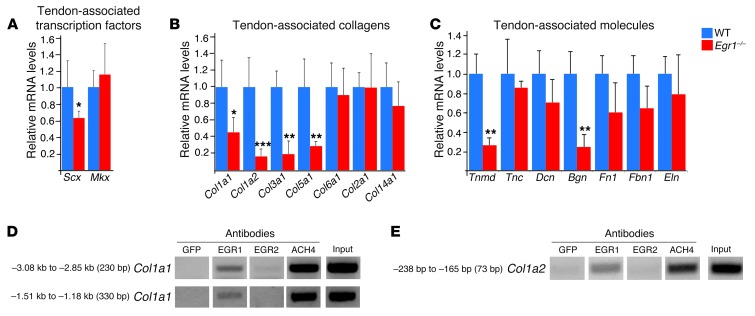
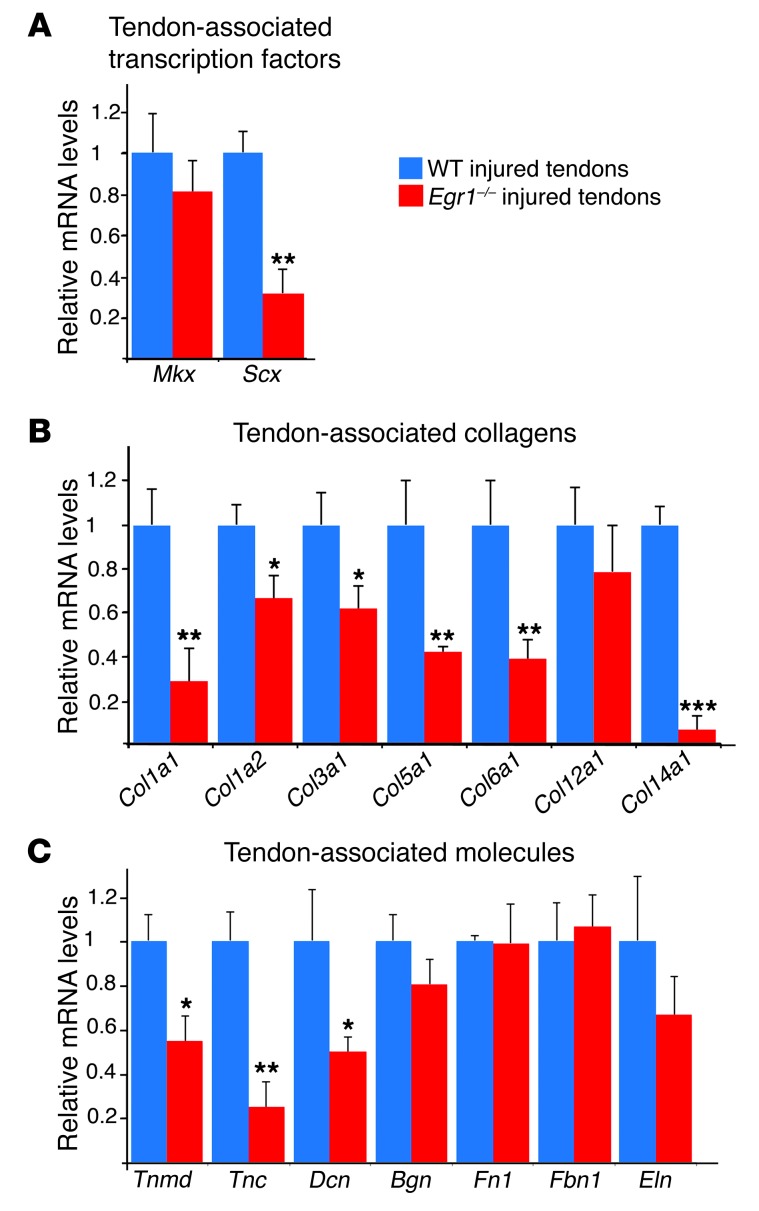
We next examined the effect of the absence of Egr1 activity on transcript expression levels of tendon-associated markers using quantitative RT-PCR (qRT-PCR) in tendons from adult mice. We observed significantly lower levels of mRNAs for the main tendon-associated transcription factor, Scx (Figure 2A), and for the 2 genes coding for the type I collagen chains, Col1a1 and Col1a2 (Figure 2B). The relative mRNA levels were also lower for other tendon-associated molecules, Col3a1, Col5a1, Tnmd, and Bgn, in Egr1-deficient tendons (Figure 2, B and C). ChIP experiments from postnatal tendons showed that EGR1 was recruited to regulatory regions of the mouse Col1a1 and Col1a2 promoters in vivo (Figure 2, D and E; full uncut gels are shown in the Supplemental Material). From these structural and molecular analyses, we conclude that EGR1 is positively involved in collagen transcription in postnatal tendons and is required for complete tendon formation.
Figure 2. Molecular analyses of tendons from Egr1–/– mutant mice.
(A–C) qRT-PCR analyses of tendon gene expression in Egr1–/– versus WT mice. The following tendon markers were analyzed: the tendon-associated transcription factors, Scx and Mkx (A); the tendon-associated collagens, Col1a1, Col1a2, Col3a1, Col5a1, Col6a1, Col12a1, and Col14a1; (B) and the tendon-associated molecules, Tnmd, Tnc, Dcn, Bgn, Fn1, Fbn1, and Eln (C). mRNA levels of tendon markers in WT tendons were normalized to 1. Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001, unpaired Student’s t tests. (D and E) ChIP assays were performed on tendons from postnatal mice with antibodies against EGR1 and EGR2 or ACH4 (acetylated histone H4) as a positive control, or with GFP antibody as a negative control. ChIP products were analyzed by PCR to study the interaction of EGR1 with the tendon regulatory regions of mouse Col1a1 and Col1a2 promoters. Primers targeting a 230-bp region flanking the tendon-specific elements TSE1 and TSE2 and targeting a 330-bp region within the tendon-specific deletion identified DNA regions immunoprecipitated by EGR1 (D). Primers targeting a 73-bp fragment of the Col1a2 promoter identified DNA regions immunoprecipitated by EGR1 (E).
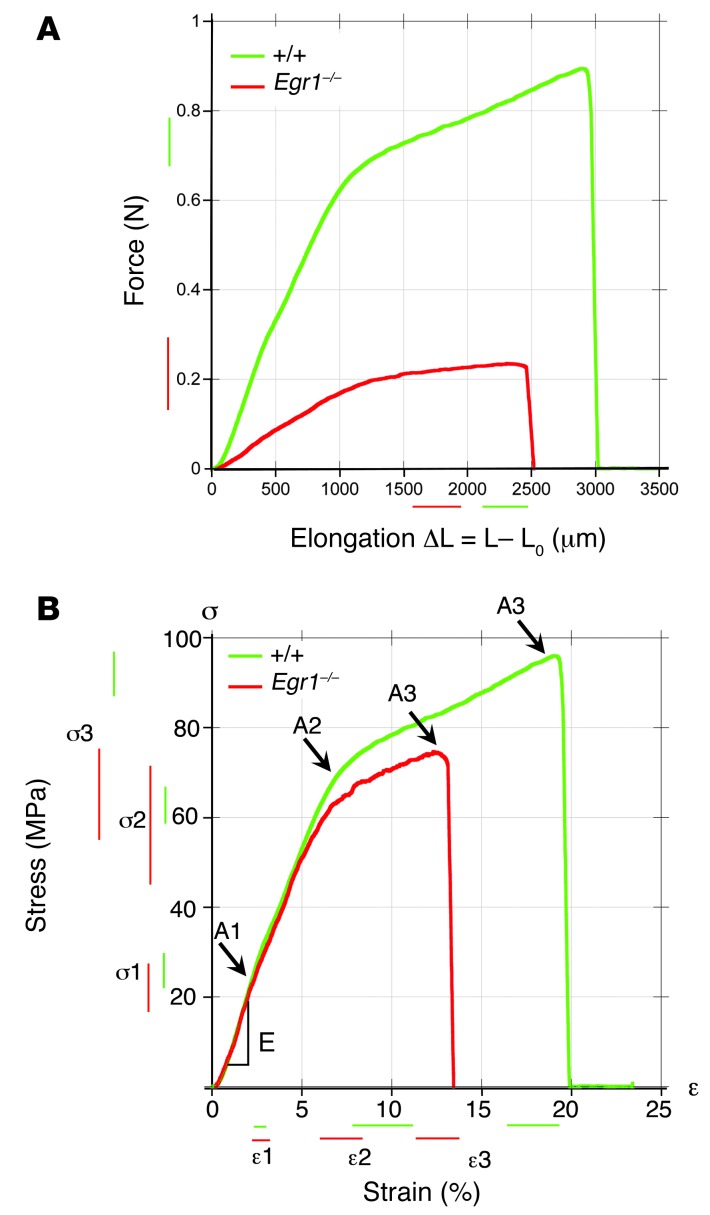
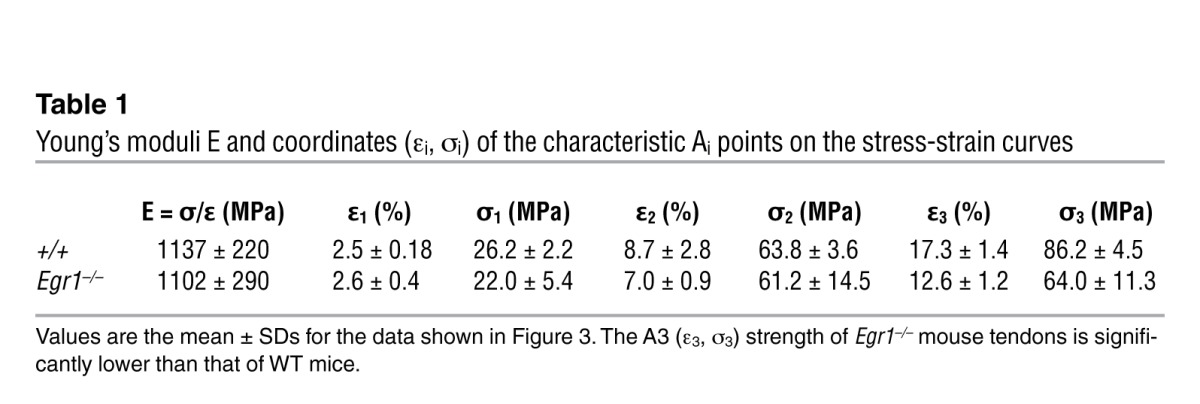
Next, we tested whether the type I collagen deficiency (reduction in Col1a1, Col1a2 transcription, collagen fiber size, and fibril packing density) in Egr1–/– tendons from adult mice altered their mechanical properties (Figure 3). The force-displacement response showed that the Egr1–/– tail tendons were much weaker than the WT tendons (Figure 3A). Because the diameters of individual tail tendons were significantly smaller in Egr1–/– mice than in WT mice (Figure 1D), the stress versus strain curves were established to compare material properties. The initial stress-strain response was found to be linear, with a slope defining the Young’s modulus (E) of the tendon, on the order of 1 MPa (Figure 3B and Table 1). Two successive changes of slopes A1 and A2 were then observed, respectively, at stress/strain coordinates (ε1, σ1) and (ε2, σ2). Finally, we noted the coordinates of the rupture point A3 (ε3, σ3), defining the strength of the tendon. We observed that the low-strain characteristics of the mechanical responses, A1 and A2, were not significantly different between WT and Egr1–/– tendons (Figure 3B and Table 1), showing that in this regime, the difference observed in the force-elongation curves was due to the smaller diameters of the Egr1–/– tendons. However, the ultimate tensile strength, characterized by the rupture point A3 (ε3, σ3), was lower for Egr1–/– tendons than for WT tendons (Table 1), consistent with the increased interfibrillar space (Figure 1K). We conclude that Egr1 gene activity is involved in the failure properties of adult mouse tendons.
Figure 3. Biomechanical properties of tail tendons deficient for Egr1.
(A) Typical force-elongation curves for WT (green) and Egr1–/– (red) mouse tail tendons. (B) Typical stress-strain curves for WT (green) and Egr1–/– (red) mouse tail tendons. These curves can be characterized by the slope E (Young’s modulus) of the linear regime at low strain, by the coordinates (ε1, σ1) and (ε2, σ2) of the 2 successive changes of slopes A1 and A2, and finally, by the coordinates (ε3, σ3) of the rupture point A3. The position and length of the bars next to the axes represent the means and SDs of these coordinates for all animals, averaged over all the tendons tested for a single animal.
Table 1.
Young’s moduli E and coordinates (εi, σi) of the characteristic Ai points on the stress-strain curves
Egr1 deficiency affects tendon gene expression during Achilles tendon healing.
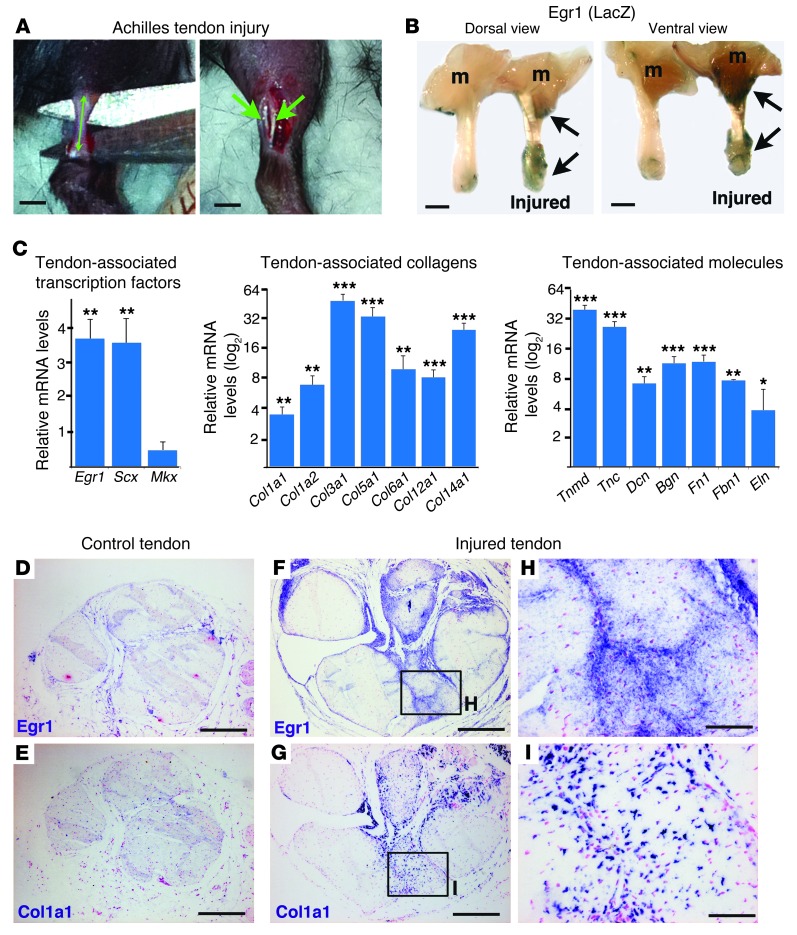
To analyze Egr1 involvement during tendon healing, we developed a mouse model of Achilles tendon injury. Achilles tendons from adult WT mice were sectioned longitudinally (Figure 4A), and the global gene response of tendon markers was analyzed 1–3 weeks after injury (Figure 4, B–I). Egr1 expression was increased in injured tendons compared with noninjured tendons 1 week after injury (Figure 4, B and C). Tendon injury also increased the relative levels of Scx mRNA, without affecting those of Mkx (Figure 4C). In addition, all the tendon-associated collagens and molecules we tested displayed a dramatic increase in mRNA levels in injured tendons compared with noninjured tendons (Figure 4C). Three weeks after injury, Egr1 and Col1a1 were still overexpressed at the sites of tendon injury compared with basal expression in the control tendons (Figure 4D). The massive upregulation of tendon gene expression after Achilles tendon injury is consistent with other studies in murine tendon injury models (26, 27). We next examined the requirement for Egr1 in the gene response during the tendon healing process. Following injury, we found that the increase in tendon gene expression was lower in Egr1-deficient tendons compared with WT tendons (Figure 5, A–C). Interestingly, Col6a1 and Col14a1 mRNA levels, which are not modified in Egr1-deficient (noninjured) tendons (Figure 2B), displayed a significant decrease in Egr1–/– versus WT tendons following injury (Figure 5B), suggesting an involvement of these 2 collagens during tendon healing downstream of Egr1. We conclude that Egr1 is required for injury-induced expression of matrix synthesis genes.
Figure 4. Upregulation of tendon gene expression following injury.
(A) Longitudinal sections parallel to the axis of Achilles tendons were made in adult mice to create a mouse model for tendon injury. (B) We observed increased LacZ expression (reflecting Egr1 expression) in Achilles tendons 1 week after tendon injury in Egr1+/– mice. m, muscle. (C) qRT-PCR analyses of tendon-associated gene expression in injured tendons versus noninjured tendons 1 week after injury. The following tendon markers were analyzed: the tendon-associated transcription factors, Scx and Mkx; the tendon-associated collagens, Col1a1, Col1a2, Col3a1, Col5a1, Col6a1, Col12a1, and Col14a1; and the tendon-associated molecules, Tnmd, Tnc, Dcn, Bgn, Fn1, Fbn1, and Eln. mRNA levels of noninjured tendons were normalized to 1. Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001, paired Student’s t tests. (D–I) Adjacent transverse sections of noninjured (D and E) and injured (F–I) Achilles tendons were hybridized with Egr1 (D, F, and H) or Col1a1 (E, G, and I) probes (blue labeling). Three weeks after injury, Egr1 and Col1a1 expression was upregulated in injured tendons at the site of injury compared with control (noninjured) tendons. Scale bars: 1 mm (A and B); 200 μm (D–G); 50 μm (H and I).
Figure 5. Egr1 is required for normal tendon gene response following mouse tendon injury.
qRT-PCR analyses of tendon-associated gene expression in Egr1–/– injured tendons versus WT injured tendons 1 week after injury. The following tendon markers were analyzed: the tendon-associated transcription factors, Scx and Mkx (A); the tendon-associated collagens, Col1a1, Col1a2, Col3a1, Col5a1, Col6a1, Col12a1, and Col14a1 (B); and the tendon-associated molecules, Tnmd, Tnc, Dcn, Bgn, Fn1, Fbn1, and Eln (C). mRNA levels of injured tendons from WT mice were normalized to 1. Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001, paired Student’s t tests.
The presence of Egr1 results in programming of MSCs toward the tendon lineage.
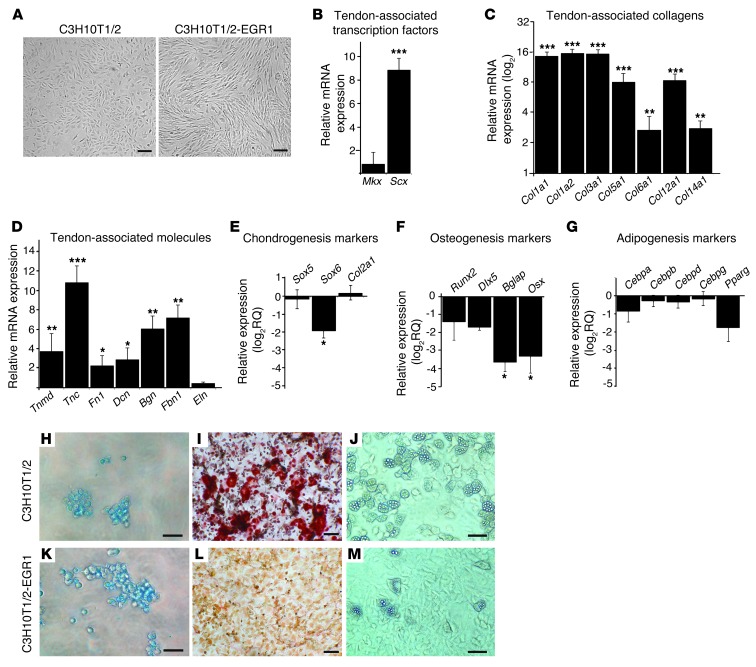
We next investigated whether forced Egr1 expression can commit MSCs to tendon differentiation. We used multipotent murine C3H10T1/2 stem cells (28). C3H10T1/2 cells stably transfected with Egr1 exhibited an elongated shape similar to that of tendon cells in culture (Figure 6A). qRT-PCR analysis revealed that overexpression of Egr1 resulted in increased expression levels of Scx (Figure 6B) and of the 2 type I collagen genes Col1a1 and Col1a2 (Figure 6C). The relative transcript levels of tendon-associated collagens, Col3a1, Col5a1, Col6a1, Col12a1, and Col14a1, were increased as well (Figure 6C). The mRNA levels of other tendon-associated molecules were also increased, especially those of Tnc (Figure 6D). Consistently, the transcript levels of the associated markers for the other mesodermal lineages, cartilage, bone, and fat, were not increased and were even downregulated in some cases in the presence of EGR1 (Figure 6, E–G). The stemness of Egr1-transfected cells was evaluated by examining the differentiation potential of C3H10T1/2-EGR1 cells compared with C3H10T1/2 cells. When stimulated toward the 3 different mesodermal lineages, C3H10T1/2 cells differentiated into chondrocytes, osteocytes, and adipocytes (Figure 6, H–J, and Supplemental Figure 3), as previously described (29). In contrast, the C3H10T1/2-EGR1 cells failed to enter the osteogenic or adipogenic differentiation pathways (Figure 6, L and M, and Supplemental Figure 3). However, the presence of Egr1 did not affect the chondrogenic differentiation pathway (Figure 6K). We conclude that Egr1 is sufficient to trigger the tendon differentiation program in mouse C3H10T1/2 cells and abolishes the capacity of C3H10T1/2 cells to differentiate into osteogenic and adipogenic lineages.
Figure 6. EGR1-producing C3H10T1/2 cells increase the expression of tendon markers and lose their capacity to differentiate into bone and fat lineages.
(A) EGR1-producing C3H10T1/2 cells showed a fibroblastic phenotype compared with C3H10T1/2 cells. EGR1-producing C3H10T1/2 cells displayed increased mRNA expression levels of Scx (B), of the all the tendon-associated collagens, Col1a1, Col1a2, Col3a1, Col5a1, Col6a1, Col12a1, and Col14a1 (C), and of the tendon-associated molecules, Tnmd, Tnc, Fn1, Dcn, Bgn, and Fbn1 (D), but no change in Mkx (B) or Eln (D) expression levels compared with C3H10T1/2 cells. Relative expression levels of mRNAs for the markers associated with chondrogenic (E), osteogenic (F), and adipogenic (G) differentiation were not increased in EGR1-producing C3H10T1/2 cells compared with C3H10T1/2 cells. Relative mRNA expression levels for Sox6 (chondrogenesis) (E), Bglap, and Osx (osteogenesis) (F) were downregulated in the presence of EGR1 compared with control C3H10T1/2 cells. Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001, unpaired Student’s t tests. (H–M) C3H10T1/2 (H–J) and C3H10T1/2-EGR1 (K–M) cells were subjected to chondrocyte (H and K), osteocyte (I and L), and adipocyte (J and M) differentiation. In contrast to C3H10T1/2 cells showing full differentiation into the 3 lineages (H–J), C3H10T1/2-EGR1 cells showed a minimal capacity to differentiate into osteocytes (L) and adipocytes (M), but appeared to retain their capacity to differentiate into chondrocytes (K). Scale bars: 200 μm (A, H–M).
Egr1 promotes the formation of in vitro–engineered tendons from MSCs.
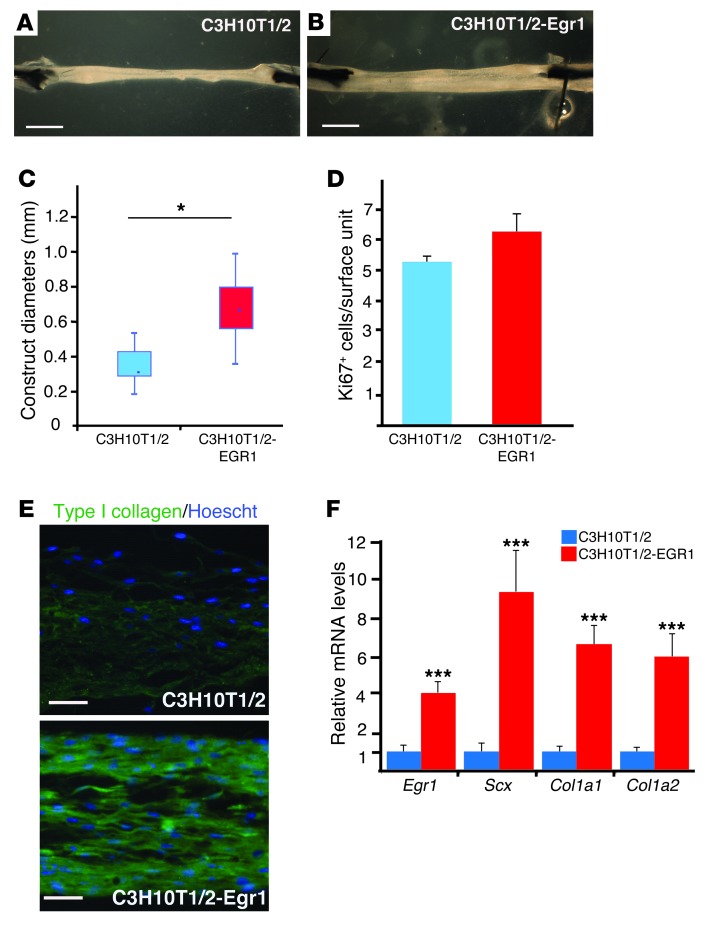
Egr1 was sufficient to trigger the expression of tendon markers in C3H10T1/2 cells (Figure 6). However, this 2D culture system did not allow the analysis of terminal tendon differentiation. Consequently, we investigated the role of Egr1 in the formation of in vitro–engineered tendons. We used a 3D fibrin-based cell culture system that promotes tendon differentiation from primary chick tendon cells and human MSCs (30–32). C3H10T1/2-EGR1 cells cultured under uniaxial tension led to thicker tendon-like structures compared with C3H10T1/2 cells (Figure 7, A and B). Although C3H10T1/2 cells produced tendon-like structures with heterogeneous diameters, diameters of constructs from Egr1-expressing cells were significantly larger compared with those made from C3H10T1/2 cells (Figure 7C). However, the number of proliferative cells did not significantly increase in the presence of Egr1 (Figure 7D), suggesting an increase in matrix synthesis rather than an increase in cell proliferation. Consistent with this result, we observed an increase in type I collagen expression in EGR1-producing constructs versus controls (Figure 7E). This was confirmed with the relative increase in transcript levels for Col1a1, Col1a2, and Scx genes in EGR1-producing constructs versus control constructs (Figure 7F). We conclude that the presence of Egr1 augments the formation of in vitro–engineered tendons from MSCs by increasing Scx and Col1a gene transcription.
Figure 7. EGR1 promotes the formation of tendon-like structures from C3H10T1/2 cells.
(A and B) C3H10T1/2 (A) or C3H10T1/2-EGR1 (B) fibrin gel constructs. The 3-week-old tendon-like constructs made with C3H10T1/2-EGR1 cells were larger than those made with C3H10T1/2 cells. (C) Diameters were significantly larger in the presence of Egr1 compared with those of controls. (D) There was no significant increase in cell proliferation based on Ki67+ cell numbers in C3H10T1/2-EGR1 tendon constructs compared with C3H10T1/2 tendon constructs. (E) Longitudinal sections of C3H10T1/2 or C3H10T1/2-EGR1 fibrin gel constructs were immunostained with type I collagen antibody. (F) An increase in the expression levels of Scx, Col1a1, and Col1a2 transcripts was observed in tendon-like constructs from C3H10T1/2-EGR1 cells compared with control constructs from C3H10T1/2 cells. *P < 0.05; ***P < 0.001, unpaired Student’s t tests. Scale bars: 1 mm (A and B); 250 μm (E).
Improvement in tendon repair using EGR1-producing MSCs in a rat model for tendon injury.
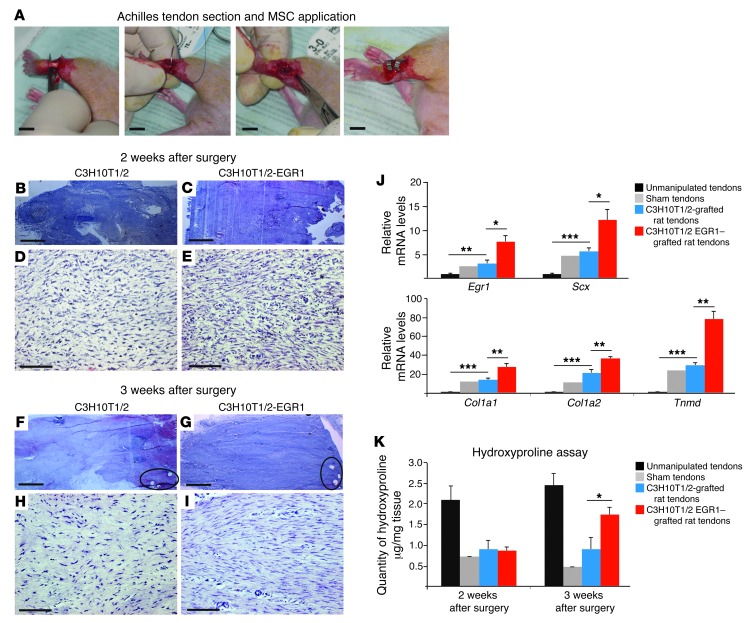
Based on the fact that Egr1 is sufficient to induce tendon differentiation in MSCs (Figure 6) and to promote the formation of in vitro tendon constructs from MSCs (Figure 7), we tested whether the presence of Egr1 could improve tendon repair. We developed a rat model of tendon injury by creating a transverse section in Achilles tendons and immediately suturing both tendon ends, then injecting control C3H10T/1/2 or C3H10T1/2-EGR1 cells (Figure 8A). C3H10T/1/2 or C3H10T1/2-EGR1 cells were implanted in sectioned/sutured Achilles tendons of nude rats. Histological analyses 1 week after cell implantation showed no obvious difference between tendons treated with C3H10T1/2 or EGR1-producing C3H10T1/2 cells (data not shown). Two weeks after cell implantation, an increase in cell density was observed in the presence of Egr1 compared with controls (Figure 8, B–E). Three weeks after implantation, the histology and cell organization of C3H10T1/2-EGR1–treated tendons more closely resembled the histology of normal tendons than that of C3H10T1/2-treated tendons (Figure 8, F–I). We also observed an increase in tendon gene expression, including Egr1, Scx, Col1a1, Col1a2, and Tnmd, in C3H10T1/2-EGR1–treated tendons versus C3H10T1/2-treated tendons 2 weeks after surgery (Figure 8J). An increase in collagen content was also observed in C3H10T1/2-EGR1–treated tendons compared with C3H10T1/2-treated tendons 3 weeks after surgery (Figure 8K). We conclude that the presence of Egr1 in MSCs improves the formation of tendon-like tissues during the process of tendon repair in a rat model of tendon injury.
Figure 8. EGR1 promotes the formation of tendon-like tissues in a rat model for tendon injury.
(A) Achilles tendons of adult nude rats were separated from the plantaris and soleus tendons. A total transverse section of the Achilles tendon was created, and both ends were immediately sutured back together with surgical sutures. C3H10T1/2 or C3H10T1/2-EGR1 cells were then implanted in the injured/sutured tendon. C3H10T1/2- and C3H10T1/2-EGR1–grafted tendons were morphologically and histologically analyzed by H&E staining of sections along the axis of the tendon, 2 (B–E) and 3 (F–I) weeks after manipulation. (F and G) Holes left by the suture points are indicated by ellipses. All the longitudinal sections (B–I) are orientated bone to the left and muscle to the right. (J) qRT-PCR analyses of tendon gene expression in nonmanipulated tendons, sham tendons (with no cell application), C3H10T1/2-grafted tendons, and C3H10T1/2-EGR1–grafted tendons 2 weeks after the operation. mRNA levels of nonmanipulated tendons were normalized to 1. (K) Collagen quantity was assessed using a hydroxyproline assay in nonmanipulated tendons, sham tendons, C3H10T1/2-grafted tendons, and C3H10T1/2-EGR1–grafted tendons 2 and 3 weeks postoperation. Error bars represent SD (J and K). *P < 0.05; **P < 0.01; ***P < 0.001, unpaired Student’s t tests. Scale bars: 1 cm (A); 1 mm (B, C, F, and G); 100 μm (D, E, H, and I).
The tendon-promoting effect of Egr1 acts partially via the TGF-β signaling pathway.
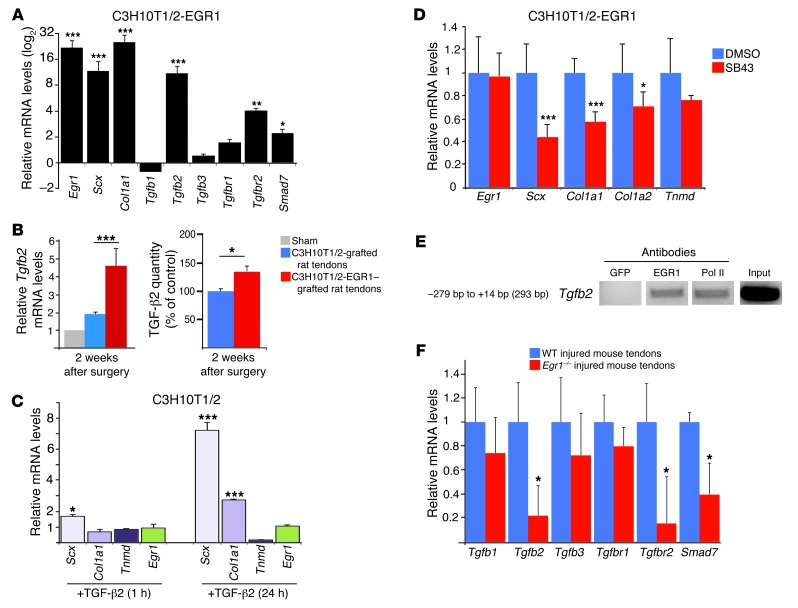
In our rat tendon injury model, we observed that the injected stem cells were not integrated into the manipulated tendons 3 weeks after surgery (Supplemental Figure 4). This absence of MSC colonization in tendons is consistent with previous reports of MSC implantation in rat tendons and in other tissues (20, 23). That MSCs were not integrated into tendons during the process of tendon repair led us to hypothesize that C3H10T1/2-EGR1 cells produced secreted factors promoting the formation of tendon-like tissues. Obvious candidates from mouse developmental studies are the TGF-β family members (33, 34). Consequently, we examined whether EGR1 is able to activate the expression of TGF-β signaling components in C3H10T1/2 cells. EGR1 increased the relative expression levels of Tgfb2 transcripts, without affecting those of Tgfb1 and Tgfb3 in C3H10T1/2 cells (Figure 9A). The relative mRNA expression levels of the receptor TgfbR2 and of the TGF-β intracellular component Smad7 were also significantly increased. The upregulation of Smad7 expression indicated that the canonical SMAD3/TGF-β signaling pathway was active in these cells, since SMAD3 is necessary and sufficient for Smad7 promoter activity through direct SMAD3 binding to the Smad7 promoter in human and murine cells (35–37). We conclude that the SMAD2/3 TGF-β signaling pathway is activated in the presence of EGR1 in MSCs. Next, we determined whether the levels of Tgfb2 mRNAs and TGF-β2 protein were modified in rat tendons grafted with C3H10T1/2-EGR1 cells versus those grafted with C3H10T1/2 cells. Two weeks after surgery, we observed an increase in Tgfb2 mRNA and TGF-β2 protein levels in rat tendons grafted with C3H10T1/2-EGR1 cells versus those grafted with control C3H10T1/2 cells (Figure 9B). We conclude that EGR1 activates Tgfb2 transcription and TGF-β2 production in MSCs using in vitro and in vivo systems.
Figure 9. Link between Egr1 and TGF-β signaling pathway components during tendon cell differentiation in postnatal tendons and injured tendons.
(A) EGR1-producing C3H10T1/2 cells displayed increased mRNA expression levels of the TGF-β signaling pathway components, Tgfb2, Tgfbr2, and Smad7, compared with control C3H10T1/2 cells. mRNA levels for each gene in the control C3H10T1/2 cells were established at 1. (B) Tgfb2 mRNA levels and TGF-β2 quantity were determined in manipulated rat tendons grafted with C3H10T1/2-EGR1 versus control C3H10T1/2 cells 2 weeks postoperation. (C) Application of human recombinant TGF-β2 in C3H10T1/2 cells led to an increase in Scx and Col1a1 expression, while that of Egr1 and Tnmd was not induced 1 or 24 hours after TGF-β2 exposure. (D) Application of a specific TGF-β inhibitor SB43 on C3H10T1/2-EGR1 cells diminished the mRNA expression levels of Scx and Col1a1 genes. mRNA levels of C3H10T1/2-EGR1 cells treated with DMSO were normalized to 1. (E) ChIP assays were performed on tendons from postnatal mice with antibodies against EGR1. ChIP products were analyzed by PCR. Primers targeting a 293-bp fragment of the Tgfb2 promoter identified DNA regions immunoprecipitated by EGR1. (F) qRT-PCR analyses of TGF-β pathway components in Egr1–/– injured tendons versus WT injured tendons 1 week after injury. mRNA levels of injured tendons from Egr1–/– and WT mice were normalized to those of Gapdh in each experiment. For qRT-PCR analyses, the error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Because Egr1 has already been described as being downstream of TGF-β1 signaling in human skin fibroblasts (38, 39), we sought to determine whether TGF-β2 could induce Egr1 expression in C3H10T/2 cells. We found, however, that Egr1 expression was not significantly modified 1, 4, 24, and 48 hours after exposure to TGF-β2 (Figure 9C and Supplemental Figure 5A). In addition, Egr1 expression was not activated 4 hours after TGF-β2 bead application in chick embryos (Supplemental Figure 5B). To determine whether TGF-β2 could mediate the EGR1 effect of increasing tendon gene expression, we analyzed the expression of the main tendon markers that were upregulated by EGR1. The mRNA expression levels of Scx and Col1a1 genes were increased after exposure to TGF-β2 (Figure 9C). However, Tnmd expression levels in C3H10T1/2 cells did not increase after exposure to TGF-β2 (Figure 9C). In addition, the blockade of TGF-β signaling in C3H10T1/2-EGR1 cells using a specific chemical inhibitor, SB43 (40), induced a diminution of mRNA levels of Scx and Col1a1 genes, while it did not affect Tnmd mRNA levels (Figure 9D). We conclude that TGF-β2 does not increase Egr1 expression in C3H10T1/2 cells and only partially mediates the EGR1 effect on tendon gene expression.
Egr1 activates Tgfb2 in postnatal mouse tendons.
The previous experiment in C3H10T1/2 and C3H10T1/2-EGR1 cells suggested that Egr1 was acting upstream of TGF-β signaling during the tendon differentiation process. We therefore performed ChIP assays to test the possibility of an in vivo EGR1 recruitment to the Tgfb2 promoter in mouse tendons. We indeed observed in vivo occupancy of EGR1 in the Tgfb2 promoter regions in postnatal mouse tendons (Figure 9E; full uncut gels are shown in the Supplemental Material). Consistently, in the absence of Egr1 activity, the relative mRNA expression levels of Tgfb2, Tgfbr2, and Smad7 were diminished in Egr1–/– injured tendons compared with WT injured tendons (Figure 9F). We conclude that EGR1 is a good candidate for activating in vivo Tgfb2 transcription in postnatal tendons in physiological and healing conditions.
Discussion
During the first month of postnatal life, there is a marked increase in collagen fibril size in mouse tendons as a consequence of fibril assembly and growth (41). Based on our findings of in vivo EGR1 binding to Col1a1 and Col1a2 promoters in tendons, combined with the decrease in Col1a1 and Col1a2 mRNA levels and the reduction of tendon diameters in Egr1–/– mice, we conclude that EGR1 is actively involved in this postnatal phase of collagen production. Defective collagen transcription in the absence of Egr1 activity leads to tendon fragility. It should be noted, however, that the molecular and biomechanical changes in tendons do not lead to any overt locomotor deficits in Egr1–/– mutant mice. This is consistent with the absence of any obvious visible phenotype in Egr1–/– mutant mice, except for that of infertility (42, 43). In nonmanipulated Egr1–/– mutant mice, only molecular changes with no overt consequences have been reported in different organs (44, 45). However, the molecular changes in tendon gene expression were amplified following tendon injury in Egr1–/– mutant mice, demonstrating a requirement for Egr1 in tendon healing. Amplification of the molecular changes in our mouse model of Achilles tendon injury is consistent with the amplified phenotypes observed in experimentally induced models for fibrosis, atherosclerosis, cardiac hypertrophy, and liver regeneration in Egr1–/– mutant mice (44, 46–51).
We have shown that Egr1 is sufficient to promote the tendon differentiation program in 2D and 3D MSC culture systems and improves the tendon-like histology of a rat model of tendon injury. This tendon-inductive activity is fully consistent with the ability of Egr1 to activate ectopic tendon gene expression in chick embryos (14). Consequently, one main conclusion of this study is that Egr1 is a potent inducer of tendon cell differentiation in vitro and in vivo. Egr1 is sufficient for the expression of a wide range of tendon-associated genes in MSCs, including the main tendon marker Scx, the main tendon collagen genes Col1a1 and Col1a2, the other tendon-associated collagens, the tendon differentiation marker Tnmd, and the secreted molecule Tgfb2. From ChIP experiments, we were able to establish the in vivo recruitment of EGR1 to the Col1a1, Col1a2, and Tgfb2 promoters, showing that EGR1 directly regulates these genes in postnatal tendons. These data are consistent with the direct activation of COL1A2 transcription by EGR1 in human skin fibroblasts (38). Although we are comparing in vivo and in vitro studies, we propose that EGR1 directly regulates Col1a gene transcription in different cell types. Whether EGR1 directly or indirectly regulates transcription of the other tendon-associated genes remains to be determined. We note that early promoter in vitro analyses highlighted EGR1 binding to the mouse Tnc and human FN1 promoters in cell culture systems (52, 53), suggesting direct EGR1 regulation of these genes. In contrast, we could not detect binding of EGR1 to the Scx promoter in mouse tendons in vivo, suggesting that EGR1 indirectly regulates Scx expression. Bgn, known to be required for normal tendon progenitor cell self-renewal and differentiation (54), is also regulated by EGR1 in MCSs and injured tendons. Mkx expression was not regulated by EGR1, consistent with the independent expression of Scx in Mkx mutant mice (12).
EGR1 is sufficient for tendon cell differentiation in vitro and in vivo and is required for complete tendon formation and healing in mice, consistent with EGR1 regulation of the transcription of a whole range of tendon genes. We established that one of the direct transcriptional targets of EGR1 is the Tgfb2 gene coding for the secreted molecule TGF-β2, which partially mediates the tendon-promoting effect of EGR1 in MSCs. This indicates that Egr1 acts upstream of Tgfb2 to drive tendon cell differentiation. In human skin or renal fibroblasts, EGR1 expression has been shown to be induced by TGF-β1 (38). However, we found no increase in Egr1 expression following TGF-β2 application in MSCs, nor after TGF-β2 application in embryonic chick limbs (Figure 9C and Supplemental Figure 5), suggesting differences between skin/renal and tendon systems or between human and mouse/chick models. TGF-β2 activates Scx and Col1a1 expression, but not that of Tnmd, suggesting that TGF-β2 alone is not sufficient to drive full tendon differentiation from MSCs. This is consistent with the finding that the TGF-β ligand is a potent inducer of cartilage differentiation and is the main component of the chondrocyte differentiation medium in MSCs, although chondrocyte differentiation also requires cell adhesion (29, 34). BMP is another signaling pathway involved in tendon cell differentiation (15, 16, 55), although the link between EGR1 and BMP remains to be established in the context of tendon lineage.
Tendon injuries are very common. Cell-based therapies involve combinations of implanted cells, scaffolds, and growth factors (1). The application of nontreated MSCs has shown encouraging, but not fully satisfactory, results for the improvement of histological or biomechanical parameters in various animal models of tendon injury (21–23). In addition, MSCs are not fully integrated into the host tissues, but appear to be important for their paracrine activity (19, 23, 56). This MSC property has led to the optimal therapy of combining MSCs with molecules that promote tendon repair (55, 57, 58). However, the question of which molecules to use still remains unanswered. Here, we present evidence that the transcription factor Egr1 acts upstream of most tendon-associated genes (transcription factors, secreted molecules, collagens, and others types of molecules) in vitro and in vivo. We believe that EGR1 orchestrates several signaling pathways, including TGF-β, to promote tendon cell differentiation in MSCs as well as tendon healing and repair. We suggest that EGR1-producing MSCs remain a good option for tendon repair.
In addition to MSCs and growth factors, mechanical stimulation is required for tendon formation and improves tendon repair (59). Interestingly, EGR1 is known to be expressed in response to mechanical stimuli, mainly via the MAPK pathways (25), and has been described as a mechanosensitive transcription factor that contributes to vascular remodeling of vein grafts in a mouse model (47). An attractive deduction from these reports is that mechanical parameters act upstream of EGR1 during tendon formation and repair and that EGR1 could be the main molecular actor by which mechanical parameters promote the tendon differentiation program. Consistent with this, TGF-β signaling, which we believe acts downstream of EGR1, has been recently shown to link mechanical forces to Scx expression in a mouse model of tendon repair (7).
Methods
Animals.
The Egr1–/– mutant mice bred in a C57BL/6 background had an insertion of a LacZ-neo cassette that inactivates the Egr1 gene (43). Male athymic nude and Wistar rats were purchased from Janvier (France). Gfp transgenic rats, originally generated by Masaru Okabe (University of Osaka, Japan), were provided by A. Chapel (IRSN, France).
Macroscopic analyses of tendons of Egr1–/– mice.
For tail tendon numbers and diameters, a piece of the proximal part of the tail (close to the body) was dissected. All individual tendons were isolated under a stereomicroscope and counted for each tail. Ten Egr1–/– and 10 WT 2- to 3-month-old mice from 10 different litters were analyzed for tail tendon numbers. The absolute number of tail tendons was highly reproducible in WT animals ranging from 58 to 64, with no differences between ages (2 to 3 months) or sex. Tail tendon diameters and the number of nuclei per unit area were measured using ImageJ software (NIH) from 2- to 4-month-old Egr1–/– mice (n = 5) and WT (n = 4) mice.
Transmission electron microscopy.
Tails or Achilles tendons from 2- to 4-month-old Egr1–/– or WT mice from 3 different litters were dissected and fixed in 2% glutaraldehyde and 0.05 M sodium phosphate buffer (pH 7.2). Following fixation, the specimens were processed as described (30). Images were recorded with a MegaView II camera (Olympus) fitted to an FEI BioTWIN transmission electron microscope. ImageJ software (NIH) was used for the measurement of collagen fibril diameters and the interfibrillar area. The collagen fibril diameters from 3 experiments were pooled to show the repartition of fibril diameters in tail tendons.
Biomechanical analysis.
Mechanical testing of mouse tail tendons was performed using a home-built setup. Specimens were bonded to the stainless steel crossheads by setting each extremity on a cyanoacrylate glue drop. The sample and crossheads were immersed in a glass Petri dish filled with PBS buffer at room temperature. The tendon’s initial diameter, d0, was determined assuming full cylindrical symmetry. Mechanical loading of the tendon was achieved by driving 1 crosshead at a constant velocity of 0.1 mm per second with a stepping motor (0.1-micron steps). The loading force F was measured with a 300-g load cell. We defined the nominal tensile stress as σ = 4F/πd02. The nominal strain is defined as e = (L–L0)/L0, where L is the distance between the glue points. The reference length L0 corresponds to the onset of a tensile-restoring force. We estimated L0 as the length above which the crimped structure of the tendon, as revealed by its characteristic birefringence pattern (60, 61), changed noticeably. Starting from a slightly loose tendon state (L < L0), we increased L incrementally until we detected a change in the pitch of the periodic crimp pattern observed between polarizers crossed at 45° of the tendon axis. We measured the nominal stress-strain curves up to rupture for 3 to 4 tendons from WT and Egr1–/– mice from 3 different litters.
Cell culture.
C3H10T1/2 cells (28) of the multipotent murine MSC line were grown in supplemented DMEM (Invitrogen). The human recombinant TGF-β2 (R&D Systems) was applied at 20 ng/ml to C3H10T1/2 cells for 1, 4, 24, or 48 hours. The SB431542 inhibitor (Selleck Chemicals) was applied at 25 mM to C3H10T1/2 cells for 24 hours. TGF-β2–treated, SB43-treated, and nontreated C3H10T1/2 cells were then fixed and processed for qRT-PCR assays to analyze tendon gene expression. Primary GFP MSCs were obtained from 2-month-old Gfp transgenic rats by flushing the bone marrow from the humerus bones and then cultured with DMEM.
Plasmid transfection.
The Egr1 coding sequence obtained from the CMV-EGR1 construct (14) was inserted into the pCDNA3.1+ vector (Invitrogen). The pCDNA-EGR1 construct or empty pCDNA vector (as a control) were transfected using Lipofectamine 2000 reagent (Invitrogen) in C3H10T1/2 cells. Stable transfectants were selected in a medium containing G418 (800 μg/ml). We isolated and expanded about 10 different clones containing the pCDNA-EGR1 construct. The Egr1 clones are referred to as C3H10T1/2-EGR1, and the C3H10T1/2 cells transfected with the empty vector are referred to as C3H10T1/2-EV. We detected no differences between the C3H10T1/2 and C3H10T1/2-EV cells in any of our experiments in terms of morphology, behavior, and gene expression. C3H10T1/2 and C3H10T1/2-EV cells were consequently used indifferently as control cells.
Chondrogenic, osteogenic, and adipogenic differentiation.
C3H10T1/2-EGR1, C3H10T1/2-EV, and C3H10T1/2 cells were stimulated for chondrogenic, osteogenic, and adipogenic differentiation as described (29). Chondrocyte differentiation was analyzed by Alcian blue staining after 26 days of culture. The osteogenic differentiation of MSCs was assayed after 16 days of culture by Alizarin red S staining. Adipogenesis was assayed 7 days afterward, with the apparition of lipid droplets in the culture.
Tendon-like constructs from MSCs.
Tendon-like structures from mouse C3H10T1/2 cells were formed as previously described from embryonic chick tendon cells (30) or from human MSCs (31, 32). After 4 to 7 days, the C3H10T1/2, C3H10T1/2-EV, and C3H10T1/2-EGR1 cells formed continuous tendon-like constructs between the 2 anchors. Each tendon construct was analyzed separately for diameter measurements, cell proliferation, or qRT-PCR. ImageJ software (NIH) was used for the measurement of construct diameters. The diameters were measured at 10 randomly and evenly spaced positions in each construct from C3H10T1/2, C3H10T1/2-EV, or C3H10T1/2-EGR1 cells. Four different C3H10T1/2-EGR1 clones were used, with at least 6 constructs made for each clone. Diameter measurements and cell proliferation numbers were pooled for constructs from C3H10T1/2 and C3H10T1/2-EV cells.
Mouse model for Achilles tendon injury.
Three- to 4-month-old mice were anesthetized by isoflurane inhalation. Left-leg Achilles tendons were manipulated, while right-leg tendons without lesions were left as controls. A 0.5-mm longitudinal full-thickness defect was made along the axis of the tendon. After injury, the skin was closed using 2-0 Mersilk sutures, and the animals were stabulated for 1 to 3 weeks. Sham-operated mice without Achilles tendon lesions were obtained after sectioning and suturing the skin. Mouse Achilles tendons were processed for qRT-PCR analyses 1 week after injury and for in situ hybridization experiments 3 weeks after injury.
Rat model for Achilles tendon injury and MSC injection.
A rat model of Achilles tendon injury was developed. Two- to 3-month-old rats were anesthetized by isoflurane inhalation. Achilles tendons of the left legs were manipulated, while Achilles tendons of the right legs without lesions were left as controls. Achilles tendons from athymic or normal rats were cut transversely to the tendon axis at equal distances between the calcaneum and gastrocnemius muscle. Freshly cultured C3H10T1/2-EGR1 cells, C3H10T1/2-EV cells, or primary rat GFP MSCs were suspended in 40 mg/ml of bovine fibrinogen (Sigma-Aldrich) and mixed with an equal volume of bovine thrombin solution (20 UI/ml; Sigma-Aldrich) to obtain a final concentration of 4 × 106 cells/100 μl. The solution was injected at the site of tendon injury. After cell injection, the skin was closed, and the rats were stabulated for 1 to 3 weeks. A sham surgery (rat Achilles tendons without xenografted cells were cut transversely and sutured) was performed and called the “sham tendon.” For experiments, rats were allowed to move immediately after surgery, and thus tension on their tendons returned to normal. At the end of the experiment, the animals were killed by CO2 inhalation. Rat Achilles tendons were processed for histological analysis 1, 2, and 3 weeks after surgery, and for hydroxyproline assay and qRT-PCR analyses 2 and 3 weeks after surgery.
β-Galactosidase activity.
β-Galactosidase activity was assessed in Achilles tendons from Egr1+/– mice 1 week after injury. Noninjured and injured tendons were fixed for 20 minutes in 4% paraformaldehyde and then incubated in X-gal staining solution for 2 to 4 hours at 37°C.
Histology, in situ hybridization, and immunohistochemistry.
Mouse tendons (from the tail or Achilles tendon) were dissected, fixed, and processed for in situ hybridization (14) or for standard H&E staining of paraffin sections. In situ hybridization analyses of mouse adult tendons were processed using the mouse Egr1 and Col1a1 digoxigenin-labeled mRNA probes, as previously described for mouse embryos (14, 62), followed by nuclear fast red staining. Rat Achilles tendons manipulated with C3H10T1/2-EV or C3H10T1/2-EGR1 cells were fixed in 4% paraformaldehyde, processed for cryostat sections, and stained with H&E. Tendon-like constructs from control C3H10T1/2 or C3H10T1/2-EGR1 cells were fixed in 4% paraformaldehyde, transverse paraffin sectioned, and immunostained with the Ki67 antibody (Ab15580; Abcam) or with the polyclonal antibody against type I collagen (Ab292; Abcam). The slides were counterstained with Hoechst.
RNA isolation, reverse transcription, and qRT-PCR.
Total RNAs were extracted from tail and Achilles tendons of adult (2- to 3-month-old) WT or Egr1–/– mice, from injured Achilles tendons from WT or Egr1–/– mice, from grafted and nongrafted rat tendons, from tendon-like constructs, or from cell cultures (C3H10T1/2, C3H10T1/2-EV, C3H10T1/2-EGR1, TGF-β2–treated C3H10T1/2 cells). qRT-PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) as previously described (14). The primer sequences used for qRT-PCR are listed in Supplemental Table 1. For the xenografting experiments, the primer sequences match the rat and mouse sequences. The results were expressed as mRNA levels corrected with Gapdh levels in each sample. For mRNA levels of tendons from Egr1–/– versus WT mice, 8 independent RNA samples originating from 3 independent litters were analyzed at least in triplicate. For mRNA levels in noninjured versus injured tendons from WT mice, 8–10 independent RNA samples originating from noninjured or injured tendons of WT mice were analyzed at least in triplicate. To determine mRNA levels in injured tendons from WT versus Egr1–/– mice, 6–8 independent RNA samples were analyzed at least in triplicate. mRNA levels in xenografted rat tendons were determined by analyzing 3–4 independent RNA samples in duplicate. For mRNA levels in C3H10T1/2-EGR1 clones versus C3H10T1/2 or C3H10T1/2-EV cells, RNA samples were prepared from at least 8 independent C3H10T1/2-EGR1 clones and analyzed at least 5 times. To determine mRNA levels in tendon-like constructs from C3H10T1/2-EGR1 versus C3H10T1/2 or C3H10T1/2-EV cells, 18 independent constructs from 3 independent C3H10T1/2-EGR1 clones and 18 constructs from C3H10T1/2 or C3H10T1/2-EV cells were analyzed at least in triplicate. RNA samples were prepared from at least from 3 independent experiments for mRNA analyses of C3H10T1/2 cultures treated with TGF-β2.
Hydroxyproline assay.
Xenografted or nongrafted rat tendons were dissected and cut into small pieces (less than 2 mm). A hydroxyproline assay kit (Sigma-Aldrich) was used to measure the production of hydroxyproline in xenografted tendons by the reaction of oxidized hydroxyproline with dimethylaminobenzaldehyde (DMAB), which results in a colorimetric product (560 nm) that is proportional to the hydroxyproline quantity. Three to 4 independent samples per condition were analyzed. Data were analyzed by a Student’s t test using Microsoft Excel. A P value less than 0.05 was considered statistically significant.
ChIP assays.
ChIP assays were performed as previously reported (62). Achilles tendons from 100 postnatal mice were homogenized using a mechanical disruption device (Lysing Matrix A, Fast Prep MP1, 3 × 30 seconds; MP Biomedicals). Eight micrograms of each of the following antibodies were used to immunoprecipitate 25 μg of sonicated chromatin: rabbit polyclonal anti-EGR1 antibody (C-19; Santa Cruz Biotechnology), anti-EGR2 antibody (Covance), antiacetylated histone H4 antibody (Upstate Biotechnology) or 3 μl of anti-RNA polymerase II (Pol II), CTD, and clone 8WG1 (Millipore). ChIP products were analyzed by PCR. Two pairs of primers were used to amplify fragments associated with tendon regulatory regions of the mouse Col1a1 proximal promoter, as previously described (14). The reverse (5′GGCTACGGGGCTTCTTAAAT3′) and forward (5′CCTCGAAAGCCTCAAAAGTG3′) pair of primers was used to amplify a 73-bp fragment (from –238 bp to –165 bp) associated with regulatory regions of the murine Col1a2 promoter. The reverse (5′GCTGCCAGCAGATAACATCA3′) and forward (5′ATGAGTGATCCCCACCATGT3′) pair of primers was use to amplify a 293-bp fragment (from –279 bp to +14 bp) associated with regulatory regions of the murine Tgfb2 promoter.
TGF-β2 immunoassay.
After gelatin-sucrose removal, 30 frozen sections of rat tendons were scraped with a scalpel blade from the slide into a tube containing 400 μl of protein lysis buffer (1X; Cell Signaling Technology) supplemented with a protease inhibitor cocktail. After incubation, samples were homogenized, subjected to sonication, centrifuged, and supernatant was collected and stored at –20°C. The quantity of total protein contained in the supernatant was determined by the Bradford method. The TGF-β2 concentration was measured in the supernatant by ELISA using the Quantikine TGF-β2 ELISA kit (R&D Systems) according to the manufacturer’s instructions. The TGF-β2 concentration was reported to total protein quantity contained in the supernatant.
Statistics.
Each data point represents the mean ± SEM for sample sizes greater than 10. SD was used when the sample size was less than 10. Images show a representative experiment that was repeated at least 3 times. The exact numbers of independent experiments are reported above. Data were analyzed using GraphPad Instat 3.0 (GraphPad Software), by a paired or unpaired Student’s t test. Statistical significance was set at *P < 0.05, **P < 0.01, and ***P < 0.001.
Study approval.
The animal studies were reviewed and approved by the animal care committee of the Comité Régional d’Ethique pour l’Expérimentation Animale N°3, Ile de France (Paris, France).
Supplementary Material
Acknowledgments
We thank Alain Chapel for the Gfp transgenic rats and Patrick Charnay for the mutant mice. We thank Sophie Gournet for illustrations and David F. Holmes for his help with the collagen fibril measurements. The work by D. Duprez was supported by the Fondation pour la Recherche Médicale (FRM) (DVO20081013480), the Agence Nationale de la Recherche (ANR) (1219 01), the CNRS, UPMC, the Association Française contre les Myopathies (AFM), and the FP6 NoE Myores. This work was a collaboration between the CNRS and AP-HP. The work by K.E. Kadler was supported by the Wellcome Trust (091840/Z/10/Z).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2013;123(8):3564–3576. doi:10.1172/JCI67521.
References
- 1.Butler DL, et al. Functional tissue engineering for tendon repair: A multidisciplinary strategy using mesenchymal stem cells, bioscaffolds, and mechanical stimulation. J Orthop Res. 2008;26(1):1–9. doi: 10.1002/jor.20456. [DOI] [PubMed] [Google Scholar]
- 2.Kadler KE, Holmes DF, Trotter JA, Chapman JA. Collagen fibril formation. Biochem J. 1996;316(pt 1):1–11. doi: 10.1042/bj3160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silver FH, Freeman JW, Seehra GP. Collagen self-assembly and the development of tendon mechanical properties. J Biomech. 2003;36(10):1529–1553. doi: 10.1016/S0021-9290(03)00135-0. [DOI] [PubMed] [Google Scholar]
- 4.Schweitzer R, et al. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128(19):3855–3866. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- 5.Murchison ND, et al. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development. 2007;134(14):2697–2708. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- 6.Pryce BA, Brent AE, Murchison ND, Tabin CJ, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev Dyn. 2007;236(6):1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- 7.Maeda T, et al. Conversion of mechanical force into TGF-beta-mediated biochemical signals. Curr Biol. 2011;21(11):933–941. doi: 10.1016/j.cub.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Docheva D, Hunziker EB, Fassler R, Brandau O. Tenomodulin is necessary for tenocyte proliferation and tendon maturation. Mol Cell Biol. 2005;25(2):699–705. doi: 10.1128/MCB.25.2.699-705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shukunami C, Takimoto A, Oro M, Hiraki Y. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev Biol. 2006;298(1):234–247. doi: 10.1016/j.ydbio.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 10.Tozer S, Duprez D. Tendon and ligament: development, repair and disease. Birth Defects Res C Embryo Today. 2005;75(3):226–236. doi: 10.1002/bdrc.20049. [DOI] [PubMed] [Google Scholar]
- 11.Schweitzer R, Zelzer E, Volk T. Connecting muscles to tendons: tendons and musculoskeletal development in flies and vertebrates. Development. 2010;137(17):2807–2817. doi: 10.1242/dev.047498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W, et al. The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol Cell Biol. 2010;30(20):4797–4807. doi: 10.1128/MCB.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito Y, et al. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc Natl Acad Sci U S A. 2010;107(23):10538–10542. doi: 10.1073/pnas.1000525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lejard V, et al. EGR1 and EGR2 involvement in vertebrate tendon differentiation. J Biol Chem. 2011;286(7):5855–5867. doi: 10.1074/jbc.M110.153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann A, et al. Neotendon formation induced by manipulation of the Smad8 signalling pathway in mesenchymal stem cells. J Clin Invest. 2006;116(4):940–952. doi: 10.1172/JCI22689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JY, et al. BMP-12 treatment of adult mesenchymal stem cells in vitro augments tendon-like tissue formation and defect repair in vivo. PLoS One. 2011;6(3):e17531. doi: 10.1371/journal.pone.0017531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alberton P, et al. Conversion of human bone marrow-derived mesenchymal stem cells into tendon progenitor cells by ectopic expression of scleraxis. Stem Cells Dev. 2012;21(6):846–858. doi: 10.1089/scd.2011.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ker ED, et al. Engineering spatial control of multiple differentiation fates within a stem cell population. Biomaterials. 2011;32(13):3413–3422. doi: 10.1016/j.biomaterials.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caplan AI. Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J Cell Physiol. 2007;213(2):341–347. doi: 10.1002/jcp.21200. [DOI] [PubMed] [Google Scholar]
- 20.Shahab-Osterloh S, et al. Mesenchymal stem cell-dependent formation of heterotopic tendon-bone insertions (osteotendinous junctions). Stem Cells. 2010;28(9):1590–1601. doi: 10.1002/stem.487. [DOI] [PubMed] [Google Scholar]
- 21.Chong AK, et al. Bone marrow-derived mesenchymal stem cells influence early tendon-healing in a rabbit achilles tendon model. J Bone Joint Surg Am. 2007;89(1):74–81. doi: 10.2106/JBJS.E.01396. [DOI] [PubMed] [Google Scholar]
- 22.Nourissat G, et al. Mesenchymal stem cell therapy regenerates the native bone-tendon junction after surgical repair in a degenerative rat model. PLoS One. 2010;5(8):e12248. doi: 10.1371/journal.pone.0012248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulotta LV, Kovacevic D, Ehteshami JR, Dagher E, Packer JD, Rodeo SA. Application of bone marrow-derived mesenchymal stem cells in a rotator cuff repair model. Am J Sports Med. 2009;37(11):2126–2133. doi: 10.1177/0363546509339582. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharyya S, Wu M, Fang F, Tourtellotte W, Feghali-Bostwick C, Varga J. Early growth response transcription factors: key mediators of fibrosis and novel targets for anti-fibrotic therapy. Matrix Biol. 2011;30(4):235–242. doi: 10.1016/j.matbio.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagel JI, Deindl E. Early growth response 1 — a transcription factor in the crossfire of signal transduction cascades. Indian J Biochem Biophys. 2011;48(4):226–235. [PubMed] [Google Scholar]
- 26.Scott A, Sampaio A, Abraham T, Duronio C, Underhill TM. Scleraxis expression is coordinately regulated in a murine model of patellar tendon injury. J Orthop Res. 2011;29(2):289–296. doi: 10.1002/jor.21220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dyment NA, et al. The relationships among spatiotemporal collagen gene expression, histology, and biomechanics following full-length injury in the murine patellar tendon. J Orthop Res. 2012;30(1):28–36. doi: 10.1002/jor.21484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reznikoff CA, Brankow DW, Heidelberger C. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res. 1973;33(12):3231–3238. [PubMed] [Google Scholar]
- 29.Juffroy O, Noël D, Delanoye A, Viltart O, Wolowczuk I, Verwaerde C. Subcutaneous graft of D1 mouse mesenchymal stem cells leads to the formation of a bone-like structure. Differentiation. 2009;78(4):223–231. doi: 10.1016/j.diff.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Kapacee Z, et al. Tension is required for fibripositor formation. Matrix Biol. 2008;27(4):371–375. doi: 10.1016/j.matbio.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Kapacee Z, Yeung CY, Lu Y, Crabtree D, Holmes DF, Kadler KE. Synthesis of embryonic tendon-like tissue by human marrow stromal/mesenchymal stem cells requires a three-dimensional environment and transforming growth factor β3. Matrix Biol. 2010;29(8):668–677. doi: 10.1016/j.matbio.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bayer ML, et al. The initiation of embryonic-like collagen fibrillogenesis by adult human tendon fibroblasts when cultured under tension. Biomaterials. 2010;31(18):4889–4897. doi: 10.1016/j.biomaterials.2010.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pryce BA, Watson SS, Murchison ND, Staverosky JA, Dünker N, Schweitzer R. Recruitment and maintenance of tendon progenitors by TGFβ signaling are essential for tendon formation. Development. 2009;136(8):1351–1361. doi: 10.1242/dev.027342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorda-Diez CI, Montero JA, Martinez-Cue C, Garcia-Porrero JA, Hurle JM. Transforming growth factors beta coordinate cartilage and tendon differentiation in the developing limb mesenchyme. J Biol Chem. 2009;284(43):29988–29996. doi: 10.1074/jbc.M109.014811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagarajan RP, Zhang J, Li W, Chen Y. Regulation of Smad7 promoter by direct association with Smad3 and Smad4. J Biol Chem. 1999;274(47):33412–33418. doi: 10.1074/jbc.274.47.33412. [DOI] [PubMed] [Google Scholar]
- 36.Brodin G, Ahgren A, ten Dijke P, Heldin CH, Heuchel R. Efficient TGF-beta induction of the Smad7 gene requires cooperation between AP-1, Sp1, and Smad proteins on the mouse Smad7 promoter. J Biol Chem. 2000;275(37):29023–29030. doi: 10.1074/jbc.M002815200. [DOI] [PubMed] [Google Scholar]
- 37.von Gersdorff G, Susztak K, Rezvani F, Bitzer M, Liang D, Böttinger EP. Smad3 and Smad4 mediate transcriptional activation of the human Smad7 promoter by transforming growth factor β. J Biol Chem. 2000;275(15):11320–11326. doi: 10.1074/jbc.275.15.11320. [DOI] [PubMed] [Google Scholar]
- 38.Chen SJ, et al. The early-immediate gene EGR-1 is induced by transforming growth factor-beta and mediates stimulation of collagen gene expression. J Biol Chem. 2006;281(30):21183–21197. doi: 10.1074/jbc.M603270200. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharyya S, et al. Smad-independent transforming growth factor-beta regulation of early growth response-1 and sustained expression in fibrosis: implications for scleroderma. Am J Pathol. 2008;173(4):1085–1099. doi: 10.2353/ajpath.2008.080382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inman GJ, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62(1):65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 41.Zhang G, et al. Development of tendon structure and function: regulation of collagen fibrillogenesis. J Musculoskelet Neuronal Interact. 2005;5(1):5–21. [PubMed] [Google Scholar]
- 42.Lee SL, et al. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1). Science. 1996;273(5279):1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- 43.Topilko P, et al. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol Endocrinol. 1998;12(1):107–122. doi: 10.1210/me.12.1.107. [DOI] [PubMed] [Google Scholar]
- 44.Saadane N, Alpert L, Chalifour LE. Altered molecular response to adrenoreceptor-induced cardiac hypertrophy in Egr-1-deficient mice. Am J Physiol Heart Circ Physiol. 2000;278(3):H796–H805. doi: 10.1152/ajpheart.2000.278.3.H796. [DOI] [PubMed] [Google Scholar]
- 45.Min IM, Pietramaggiori G, Kim FS, Passegué E, Stevenson KE, Wagers AJ. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;2(4):380–391. doi: 10.1016/j.stem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 46.Liao Y, Shikapwashya ON, Shteyer E, Dieckgraefe BK, Hruz PW, Rudnick DA. Delayed hepatocellular mitotic progression and impaired liver regeneration in early growth response-1-deficient mice. J Biol Chem. 2004;279(41):43107–43116. doi: 10.1074/jbc.M407969200. [DOI] [PubMed] [Google Scholar]
- 47.Wu M, et al. Essential roles for early growth response transcription factor Egr-1 in tissue fibrosis and wound healing. Am J Pathol. 2009;175(3):1041–1055. doi: 10.2353/ajpath.2009.090241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X, et al. Mechano-sensitive transcriptional factor Egr-1 regulates insulin-like growth factor-1 receptor expression and contributes to neointima formation in vein grafts. Arterioscler Thromb Vasc Biol. 2010;30(3):471–476. doi: 10.1161/ATVBAHA.109.184259. [DOI] [PubMed] [Google Scholar]
- 49.Pritchard MT, Nagy LE. Hepatic fibrosis is enhanced and accompanied by robust oval cell activation after chronic carbon tetrachloride administration to Egr-1-deficient mice. Am J Pathol. 2010;176(6):2743–2752. doi: 10.2353/ajpath.2010.091186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albrecht C, et al. Egr-1 deficiency in bone marrow-derived cells reduces atherosclerotic lesion formation in a hyperlipidaemic mouse model. Cardiovasc Res. 2010;86(2):321–329. doi: 10.1093/cvr/cvq032. [DOI] [PubMed] [Google Scholar]
- 51.Bhattacharyya S, et al. Egr-1 induces a profibrotic injury/repair gene program associated with systemic sclerosis. PLoS One. 2011;6(9):e23082. doi: 10.1371/journal.pone.0023082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Copertino DW, Edelman GM, Jones FS. Multiple promoter elements differentially regulate the expression of the mouse tenascin gene. Proc Natl Acad Sci U S A. 1997;94(5):1846–1851. doi: 10.1073/pnas.94.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu C, Yao J, de Belle I, Huang RP, Adamson E, Mercola D. The transcription factor EGR-1 suppresses transformation of human fibrosarcoma HT1080 cells by coordinated induction of transforming growth factor-β1, fibronectin, and plasminogen activator inhibitor-1. J Biol Chem. 1999;274(7):4400–4411. doi: 10.1074/jbc.274.7.4400. [DOI] [PubMed] [Google Scholar]
- 54.Bi Y, et al. Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med. 2007;13(10):1219–1227. doi: 10.1038/nm1630. [DOI] [PubMed] [Google Scholar]
- 55.Pelled G, et al. Smad8/BMP2-engineered mesenchymal stem cells induce accelerated recovery of the biomechanical properties of the Achilles tendon. J Orthop Res. 2012;30(12):1932–1939. doi: 10.1002/jor.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.da Silva Meirelles L, Fontes AM, Covas DT, Caplan AI. Mechanisms involved in the therapeutic properties of mesenchymal stem cells. Cytokine Growth Factor Rev. 2009;20(5–6):419–427. doi: 10.1016/j.cytogfr.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 57.Gulotta LV, Kovacevic D, Montgomery S, Ehteshami JR, Packer JD, Rodeo SA. Stem cells genetically modified with the developmental gene MT1-MMP improve regeneration of the supraspinatus tendon-to-bone insertion site. Am J Sports Med. 2010;38(7):1429–1437. doi: 10.1177/0363546510361235. [DOI] [PubMed] [Google Scholar]
- 58.Gulotta LV, Kovacevic D, Packer JD, Deng XH, Rodeo SA. Bone marrow-derived mesenchymal stem cells transduced with scleraxis improve rotator cuff healing in a rat model. Am J Sports Med. 2011;39(6):1282–1289. doi: 10.1177/0363546510395485. [DOI] [PubMed] [Google Scholar]
- 59.Killian ML, Cavinatto L, Galatz LM, Thomopoulos S. The role of mechanobiology in tendon healing. J Shoulder Elbow Surg. 2012;21(2):228–237. doi: 10.1016/j.jse.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rigby BJ, Hirai N, Spikes JD, Eyring H. The mechanical properties of rat tail tendon. J Gen Physiol. 1959;43(2):265–283. doi: 10.1085/jgp.43.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Diamant J, Keller A, Baer E, Litt M, Arridge RG. Collagen; ultrastructure and its relation to mechanical properties as a function of ageing. Proc R Soc Lond B Biol Sci. 1972;180(60):293–315. doi: 10.1098/rspb.1972.0019. [DOI] [PubMed] [Google Scholar]
- 62.Havis E, et al. Sim2 prevents entry into the myogenic program by repressing MyoD transcription during limb embryonic myogenesis. Development. 2012;139(11):1910–1920. doi: 10.1242/dev.072561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.