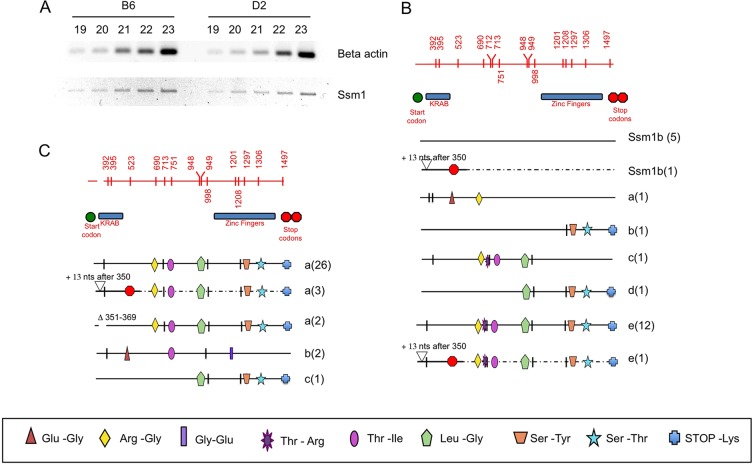
Fig. 3.
Expression of Ssm1 in B6 and D2 ESCs. (A) RT-PCR of beta actin and Ssm1 in B6 and D2 undifferentiated ESCs. The number of PCR cycles is indicated. (B) Ssm1 mRNAs in B6 ESCs. Each horizontal line represents the mRNA sequence of each member of the Ssm1 family amplified. Numbers in parentheses indicate the number of bacterial clones for each sequence. The region shown is coding sequence of Ssm1 from nt 276 to nt 1499 (supplementary material Fig. S2). Lines labeled Ssm1b indicate sequences matching the genomic sequence of Ssm1b and hence considered the Ssm1b cDNA. Lines marked a, b, c, d and e represent the other Ssm1 family sequences. Alternative splice forms of the same sequence with either addition (+13 or +39) or deletion (Δ) of nucleotides are labeled with the same letter as the original sequence. These changes lie within the translated RNA. In some splice forms the addition of 13 nt leads to a STOP codon 153 nt further 3′. Vertical marks indicate silent SNPs differing from the Ssm1b sequence. SNPs causing amino acid changes are marked with a symbol with the actual amino acid changes depicted beneath. Red top line displays the Ssm1 region with vertical marks for all SNPs with nucleotide positions indicated. The positions of the KRAB (nt 357-477, supplementary material Fig. S2) and ZF (nt 966-1448) domains and the start (nt 276-278) and STOP (nt 1497-1499) codons are also depicted. (C) Expression of Ssm1 in D2 ESCs. There is no expression of Ssm1b in the D2 ESCs.