Abstract
Transcription factors and microRNAs (miRNAs) are two important classes of trans-regulators in differential gene expression. Transcription factors occupy cis-regulatory motifs in DNA to activate or repress gene transcription, whereas miRNAs specifically pair with seed sites in target mRNAs to trigger mRNA decay or inhibit translation. Dynamic spatiotemporal expression patterns of transcription factors and miRNAs during development point to their stage- and tissue-specific functions. Recent studies have focused on miRNA functions during development; however, much remains to explore regarding how the expression of miRNAs is initiated and how dynamic miRNA expression patterns are achieved by transcriptional regulatory networks at different developmental stages. Here, we focused on the identification, regulation and function of miRNAs during the earliest stage of Drosophila development, when the maternal-to-zygotic transition (MZT) takes place. Eleven miRNA clusters comprise the first set of miRNAs activated in the blastoderm embryo. The transcriptional activator Zelda is required for their proper activation and regulation, and Zelda binding observed in genome-wide binding profiles is predictive of enhancer activity. In addition, other blastoderm transcription factors, comprising both activators and repressors, the activities of which are potentiated and coordinated by Zelda, contribute to the accurate temporal and spatial expression of these miRNAs, which are known to function in diverse developmental processes. Although previous genetic studies showed no early phenotypes upon loss of individual miRNAs, our analysis of the miR-1; miR-9a double mutant revealed defects in gastrulation, demonstrating the importance of co-activation of miRNAs by Zelda during the MZT.
Keywords: MZT, Zelda, Co-activation, Gene network, MicroRNA
INTRODUCTION
Since the discovery that lin-4 encodes a non-coding RNA (Lee et al., 1993), hundreds of microRNAs (miRNAs) have been discovered in plants, animals, single-celled eukaryotes and viruses (reviewed by Carrington and Ambros, 2003; Bartel, 2004; Kozomara and Griffiths-Jones, 2011). As inhibitors of target mRNAs, miRNAs have been shown to function in many biological processes, including development, metabolism and disease (reviewed by He and Hannon, 2004; Flynt and Lai, 2008; Mendell and Olson, 2012; Ameres and Zamore, 2013). Computational methods, classic genetic experiments and high-throughput genomic studies continue to uncover targets of miRNAs (Rajewsky, 2006; Bartel, 2009; Thomson et al., 2011). With the ever-expanding knowledge of miRNAs, their function is now seen as an important second layer of gene regulation after transcription factors in cis-trans regulatory networks (Martinez and Walhout, 2009), particularly since they share similar strategies in utilizing a small number of nucleotides to simultaneously regulate a large number of targets in a short time span.
The Drosophila model system has provided many insights into the biogenesis and functions of miRNAs. The 238 annotated miRNAs in Drosophila (miRBase; Kozomara and Griffiths-Jones, 2011) were identified by classic forward genetics, deep-sequencing and computational predictions (Berezikov et al., 2006, 2011; Sokol, 2008). Methods such as northern analysis, RT-qPCR and in situ hybridization have provided meaningful information about the level, timing and tissue-specific expression of miRNAs (Sempere et al., 2003; Aravin et al., 2003; Leaman et al., 2005; Aboobaker et al., 2005; Ruby et al., 2007).
Similar to protein-coding genes, miRNA genes are regulated by sophisticated spatial and temporal signals to ensure their proper production in specific cell types. Sokol and Ambros (2005) showed that the muscle-specific transcription factors Twist (Twi) and Mef2 are key activators of miR-1 in Drosophila. Genomic studies have also identified regulators of miRNAs, such as Dorsal (Dl), c-Myc (Diminutive – FlyBase) and Ecdysone (Zeitlinger et al., 2007; Qian et al., 2011; Sempere et al., 2003). However, for many miRNAs, particularly those differentially expressed across developmental stages, the regulatory networks that control their transcription remain unknown. We are interested in the gene network that regulates miRNA functions during the maternal-to-zygotic transition (MZT), a time when developmental control is transferred from maternal products preloaded into the egg to the embryo's own genome (reviewed by Schier, 2007), which in Drosophila is activated ∼1 hour after fertilization. During the MZT, thousands of maternal RNAs are degraded and hundreds of newly synthesized RNAs appear (Tadros and Lipshitz, 2009); thus, the MZT represents a major reprogramming event of the early transcriptome (Giraldez, 2010). Previously, we reported that the zinc-finger transcription factor Zelda (Vielfaltig – FlyBase) plays a key role during the MZT in Drosophila, collectively activating batteries of genes involved in early developmental processes, such as sex determination, cellularization and axis patterning (Liang et al., 2008; Nien et al., 2011). Interestingly, Zelda also activates the miR-309 cluster of eight miRNAs (Liang et al., 2008), which is involved in the clearance of many maternally loaded mRNAs (Bushati et al., 2008). Since Zelda plays such an extensive role in zygotic genome activation, possibly as a pioneer factor to prime genes for transcriptional activation (Nien et al., 2011; Harrison et al., 2011), we investigated the possibility that Zelda activates the miRNAs expressed during the MZT.
Here, we identified a group of miRNAs (11 clusters) that are zygotically expressed in cellular blastoderm embryos, and showed that Zelda regulates all 11. We located the enhancers of several miRNAs by virtue of Zelda ChIP binding, and demonstrated that Zelda binding sites, also known as CAGGTAG sites (De Renzis et al., 2007) or TAGteam sites (Bosch et al., 2006), in these enhancers are essential for proper activation. We further showed that anteroposterior (AP) and dorsoventral (DV) patterning factors work together with Zelda to ensure timely and robust transcriptional activation of these miRNAs, contributing to their accurate spatial expression patterns. The reduced and disrupted miRNA expression seen in zelda mutants affects their downstream functions in maternal mRNA degradation, cell death gene repression and Hox gene regulation. We observed ventral midline defects during gastrulation in miR-1; miR-9a double mutants, which was not seen in either single mutant, suggesting that the coordinated activation of miRNAs by Zelda is crucial for their combinatorial function. Our analysis offers a systems-level view and understanding of the early gene network. Zelda sits as a major hub in the network, globally activating both protein-coding and non-coding genes, thereby orchestrating the early developmental processes.
RESULTS
Identification of blastoderm-expressed miRNAs by transcriptome analysis
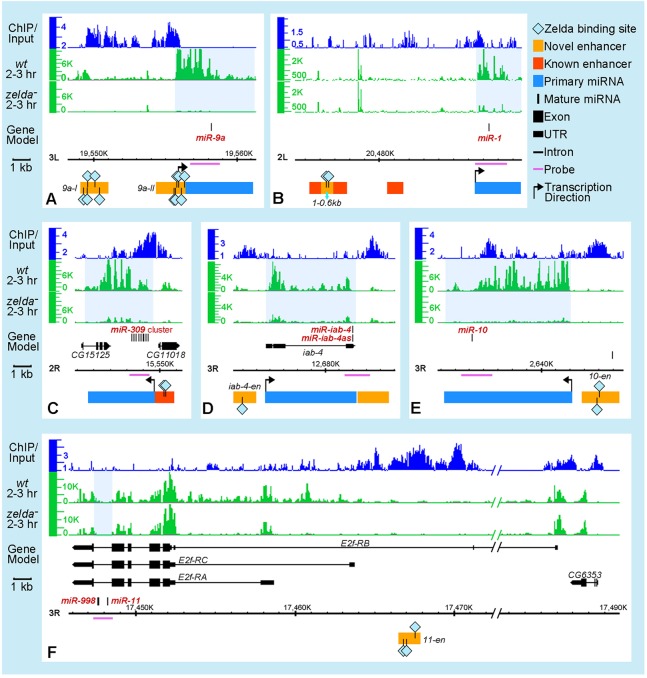
Previous studies showed that miRNAs are expressed across different developmental stages (Aravin et al., 2003; Aboobaker et al., 2005; Ruby et al., 2007; Graveley et al., 2011). In order to focus on the set of miRNAs expressed during the MZT, and to determine which are regulated by Zelda, we examined the transcription profiles of all 188 annotated Drosophila miRNA loci (comprising 238 miRNAs) in our published datasets from 2- to 3-h wild-type and zelda mutant embryos, together with Zelda ChIP profiles (Nien et al., 2011). In total, ten loci (21 miRNAs) were expressed in 2- to 3-h embryos (Fig. 1; supplementary material Fig. S1 and Table S1). One locus is transcribed in both directions, yielding two miRNAs: miR-iab-4 and miR-iab-4as (or miR-iab-8; Ronshaugen et al., 2005; Bender, 2008; Stark et al., 2008; Tyler et al., 2008); thus, 11 miRNA clusters are transcribed in the blastoderm embryo. Strikingly, Zelda binding was observed in regions close to all of them, and most were downregulated in zelda mutants compared with wild type (Fig. 1; supplementary material Fig. S1 and Table S1).
Fig. 1.
Zelda binds upstream of miRNA transcription units and regulates their expression. Integrated Genome Browser (IGB) (Nicol et al., 2009) views are shown for six Drosophila miRNA loci (A-F; miRNAs shown as black vertical lines in the gene model; genome version BDGP R5/dm3). Tracks above gene models represent data from Nien et al. (Nien et al., 2011) as follows: Zelda ChIP binding in 1- to 2-h wild-type (wt) embryos (top, blue), RNA expression profiles of 2- to 3-h wt (middle, green) and zelda mutant (bottom, green) embryos. Highlighted below the gene models are: pri-miRNA transcription units (blue rectangles) estimated from the wt expression track, previously identified miRNA enhancers (red boxes) (Biemar et al., 2005; Zeitlinger et al., 2007), and newly identified enhancers from this study (orange boxes). Zelda binding sites (light blue diamonds) within enhancers are indicated. The horizontal purple lines under the gene models delineate the probes used to detect pri-miRNA transcripts in embryos. Light blue shading on the expression tracks indicates intergenic or intronic regions harboring the miRNAs that are downregulated in zelda mutants. Note that miR-11 is located within an intron of the E2f-RC isoform (F), and Zelda-bound regions lie upstream of E2f-RB.
Coordinated activation of early zygotic miRNAs by Zelda
We further investigated the roles of Zelda in regulating miRNAs by comparing the expression patterns of miRNAs in wild-type and zelda mutant embryos using in situ hybridization, which offers finer temporal and spatial resolution than transcriptome analysis. We designed 1-2 kb probes to target the primary miRNA (pri-miRNA) transcripts, delineated from our transcriptome data (Fig. 1; supplementary material Tables S1 and S2). For miRNAs located within introns of protein-coding genes (miR-11 in E2f, miR-92a in jigr1, miR-2a-1 in spi, and miR-965 in kis; Fig. 1F; supplementary material Fig. S1), we used intronic sequences centered on the mature miRNA (supplementary material Tables S1 and S2). The presence of signal as nuclear dots is indicative of nascent transcripts, which distinguishes zygotic from maternal transcripts, as all four host genes are maternally loaded. Nuclear dots were observed in blastoderm embryos for all 11 miRNAs (Fig. 2). In addition, we tested the other 20 miRNAs known to be expressed in 0- to 6-h embryos (Aravin et al., 2003; Ruby et al., 2007), and, as predicted, we did not observe nuclear dots with any of these probes (supplementary material Table S1). Thus, we are confident that a total of 11 miRNA clusters are transcribed in blastoderm embryos.
Fig. 2.
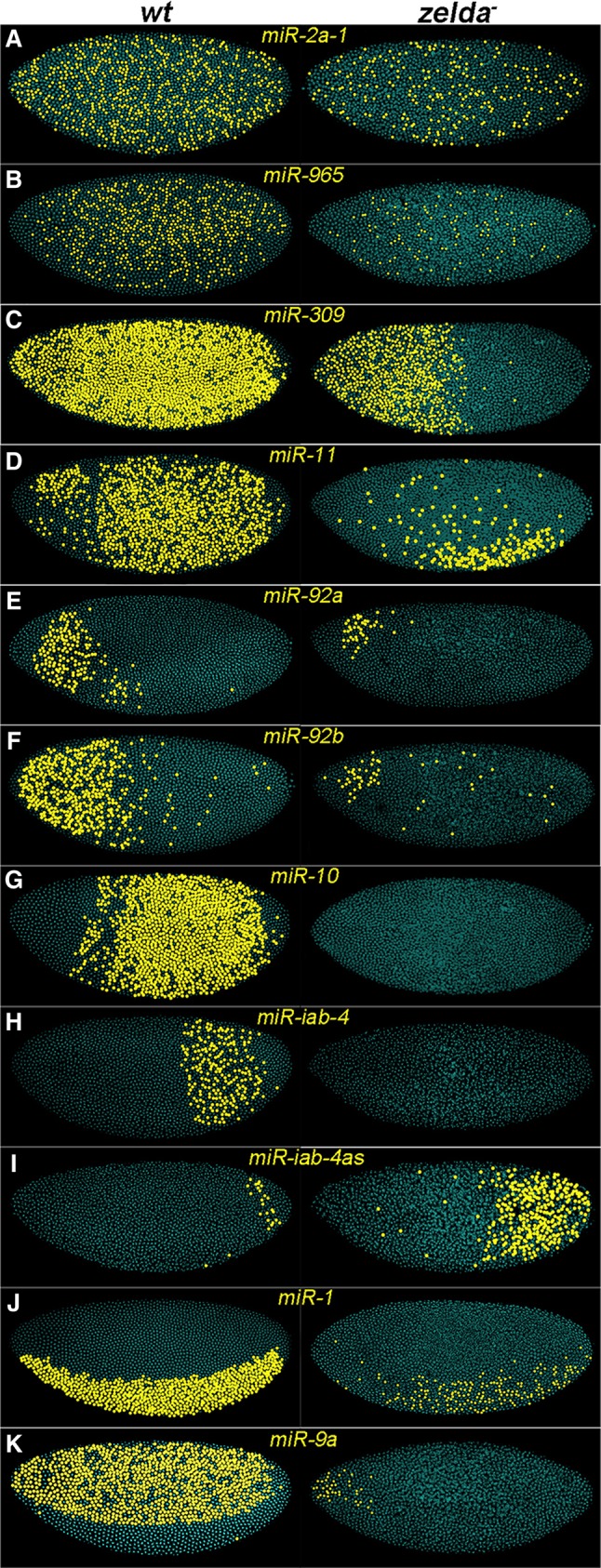
Zelda is required for proper miRNA activation. Pseudocolored confocal FISH images (see Materials and Methods) of wild-type (wt, left) and zelda mutant (right) embryos hybridized with RNA probes against pri-miRNAs (A-K, as indicated). Yellow dots represent nuclei with nascent transcripts. Embryos are oriented anterior to the left and dorsal up. Pri-miRNAs show diverse expression patterns in wt embryos, and these are affected in zelda mutants.
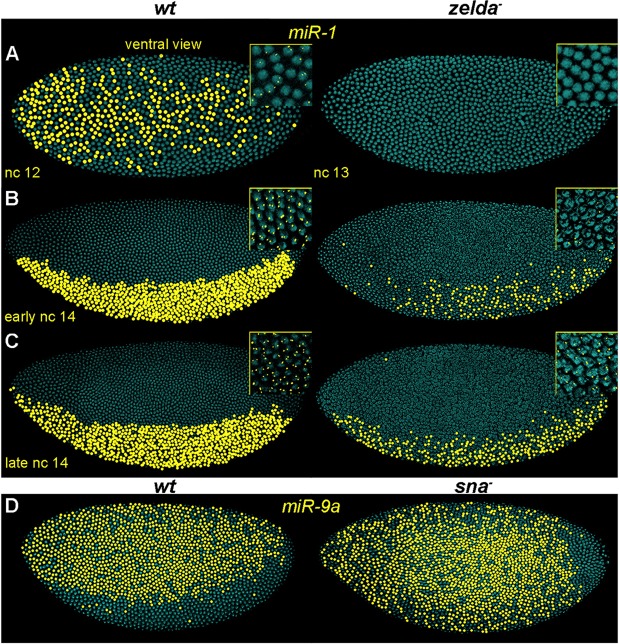
Three miRNAs displayed ubiquitous expression (miR-2a-1, miR-965 and miR-309; Fig. 2A-C), whereas the remaining eight were spatially restricted along the AP or DV axes (miR-11, miR-92a, miR-92b, miR-10, miR-iab-4, miR-iab-4as, miR-1 and miR-9a; Fig. 2D-K). All 11 miRNAs were affected in zelda mutants, with a range of defects. Some were absent, whereas others were either reduced or became spatially modified (Fig. 2, right). In addition to these changes, we observed delays in the timing of transcriptional onset. For example, miR-1 nascent transcripts were first detected in nuclear cycle (nc) 12 in the ventral region of the embryo (Fig. 3A, left). In zelda mutants, miR-1 was not detected until nc 14, and the expression was sporadic compared with wild type (Fig. 3A-C), indicating that Zelda is required for timely and robust activation of miR-1.
Fig. 3.
Patterning factors spatially regulate miRNA expression. Pseudocolored confocal FISH images of embryos hybridized with pri-miR-1 (A-C) or pri-miR-9a (D) probes. (A-C) Wild-type (left) and zelda mutant (right) embryos; insets show the actual FISH signal localized in nuclei. Note that the expression of miR-1 was first detected in nuclear cycle (nc) 12 in wt (A, left), but was not detected until early nc 14 in zelda mutants and the expression was also less robust (B, right). (D) miR-9a expression expands ventrally in sna mutants (right), indicating that Sna represses miR-9a in the presumptive mesoderm.
Zelda and the patterning factors establish precise miRNA expression domains
The fact that Zelda is expressed uniformly throughout the embryo (Liang et al., 2008; Nien et al., 2011; Kanodia et al., 2012) but affects the spatially restricted miRNAs prompted us to investigate additional regulatory inputs from the AP and DV patterning factors. Two miRNAs are expressed in complementary domains along the DV axis: miR-1 in the mesoderm and miR-9a in the ectoderm. Two key factors function in mesoderm specification, namely the activator Twi and the repressor Snail (Sna) (Leptin, 1991), and it is known that Twi binds and activates miR-1 (Sokol and Ambros, 2005; Zeitlinger et al., 2007). In zelda mutants, twi activation is delayed (Nien et al., 2011), suggesting that the loss of robust expression of miR-1 is due to both lack of direct Zelda input and reduced Twi-mediated activation.
By contrast, Sna is a likely repressor of miR-9a transcription in the mesoderm since it is known to inhibit the activation of many ectoderm genes (Boulay et al., 1987; Kosman et al., 1991; Leptin, 1991). Although miR-9a is initially ubiquitously expressed at nc 12 (data not shown), transcripts are no longer detected in the presumptive mesoderm during nc 13-14 (Fig. 3D, left), the time when the Sna protein level increases. We examined miR-9a in sna mutants and observed ectopic expression of miR-9a in the mesoderm (Fig. 3D, right). This repression by Sna is likely to be a direct effect, since there are four predicted Sna binding sites (ACAGGTAG, TCACCTGT, TCAGGTGG, CACCTTGC; ClusterDraw; Papatsenko, 2007) within the Zelda-bound region upstream (∼7 kb) of miR-9a.
Repression is also the probable mechanism that regulates Hox cluster miRNAs along the AP axis (Fig. 2G-I). For example, miR-10, which lies between Dfd and Scr in the Antennapedia (Antp) complex, is expressed in a posterior domain (∼35-85% embryo length; Fig. 2G; supplementary material Fig. S2A). In bicoid (bcd) mutant embryos, miR-10 expression expands anteriorly to cover the entire embryo (supplementary material Fig. S2B), and when Bcd is overexpressed, as in 6× bcd embryos (Ochoa-Espinosa et al., 2009), miR-10 expression moves posteriorly and becomes restricted to ∼60-85% of the embryo length (supplementary material Fig. S2C). Thus, activation by Zelda and repression by Bcd-dependent anterior repressors set the miR-10 domain. One such repressor is possibly Hunchback (Hb), since it acts mainly as a repressor of posterior gap genes in the anterior region, and its expression domain is complementary to that of miR-10 (Hülskamp et al., 1990). Similarly, miR-iab-4 and miR-iab-4as, which are expressed posteriorly (Fig. 2H,I), are derepressed in the head region of bcd mutants (data not shown).
Taken together, these results suggest that repressor activity is essential to prevent the expression of Hox cluster miRNAs in anterior regions. Given that these miRNAs are involved in regulating other Hox genes (Ronshaugen et al., 2005; Bender, 2008; Stark et al., 2008; Tyler et al., 2008; Gummalla et al., 2012), it is important that their borders of expression are precisely established. This is achieved by the coordinated activity of uniformly distributed Zelda and spatially localized AP repressors.
Interestingly, miR-iab-4 and miR-iab-4as are transcribed from opposite strands of the same locus (Ronshaugen et al., 2005; Bender, 2008; Stark et al., 2008; Tyler et al., 2008), but they are regulated differently. Although both are expressed posteriorly, miR-iab-4 is much broader, and in zelda mutants miR-iab-4 disappears, whereas miR-iab-4as expands anteriorly (compare Fig. 2H,I). Zelda is likely to regulate miR-iab-4as indirectly by ensuring the proper localization of AP repressors. It is known that in zelda mutants, the gap gene repressor domains shift and/or expand (Nien et al., 2011), which results in a cascade effect on downstream targets including miRNAs.
Patterning factors also contribute to ubiquitously expressed miRNAs
We expected to see the ubiquitously expressed miR-309 transcripts disappear in zelda mutants, but they persisted in the anterior region, in an hb-like domain (Fig. 2C). In the absence of Bcd, miR-309 is initially ubiquitous as in wild type, indicating that the onset of miR-309 expression is mediated by Zelda; however, shortly thereafter, transcripts are lost anteriorly (supplementary material Fig. S3A,B), probably owing to the lack of Bcd-dependent activation. In addition, both Zelda and Bcd exert regulatory activity on the known miR-309 enhancer region. This enhancer drives the expression of a lacZ reporter gene in a similar pattern to endogenous miR-309 (supplementary material Fig. S3C) (Biemar et al., 2005). In the absence of Zelda, lacZ is only expressed in the anterior region (supplementary material Fig. S3D), resembling the endogenous expression pattern of miR-309 in zelda mutants (Fig. 2C, right). In the absence of Bcd, lacZ expression is decreased in the anterior region at the blastoderm stage, and later becomes restricted posteriorly (supplementary material Fig. S3E). These observations suggest that the uniform miR-309 pattern is composed of multiple overlapping patterns regulated by the combinatorial action of Zelda and Bcd-dependent factors. This type of multifactor regulation is also seen for miR-11, the expression of which is observed in a discrete ventral domain in the absence of Zelda (Fig. 2D).
Using Zelda binding profiles to locate miRNA enhancers
Previous studies have shown that Zelda binds to enhancers of many known blastoderm-expressed genes (Nien et al., 2011; Harrison et al., 2011); hence, we hypothesized that enhancers of early miRNAs could be located by searching for Zelda-bound regions close to miRNA transcription units. First, we noticed that Zelda binds to previously defined enhancers of miR-1 (Zeitlinger et al., 2007) and miR-309 (Biemar et al., 2005) (red rectangles in Fig. 1B,C). Next, we tested Zelda-bound regions that lie upstream of the miR-10, miR-iab-4, miR-11, miR-9a and miR-92a transcription units (see Fig. 1) in transgenic reporter assays. Reporter gene expression recapitulated endogenous miRNA expression in all but one case, miR-92a, for which the tested fragment directed a different pattern and could be the enhancer of a nearby gene (Fig. 4A-C and Fig. 5C; data not shown for miR-92a). These results suggest that Zelda binding can be used to pinpoint bona fide enhancers of early miRNAs.
Fig. 4.
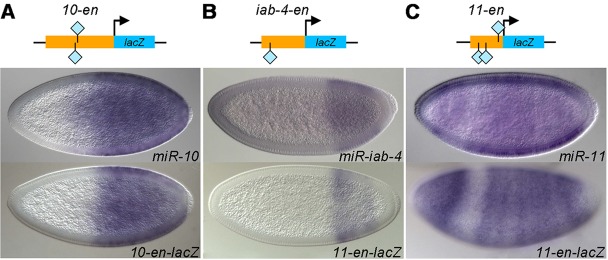
Identification of novel enhancers of miRNAs. (Above) Schematic representation of the transgenic constructs, with enhancer fragments (orange rectangles) from miR-10 (A), miR-iab-4 (B) and miR-11 (C) fused to a lacZ reporter (blue rectangles). Light blue diamond represents Zelda binding site. (Below) Reporter expression (bottom row of embryos) recapitulates the endogenous miRNA pattern (top row of embryos).
Fig. 5.
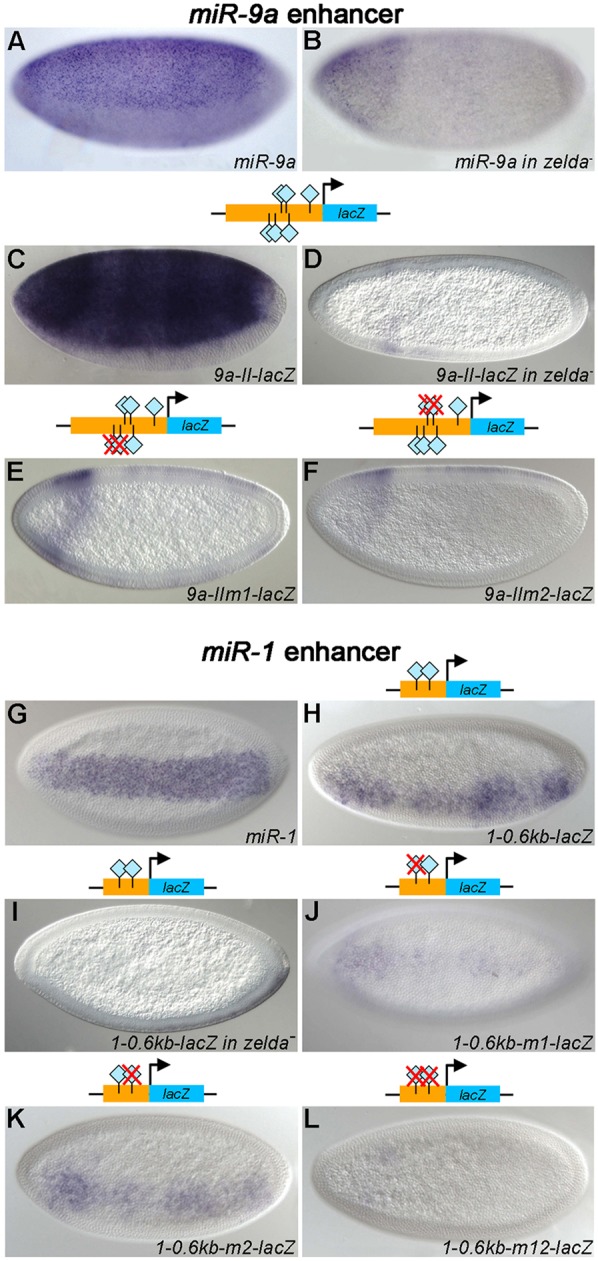
Zelda binding sites in miRNA enhancers are crucial for transcriptional activation. Embryos were hybridized with RNA probes against pri-miR-9a (A,B), pri-miR-1 (G) or lacZ (C-F,H-L). Embryos are in lateral (A-F) or ventral/ventrolateral (G-L) view. Transgenic embryos carry wild-type or mutated versions (red Xs) of the miR-9a (C-F) and miR-1 (I-L) enhancers (see genomic locations in Fig. 1). Mutation of Zelda binding sites affected reporter expression in all cases (E,F,J-L), indicating that Zelda activates miR-9a and miR-1 directly by binding to their enhancers.
Interestingly, miR-11 resides in the fifth intron of the RC isoform of the E2f gene (see Fig. 1F), which is the isoform present in blastoderm embryos. Because E2f is maternally supplied (Thomsen et al., 2010), zygotic transcripts can be distinguished only by detecting hybridization signal over introns, which is clearly reduced in zelda mutants, whereas the E2f exonic signal appears little affected (Fig. 1F). These results are consistent with the idea that Zelda regulates zygotic activation of the E2f-RC isoform, and therefore miR-11.
In our search for miR-9a enhancers, we tested two fragments, 9a-I and 9a-II (Fig. 1A), both of which drove lacZ in a miR-9a-like ectodermal pattern that was lost in the absence of Zelda (Fig. 5C,D; data not shown for 9a-I). However, the 9a-I enhancer gave a somewhat stripey pattern along the AP axis (data not shown). Although not fully redundant, both enhancers might be required for robust and accurate expression of miR-9a, as has been shown for the primary and shadow enhancers of sna and hb (Perry et al., 2010, 2011).
Zelda binding sites in the miR-1 and miR-9a enhancers are required for proper expression
To demonstrate that Zelda binding sites in the miRNA enhancers are essential for activation, we mutated the sites in the miR-9a-II and miR-1 enhancers. The miR-9a-II enhancer contains six Zelda binding sites. Mutation of the two distalmost Zelda binding sites (9a-IIm1) resulted in loss of lacZ expression except in the head region (Fig. 5E), which is similar to endogenous miR-9a expression in zelda mutants (Fig. 5B). Mutation of the middle two Zelda binding sites caused a similar effect (Fig. 5F), indicating that both sets of sites are required for transcriptional activation.
For miR-1, we further dissected the known enhancer identified by Zeitlinger et al. (Zeitlinger et al., 2007), which was located by virtue of Twi and Dl binding profiles. An internal 0.6 kb fragment that contains two Zelda binding sites directed a ventral miR-1-like pattern (Fig. 5G,H), which was lost in the zelda mutant background (Fig. 5I). Mutation of the first or second binding site, or both, caused reduced lacZ expression (Fig. 5J-L), indicating that both sites are important for miR-1 activation in blastoderm embryos. We also examined reporter gene expression in older embryos, since miR-1 is expressed throughout the mesodermal tissue in stage 10 embryos (Aboobaker et al., 2005). All four reporters directed late expression, including the mutated forms (data not shown), demonstrating that miR-1 expression at stage 10 is not regulated by Zelda.
Zelda-activated miRNAs function in diverse processes during the MZT
Our results show that Zelda is central to the miRNA early transcription network, ensuring the timely, robust and spatially precise activation of miRNAs, which in turn might be important for the proper inhibition of their targets. Since activation of all but one miRNA is substantially downregulated in zelda mutants, we reasoned that downstream targets of miRNAs might be included in the set of transcripts upregulated in zelda mutants. Indeed, several genes in this set have previously been verified as targets of miRNAs including miR-309 (Bushati et al., 2008), miR-11 and the miR-2 family (Leaman et al., 2005), and miR-iab-4/4as (Ronshaugen et al., 2005; Bender, 2008; Stark et al., 2008; Tyler et al., 2008).
The miR-309 cluster of miRNAs mediates the degradation of ∼138 maternally loaded RNAs (Bushati et al., 2008). In the absence of Zelda, 774 genes were upregulated in 2- to 3-h embryos (Nien et al., 2011), and 534 (61.6%) of these can be classified as unstable maternal mRNAs (supplementary material Table S3 and Fig. S4) (Thomsen et al., 2010) and thus represent mRNAs that are normally degraded in wild type, but persist in zelda mutants during the MZT. Degradation of 434 (81%) of these unstable maternal mRNAs depends on zygotically derived factors (Thomsen et al., 2010), 125 of which are upregulated in miR-309 mutants (Bushati et al., 2008). Importantly, of 29 experimentally verified miR-309 targets (Bushati et al., 2008), 15 are significantly upregulated in zelda mutants (supplementary material Table S3). One possible reason that not all of the genes upregulated in miR-309 mutants were upregulated in zelda mutants could relate to the residual anterior miR-309 function in zelda mutants (Fig. 2C, right).
Besides maternal genes, the upregulated genes in zelda mutants also include zygotic genes, some of which are also verified miRNA targets. For example, two key regulators of Drosophila programmed cell death, reaper (rpr) and head involution defective (hid, also known as Wrinkled), were upregulated in zelda mutants (by >1.5-fold, P<0.05; supplementary material Table S3; Fig. 6A,B). Both rpr and hid are verified targets of miR-11, miR-2 and of miR-6, which is a member of the miR-309 complex, in mid-stage embryos (Leaman et al., 2005; Truscott et al., 2011; Ge et al., 2012). In early wild-type embryos, rpr is expressed in two stripes in the anterior region (Fig. 6A), in a domain complementary to that of miR-11 (Fig. 2D and Fig. 4C). In zelda mutants, rpr is ectopically expressed posteriorly, except in the ventral region (Fig. 6A) where miR-11 remains (Fig. 2D), indicating that miR-11 might directly regulate rpr in blastoderm embryos as it does in older embryos. hid transcripts are not normally observed before gastrulation, but appear earlier in zelda mutants (Fig. 6B).
Fig. 6.
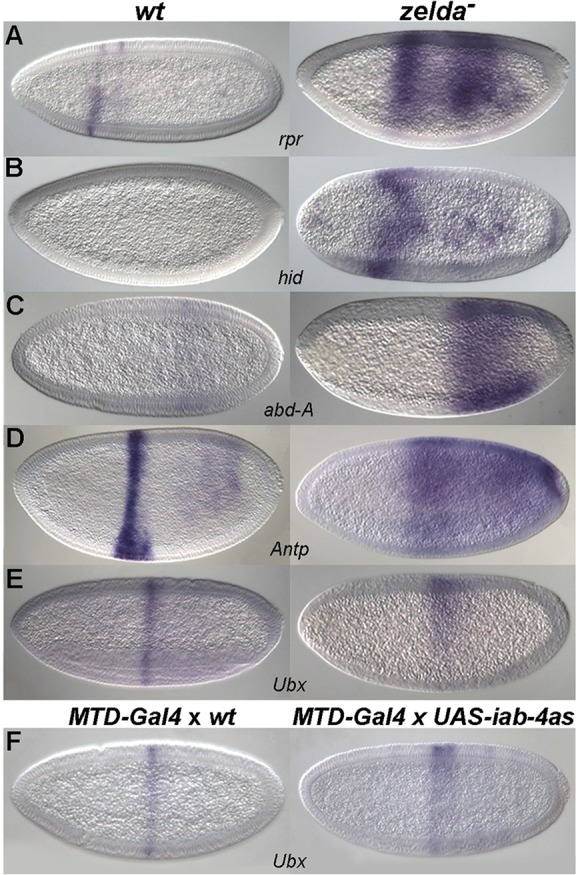
miRNA target expression is affected in zelda mutants. Wild-type (left) and zelda mutant (right) embryos were hybridized with RNA probes as indicated. Expression of miRNA targets rpr (A), hid (B), abd-A (C), Antp (D) and Ubx (E) is upregulated in zelda mutants. The posterior expansion of Ubx (E) in zelda mutants is partially due to the upregulation of miR-iab-4as (see Fig. 2I), since a similar effect was observed when miR-iab-4as was ectopically expressed (F, right).
Other examples of zygotic genes targeted by early miRNAs include Hox genes. abd-A is a direct target of miR-iab-4as (Bender, 2008; Gummalla et al., 2012), and Ubx was verified to be a target of both miR-iab-4 and miR-iab-4as (Ronshaugen et al., 2005; Bender, 2008; Stark et al., 2008; Tyler et al., 2008). Antp is predicted to be a target of miR-iab-4 and miR-iab-4as (TargetScanFly; Ruby et al., 2007). Interestingly, abd-A, Ubx and Antp are all upregulated in zelda mutant embryos, and their expression domains expand posteriorly (Fig. 6C-E; supplementary material Table S3 and Fig. S5), which might in part be due to the misregulation of miRNA activity in posterior regions (Fig. 2H,I). One piece of evidence to support this idea is the posterior expansion of Ubx when miR-iab-4as is ectopically expressed (Fig. 6F; see Materials and Methods), an effect also seen in zelda mutants (Fig. 6E).
Spatially restricted expression of Hox genes relies on multiple regulatory inputs (reviewed by McGinnis and Krumlauf, 1992; Gellon and McGinnis, 1998; Yekta et al., 2008), including cross-regulation among Hox genes, particularly 'posterior prevalence', whereby the posterior Hox genes repress the more anterior ones. Interestingly, the miRNAs embedded in the Hox complexes follow this rule, as they repress Hox genes expressed more anteriorly (Yekta et al., 2008). Finally, long intergenic non-coding RNAs (lincRNAs) transcribed from the Hox complexes have recently been identified as regulators of Hox genes by interfering with their transcription; for example, bithoraxoid (bxd) RNAs repress Ubx (Petruk et al., 2006), whereas the iab-8 RNAs, comprising lincRNA and miR-iab-4as, repress abd-A (Gummalla et al., 2012). The expression level of bxd is downregulated in zelda mutant embryos (blue shading in supplementary material Fig. S5A), which might also contribute to the derepression of Ubx in the absence of Zelda. Transcription in the iab-8 region is upregulated in zelda mutants (red shading in supplementary material Fig. S5A), but how this affects abd-A regulation is complicated by the presence of other non-coding RNAs within the long, 92 kb iab-8 transcript, including iab-4 and CR43617 (supplementary material Fig. S5A) (FlyBase; Tweedie et al., 2009), which are downregulated in zelda mutants (blue shading in supplementary material Fig. S5A). In the Antp complex, CR44931, a lincRNA that lies between ftz and Antp, is absent in zelda mutants (blue shading in supplementary material Fig. S5B). Thus, Zelda regulation of the Hox genes is multilayered, involving miRNAs and lincRNAs, as well as indirect effects on the upstream patterning factors in the segmentation hierarchy that regulates transcription of the Hox genes (Nien et al., 2011).
Co-activation of miRNAs by Zelda is crucial for normal development
So far, our results indicate that Zelda coordinately activates a set of miRNAs, and in turn these miRNAs inhibit sets of downstream target RNAs. One question is whether it is significant that these particular miRNAs are co-expressed together in the blastoderm embryo. Of the early expressed miRNAs, loss-of-function mutations have been made in miR-309 (Bushati et al., 2008) as mentioned above, miR-1 (Sokol and Ambros, 2005), miR-9a (Li et al., 2006), miR-11 (Truscott et al., 2011), miR-iab-4/4as (Bender, 2008), miR-10 (Lemons et al., 2012) and miR-92a (Chen et al., 2012,b). Curiously, none of these mutants displayed early morphological phenotypes despite the fact that miRNA targets for some were identified and verified. In most cases, mutants survived to adulthood, suggesting that, individually, these miRNAs have subtle effects at best. However, as previously noted by many investigators, miRNAs often function redundantly or have small contributions by themselves (reviewed by Smibert and Lai, 2008). Hence, knocking out combinations of miRNAs may have more dramatic effects. For example, Ge et al. (Ge et al., 2012) reported that the miR-6; miR-11 double mutant exhibits a strong reduction in survival to adulthood compared with either of the single mutants, and that this phenotype was due to elevated apoptosis activity as a result of decreased degradation of the targets rpr and hid in mid-stage embryos.
To further study the significance of the coordinated expression of miRNAs, we examined embryos doubly mutant for miR-1 and miR-9a, which are directly regulated by Zelda (Fig. 5). The double mutants are embryonic lethal and display gastrulation defects during ventral furrow formation (Fig. 7B, arrows; data not shown). Normally, rhomboid (rho), a component of EGFR signaling, is expressed in ventrolateral stripes in the blastoderm (Fig. 7A, left), which refine and then progressively meet at the ventral midline as the ventral furrow invaginates (Fig. 7B,C, left). Later, the rho-labeled cells form a single straight stripe along the ventral midline that persists through germband extension, marking the mesectoderm (Fig. 7D,E, left). In the miR-1; miR-9a double mutant, the progression of ventral-midline closure is desynchronized, with lagging gaps, eventually forming a disorganized midline (Fig. 7B, red arrows). In addition, ectopic expression of rho was observed in cells adjacent to midline cells (Fig. 7C, pink arrowheads), and the organization of rho-expressing cells in the tracheal pits is also abnormal, as shown by the larger clumps of rho-expressing cells (Fig. 7E, right, black arrow) and the lack of even spacing between the pits. These phenotypes were not observed in either of the two single mutants (data not shown), suggesting that proper gastrulation movements require the combined activity of the miR-1 and miR-9a miRNAs. Although miR-1 is expressed in the mesoderm and miR-9a in the ectoderm (Fig. 2J,K), they might influence signaling at the boundary where the mesectoderm forms.
Fig. 7.
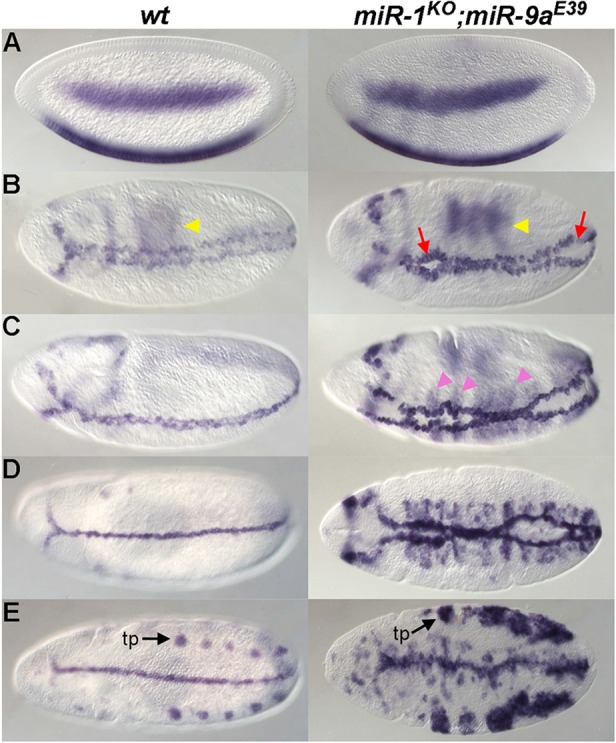
miR-1; miR-9a double mutants display ventral midline defects. Wild-type (left) and miR-1; miR-9a double-mutant (right) embryos of increasing age (A-E) were hybridized with rho probes. Yellow arrowheads in B indicate dorsal expression of rho. In the nc 14 embryo (A), rho expression is detected in two ventrolateral stripes, which come together during ventral furrow formation in stage 8-10 embryos (B-D), forming the ventral midline. rho is expressed in tracheal pits (tp, black arrows) after stage 11 (E). miR-1; miR-9a double-mutant embryos are defective in synchronized ventral midline closure, as anterior and posterior gaps are visible (B, red arrows). Ectopic rho expression is observed along the midline (C, pink arrowheads).
DISCUSSION
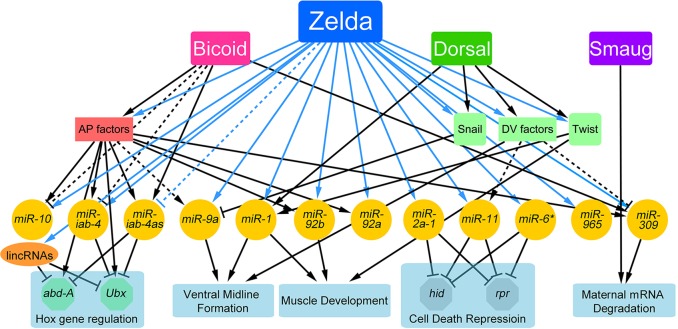
In this study we identified the set of miRNAs expressed in 2- to 3-h Drosophila embryos, a time when the MZT is well underway and the fate map of the embryo is being established (Figs 1 and 2; supplementary material Fig. S1). These early expressed miRNAs are regulated globally by Zelda, both directly via binding to cis-regulatory enhancers and indirectly by affecting the expression of additional transcriptional regulators (Figs 2-5; supplementary material Figs S2 and S3). Together with previously published data on specific miRNAs, we were able to integrate the early miRNAs, their upstream regulators and downstream targets into the early gene network (Fig. 8). In addition, our results emphasize the importance of co-activation of miRNAs by Zelda, which ensures their combined activity.
Fig. 8.
miRNAs complete the early gene network. Protein-coding genes and miRNAs are nodes in this network. Arrows indicate positive regulation (transcriptional activation or promotion); T bar represents negative regulation (transcriptional repression, or repression/destabilization via the 3′UTR); and dashed lines indicate putative connections. Zelda is a central hub in the early gene regulatory network (Nien et al., 2011), working together with maternal morphogens and newly transcribed zygotic transcription factors to ensure accurate spatial and temporal expression of miRNAs. Zelda links the interplay between miRNAs and transcription factors by regulating the transcription of both. *miR-6 is one of the eight miRNAs in the miR-309 cluster.
Zelda activates the blastoderm-specific miRNAs
Using our expression profiling data from 2- to 3-h wild-type and zelda mutant embryos (Nien et al., 2011), we identified blastoderm-specific pri-miRNA transcription units, which included seven intergenic and four intronic miRNAs (clusters) (Fig. 1; supplementary material Fig. S1). Since the expression levels of all 11 miRNAs were affected in zelda mutants (Fig. 2), we were able to better distinguish blastoderm-specific isoforms, particularly in the case of intronic miRNAs, such as miR-11, which resides in an intron of E2f (Fig. 1F). Moreover, maternal E2f expression could be differentiated from zygotic expression by observing the intronic signal, which was clearly downregulated in zelda mutants (Fig. 1F).
Precise miRNA expression patterns are established by the early gene network
The early miRNAs exhibit strikingly different expression patterns (Fig. 2), and it is noteworthy that, similar to the protein-coding targets of Zelda, two different strategies are used to regulate these miRNAs. Some miRNAs, such as miR-9a, were completely abolished in zelda mutants, indicating that Zelda is their sole activator, whereas others, such as miR-1, were affected temporally and/or spatially (Fig. 2J and Fig. 3A-C), indicating that Zelda works together with other factors to establish robust and precise domains of expression. For example, miR-1 downregulation in zelda mutants is likely to be due to the cumulative effect of loss of direct inputs from Zelda and the delayed expression of twi that occurs in zelda mutants (Nien et al., 2011). Thus, the effect on miR-1 is the result of a breakdown in the Zelda-Twi-miR-1 feedforward loop (Fig. 8).
Zelda binding marks miRNA enhancers
Cis-regulatory modules/enhancers of miRNAs have been predicted based on the presence of transcription factor binding (Zeitlinger et al., 2007; The modENCODE Consortium et al., 2010) or specific chromatin marks (Ozsolak et al., 2008), and verified in only some cases. For example, two regions upstream of miR-1 that bind Twi/Dl were shown to drive a miR-1-like expression pattern (Zeitlinger et al., 2007). We reasoned that we could locate enhancers of all early miRNAs by simply looking for Zelda-bound regions upstream of the pri-miRNA transcription units, especially since Zelda is a global activator during the MZT. This approach worked well; eight of nine enhancers recapitulated endogenous-like expression (Figs 4 and 5). Mutation of Zelda binding sites in enhancers further demonstrated direct Zelda input. As proof of principle, we assayed enhancers of two genes, miR-9a and miR-1, and showed that mutation of the Zelda binding sites had the same effect as eliminating Zelda in trans (Fig. 5). These results indicate that Zelda directly regulates the early expressed miRNAs, often in conjunction with other transcription factors, many of which are also regulated by Zelda (Nien et al., 2011). Zelda is a major hub in the early network, establishing multiple feedforward loops and closely linking the transcription factors and miRNAs expressed in this stage (Fig. 8).
Zelda plays important roles in both hallmark events of the MZT
The MZT is a key event during the development of an organism, whereby the transcriptome is reprogrammed in the first few hours of development. This requires the clearance of previous information (maternal mRNA degradation) and the initiation of a new program (zygotic genome activation). The maternal mRNA degradation machinery comprises both maternally derived and zygotically derived pathways (Bashirullah et al., 1999; Tadros and Lipshitz, 2009). In Drosophila, Smaug (Smg), a maternally loaded RNA-binding protein, is central to the mRNA clearance pathway. By recruiting the CCR4-NOT deadenylation complex, Smg destabilizes two-thirds of the maternal mRNAs that undergo degradation (i.e. that are unstable) upon egg activation (Tadros et al., 2007). By contrast, miR-309 is a key component of the zygotically derived pathway to clear mRNAs (Bushati et al., 2008). When analyzing the maternal RNAs upregulated in zelda mutants, we noted that 81% of them (434) depend on zygotic degradation pathways (supplementary material Fig. S4B); 125 of the 434 genes are also upregulated in miR-309 mutants (Bushati et al., 2008) (supplementary material Fig. S4C), indicating that Zelda, by activating miR-309, is involved in maternal RNA degradation (Fig. 8). Therefore, Zelda plays important roles in both of the hallmark events of the MZT. Interestingly, the miR-309 targets account for only ∼30% of the unstable maternal RNAs upregulated in zelda mutants (supplementary material Fig. S4B, classes III and IV), and another 14% are putative targets of the other early miRNAs, indicating that Zelda might activate additional zygotic pathways to mediate maternal mRNA degradation.
Coordinated miRNA activation by Zelda is important for in vivo function
Several miRNAs, in addition to miR-309, have been shown to target specific mRNAs in the early embryo; for example, miR-iab-4 and miR-iab-4as target Hox genes (Ronshaugen et al., 2005; Bender, 2008; Stark et al., 2008; Tyler et al., 2008). However, although each of the miRNAs is predicted to target hundreds of genes (Bartel, 2009), in many cases the individual miRNA loss-of-function phenotypes are relatively mild (reviewed by Smibert and Lai, 2008). There are several explanations for this phenomenon: (1) the miRNA does not function at the time that it is expressed, but might function later; (2) miRNAs ‘fine-tune’ the expression levels of their target genes, which might not be reflected in obvious phenotypes when they are mutated; and (3) miRNA functions are redundant, such that knockdown of one miRNA may be compensated by another.
To better address the functions of miRNAs, investigators have used several genetic approaches: gain-of-function assays (Bejarano et al., 2012), using sensitized genetic backgrounds (Brenner et al., 2010), and assaying double mutants (Ge et al., 2012). For example, the miR-6; miR-11 double mutant exhibits increased apoptosis, leading to lower survival rates compared with either of the single miRNA mutants (Ge et al., 2012). Using a similar approach, we observed fully penetrant gastrulation defects in miR-1; miR-9a double mutants (Fig. 7). Neither single mutant is embryonic lethal, nor shows any sign of ventral furrow defects; however, miR-1 mutants are larval lethal and display muscle defects (Sokol and Ambros, 2005), while miR-9a mutants show wing margin defects in adulthood (Li et al., 2006). Importantly, the double-mutant phenotype is the earliest phenotype seen for any known miRNA, or combination of miRNAs, thus far tested. These results support the idea that co-activation of miRNAs by Zelda is required for normal development.
The miR-1; miR-9a double-mutant phenotype resembles, to some extent, the ventral furrow defects observed in RhoGEF2 loss-of-function mutants (Barrett et al., 1997). Rho signaling is involved in the cell shape changes associated with ventral furrow invagination, and loss of Rho signaling results in very disorganized invagination (Barrett et al., 1997). Curiously, miR-1 and miR-9a are both predicted to target RhoGAP68F, a negative regulator of Rho signaling (Sanny et al., 2006). RhoGAP68F is maternally loaded and cleared during the MZT (Thomsen et al., 2010). It is possible that the gastrulation phenotype of the miR-1; miR-9a double mutant is caused in part by excess activity of RhoGAP68F. Although we did not see any obvious upregulation of RhoGAP68F transcripts in miR-1; miR-9a mutant embryos by in situ hybridization (data not shown), it is possible that subtle upregulation of RhoGAP68F, combined with effects on other predicted targets, all contribute to the gastrulation defects observed in the double mutant.
The co-activation of groups of miRNAs by master regulators such as Zelda may be crucial for miRNA activity during development, as revealed by the severe gastrulation phenotype of the miR-1; miR-9a double mutant. Such coordinated activation of miRNAs might also occur at later stages in development in tissues in which Zelda is expressed, such as the central nervous system (Liang et al., 2008). In the future, various combinations of mutations in miRNA genes that are co-regulated by Zelda, or other key factors, might unveil additional functions of miRNAs across development stages.
MATERIALS AND METHODS
Drosophila genetics
The y1w67c23 strain was used as wild-type (wt) control. The zld294 allele was used to obtain zelda germline clones as previously described (Liang et al., 2008). Other alleles used include bcdE1, sna1, gd7 and MTD-gal4 (31777, Bloomington Drosophila Stock Center). bcd+5 bcd+8/bcd+5 bcd+8; vasPD exuPJ/vasPD exuPJ (6× bcd; Ochoa-Espinosa et al., 2009) was used to overexpress Bcd. P{attP.w+.attP} JB38F (38F1; 27388, Bloomington Drosophila Stock Center) was used for transgene injection. The UAS-miRNA fly lines were generously supplied by Eric Lai (Bejarano et al., 2012). miR-1; miR-9a double mutants were obtained by crossing miR-1KO/SM6B, eve-lacZ (Sokol and Ambros, 2005) and miR-9aE39/TM3B, hb-lacZ (Li et al., 2006) flies. Homozygous miR-1; miR-9a double-mutant embryos were identified by the absence of lacZ expression.
Mutagenesis and enhancer testing
Candidate enhancer fragments were chosen by identifying regions near transcription units of miRNAs that overlapped with Zelda binding peaks and contained clusters of Zelda binding sites (limited to eight CAGGTAG and related sites; Nien et al., 2011). DNA fragments were amplified using wt genomic DNA as template and the Expand High Fidelity PCR System (Roche). Forward primers for amplification of candidate enhancers contain BglII (or BamHI) restriction sites and reverse primers contain AscI restriction sites. Amplified fragments were digested with BglII (or BamHI) and AscI and inserted into the piB-HC-lacZ vector (Chen et al., 2012a). Nucleotide changes in the Zelda binding sites were introduced by PCR mutagenesis. Primer sets are listed in supplementary material Table S2. Constructs were injected into embryos carrying the 38F1 landing site (Bateman et al., 2006; Bischof et al., 2007) to generate transgenic flies. At least three transformant lines for each construct were analyzed.
RNA probes and in situ hybridization
Primers for miRNA probe design are listed in supplementary material Table S2. In situ hybridization was performed as previously described (Liang et al., 2008; Nien et al., 2011). Embryos were stained with DAPI (1 mg/ml; Sigma-Aldrich) following hybridization.
Confocal image acquisition and image processing
Fluorescence in situ hybridization (FISH) images were acquired using a Leica TCS SP5 confocal microscope. z-sections containing pixel intensities higher than the median intensity of all pixels were selected for analysis. For each position in the x-y plane, the pixel with the strongest intensity across all z-sections was defined as the intensity value for that x-y position. All z-sections from the DAPI channel were processed to generate clear images of nuclei by customized Matlab scripts (Nien et al., 2011). Every FISH signal was assigned to the closest nucleus, when the distance between a FISH signal and the center of a nucleus was less than 1.5× the radius of the nucleus. The assigned nuclei were considered to show expression and were pseudocolored. Images were then further prepared using ImageJ (Abramoff et al., 2004).
miRNA target prediction
PicTar/doRiNA (Grün et al., 2005; Anders et al., 2012) and TargetScanFly (Ruby et al., 2007) were used for miRNA target predictions.
Supplementary Material
Acknowledgements
We thank Nicholas Sokol for the miR-1KO fly stock; Fen-Biao Gao for miR-9aE39; Steve Small for bcdE1 and the piB-HC-lacZ vector; Eric Lai for the UAS-miRNA lines; Bomyi Lim for the miR-2a-1 RNA probe; Kris Gunsalus for helpful discussions regarding miRNA targets; Nikolai Kirov and Yujia Sun for many insightful discussions and comments on the manuscript; and Josephine DeLuca and Utkarsh Patel for help with the transgenic studies.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
S.F. and C.R. designed the experiments and prepared the manuscript. S.F. performed the transgenic reporter assays and genetic experiments. C.-Y.N. and H.-L.L. analyzed the expression profiling data. C.-Y.N. processed the FISH data for false coloring.
Funding
This work was supported by a grant from the National Institutes of Health [GM63024]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.108118/-/DC1
References
- Aboobaker A. A., Tomancak P., Patel N., Rubin G. M., Lai E. C. (2005). Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc. Natl. Acad. Sci. U.S.A. 102, 18017-18022 10.1073/pnas.0508823102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramoff M. D. M., Paulo J., Ram S. J. (2004). Image processing with ImageJ. Biophotonics Int. 11, 36-42 [Google Scholar]
- Ameres S. L., Zamore P. D. (2013). Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 14, 475-488 10.1038/nrm3611 [DOI] [PubMed] [Google Scholar]
- Anders G., Mackowiak S. D., Jens M., Maaskola J., Kuntzagk A., Rajewsky N., Landthaler M., Dieterich C. (2012). doRiNA: a database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res. 40, D180-D186 10.1093/nar/gkr1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A. A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. (2003). The small RNA profile during Drosophila melanogaster development. Dev. Cell 5, 337-350 10.1016/S1534-5807(03)00228-4 [DOI] [PubMed] [Google Scholar]
- Barrett K., Leptin M., Settleman J. (1997). The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell 91, 905-915 10.1016/S0092-8674(00)80482-1 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281-297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215-233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirullah A., Halsell S. R., Cooperstock R. L., Kloc M., Karaiskakis A., Fisher W. W., Fu W., Hamilton J. K., Etkin L. D., Lipshitz H. D. (1999). Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J. 18, 2610-2620 10.1093/emboj/18.9.2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Lee A. M., Wu C.-t. (2006). Site-specific transformation of Drosophila via φC31 integrase-mediated cassette exchange. Genetics 173, 769-777 10.1534/genetics.106.056945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F., Bortolamiol-Becet D., Dai Q., Sun K., Saj A., Chou Y.-T., Raleigh D. R., Kim K., Ni J.-Q., Duan H., et al. (2012). A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development 139, 2821-2831 10.1242/dev.079939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W. (2008). MicroRNAs in the Drosophila bithorax complex. Genes Dev. 22, 14-19 10.1101/gad.1614208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Cuppen E., Plasterk R. H. A. (2006). Approaches to microRNA discovery. Nat. Genet. 38, S2-S7 10.1038/ng1794 [DOI] [PubMed] [Google Scholar]
- Berezikov E., Robine E., Samsonova A., Westholm J. O., Naqvi A., Hung J.-H., Okamura K., Dai Q., Bortolamiol-Becat D., Martin R., et al. (2011). Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 21, 203-215 10.1101/gr.116657.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemar F., Zinzen R., Ronshaugen M., Sementchenko V., Manak J. R., Levine M. S. (2005). Spatial regulation of microRNA gene expression in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 102, 15907-15911 10.1073/pnas.0507817102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312-3317 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J. R. t., Benavides J. A., Cline T. W. (2006). The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133, 1967-1977 10.1242/dev.02373 [DOI] [PubMed] [Google Scholar]
- Boulay J. L., Dennefeld C., Alberga A. (1987). The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. Nature 330, 395-398 10.1038/330395a0 [DOI] [PubMed] [Google Scholar]
- Brenner J. L., Jasiewicz K. L., Fahley A. F., Kemp B. J., Abbott A. L. (2010). Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr. Biol. 20, 1321-1325 10.1016/j.cub.2010.05.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Stark A., Brennecke J., Cohen S. M. (2008). Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 18, 501-506 10.1016/j.cub.2008.02.081 [DOI] [PubMed] [Google Scholar]
- Carrington J. C., Ambros V. (2003). Role of microRNAs in plant and animal development. Science 301, 336-338 10.1126/science.1085242 [DOI] [PubMed] [Google Scholar]
- Chen H., Xu Z., Mei C., Yu D., Small S. (2012a). A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell 149, 618-629 10.1016/j.cell.2012.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Liang S., Zhao Y., Han Z. (2012b). miR-92b regulates Mef2 levels through a negative-feedback circuit during Drosophila muscle development. Development 139, 3543-3552 10.1242/dev.082719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Renzis S., Elemento O., Travzoie S., Wieschaus E. F. (2007). Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryos. PLoS Biol. 5, e117 10.1371/journal.pbio.0050117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt A. S., Lai E. C. (2008). Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 9, 831-842 10.1038/nrg2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W., Chen Y.-W., Weng R., Lim S. F., Buescher M., Zhang R., Cohen S. M. (2012). Overlapping functions of microRNAs in control of apoptosis during Drosophila embryogenesis. Cell Death Differ. 19, 839-846 10.1038/cdd.2011.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellon G., McGinnis W. (1998). Shaping animal body plans in development and evolution by modulation of Hox expression patterns. BioEssays 20, 116-125 [DOI] [PubMed] [Google Scholar]
- Giraldez A. J. (2010). MicroRNAs, the cell's Nepenthe: clearing the past during the maternal-to-zygotic transition and cellular reprogramming. Curr. Opin. Genet. Dev. 20, 369-375 10.1016/j.gde.2010.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carison J. W., Duff M. O., Landolin J. M., Yang L., Artieri C. G., van Baren M. J., Boley N., Booth B. W., et al. (2011). The developmental transcriptome of Drosophila melanogaster. Nature 471, 473-479 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D., Wang Y.-L., Langenberger D., Gunsalus K. C., Rajewsky N. (2005). microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 1, e13 10.1371/journal.pcbi.0010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummalla M., Maeda R. K., Castro Alvarez J. J., Gyurkovics H., Singari S., Edwards K. A., Karch F., Bender W. (2012). abd-A regulation by the iab-8 noncoding RNA. PLoS Genet. 8, e1002720 10.1371/journal.pgen.1002720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. M., Li X.-Y., Kaplan T., Botchan M. R., Eisen M. B. (2011). Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7, e1002266 10.1371/journal.pgen.1002266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522-531 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- Hülskamp M., Pfeifle C., Tautz D. (1990). A morphogenetic gradient of hunchback protein organizes the expression of the gap genes Krüppel and knirps in the early Drosophila embryo. Nature 346, 577-580 10.1038/346577a0 [DOI] [PubMed] [Google Scholar]
- Kanodia J. S., Liang H.-L., Kim Y., Lim B., Zhan M., Lu H., Rushlow C. A., Shvartsman S. Y. (2012). Pattern formation by graded and uniform signals in the early Drosophila embryo. Biophys. J. 102, 427-433 10.1016/j.bpj.2011.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D., Ip Y., Levine M., Arora K. (1991). Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science 254, 118-122 10.1126/science.1925551 [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152-D157 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman D., Chen P. Y., Fak J., Yalcin A., Pearce M., Unnerstall U., Marks D. S., Sander C., Tuschl T., Gaul U. (2005). Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 121, 1097-1108 10.1016/j.cell.2005.04.016 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843-854 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Lemons D., Paré A., McGinnis W. (2012). Three Drosophila Hox complex microRNAs do not have major effects on expression of evolutionarily conserved Hox gene targets during embryogenesis. PLoS ONE 7, e31365 10.1371/journal.pone.0031365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leptin M. (1991). twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 5, 1568-1576 10.1101/gad.5.9.1568 [DOI] [PubMed] [Google Scholar]
- Li Y., Wang F., Lee J.-A., Gao F.-B. (2006). MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 20, 793-2805 10.1101/gad.1466306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H.-L., Nien C.-Y., Liu H.-Y., Metzstein M. M., Kirov N., Rushlow C. (2008). The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400-403 10.1038/nature07388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N. J., Walhout A. J. M. (2009). The interplay between transcription factors and microRNAs in genome-scale regulatory networks. BioEssays 31, 435-445 10.1002/bies.200800212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W., Krumlauf R. (1992). Homeobox genes and axial patterning. Cell 68, 283-302 10.1016/0092-8674(92)90471-N [DOI] [PubMed] [Google Scholar]
- Mendell J. T., Olson E. N. (2012). MicroRNAs in stress signaling and human disease. Cell 148, 1172-1187 10.1016/j.cell.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol J. W., Helt G. A., Blanchard S. G., Raja A., Loraine A. E. (2009). The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25, 2730-2731 10.1093/bioinformatics/btp472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nien C.-Y., Liang H.-L., Butcher S., Sun Y., Fu S., Gocha T., Kirov N., Manak J. R., Rushlow C. (2011). Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 7, e1002339 10.1371/journal.pgen.1002339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Espinosa A., Yu D., Tsirigos A., Struffi P., Small S. (2009). Anterior-posterior positional information in the absence of a strong Bicoid gradient. Proc. Natl. Acad. Sci. U.S.A. 106, 3823-3828 10.1073/pnas.0807878105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F., Poling L. L., Wang Z., Liu H., Liu X. S., Roeder R. G., Zhang X., Song J. S., Fisher D. E. (2008). Chromatin structure analyses identify miRNA promoters. Genes Dev. 22, 3172-3183 10.1101/gad.1706508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatsenko D. (2007). ClusterDraw web server: a tool to identify and visualize clusters of binding motifs for transcription factors. Bioinformatics 23, 1032-1034 10.1093/bioinformatics/btm047 [DOI] [PubMed] [Google Scholar]
- Perry M. W., Boettiger A. N., Bothma J. P., Levine M. (2010). Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 20, 1562-1567 10.1016/j.cub.2010.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M. W., Boettiger A. N., Levine M. (2011). Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 108, 13570-13575 10.1073/pnas.1109873108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S., Sedkov Y., Riley K. M., Hodgson J., Schweisguth F., Hirose S., Jaynes J. B., Brock H. W., Mazo A. (2006). Transcription of bxd noncoding-RNAs promoted by Trithorax represses Ubx in cis by transcriptional interference. Cell 127, 1209-1221 10.1016/j.cell.2006.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Zhang Z., Liang J., Ge Q., Duan X., Ma F., Li F. (2011). The full-length transcripts and promoter analysis of intergenic microRNAs in Drosophila melanogaster. Genomics 97, 294-303 10.1016/j.ygeno.2011.02.004 [DOI] [PubMed] [Google Scholar]
- Rajewsky N. (2006). MicroRNA target predictions in animals. Nat. Genet. 38, S8-S13 10.1038/ng1798 [DOI] [PubMed] [Google Scholar]
- Ronshaugen M., Biemar F., Piel J., Levine M., Lai E. C. (2005). The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 19, 2947-2952 10.1101/gad.1372505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J. G., Stark A., Johnston W. K., Kellis M., Bartel D. P., Lai E. C. (2007). Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17, 1850-1864 10.1101/gr.6597907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanny J., Chui V., Langmann C., Pereira C., Zahedi B., Harden N. (2006). Drosophila RhoGAP68F is a putative GTPase activating protein for RhoA participating in gastrulation. Dev. Genes Evol. 216, 543-550 10.1007/s00427-006-0067-6 [DOI] [PubMed] [Google Scholar]
- Schier A. F. (2007). The maternal-zygotic transition: death and birth of RNAs. Science 316, 406-407 10.1126/science.1140693 [DOI] [PubMed] [Google Scholar]
- Sempere L. F., Sokol N. S., Dubrovsky E. B., Berger E. M., Ambros V. (2003). Temporal regulation of microRNAs expression in Drosophila melanogaster mediated by hormonal signals and Broad-Complex gene activity. Dev. Biol. 259, 9-18 10.1016/S0012-1606(03)00208-2 [DOI] [PubMed] [Google Scholar]
- Smibert P., Lai E. C. (2008). Lessons from microRNA mutants in worms, flies and mice. Cell Cycle 7, 2500-2508 10.4161/cc.7.16.6454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol N. (2008). An overview of the identification, detection, and functional analysis of Drosophila microRNAs. In Drosophila: Methods and Protocols (ed. Dahmann C.), pp. 319-334 Totowa, NJ: Humana Press; [DOI] [PubMed] [Google Scholar]
- Sokol N. S., Ambros V. (2005). Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 19, 2343-2354 10.1101/gad.1356105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Bushati N., Jan C. H., Kheradpour P., Hodges E., Brennecke J., Bartel D. P., Cohen S. M., Kellis M. (2008). A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 22, 8-13 10.1101/gad.1613108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W., Lipshitz H. D. (2009). The maternal-to-zygotic transition: a play in two acts. Development 136, 3033-3042 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Tadros W., Goldman A. L., Babak T., Menzies F., Vardy L., Orr-Weaver T., Hughes T. R., Westwood J. T., Smibert C. A., Lipshitz H. D. (2007). SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev. Cell 12, 143-155 10.1016/j.devcel.2006.10.005 [DOI] [PubMed] [Google Scholar]
- The modENCODE Consortium, Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., Eaton M. L., Landolin J. M., Bristow C. A., Ma L., et al. (2010). Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787-1797 10.1126/science.1198374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen S., Anders S., Janga S. C., Huber W., Alonso C. (2010). Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol. 11, R93 10.1186/gb-2010-11-9-r93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson D. W., Bracken C. P., Goodall G. J. (2011). Experimental strategies for microRNA target identification. Nucleic Acids Res. 39, 6845-6853 10.1093/nar/gkr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott M., Islam A. B. M. M. K., Lopez-Bigas N., Frolov M. V. (2011). MiR-11 limits the proapoptotic function of its host gene, dE2f1. Genes Dev. 25, 1820-1834 10.1101/gad.16947411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S., Ashburner M., Falls K., Leyland P., McQuilton P., Marygold S., Millburn G., Osumi-Sutherland D., Schroeder A., Seal R., et al. (2009). FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37, D555-D559 10.1093/nar/gkn788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler D. M., Okamura K., Chung W.-J., Hagen J. W., Berezikov E., Hannon G. J., Lai E. C. (2008). Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 22, 26-36 10.1101/gad.1615208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S., Tabin C. J., Bartel D. P. (2008). MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat. Rev. Genet. 9, 789-796 10.1038/nrg2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J., Zinzen R. P., Stark A., Kellis M., Zhang H., Young R. A., Levine M. (2007). Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 21, 385-390 10.1101/gad.1509607 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.