Abstract
The HIV epidemic in Cameroon is marked by a broad genetic diversity dominated by Circulating Recombinant Forms (CRFs). Studies performed more than a decade ago in urban settings of Southern Cameroon revealed a dominance of the CRF02_AG and clade A variants in >90% of the infected subjects; however, little is known about the evolving viral variants circulating in this region. To document circulating HIV viral diversity, four regions of the viral genome (gag, PR, reverse transcriptase, env) in 116 HIV-1 positive individuals in Limbe, Southern Cameroon, were PCR-amplified. Sequences obtained at the RT and protease regions were analyzed for mutations that conferred drug resistance using the Stanford Drug Resistance Database. The present study reveals a broad genetic diversity characterized by several unique recombinant forms (URF) accounting for 36% of infections, 48.6% of patients infected with CRF02_AG, and the emergence of CRF22_01A1 in 7.2% of patients. Three out of 15 (20%) treated patients and 13 out of 93 (13.9%) drug naïve patients harbor drug resistance mutations to RT inhibitors, while 3.2% of drug naïve patients harbor drug resistance mutations associated with protease inhibitors. The high proportion (13.9%) of drug resistance mutations among the drug naïve patients reveals the ongoing transmission of these viruses in this region of Cameroon and highlights the need for drug resistance testing before starting treatment for patients infected with HIV-1.
Keywords: Cameroon, Genetic Diversity, Unique Recombinant Forms, Drug Resistance Mutations
Introduction
In Cameroon, the first HIV/AIDS case was reported in 1985 [Mbopi Keou et al., 1998]. By 1995, a marked epidemic was documented, with average rates that have ranged between 4-6% over the past ten years [Cameroon, 2010]. Studies carried out in Cameroon's major cities (2007) indicated high infection rates of 40% to 50% among high risk groups, such as commercial sex workers and long distance truck drivers [UNAIDS/WHO, 2007]. Despite the low infection rate among the general population, the genetic diversity of the viruses infecting individuals in Cameroon is broad [Nkengasong et al., 1994; Carr et al., 1998; Fonjungo et al., 2000; Nyambi et al., 2002; Zhong et al., 2002; Zhong et al., 2003; Konings et al., 2006a; Brennan et al., 2008; Carr et al., 2010; Powell et al., 2010a; Ragupathy et al., 2011; Ceccarelli et al., 2012], though much is not known on the evolution of this diversity. The HIV epidemic in Cameroon has since become more complex, with numerous divergent subtypes, sub-subtypes and recombinant forms of HIV-1 group M viruses [Powell et al., 2007; Carr et al., 2010; Powell et al., 2010a; Ragupathy et al., 2011; Ceccarelli et al., 2012; Tongo et al., 2013]. Reports during the early stage of this epidemic (1994-2004) show a viral population dominated by non-recombinant subtypes such as A (A1, A2), C, D, F (F1, F2), G, H, J, K and a few less complex recombinants forms, such as CRF02_AG and CRF01_AE. In particular, previous studies in several urban settings in Southern Cameroon including the city of Limbe in 1990 revealed the dominance of HIV-1 subtype A and CRF02_AG variants in the general population [Nyambi et al., 2002], which accounted for >90% of the subtype infection [Nyambi et al., 2002]. Even recent studies in rural villages of Cameroon revealed that CRF02_AG accounted for 65% to 75% of HIV infections [Carr et al., 2010; Powell et al., 2010a; Ragupathy et al., 2011] as well as other recombinant variants including CRF01_AE, CRF22_01A1, CRF09_cpx, CRF18_cpx, CRF19_cpx, CRF36_cpx, CRF37_cpx [Nyambi et al., 2002; Konings et al., [2006 a,b]; Brennan et al., 2008; Ndembi et al., 2008; Carr et al., 2010; Powell et al., [2010 a,b]]. The genetic diversity of HIV-1 virus poses a major challenge for diagnosis, treatment, and vaccine development as new recombinant forms arise continuously in different regions of Cameroon and globally.
In 2008, Cameroon adapted the World Health Organization (WHO) guidelines for the treatment of persons with HIV/AIDS in income constrained countries [WHO, 2008]. These guidelines stipulate that first-line Highly Active Antiretroviral Therapy (HAART) should be composed of two nucleoside reverse transcriptase inhibitors (NRTI) and one non-nucleoside reverse transcriptase inhibitor (NNRTI). This fixed-dose medication is comprised of 3 generic drugs (Lamivudine, Stavudine, Nevirapine) known as Triomune and is administered along with additional monitoring of CD4 T-cell counts, when available [Gilks et al., 2006; WHO, 2008]. Most of the patients who are on treatment are 20-39 years old, which is the age group most affected by HIV/AIDS in Cameroon [Cameroon, 2011]. These patients obtain care and treatment from any of the 151 treatment centers in the country [Cameroon, 2011]. Many of these patients with HIV have benefited from HAART, which has resulted in marked viral suppression, reduced transmission, morbidity and mortality among patients with advanced infection [Hammer et al., 1996; Palella et al., 1998]. It would be important to monitor the diversity and evolution of HIV strains in these different treatment centers in Cameroon and to monitor the emergence of drug resistant strains as this might impact treatment strategies in this population.
Anti-HIV drugs were developed in Western countries based on HIV-1 subtype B models [Butler et al., 2007] and are now used in developing countries such as Cameroon, where broad genetic diversity exists. Because viruses bear different sequences in their respective regions and have different mutation rates, anti-HIV drugs developed for subtype B viruses may not be as effective against the diverse non-B subtype viruses present in Cameroon. Therefore, studies are needed to carefully monitor the efficacy of these drugs on different HIV-1 subtype infected patients. The natural resistance of different viruses in drug naïve individuals, as well as the tendency of developing drug resistance after treatment are major concerns when intiating therapy [Butler et al., 2007]. Thus, proper monitoring of HIV/AIDS patients and treatment strategies is imperative.
Data on drug resistance development in Cameroon are sparse. However, some studies identified mutations in viruses isolated from patients on treatment that confer drug resistance to NRTIs and NNRTIs with a range of 16.4% to 84.4%, as well as mutations in drug naïve individuals, ranging from 8.2% to 44% [Laurent et al., 2006; Burda et al., 2010; Ragupathy et al., 2011; Ceccarelli et al., 2012]. Because new HIV-1 viral variants emerge due to recombination with well documented or existing recombinant forms or from entirely new viral variants, it is important to continue to evaluate the evolution of HIV-1 genetic diversity and the effectiveness of anti-retroviral therapy in different populations of the world.
Materials and Methods
Ethical considerations
This study was performed in accordance with the guidelines of the Helsinki Declaration and was approved by the Institutional Ethical Review Board of New York University School of Medicine, New York, USA. Written informed consent was obtained from all the participants.
Study Site and Subjects
The Limbe Regional Hospital in the South West Region of Cameroon is one of the major hospitals and one of the main HIV treatment centers in Cameroon. HIV infected patients seen at the treatment center of this hospital come from different urban and rural areas of the South West Region of Cameroon. Thus, the results of studies conducted here would give a fair representation of the genetic diversity in this city and in parts of Southern Cameroon. Notably, Limbe is located on the coast line on the Atlantic Ocean and is a major touristic city due to its proximity to many natural attractions. Thus, understanding the distribution and evolution of HIV diversity in this region of Cameroon is crucial.
Whole blood samples were collected from 116 recruited subjects at the Limbe Regional Hospital from January to October 2010 and shipped to NYU School of Medicine. Subjects were mostly residents of Limbe and neighboring towns and villages, men and women of all income levels, and with educational background ranging from primary school to university level. The age range was 14 to 67 years (mean age of 27 years) at the time of enrollment in the study. Participants included those that were drug naïve (n=100) and those on antiretroviral therapy (n=16). All patients receiving HAART obtained the drugs free of charge through the Ministry of Public Health, when their CD4 T-cell count was ≤ 350cells/mm3. Informed consent and ethical clearance were obtained for the blood donation of all individuals (LB001-1 to LB116-1) studied.
RNA Extraction, Polymerase Chain reaction and Sequencing
Plasma was obtained by Ficoll-hypaque gradient centrifugation. Viral RNA was extracted from plasma using the QIAamp Viral RNA Mini kit (Qiagen, Valencia, CA) followed by reverse transcription and nested PCR using SuperScript One-Step RT-PCR system and Platinum PCR supermix (Life Technologies, Carlsbad, CA), following manufacturer's instructions. Four fragments of the genome were amplified (fragments of gag, PR, RT, and env genes). The primers used are listed (Table 1) [Konings et al., 2004]. Thermocycling conditions for the first-round RTPCR were as follows: 50°C for 30 min, 94C° for 2 min, followed by 40 cycles at 94°C for 15 sec, 50°C for 30 sec, and 68°C for 1 min, and a final extension step at 72°C for 7 min. A 2 µlaliquot of the first-round PCR was used for second round PCR. Conditions for the nested PCR were as follows: 94°C for 2 min, followed by 35 cycles at 94°C for 15 sec, 50°C for 30 sec, and 68°C for 30 sec. PCR products were run on a 1.5% agarose gel, and DNA fragments of the expected size (2ul of PCR product + 14ul of nuclease free water) along with the forward and reverse primers were sent for sequencing to Macrogen (New York, USA) using the capillary electrophoresis sequencing method with the 3730xl DNA Analyzer Capillary Array from Life Technology Scientific.
Table 1.
PCR primers used for amplification of HIV-1 genomic material in patient plasma Locations of the primers are based on the HxB2 numbering engine
| Primer Name | HxB2 Location | Sequence 5′- 3′ |
|---|---|---|
| HIG777 | 1231-1255 | TCACCTAGAACTTTGAATGCATGGG |
| HIP202 | 2328-2352 | CTAATACTGTATCATCTGCTCCTGT |
| H1gag1584 | 1577-1595 | AAAGATGGATAATCCTGGG |
| G17 | 2017-2040 | TCCACATTTCCAACAGCCCTTTTT |
| NYUPOL6 | 2114 - 2132 | AGGGAAGGCCAGGGAATTT |
| NYUPOL7 | 2124 - 2144 | AGGAAATTTTCCTCAGAGCAG |
| NYUPOL8 | 2634 - 2615 | CTTCTGTCAATGGCCATTGT |
| NYUPOL9 | 2241 - 2264 | TCCTTTAACTTCCCTCAAATCACT |
| NYUPOL10 | 2577 - 2556 | CTGGCACGGTTTCAATAGGACT |
| ED5 | 6557-6582 | ATGGGATCAAAGCCTAAAGCCATGTG |
| ED12 | 7782-7811 | AGTGCTTCCTGCTGCTCCCAAGAACCCAAG |
| ES7 | 6983-7021 | TGTAAAACGACGGCCAGTCTGTTAAATGGCAGTCTAGC |
| ES8 | 7648- 7686 | CAGGAAACAGCTATGACCCACTTCTCCAATTGTCCCTCA |
| Env1 | 6949-6976 | TCAGCACAGTACAATGTACACATGGAAT |
| Env2 | 7784-7810 | GTGCTTCCTGCTCCCAAGAACCCA |
| Env3 | 7003-7025 | TGTTAAATGGCAGTCTAGCAGAA |
| Env4 | 7656-7678 | TTATATAATTCACTTCTCCAATT |
| RTPOL1F | 2475-2498 | GTATTAGTAGGACCTACACCTGTC |
| RTPOL1R | 4203-4225 | ACCTTCCTGATTCCATTACTGAC |
| RTPOL2F | 2584-2605 | TAAAGCCAGGAATGGATGGCCC |
Phylogenetic Analysis
All sample sequences were automatically aligned with reference sequences of all known HIV-1 group M subtypes, sub-subtypes (A1, A2, B, C, D, F1, F2, G, H J, and K) and circulating recombinant forms (CRFs) from the Los Alamos database. (www.hiv-web.lanl.gov), using the CLUSTAL X [Thompson et al., 1997] alignment software with minor manual adjustments. Phylogenetic analyses were conducted using the MEGA version 3.1 software package [Kumar et al., 2004], with pairwise evolutionary distances estimated by using Kimura's two-parameter method. Phylogenetic trees were constructed for each sample for each gene and amplified by the neighbor-joining method. The reliability of topologies was estimated by performing bootstrap analysis (1000 replicates) [Kimura, 1980]. Clustering of sequences with a bootsrap value of more than 70% was considered significant for defining a subtype.
Drug-Resistance Genotyping
The RT and protease DNA sequences were analyzed for drug-resistance mutations using the Stanford University HIV database genotypic resistance interpretation algorithms (http://hivdb.standord.edu/). This program identifies documented drug-resistance mutations in user-entered sequences and infers the level of resistance to NRTIs and NNRTIs and protease inhibitors.
All the sequences were submitted to the GenBank with accession numbers KF540274-KF540377, KF576407-KF576512, KF576513-KF576620, and KF576329-KF576406
Results
Overall genetic diversity
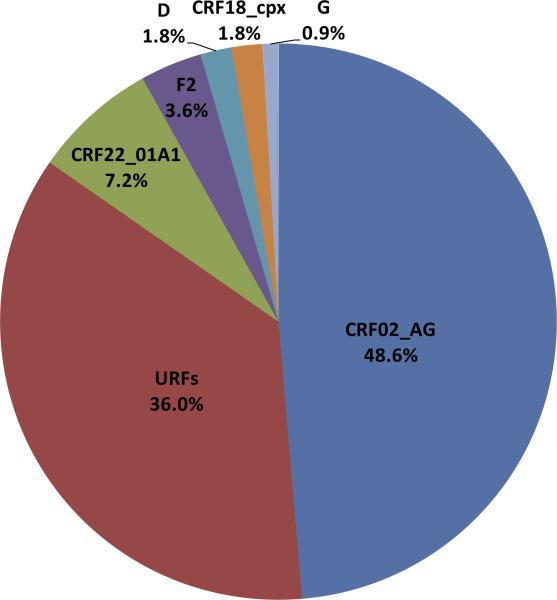
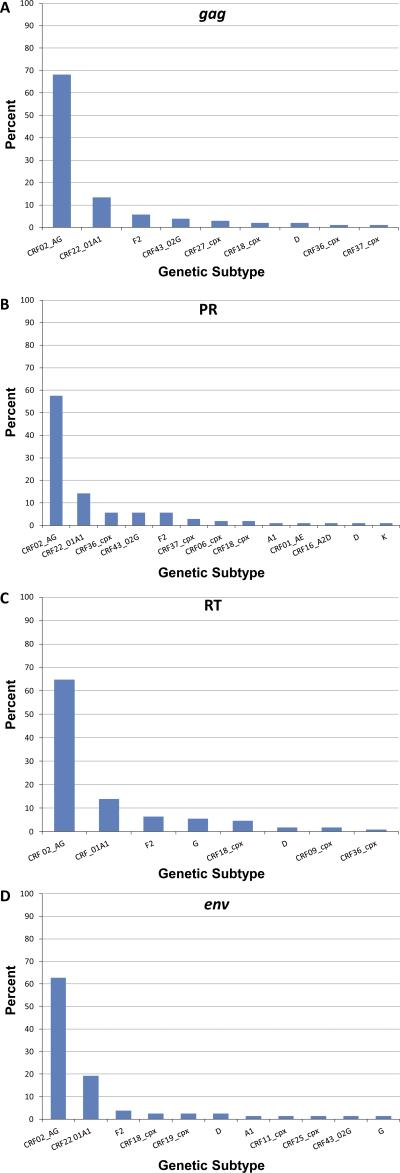
To characterize the viruses circulating in HIV-1 positive individuals seen at the HIV treatment center in Limbe, South West region of Cameroon, 116 samples were amplified by nested PCR in four regions (gag, PR, RT, and env). Sequences of at least one region were successfully obtained for 96% of all samples (111/116). CRF02_AG was the most predominant strain, in 48.6% (54/111) of the study population. URFs made up the second biggest group with 36% (40/111) followed by CRF22_01A1 at 7.2% (8/111), subtype F2 at 3.6% (4/111), subtype D at 1.8% (2/111), CRF18_cpx at 1.8% (2/111), and subtype G at 0.9% (1/111) (Figure 1). Representative subtypes from study subjects and the subtype designations for all the study subjects based on genomic regions analyzed are shown in supplementary Figure 1 and supplementary Table 1. Based on phylogenetic analysis of gag, PR, RT, and env fragments, five (sub) subtypes were detected including A1, D, F2, G, and K and 14 CRFs (CRF01_AE, CRF02_AG, CRF06_cpx, CRF09_cpx, CRF11_cpx, CRF16_A2D, CRF18_cpx, CRF19_cpx, CRF22_01A1, CRF25_cpx, CRF27_cpx, CRF36_cpx, CRF37_cpx, CRF43_02G) (Figures 2A-D).
Figure 1.
HIV-1 group M genetic diversity detected among patients infected with HIV-1 in Limbe, Cameroon. The genetic subtype is defined based on phylogenetic analysis of the sequences of four viral genetic regions (gag, PR, RT, and env). URF, Unique Recombinant Form; URF is defined when one or more region is a discordant subtype
Figure 2.
Proportions of each HIV-1 group M sub-subtype and CRF identified from sequences amplified in (A) gag, (B) PR, (C) RT, and (D) env region of HIV-1 strains from patients in Limbe, Cameroon. Classifications are based on phylogenetic analysis of the sequences of each genetic region. CRF, circulating recombinant form.
Genetic diversity by HIV-1 genomic regions
Genetic diversity in gag: Amplification was successful for 90% (104/116) of the specimens in gag. CRF02_AG was the most predominant subtype detected in gag (68.3%, 71/104), followed by CRF22_01A1 (13.5%, 14/104), subtype F2 (5.8%, 6/104), CRF43_02G (3.8%, 4/104), CRF27_cpx (2.9%, 3/104), CRF18_cpx (1.9%, 2/104), subtype D (1.9%, 2/104), CRF36_cpx (1%, 1/104), and CRF37_cpx (1%, 1/104) (Figure 2A).
Genetic diversity in PR
Sequences of the PR region were obtained for 91% (106/116) of samples. (Sub) subtypes identified included A1 (0.9%, 1/106), F2 (5.7%, 6/106), D (0.9%, 1/106), and K (0.9%, 1/106). CRFs detected at the PR region were: CRF01_AE (0.9%, 1/106), CRF02_AG (57.5%, 61/106), CRF06_cpx (1.9%, 2/106), CRF16_A2D (0.9%, 1/106), CRF18_cpx (1.9%, 2/106), CRF22_01A1 (14.2%, 15/106), CRF36_cpx (5.7%, 6/106), CRF37_cpx (2.8%, 3/106), and CRF43_02G (5.7%, 6/106) (Figure 2B).
Genetic diversity in RT
In 108 of 116 patients (93%), the RT region was amplified successfully via nested PCR, and the following (sub) subtypes and CRFs were identified: CRF02_AG (64.8%, 70/108), CRF22_01A1 (13.9%, 15/108), F2 (6.5%, 7/108), G (5.6%, 6/108), CRF18_cpx (4.6%, 5/108), D (1.9%, 2/108), CRF09_cpx (1.9%, 2/108), and CRF36_cpx (0.9%, 1/108) (Figure 2C).
Genetic diversity in env
Sequences were successfully obtained for 78/116 (67.2%) specimens in env. CRF02_AG was the most predominant subtype (62.8%, 49/78), followed by CRF22_01A1 (19.2%, 15/78), subtype F2 (3.8%, 3/78), subtype D (2.6%, 2/78), CRF18_cpx (2.6%, 2/78), CRF19_cpx (2.6%, 2/78), subtype G (1.3%, 1/78), subtype A1 (1.3%, 1/78), CRF11_cpx (1.3%, 1/78), CRF25_cpx (1.3%, 1/78), CRF43_02G (1.3%, 1/78) (Figure 2D).
Composition of the URFs
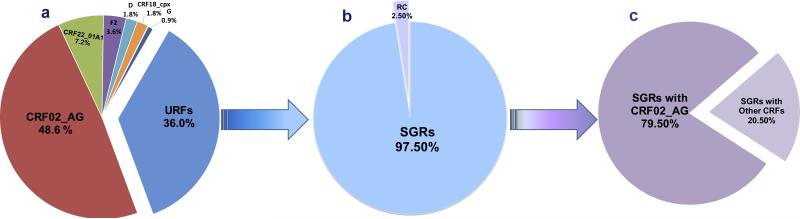
To begin to gain an understanding of the proportion of URFs in this study population, specimens with successful amplification of at least two genomic regions were further analyzed; in this case, 93.1% (108/116) of specimens at more than one region were analyzed. Thus, the different genes that were amplified, sequenced, and phylogenetically subtyped were evaluated for concordant or discordant combinations of subtypes. From this analysis, 36% of the specimens (40/111) were found to be URFs, of which 39/40 included one or more fragments of a previously identified CRF (Figure 3). These 39 specimens represent the second generation recombinant (SGR) infections in this study. Of note, 31 of the 39 SGRs contained fragments of CRF02_AG (Figure 3), and 15 of the 39 SGRs carried CRF22_01A1.
Figure 3.
HIV-1 subtype analysis in patients in Limbe, Cameroon reveals proportion of a) diverse HIV-1 subtypes and URFs, b) SGRs, and c) SGRs containing CRF02_AGs. URF, Unique Recombinant Form. Most of the URFs reveal recombination with CRFs and in particular, CRF02_AG. URFs comprising portions of CRFs are referred to as Second Generation Recombinants (SGRs). The definition of the URF is based on phylogenetic analysis of the sequences of four viral genetic regions (gag, PR, RT, and env) which reveal discordant subtype in at least one gene region.
Drug Resistance Testing for HAART and treatment naïve patients
The 108 sequences obtained from the RT region were used to identify mutations that conferred drug resistance to NRTIs and NNRTIs. Of 108 patients whose HIV-1 RT sequences were amplified successfully, 15 were on treatment and 93 were drug naive. The majority of patients received NRTI/NNRTI standard combinations consisting of Lamivudine (brand name Epivir), Stavudine (brand name Zerit), and Nevirapine (brand name Viramune); this combination is known as Triomune. One patient received Lamivudine (3TC), one received Duovir (a combination of Zidovudine or Azidothymidine (brand name Retrovir) and Lamivudine) plus Efavirenz, and two patients received Stocrin (a brand name for Efavirenz) (Table 2). Three patients (20%) harbored mutations associated with NRTIs (1 patient, 6.7%) and NNRTIs (2 patients, 13%). The identified mutations were K101N, K101Q, and V118I (Table 2). The mutation K101N is an uncommon NNRTI-associated mutation that, in combination with other NNRTI-resistance mutations, has been linked to a low-level decrease in drug susceptibility [Vergne et al., 2006]. Similarly, mutation K101Q by itself does not cause major drug resistance but contributes to reduced responses to Nevirapine (NVP), Efavirenz (EFV), and Etravirine (ETR) when present with other NNRTI-resistance mutations (http://hivdb.stanford.edu/). NRTI mutation V118I is predicted to decrease efficacy of Abacavir (ABC), Zidovudine (AZT), Stavudine (D4T), Didanosine (DDI), and Tenofovir (TDF) (Table 2). Four of the 15 patients on HAART harbored viruses with other polymorphisms: V179I (2 patients), K238R (1 patient), and V90I (1 patient).
Table 2. Drug resistance mutations found in HIV-1 infected patients on HAART.
NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor. The mutations that cause drug resistance are shown. The subtype designation is based on analysis of the RT region and is shown in Supplemental Table 1.
| Mutations | Drugs resistance | Subtype | ||||
|---|---|---|---|---|---|---|
| Sample | Current HAART | NRTI | NNRTI | NRTI | NNRTI | |
| LB046-1 | Lamivudine | CRF02_AG | ||||
| LB047-1 | Triomune | K101Q | NVP, EFV, ETR | CRF02_AG | ||
| LB051-1 | Duovir + Efavirenz | CRF02_AG | ||||
| LB075-1 | Triomune | CRF02_AG | ||||
| LB079-1 | Stocrin | G | ||||
| LB082-1 | Triomune | CRF22_01A1 | ||||
| LB083-1 | Stocrin | CRF02_AG | ||||
| LB084-1 | Triomune | CRF02_AG | ||||
| LB085-1 | Triomune | V118I | ABC, AZT, TDF, D4T,DDI | CRF22_01A1 | ||
| LB086-1 | Triomune | K101N | CRF22_01A1 | |||
| LB091-1 | Triomune | CRF02_AG | ||||
| LB092-1 | Triomune | CRF22_01A1 | ||||
| LB095-1 | Triomune | CRF02_AG | ||||
| LB096-1 | Triomune | CRF02_AG | ||||
| LB099-1 | Triomune | CRF02_AG | ||||
Of the 93 drug naïve patients studied, 13 (13.9%) were infected with viruses that harbored one or more NRTI and/or NNRTI-associated mutations (Table 3). Seven out of 93 patients (7.5%) harbored viruses with mutations associated with NRTIs, three patients (3.2%) harbored viruses with NNRTI-associated mutations, and three (3.2%) had mutations associated with both NRTI and NNRTI (Table 3). The following NRTI-associated mutations were detected: E40F, M41L, T69S, K70R, L74V, Y115F, V118I, M184V, and K219Q. NNRTI-associated mutations included K101N, K103N, K103Q, E138K, and Y188C.
Table 3. Drug resistance mutations in drug-naive HIV-1 infected patients.
NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor. The mutations that cause drug resistance are shown. The subtype designation is based on analysis of the RT region and is shown in Supplemental Table 1.
| Mutations | Drug Resistance | ||||
|---|---|---|---|---|---|
| Sample | NRTI | NNRTI | NRTI Drug | NNRTI Drug | Based on RT |
| LB015-1 | L74V | DDI, ABC | CRF02_AG | ||
| LB025-1 | V118I | E138K | ABC,AZT,D4T, DDI, TDF | ETR,DLV,NVP, EFV | CRF09_cpx |
| LB061-1 | Y188C | NVP,EFV | CRF02_AG | ||
| LB063-1 | M41L | AZT, D4T | CRF22_01A1 | ||
| LB064-1 | K219Q | AZT, D4T | CRF22_01A1 | ||
| LB070-1 | T69S, L74V, Y115F, V118I | DDI, ABC, TDF | CRF02_AG | ||
| LB071-1 | M184V | 3TC,FTC,DDI,ABC | CRF22_01A1 | ||
| LB087-1 | K103Q | F2 | |||
| LB097-1 | E40F, M41L, K70R | AZT,D4T,DDI,ABC,TDF | CRF02_AG | ||
| LB098-1 | K219Q | K103N | AZT.D4T | EFV, NVP | CRF02_AG |
| LB102-1 | Y115F | K101N | ABC, TDF | CRF18_cpx | |
| LB106-1 | M41L | AZT,D4T | CRF02_AG | ||
| LB109-1 | K103N | EFV, NVP | CRF02_AG | ||
Patients infected with viruses carrying one NRTI mutation included LB015-1, LB063-1, LB070-1, LB071-1 and LB106-1. These mutations either conferred resistances to ABC, DDI, AZT, D4T, 3TC, and Emtricitabine (FTC) (L74V, M41L, and M184V) or are predicted to decrease efficacy of ABC, AZT, D4T, DDI, and TDF (V118I). Two patients (LB064-1, and LB097-1) harbored viruses with more than one NRTI associated mutation. Four NRTI associated mutations in the virus infecting LB064-1 (T69S, L74V, Y115F, and K219Q) conferred drug resistance to ADC, DDI and TDF. A total of 3 NRTIs associated mutations were found in the virus harbored by patient LB097-1 (E40F, M41L, and K70R). The combination of these mutations was predicted to confer resistance to five NRTI drugs (AZT, D4T, DDI, ABC, and TDF). Three patients infected with viruses carried one of the following NNRTI mutations (K103N, K103Q, or Y188C). K103N and Y188C confer resistance to the drugs NVP and EFV, while K103Q, when present with other NNRTI-resistance mutations, contributes to reduced efficacy of NVP, EFV and ETR, but is not associated with drug resistance in this patient. Three patients harbored viruses with mutations associated with resistance to both NRTI and NNRTI (ABC, AZT, D4T, DDI, TDF, EFV, ETR, DLV, and NVP) (Table 3). Nineteen (20%) drug naïve patients harbored viruses with the following polymorphisms: V179I (5 patients), E138A (5 patients), V90I (6 patients), A98S (1 patient), V106I (1 patient), and K103R (1 patient).
Analyses for protease mutations
Sequences were next analyzed to determine the presence of mutations that confer resistance to protease inhibitors if such drugs were to be administered to any of the study subjects. Thus, 106 of the (protease naïve) 116 patients (91%) whose protease sequences that were amplified successfully, were subjected to the Stanford University HIV database genotypic resistance interpretation algorithms to determine pre-existing mutations that could cause resistance. The analyses identified 3 (2.5%) of the 116 patient samples that harbored viruses with protease mutations: Q58E mutation, which reduces efficacy of Nelfinavir (NFV) and Tipranavir,(TPV/r), L89V mutation, which causes resistance to NFV, and L33F mutation, which causes resistance to Fosamprenavir (FPV/r) and NFV. All three patients were drug naïve 3/93 (3.2%), with two of three harboring either an NRTI or NNRTI mutation.
Discussion
Previous studies conducted in major cities in Cameroon and in particular at the Limbe Regional Hospital reported HIV-1 CRF02_AG as the dominant variant accounting for 90% in patients from 7 major cities in Southern Cameroon. In fact, the CRF02_AG strain, which is a complex mosaic virus with alternating subtype A and G sequences [Carr et al., 1998; Tscherning-Casper et al., 2000], has consistently been found to be the most predominant in the cities and rural villages of Cameroon [Fonjungo et al., 2000; Nyambi et al., 2002; Zhong et al., 2002; Zhong et al., 2003; Konings et al., 2006a; Ndongmo et al., 2006; Powell et al., 2010a]. Reports from studies in urban areas in Cameroon revealed its prevalence to range from 58% to 75% in gag, pol, env genes and gp41 genes [Konings et al., 2006a; Carr et al., 2010; Ragupathy et al., 2011; Ceccarelli et al., 2012]. Similarly, in some rural areas, prevalence of CRF02_AG has been described to range from 60% to 67% [Konings et al., 2004; Konings et al., 2006a; Carr et al., 2010].
This study reveals that the CRF02_AG variant accounts for only 49% of the infections among a sample of patients in Limbe, Cameroon who attend the Regional Hospital (Figure 1). This low proportion (49%) of CRF02_AG observed could be the result of the in-depth analysis of four different gene regions (gag, PR, RT, and env), which would discriminate URFs from CRF02_AG. However, if only one gene region is analyzed, it would reveal a CRF02_AG prevalence rate ranging from 58% in PR to 68% in gag (Figure 2). Therefore, in-depth analyses analyzing several gene regions or full length sequences are needed to better understand the evolutionary dynamics of the HIV-1 epidemic.
Of interest was the second biggest group detected in this study – URFs – accounting for 36% of infections (Figure 1). A huge percentage (79.5%) of these URFs was SGRs that contained CRF02_AG in one or more of the four analyzed genetic regions (Figure 3). Several possibilities or explanations could account for this high proportion of URFs. First, it is possible that, through superinfection and recombination with CRF02_AG, new SGRs could be emerging within the population studied. It is also possible that these URFs are evolutionary relics [Carr et al., 2010] or a result of recombination events that led to the emergence of viruses with varying fitness and transmissibility. A more detailed analysis is needed to differentiate URFs that may represent ancient variants from those that are newly emerging as well; their fitness and transmissibility should also be studied to determine evolutionary pathways of these viruses. Indeed, different replicative fitness has been reported for the parental subtypes A and G versus the recombinant CRF02_AG [Konings et al., 2006b]; furthermore, other studies have shown URFs with higher replicative kinetics than other viruses [Konings et al., 2006a; Carr et al., 2010; Ragupathy et al., 2011; Ceccarelli et al., 2012]. If superinfection and recombination accounts for the high proportion of URFs reported in this study, it will therefore continue to enable the genetic diversity, and the evolution and emergence of new virus strains; thus, studies to monitor the evolution of viral diversity in such a population are needed. Tourism and entry of migrants from other regions with diverse viruses [Nyambi et al., 2002; Konings et al., 2006a; Torimiro et al., 2009; Powell et al., 2010a] may traffic HIV viruses into the city, fueling the epidemic through superinfection and recombination. Some viral variants may have higher replicative fitness or transmissibility or may be dominant in a population due to founder effect. For example, the predominance of CRF02_AG suggests that this virus strain may be well adapted in the Cameroonian population due to a founder effect [Njai et al., 2004] or has some biologic advantages such as a higher replicative fitness and modification of tropism over other co-circulating strains [Montavon et al., 2000; Fischetti et al., 2004; Sarr et al., 2005; Konings et al., 2006a; Njai et al., 2006].
In this study, (sub)subtypes that were concordant in all four genetic regions analyzed, referred to here as “pure” subtypes, were detected with low prevalence and accounted for only 3/111 (2.7%) of infections. Before the use of antiretroviral drugs, the AIDS epidemic was mostly characterized by non-recombinant subtypes [Takehisa et al., 1998; Nyambi et al., 2002; Konings et al., 2004; Torimiro et al., 2009]. For example, subtype A, which was reported to be the most predominant subtype in Cameroon during initial studies [Nkengasong et al., 1994], was not found in any of the patient sequences, and only one (sub) subtype A1 was identified. This suggests pure subtype A may be rare or absent in this population. However, since only 4 genomic fragments were studied, it is possible that some of the infections considered to be caused by “pure” (sub) subtypes or CRFs are actually caused by URFs, and the real frequency of URFs and SGRs in this study is higher. This study did not identify non-group M subtype infections, which could be either due to the inability of the primers used to amplify such infections or the lack of such infections in this population.
In 2007, the Ministry of Public Health declared free drugs to all eligible individuals, leading to increased use of ARVs in Cameroon. Being a resource constrained country where regular drug supply and patient follow-up may not be as developed as in resource rich countries, this increase in ARV therapy could have resulted in the emergence and transmission of drug resistant strains. In Europe and the United States, routine genotyping for newly diagnosed patients has been shown to be cost effective in managing and treating HIV infections. While this approach at the individual level may be cost prohibitive for resource limited countries, an approach at the population level through field studies such as that described in the present study may be an option to include in HIV treatment and management programs. Taken together, there is a need to monitor the emergence of drug resistant strains in the population to assess the efficacy of drugs commonly used in this region and in regions where drug resistance mutations are common. Interestingly, 13.9% of drug naïve patients have mutations associated with RTIs (Table 3) and 3.2% have mutations associated with protease inhibitors. In cities in Southern Cameroon, 5 of 21 drug naïve patients (24%) in the North West and South West regions harbored viruses with drug resistant mutations, while 18 of 43 drug naive patients (42%) in three different cities (Bamenda, Buea, Limbe) and a few strains found in villages had similar mutations [Burda et al., 2010; Ragupathy et al., 2011]. The high level of drug resistance among the drug naïve population suggests that drug resistance viruses are transmitted in these communities, which poses a major threat to the success of HIV treatment programs.
In this sample population in Southern Cameroon, it revealed a lower CRF02_AG prevalence, an emerging population of URFs bearing the genes of CRF02_AG, and the presence of several drug resistant mutations among drug naïve patients and patients on first and second line of HAART. These studies highlight the need to establish a monitoring system to detect drug resistant mutations before the initiation of HAART and to introduce third line treatment for patients with treatment failure from first and second line therapy.
Supplementary Material
Acknowledgments
Sources of Support: This work was supported by funds from the Department of Veterans Affairs (Merit Review Award and the Research Enhancement Program) and from grants AI083142 from the National Institute of Allergy and Infectious Diseases (NIAID), CA153726 from the National Cancer Institute (NCI), TW001409 from Fogarty International Center (FIC).
The authors are grateful to the individuals who have donated their blood for these studies. The authors wish to acknowledge the continued support of the Ministry of Public Health, Cameroon. The findings and conclusions presented in this manuscript are those of the authors and do not necessarily reflect those of the FDA.
References
- Brennan CA, Bodelle P, Coffey R, Devare SG, Golden A, Hackett J, Jr., Harris B, Holzmayer V, Luk KC, Schochetman G, Swanson P, Yamaguchi J, Vallari A, Ndembi N, Ngansop C, Makamche F, Mbanya D, Gurtler LG, Zekeng L, Kaptue L. The prevalence of diverse HIV-1 strains was stable in Cameroonian blood donors from 1996 to 2004. J Acquir Immune Defic Syndr. 2008;49:432–439. doi: 10.1097/QAI.0b013e31818a6561. [DOI] [PubMed] [Google Scholar]
- Burda ST, Viswanath R, Zhao J, Kinge T, Anyangwe C, Tinyami ET, Haldar B, Powell RL, Jarido V, Hewlett IK, Nyambi PN. HIV-1 reverse transcriptase drug-resistance mutations in chronically infected individuals receiving or naive to HAART in Cameroon. J Med Virol. 2010;82:187–196. doi: 10.1002/jmv.21677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler IF, Pandrea I, Marx PA, Apetrei C. HIV genetic diversity: biological and public health consequences. Curr HIV Res. 2007;5:23–45. doi: 10.2174/157016207779316297. [DOI] [PubMed] [Google Scholar]
- Cameroon MoPH Report of the first semester activities for the fight against HIV/AIDS in Cameroon. 2011 [Google Scholar]
- Cameroon NAcCCTG The impact of HIV and AIDS in Cameroon through 2020. 2010 [Google Scholar]
- Carr JK, Salminen MO, Albert J, Sanders-Buell E, Gotte D, Birx DL, McCutchan FE. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247:22–31. doi: 10.1006/viro.1998.9211. [DOI] [PubMed] [Google Scholar]
- Carr JK, Wolfe ND, Torimiro JN, Tamoufe U, Mpoudi-Ngole E, Eyzaguirre L, Birx DL, McCutchan FE, Burke DS. HIV-1 recombinants with multiple parental strains in low-prevalence, remote regions of Cameroon: evolutionary relics? Retrovirology. 2010;7:39. doi: 10.1186/1742-4690-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli L, Salpini R, Moudourou S, Cento V, Santoro MM, Fokam J, Takou D, Nanfack A, Dori L, Torimiro J, Sarmati L, Andreoni M, Perno CF, Colizzi V, Cappelli G. Characterization of drug resistance mutations in naive and ART-treated patients infected with HIV-1 in Yaounde, Cameroon. J Med Virol. 2012;84:721–727. doi: 10.1002/jmv.23244. [DOI] [PubMed] [Google Scholar]
- Fischetti L, Opare-Sem O, Candotti D, Lee H, Allain JP. Higher viral load may explain the dominance of CRF02_AG in the molecular epidemiology of HIV in Ghana. AIDS. 2004;18:1208–1210. doi: 10.1097/00002030-200405210-00017. [DOI] [PubMed] [Google Scholar]
- Fonjungo PN, Mpoudi EN, Torimiro JN, Alemnji GA, Eno LT, Nkengasong JN, Gao F, Rayfield M, Folks TM, Pieniazek D, Lal RB. Presence of diverse human immunodeficiency virus type 1 viral variants in Cameroon. AIDS Res Hum Retroviruses. 2000;16:1319–1324. doi: 10.1089/08892220050117087. [DOI] [PubMed] [Google Scholar]
- Gilks CF, Crowley S, Ekpini R, Gove S, Perriens J, Souteyrand Y, Sutherland D, Vitoria M, Guerma T, De Cock K. The WHO public-health approach to antiretroviral treatment against HIV in resource-limited settings. Lancet. 2006;368:505–510. doi: 10.1016/S0140-6736(06)69158-7. [DOI] [PubMed] [Google Scholar]
- Hammer SM, Katzenstein DA, Hughes MD, Gundacker H, Schooley RT, Haubrich RH, Henry WK, Lederman MM, Phair JP, Niu M, Hirsch MS, Merigan TC. A trial comparing nucleoside monotherapy with combination therapy in HIV-infected adults with CD4 cell counts from 200 to 500 per cubic millimeter. AIDS Clinical Trials Group Study 175 Study Team. N Engl J Med. 1996;335:1081–1090. doi: 10.1056/NEJM199610103351501. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Konings FA, Haman GR, Xue Y, Urbanski MM, Hertzmark K, Nanfack A, Achkar JM, Burda ST, Nyambi PN. Genetic analysis of HIV-1 strains in rural eastern Cameroon indicates the evolution of second-generation recombinants to circulating recombinant forms. J Acquir Immune Defic Syndr. 2006a;42:331–341. doi: 10.1097/01.qai.0000219784.81163.2e. [DOI] [PubMed] [Google Scholar]
- Konings FA, Burda ST, Urbanski MM, Zhong P, Nadas A, Nyambi PN. Human immunodeficiency virus type 1 (HIV-1) circulating recombinant form 02_AG (CRF02_AG) has a higher in vitro replicative capacity than its parental subtypes A and G. J Med Virol. 2006b;78:523–534. doi: 10.1002/jmv.20572. [DOI] [PubMed] [Google Scholar]
- Konings FA, Zhong P, Agwara M, Agyingi L, Zekeng L, Achkar JM, Ewane L, Saa, Afane Ze E, Kinge T, Nyambi PN. Protease mutations in HIV-1 non-B strains infecting drug-naive villagers in Cameroon. AIDS Res Hum Retroviruses. 2004;20:105–109. doi: 10.1089/088922204322749558. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Laurent C, Kouanfack C, Vergne L, Tardy M, Zekeng L, Noumsi N, Butel C, Bourgeois A, Mpoudi-Ngole E, Koulla-Shiro S, Peeters M, Delaporte E. Antiretroviral drug resistance and routine therapy, Cameroon. Emerg Infect Dis. 2006;12:1001–1004. doi: 10.3201/eid1206.050860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbopi Keou FX, Mbu R, Mauclere P, Andela A, Tetanye E, Leke R, Chaouat G, Barre-Sinoussi F, Martin P, Belec L. Antenatal HIV prevalence in Yaounde, Cameroon. Int J STD AIDS. 1998;9:400–402. doi: 10.1258/0956462981922485. [DOI] [PubMed] [Google Scholar]
- Montavon C, Toure-Kane C, Liegeois F, Mpoudi E, Bourgeois A, Vergne L, Perret JL, Boumah A, Saman E, Mboup S, Delaporte E, Peeters M. Most env and gag subtype A HIV-1 viruses circulating in West and West Central Africa are similar to the prototype AG recombinant virus IBNG. J Acquir Immune Defic Syndr. 2000;23:363–374. doi: 10.1097/00126334-200004150-00001. [DOI] [PubMed] [Google Scholar]
- Ndembi N, Abraha A, Pilch H, Ichimura H, Mbanya D, Kaptue L, Salata R, Arts EJ. Molecular characterization of human immunodeficiency virus type 1 (HIV-1) and HIV-2 in Yaounde, Cameroon: evidence of major drug resistance mutations in newly diagnosed patients infected with subtypes other than subtype B. J Clin Microbiol. 2008;46:177–184. doi: 10.1128/JCM.00428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndongmo CB, Pieniazek D, Holberg-Petersen M, Holm-Hansen C, Zekeng L, Jeansson SL, Kaptue L, Kalish ML. HIV genetic diversity in Cameroon: possible public health importance. AIDS Res Hum Retroviruses. 2006;22:812–816. doi: 10.1089/aid.2006.22.812. [DOI] [PubMed] [Google Scholar]
- Njai FH, van der Auwera G, Ngong CA, Heyndrickx L, Sawadago S, Whittle H, Nyambi P, Colebunders R, van der Groen G, Janssens W. Development, evaluation and validation of an oligonucleotide probe hybridization assay to subtype human immunodeficiency virus type 1 circulating recombinant from CRF02_AG. J Clin Microbiol. 2004;42:1428–1433. doi: 10.1128/JCM.42.4.1428-1433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njai HF, Gali Y, Vanham G, Clybergh C, Jennes W, Vidal N, Butel C, Mpoudi-Ngolle E, Peeters M, Arien KK. The predominance of Human Immunodeficiency Virus type 1 (HIV-1) circulating recombinant form 02 (CRF02_AG) in West Central Africa may be related to its replicative fitness. Retrovirology. 2006;3:40. doi: 10.1186/1742-4690-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkengasong JN, Janssens W, Heyndrickx L, Fransen K, Ndumbe PM, Motte J, Leonaers A, Ngolle M, Ayuk J, Piot P, et al. Genotypic subtypes of HIV-1 in Cameroon. AIDS. 1994;8:1405–1412. doi: 10.1097/00002030-199410000-00006. [DOI] [PubMed] [Google Scholar]
- Nyambi P, Heyndrickx L, Vereecken K, Burda S, De Houwer K, Coppens S, Urbanski M, Williams C, Ndumbe P, Janssens W. Predominance of infection with HIV-1 circulating recombinant form CRF02_AG in major Cameroonian cities and towns. AIDS. 2002;16:295–296. doi: 10.1097/00002030-200201250-00022. [DOI] [PubMed] [Google Scholar]
- Palella FJ, Jr., Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- Powell R, Barengolts D, Mayr L, Nyambi P. The Evolution of HIV-1 Diversity in Rural Cameroon and its Implications in Vaccine Design and Trials. Viruses. 2010a;2:639–654. doi: 10.3390/v2020639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell RL, Lezeau L, Kinge T, Nyambi PN. Longitudinal quasispecies analysis of viral variants in HIV type 1 dually infected individuals highlights the importance of sequence identity in viral recombination. AIDS Res Hum Retroviruses. 2010b;26:253–264. doi: 10.1089/aid.2009.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell RL, Zhao J, Konings FA, Tang S, Nanfack A, Burda S, Urbanski MM, Saa DR, Hewlett I, Nyambi PN. Identification of a novel circulating recombinant form (CRF) 36_cpx in Cameroon that combines two CRFs (01_AE and 02_AG) with ancestral lineages of subtypes A and G. AIDS Res Hum Retroviruses. 2007;23:1008–1019. doi: 10.1089/aid.2006.0289. [DOI] [PubMed] [Google Scholar]
- Ragupathy V, Zhao J, Wood O, Tang S, Lee S, Nyambi P, Hewlett I. Identification of new, emerging HIV-1 unique recombinant forms and drug resistant viruses circulating in Cameroon. Virol J. 2011;8:185. doi: 10.1186/1743-422X-8-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarr AD, Eisen G, Gueye-Ndiaye A, Mullins C, Traore I, Dia MC, Sankale JL, Faye D, Mboup S, Kanki P. Viral dynamics of primary HIV-1 infection in Senegal, West Africa. J Infect Dis. 2005;191:1460–1467. doi: 10.1086/429409. [DOI] [PubMed] [Google Scholar]
- Takehisa J, Zekeng L, Ido E, Mboudjeka I, Moriyama H, Miura T, Yamashita M, Gurtler LG, Hayami M, Kaptue L. Various types of HIV mixed infections in Cameroon. Virology. 1998;245:1–10. doi: 10.1006/viro.1998.9141. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongo M, Martin DP, Zembe L, Mpoudi-Ngole E, Williamson C, Burgers WA. Characterization of HIV-1 gag and nef in Cameroon: further evidence of extreme diversity at the origin of the HIV-1 group M epidemic. Virol J. 2013;10:29. doi: 10.1186/1743-422X-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torimiro JN, D'Arrigo R, Takou D, Nanfack A, Pizzi D, Ngong I, Carr JK, Joseph FP, Perno CF, Cappelli G. Human immunodeficiency virus type 1 intersubtype recombinants predominate in the AIDS epidemic in Cameroon. New Microbiol. 2009;32:325–331. [PubMed] [Google Scholar]
- Tscherning-Casper C, Dolcini G, Mauclere P, Fenyo EM, Barre-Sinoussi F, Albert J, Menu E. Evidence of the existence of a new circulating recombinant form of HIV type 1 subtype A/J in Cameroon. The European Network on the Study of In Utero Transmission of HIV-1. AIDS Res Hum Retroviruses. 2000;16:1313–1318. doi: 10.1089/08892220050117078. [DOI] [PubMed] [Google Scholar]
- UNAIDS/WHO AIDS Epidemic updates. 2007 [Google Scholar]
- Vergne L, Stuyver L, Van Houtte M, Butel C, Delaporte E, Peeters M. Natural polymorphism in protease and reverse transcriptase genes and in vitro antiretroviral drug susceptibilities of non-B HIV-1 strains from treatment-naive patients. J Clin Virol. 2006;36:43–49. doi: 10.1016/j.jcv.2006.01.012. [DOI] [PubMed] [Google Scholar]
- WHO Towards universal access: Scaling up priority HIV/AIDS interventions in the Health Sector. 2008 [Google Scholar]
- Zhong P, Burda S, Urbanski M, Kenfack H, Tongo M, Heyndrickx L, Nanfack A, Shang J, Agyingi L, Zolla-Pazner S, Zekeng L, Nyambi P. HIV type 1 group M clades infecting subjects from rural villages in equatorial rain forests of Cameroon. J Acquir Immune Defic Syndr. 2002;31:495–505. doi: 10.1097/00126334-200212150-00007. [DOI] [PubMed] [Google Scholar]
- Zhong P, S BU, Konings F, Urbanski M, Ma L, Zekeng L, Ewane L, Agyingi L, Agwara M, Saa, Afane ZE, Kinge T, Zolla-Pazner S, Nyambi P. Genetic and biological properties of HIV type 1 isolates prevalent in villagers of the Cameroon equatorial rain forests and grass fields: further evidence of broad HIV type 1 genetic diversity. AIDS Res Hum Retroviruses. 2003;19:1167–1178. doi: 10.1089/088922203771881284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.