Abstract
Despite recent advances in the understanding of ethanol’s biological action, many of the molecular targets of ethanol and mechanisms behind ethanol’s effect on behavior remain poorly understood. In an effort to identify novel genes, the products of which regulate behavioral responses to ethanol, we recently identified a mutation in the dtao gene that confers resistance to the locomotor stimulating effect of ethanol in Drosophila. dtao encodes a member of the Ste20 family of serine/threonine kinases implicated in MAP kinase signaling pathways. In the present study, we report that conditional ablation of the mouse dtao homolog, Taok2, constitutively and specifically in the nervous system, results in strain-specific and overlapping alterations in ethanol-dependent behaviors. These data suggest a functional conservation of dtao and Taok2 in mediating ethanol’s biological action and identify Taok2 as a putative candidate gene for ethanol use disorders in humans.
Keywords: TAOK2, MAP Kinase, ethanol sensitivity, ethanol consumption, conditioned place preference
Introduction
An estimated 12.5% of the U.S. population suffers from alcohol dependence (Hasin et al. 2007), exacting immense individual and societal costs. Despite the high prevalence of alcohol use disorders, the biological effects of ethanol remain poorly understood. In contrast to many drugs of abuse, ethanol acts on multiple biological targets, including neurotransmitter systems, cellular membrane components and intracellular signaling pathways (Buck and Harris, 1991; Chandler et al. 2003; Harris and Hitzmann, 1981; Moonat et al. 2010; Newton and Messing, 2006).
Emerging evidence suggests that mitogen activated protein kinase (MAPK) signaling cascades may play an important role in the action of ethanol and other drugs of abuse (Arror and Shukla, 2004; Corominas et al. 2007; Herbert and O’Callaghan, 2000). MAPK signaling transduces such diverse extracellular stimuli as growth factors, cytokines, and environmental stressors via a three-tiered intracellular signaling pathway to induce changes in gene transcription, cellular morphology, differentiation, proliferation and death (Krishna and Narang, 2008; Seger and Krebs, 1995). Specifically, MAP kinase kinase kinases (MAP3Ks) phosphorylate and activate MAP2Ks, which in turn phosphorylate and activate three classes of MAP kinases: p38, ERK, and JNK (Davis, 1994; Sugden and Clerk, 1997; Takeda and Ichijo, 2002). Meta-analysis of expression studies has revealed that brain transcript levels of various MAPK signaling components are altered in response to ethanol, cocaine, and other drugs of abuse (Daniels et al. 2002; Freeman et al. 2010; Hassan et al. 2003; Konu et al. 2001). Furthermore, ethanol treatment increases phosphorylation and activation of p38, ERK and JNK in both liver and brain (Aroor and Shukla, 2004; Aroor et al. 2010; Jung et al. 2010; Ku et al. 2007).
In a screen for Drosophila melanogaster mutants with abnormal ethanol-induced hyperactivity, we recently identified a loss-of-function mutation in the dtao gene that almost completely abolishes flies’ hyperactivity response to ethanol (King et al. 2011). dtao encodes a putative MAP3K of the GCK-VIII subfamily of Ste20p (sterile 20 protein) kinases (Dan et al. 2001). These proteins, also known as TAO (thousand-and-one amino acid) kinases, are characterized by a highly conserved serine/threonine kinase domain, which regulates MAP signaling (Chen and Cobb, 2001; Hutchison et al. 1998; Tassi et al. 1999; Yasuda et al. 2007). In addition to their catalytic function, TAO kinases regulate cytoskeletal organization through interaction with actin and tubulin, via a structurally divergent C-terminal tail region (Johne et al. 2008; Mitsopoulos et al. 2003; Moore et al. 2000; Timm et al. 2003; Zihni et al. 2006).
In mammals, dtao is represented by three orthologous genes: Taok1, Taok2 and Taok3 (Yustein et al. 2003). TAOK2, in addition to putative roles in the regulation of actin and tubulin dynamics, regulates neuronal plasticity via activation of p38 and trafficking of cell adhesion molecules arcadlin and N-cadherin (Yamagata et al. 1999; Yasuda et al. 2007), indicating that it may also regulate neuroadaptive changes associated with ethanol exposure. Based on these observations, we hypothesized that disruption of the Taok2 gene in mice would alter the behavioral response to ethanol.
Methods
Generation of Taok2tm1fl mice
The Taok2tm1 targeting construct was generated using an approach similar to that reported by Aoyama et al. 2005. Briefly, a C57BL/6J-derived bacterial artificial chromosome clone (RP23-142A14) comprising the Taok2 gene was obtained from Invitrogen (Carlsbad, CA, USA), digested with BamHI and BsrBI (New England Biolabs, Ipswich, MA, USA) and shotgun subcloned into the BamHI/SmaI sites of vector pBSDT-AII (Aoyama et al. 2005). Plasmid-transformed E.coli colonies were screened by PCR using the following primers: forward: 5’GCTGAGGCTACCTCCTCCTT, reverse: 5’TGCTGCTTATGCAGTTGGAC to identify an 8.7 Kb genomic clone containing exons 1–7 of Taok2 and flanking intronic sequence. DNA from insert-containing plasmids was incubated with plasmid pMODloxZeoΔamp3 (Aoyama et al. 2005) in the presence of EZ:Tn transposase (Epicentre Biotechnologies, Madison, WI, USA) to introduce two lox P sites flanking a zeocin selectable marker. The position and orientation of this insertion was determined by sequencing. Plasmid DNA from this clone was incubated with Cre recombinase (New England Biolabs) to remove the zeocin selectable marker and combine the two lox P sites into a single site. To introduce a second lox P site into the construct, plasmid DNA from this clone was incubated with plasmid pGPS21loxFRTNeo (Aoyama et al. 2005) in the presence of TnsABC transposase (New England Biolabs). pGPS21loxFRTNeo contains two Tn7-derived transposable elements flanking two lox P sites, two FRT sites and the neomycin phosphotransferase gene (Neo) placed downstream of the EM7 and PGK promotors to confer kanamycin and G418 resistance in E. coli and mouse embryonic stem (ES) cells, respectively. Removal of the Neo cassette and one of the lox P sites may be performed either in ES cells or mice with Flpe-mediated recombination. Position and orientation of the insertion into the genomic fragment was determined by sequencing, and a clone with both lox P sites in the same orientation was selected for electroporation in ES cells. Sequence analysis of the entire insert of the resulting targeting construct verified insertion of lox P sites within the first and seventh introns of the Taok2 gene and confirmed that the remaining genomic sequence was left intact. Mouse ES cells and transgenic mice targeting Taok2 were generated using standard methods by the Transgenic Core Facility at the Ernest Gallo Clinic and Research Center (EGCRC, Bradley 1987, Hogan et al.1994). Briefly, DNA from plasmids containing the Taok2-targeted locus was linearized at the PmeI (New England Biolabs) site in the multiple cloning site of the vector and electroporated into C57BL/6-derived ES cells (Primogenix, Laurie, MO, USA). PCR analysis was performed on the resulting G418-resistent clones using a forward primer spanning part of the PGK promotor (5’-GGGGAACTTCCTGACTAGGG) and a reverse primer recognizing mouse genomic sequence immediately 3’ of that included in the targeting construct (5’-AGGGCCTAGGGCAAAATAGA) to assess homologous recombination. Prior to blastocyst injection, the integrity of these targeted ES cell clones was verified by sequence analysis of the targeted region. Cells were injected into C57BL/6Jtyrc-2J-derived blastocysts (Jackson Laboratory, Bar Harbor, ME, USA) and implanted in pseudopregnant CD1 dams to generate chimeric mice. C57BL/6Jtyrc-2Jmice carry a recessive point mutation in the tyrosinase gene resulting in a white coat color, allowing the distinction of targeted cells that confer a black coat color. Chimeric pups obtained from these lines were crossed with C57BL/6Jtyrc-2Jmice to test for germline transmission and stabilization of the line, designated, Taok2tm1. Subsequently, Taok2tm1/+ mice were crossed with Flpe mice (Jackson Laboratory) to remove the Neo marker and yield the Taok2tm1fl allele.
Mouse crosses
Taok2tm1fl/+ mice were intercrossed to generate Taok2tmfl/fl homozygous animals for subsequent crosses. Additionally, Taok2tm1fl/+ mice were mated with B6.Cg-Tg(Nes-cre)1Kln/J/+ mice (Jackson Laboratory). B6.Cg-Tg(Nes-cre)1Kln/J/+ mice, abbreviated “Nes-cre/+”, express CRE recombinase in the nervous system under control of the rat nestin promotor (Tronche et al.1999). Taok2tm1fl/+;Nes-cre/+ mice were crossed to Taok2tm1fl/fl mice to generate Taok2tm1fl/fl;Nes-cre/+ mice and littermate controls (Taok2tm1fl/+ mice) for expression analysis and behavioral experiments. In the course of analyzing these crosses, we observed “leaky” germline expression of CRE recombinase in a small subset of progeny. These animals were bred to C57BL/6J mice to generate Taok2+/− mice, which were subsequently intercrossed to generate Taok2−/− mice and controls for behavioral experiments.
Genotypic Analysis
Mice were genotyped using DNA isolated from tail biopsies using standard protocols. The following primers were used to genotype progeny from the above described crosses by PCR analysis: forward primer 5’-CCAAGGACCAGACATCCACT and reverse primer 5’-ACCAGTCCTCGTTTTTGCTG were used to detect presence of the wild-type and Taok2tm1fl alleles; forward primer 5’-GATGCAACGAGTGATGAG and reverse primer 5’-TCGGCTATACGTAACAGG were used to detect presence of the Nes-cre transgene; forward primer 5’-TGCAGGGTTTGACAGTTGAT and reverse primer 5’-ACTCTGCCTCAGGAGTCCAA were used to detect presence of the null allele.
Immunohistochemistry
Mice were deeply anesthetized with 100mg/kg of Euthasol® (Virbac, Forth Worth, TX, USA) and intracardially perfused with 0.9% NaCl, followed by 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO) in phosphate buffer, (PB) pH 7.4. Brains were removed, post-fixed overnight in the same fixative at 4 °C, incubated in 30% sucrose for 48 hours for cryoprotection. Frozen sagittal, 50 µm-thick sections were cut on a cryostat (Leica Instruments, Nussloch, Germany). Free-floating sections were incubated in 3% hydrogen peroxide in phosphate-buffered saline (PBS) for 10 min, then in 50% alcohol for 10 min, rinsed in PBS, incubated in 10% normal donkey serum in PBS (Jackson ImmunoResearch, West Grove, PA, USA) for 30 min, and incubated overnight in the goat polyclonal antibody recognizing TAOK2 (sc-47447, Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:1,000). On the next day sections were rinsed in PBS incubated in 2% normal donkey serum in PBS for 10 min and incubated in biotinylated donkey anti-goat (1:500, Jackson ImmunoResearch) for 3 hours, rinsed in PBS and incubated in ExtrAvidin-peroxidase complex (1:2,500, Sigma-Aldrich, St. Louis, MO, USA) for 2 hours. Peroxidase was histochemically visualized with diaminobenzidine (Sigma-Aldrich). Sections were mounted on gelatin-coated slides, air-dried, dehydrated in graded alcohols, cleared with xylene, and mounted with DPX mounting media (Sigma-Aldrich). Images were taken examined in a Nikon Eclipse E600 microscope equipped with Spot 2 CCD camera (Diagnostic Instruments, Sterling Heights, MI, USA). In control experiments the omission of primary antibody resulted in lack of immunostaining.
Quantitative PCR (QPCR)
Total RNA was isolated from whole brain tissue using Trizol® reagent (Invitrogen) according to the manufacturer’s instructions and was treated with RNase-free DNase (Promega, Madison, WI, USA) to remove genomic DNA contamination. cDNA was synthesized from 1 µg of total RNA using reverse transcription reagents from Applied Biosystems (Foster City, CA, USA). Following synthesis, cDNA was diluted 1:10 in water. TaqMan QPCR was performed using standard thermal cycling conditions on an ABI PRISM 7900 Sequence Detection System (Applied Biosystems). Amplification reactions contained 5 µl of cDNA template, 1x Universal PCR Master Mix, 100 nM each of forward and reverse primers, and 200 nM of FAM-labeled probe in a final volume of 10 µl. The Taok2 5’probe and primers set spanning exons 4–5 was Applied Biosystems catalog # Mm001139118. Rodent GAPDH probe and primers (Applied Biosystems) were used as a control for the PCR reactions. Data was analyzed using the comparative Ct method (Applied Biosystems user bulletin #2).
Western Analysis
Protein extracts were isolated from whole brain tissue in RIPA buffer using standard techniques. 10 µg protein samples were electrophoresed on NuPAGE 4–12% SDS-polyacrylamide gels (Invitrogen) and transferred to PVDF membranes (Invitrogen). Primary antibodies included goat anti-TAOK2 (sc-47447, Santa Cruz Biotechnology, Inc) 1:500 and mouse anti-GAPDH (Fisher Scientific, Pittsburgh, PA, USA), 1:5000. Western blots were incubated with either HRP-linked donkey anti-goat or HRP-linked sheep anti-mouse secondary antibody (GE Healthcare, Biosciences, Piscataway, NJ, USA), 1:5000, and processed with ECL Plus Western Blotting Detection System (GE Healthcare, Biosciences). Chemiluminescence was visualized and quantified using the Storm 660 phosphorimager system (Molecular Dynamics, Sunnyvale, CA, USA).
Behavioral Testing
Behavioral testing was performed on male mice (to avoid potential behavioral effects of the estrus cycle on behavior) that were 3–6 months of age at time of testing. Behavior was performed as follows: group 1 (Taok2−/− and control animals): open field activity, stationary dowel test, LORR test and drinking in the dark with one week rest period between each behavioral test. Group 2 (Taok2−/− and control animals): ethanol metabolism and clearance. Group 3: (Taok2−/− and control animals): ethanol CPP. Group 4: (Taok2fl/fl:Nes-cre and control mice): LORR test. Group 5: (Taok2fl/fl:Nes-cre and control mice): drinking in the dark. Group 6: (Taok2fl/fl:Nes-cre and control mice): ethanol metabolism and clearance. All animal protocols were approved by the EGCRC institutional animal care and use committee.
Stationary dowel test of acute functional tolerance (AFT)
A stationary wooden dowel (1.5 cm diameter and 30-cm long) was suspended between two Plexiglas walls 50 cm above a cushioned surface. Mice were trained to remain on the stationary dowel for 5 minutes. Each mouse was then given an intraperitoneal injection of 1.5 g/kg of ethanol (10% w/v in saline) and placed on the dowel. At this dose, subjects were unable to balance on the dowel. After a subject fell from the dowel, it was retested for regain of balance at 5 minutes intervals until it was able to remain on the dowel for 1 minute. Immediately following regain of balance, a blood sample was taken by tail vein puncture (BEC1), the mouse was given a booster injection of 1.5 g/kg ethanol and placed back on the dowel. The time at which the second period of ataxia began was recorded and when the subject fell from the dowel. Recovery was again tested at 5 minute intervals until the subject was able to remain on the dowel for 1 minute, at which point a second blood sample was taken (BEC2). AFT was calculated as the difference between BEC2 and BEC1. The rate at which AFT developed was calculated as (BEC2-BEC1)/duration of ataxia following the booster injection.
Loss-of-righting reflex (LORR) assay
Ethanol (10% v/v in saline) was administered IP at a dose of 4g/kg. After injection, mice were placed on their backs and tested for loss of the righting reflex. The mouse was judged to have lost the righting reflex at the time when it could not right itself three times within 30 seconds. When the animal was able to right itself three times within 30 seconds it was deemed to have recovered. The duration of the loss of the righting reflex was calculated as the difference between when the reflex was lost and when it was recovered. For Taok2−/− and control mice, blood ethanol content at time of recovery was measured by tail vein bleed.
Drinking in the dark (DID) assay
Oral alcohol self-administration was examined using a limited access, drinking in the dark assay. One week prior to data collection, mice were singly housed and transferred to a reverse light-dark cycle. On day 8 of the experiment, water intake (ml) was measured for the 4-hour period beginning 4 hours after lights off and body weight (g) was recorded. Subsequently, mice were given access to a single bottle of 20%w/v ethanol during the same time of day, on alternate days for 10 days (i.e., 5 days of data collection) and ethanol consumption was calculated. For Taok2−/− and control mice, water consumption was also measured on alternate days during the same 4 hour time period for the duration of the study.
Two-bottle choice assay for taste preference
Taste preference for saccharin (sweet) and quinine (bitter) was examined using a two-bottle choice protocol. Mice were singly housed for 4 days in double-grommet cages with continuous access to two water bottles under standard 12 hour light/12 hour dark conditions. On day 5, the left water bottle was replaced with a 0.06% w/v solution saccharin in water. On the following day, the amount of saccharin solution and water consumed was determined and the position of the bottles was switched. The next day, saccharin and water consumption were again determined and the procedure was repeated using a 0.03 mM quinine solution (in water) and water.
Conditioned place preference assay and ethanol-induced locomotor stimulation
Ethanol-induced conditioned place preference (CPP) was performed using an apparatus (Med Associates, St. Albans, VT, USA) consisting of two open field chambers (each 27.3×27.3 cm) separated by a central guillotine door and characterized by custom acrylic floors that are contextually distinct. Ethanol-naïve mice were allowed free access to both chambers of the CPP apparatus for 30 minutes on day 1 of the experiment to habituate to the apparatus, assess general activity level and initial side-preference. Subjects were then assigned to either of two experimental groups in a counterbalanced design in which ethanol was paired with the right chamber or left chamber. On day 2, subjects received either an intraperitoneal injection of 2g/kg ethanol or saline prior to placement in one of the two chambers for 5 minutes. On day 3, subjects that received an ethanol injection on day 2 were given a saline injection and allowed access to the opposite chamber for 5 minutes. Subjects were thus trained with alternating ethanol and saline treatments for a total of 8 days. On the test day, subjects were allowed free access to both chambers for 30 minutes, during which the time spent in each chamber was recorded. For baseline activity and ethanol-induced locomotor stimulation, the horizontal activity of subjects on the first day of saline treatment (either day 2 or day 3) was compared to activity on the first day of ethanol treatment (day 2 or day 3).
Open field activity
Locomotor activity measurements were performed in Plexiglas locomotor activity chambers (43 cm X 43 cm, Med Associates, St. Albans, VT, USA), located in sound-attenuated cubicles equipped with a 2.8 watt house light and exhaust fans to mask external noise. The chambers contained two sets of 16 pulse-modulated infrared photobeams on opposite walls to record x, y ambulatory movements and are computer interfaced for data sampling at 100 ms resolution. Mice were placed in chambers for 60 minutes and the distance traveled (cm) was measured in 5 minute bins.
Ethanol metabolism and clearance
Ethanol-naive mice were injected with 4g/kg of ethanol and tail blood samples (10 µl) were obtained at 10, 30, 60, 90, 120 and 180 minutes to measure blood alcohol levels. Blood ethanol content was assessed from serum using the Analox AM1 Analyzer (Analox Instruments, North Yorkshire, UK).
Statistical Analysis
Data were analyzed using SigmaStat 3.1 software (Systat Software, San Jose, CA, USA) with appropriate post-hoc comparisons performed as indicated by SigmaStat. Statistical tests were performed as follows: mRNA and protein quantification in Taok2tm1fl/fl;Nes-cre/+ mice was normalized to control littermates and GAPDH signal and compared by student’s t-test . Water consumption data for Taok2tm1fl/fl;Nes-cre/+ and control mice were analyzed by student’s t-test and for Taok2−/− and control mice during DID experiments by 2-way repeated-measures ANOVA for genotype X day. AFT and LORR data were analyzed by student’s t-test. Ethanol consumption data during DID experiments were analyzed by 2-way repeated-measures ANOVA for genotype X day. Similarly, ethanol clearance data were analyzed by 2-way repeated-measures ANOVA for genotype X time. CPP data were analyzed by 2-way repeated measures ANOVA for genotype X time spent in the ethanol-paired chamber on day 1 (habituation) and day 10 (test). Open field activity data were analyzed by 2-way repeated-measures ANOVA for genotype X time. Ethanol-induced locomotor stimulation data were also analyzed by 2-way repeated measures ANOVA for genotype X treatment. Data are presented as mean ± standard error of the mean.
Results
Generation and characterization of Taok2−/− and Taok2tm1fl/fl;Nes-cre mice
To generate a Taok2 allele in which the gene is conditionally disrupted, we used a modified approach described by Aoyama et al. 2005 and Seong et al. 2004. This approach simplifies construction of a targeting vector by the insertion of lox P sites and selectable markers in a region of interest using in vitro transposition (Aoyama et al. 2005). The targeting construct was made using C57BL/6J-derived genomic DNA, a strain for which fully sequenced bacterial artificial clones (BACs) are commercially available, obviating the need to screen a BAC library and facilitating in silico design of the targeting construct. The resulting C57BL/6J-derived embryonic stem cells were injected into C57BL/6Jtyr-derived blastocysts. C57BL/6Jtyr mice carry a recessive mutation in the tyrosinase gene that results in a white coat color, allowing for the identification of C57BL/6JTaok2tm1fl; C57BL/6Jtyr chimeric pups and germline transmission of the Taok2tm1fl allele in the subsequent generation. An important advantage of this approach over standard mouse transgenesis methods is that the targeted allele is generated on an isogenic background as opposed to a mixed C57BL/6J X 129-derived background, which may complicate phenotypic analysis (Seong et al. 2004).
We targeted the kinase domain of the Taok2 gene by insertion of lox P sites within introns 1 and 7 to generate the Taok2tm1 allele. In the presence of FLPe recombinase, the neomycin resistance selectable marker was removed to yield the Taok2tm1fl allele (Fig. 1a). This allele, in the presence of a CRE recombinase-expressing transgene, is expected to excise sequences containing exons 2–7 of the Taok2 locus to generate the deleted or Taok2 null allele (Taok2−/−, Fig. 1a).
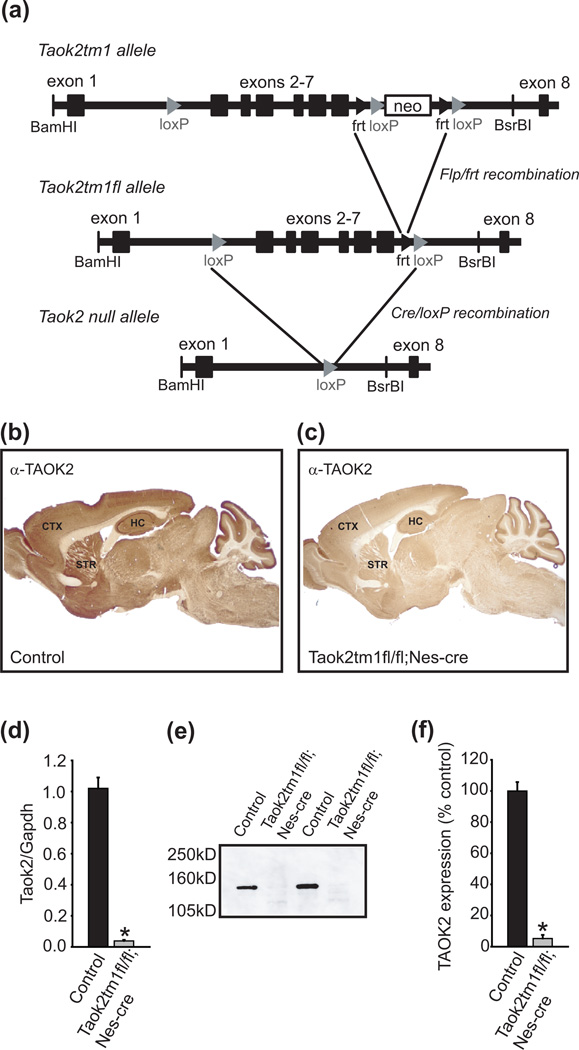
Figure 1. Targeted disruption of the Taok2 gene in mice.
(a) Schematic of the conditionally targeted Taok2 allele. The Taok2tm1 allele was generated by introduction of lox P sites flanking exons 2 though 7 of the Taok2 gene. Within intron 7 is a neomycin resistance gene flanked by lox P and frt sites. Upon crossing this line to mice expressing Flpe recombinase, the neomycin selectable marker is removed along with one of the lox P sites to generate the Taok2tm1fl allele. In the presence of Cre recombinase, the remaining lox P sites are recombined into a single lox P site, removing exons 2 though 7 and the translational start site to generate the Taok2tm1Δ allele. (b) Brain sagittal section of a control mouse shows strong expression of TAOK2 by immunohistological analysis in all areas, especially in cortex, hippocampus and striatum. (c) Immunohistological analysis shows reduced TAOK2 expression in Taok2tm1fl/fl;Nes-cre brain. (d) Quantitative PCR analysis of total mouse brain extracts shows near absence of Taok2 transcript in Taok2tm1fl/fl;Nes-cre brain (n = 4) compared to control mice (n = 2). (e) Western blot showing comparative loss of TAOK2 protein in total brain lysates of Taok2tm1fl/fl;Nes-cre mice relative to controls. (f) Quantification of anti-TAOK2 signal from (e) comparing Taok2tm1fl/fl;Nes-cre mice (n = 2) and controls (n = 2). Lox P sites are denoted by grey chevrons, frt sites by black chevrons. Neo = neomycin resistance gene. The positions of restriction sites BamH1 and BsrB1 used in the generation of the targeting construct are indicated. CTX = cortex; HC = hippocampus; STR = striatum. Error bars are mean ± SEM. Asterisks indicate level of significance (* P < 0.05).
To disrupt the Taok2 gene specifically in the nervous system, we crossed Taok2tm1fl/fl mice with B6.Cg-Tg(Nes-cre)1Kln/J/+ mice (abbreviated Nes-cre/+). Nes-cre/+ mice express CRE recombinase under control of the rat nestin promotor (Tronche et al. 1999). Taok2tm1fl/fl;Nes-cre/+ mice were viable and normal in appearance. In addition, gross brain morphology of Taok2tm1fl/fl;Nes-cre/+ mice, as determined by immunohistological analysis, was indistinguishable from that of control animals (Fig. 1b,c). In wild-type mice, we observed abundant TAOK2 expression throughout the brain, including cortex, striatum and hippocampus (Fig. 1b). In contrast, we confirmed near absence of TAOK2 protein in brains of Taok2tm1fl/fl;Nes-cre/+ mice (Fig. 1c).
We assessed the efficiency of Nes-cre mediated Taok2 transcript disruption in brains of Taok2tm1fl/fl;Nes-cre/+ mice by quantitative PCR (qPCR). QPCR analysis using a 5’ primer/probe set internal to the lox P sites revealed nearly 100% excision of the intervening sequence in Taok2tm1fl/fl;Nes-cre/+ mice compared to control animals (t(4) = 22.40, P < 0.001, Fig. 1d). These data indicate that the Nes-cre transgene excises exons 2–7 with high efficiency in brains of Taok2tm1fl/fl;Nes-cre/+ mice.
In addition to disrupting most of the kinase domain, deletion of exons 2–7 is also expected to remove the translational start site of the Taok2 transcript. To determine if the Nes-cre transgene effectively knocks down TAOK2 protein in brains of Taok2tm1fl/fl;Nes-cre/+ mice, we performed Western blot analysis using a polyclonal antibody raised against a peptide mapping to an internal region of the human TAOK2 protein and cross-reacting to mouse TAOK2. This analysis showed nearly complete loss of TAOK2 protein in brain tissue isolated from Taok2tm1fl/fl;Nes-cre/+ mice (t(2) = −15.38, P < 0.01) and absence of an aberrantly translated product (Fig. 1e,f). These data suggest that Taok2tm1fl/fl;Nes-cre/+ mice are effectively null for the TAOK2 protein in brain tissue.
Behavioral characterization
A loss-of-function mutation in the Drosophila homolog of Taok2, dtao, confers resistance to the locomotor activating effects of ethanol (King et al. 2011). To determine if loss of Taok2 in the mouse alters ethanol-related behaviors, we tested Taok2-deficient and control mice for ethanol-induced ataxia, acute functional tolerance, sedation, ethanol consumption, conditioned place preference and ethanol-induced hyperactivity.
To assess sensitivity to the ataxic affects of ethanol and acute functional tolerance, we tested Taok2−/− and control mice in the stationary dowel assay. Taok2−/− mice recovered their balance more quickly from both the first (t(19) = −2.49, P < 0.05, Fig. 2a) and second (t(19) = −2.16, P < 0.05, Fig. 2c) injections of ethanol in this assay, suggesting that they may be acutely resistant to the ataxic effects of ethanol. To further characterize the acute response of Taok2−/− mice to ethanol in this assay, we measured blood ethanol content (BEC) at the time of recovery from both injections and observed that Taok2−/− mice exhibited significantly reduced BEC upon recovery from the first ethanol injection (t(19) = −3.06, P < 0.05, Fig. 2b), whereas BEC values in null mice were comparable to control animals following recovery from the second ethanol injection (t(19) = −0.83, P = 0.42, Fig. 2d). These data indicate a trend toward increased acute functional tolerance to the ataxic effects of ethanol in Taok2−/− mice (t(19) = 1.94, P = 0.07, Fig. 2e).
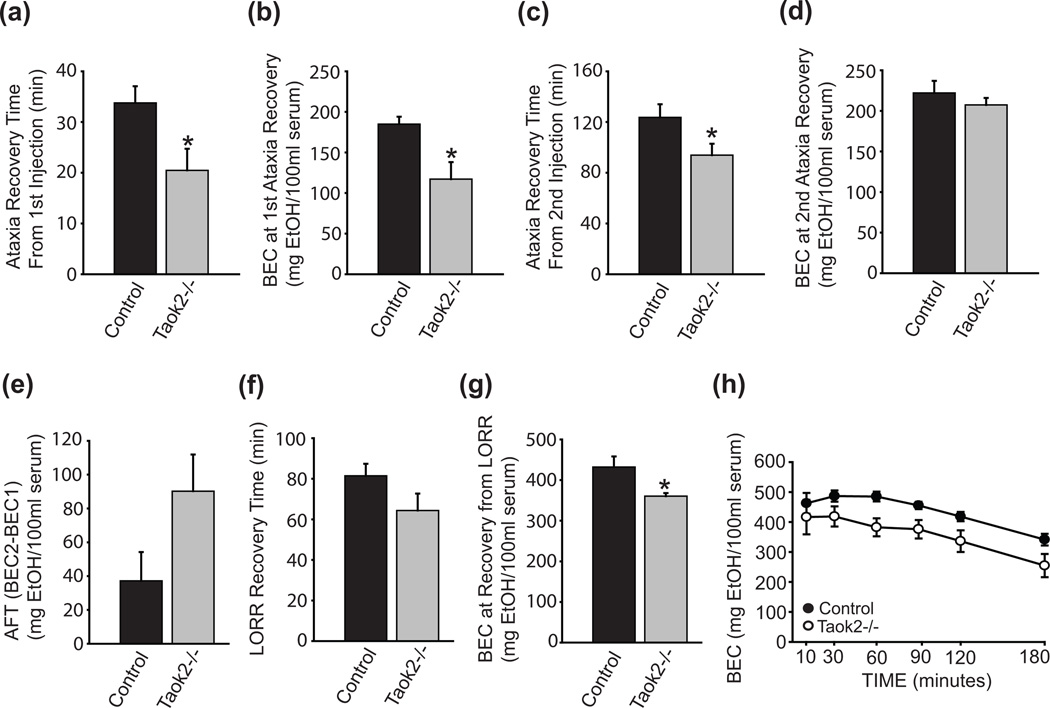
Figure 2. Constitutive loss of Taok2 in mice confers resistance to the acute ataxic and sedative effects of ethanol.
(a) Taok2−/− mice (n = 10) recover more quickly from the ataxic effects of an initial 1.5g/kg dose ethanol than control mice (n = 11). (b) BEC1 is reduced in Taok2−/− mice upon recovery from the ataxic effects of ethanol. (c) Taok2−/− mice recover more quickly from the ataxic effects of a second 1.5g/kg injection of ethanol (d) BEC2 is comparable among control and Taok2−/− mice upon recovery from a second injection of ethanol. (e) Taok2−/− mice show a trend toward increased ethanol-induced AFT. (f) Taok2−/− mice show a trend toward resistance to the sedative effects of ethanol in the LORR assay. (g) BEC levels are significantly decreased upon recovery of the ethanol-induced loss of righting reflex in Taok2−/− mice compared to control subjects. (h) Altered metabolism of a 4g/kg dose of ethanol is observed in Taok2−/− mice (n = 7) compared to control animals (n = 8). Error bars are mean ± SEM. Asterisks indicate level of significance (* P < 0.05).
We next assessed Taok2−/− mice for sensitivity to the sedating effects of ethanol in the loss-of righting-reflex (LORR) assay. After administration of a 4g/kg injection of ethanol intraperitoneally, we measured the time it took for subjects to recover the righting reflex. Compared with controls, Taok2−/− mice exhibited a trend toward shorter recovery time from ethanol-induced sedation (t(19) = −1.69, P = 0.11, Fig. 2f). At the time of recovery, Taok2−/− mice showed reduced BEC (t(19) = −2.54, P < 0.05, Fig. 2g).
The observation that Taok2−/− mice recover more quickly with reduced BEC levels from both ataxia and sedation-inducing doses of ethanol suggested that Taok2−/− mice may absorb and/or metabolize ethanol more quickly than controls. To address this possibility, we measured BEC levels in Taok2−/− and control mice following a 4g/kg intraperitoneal dose ethanol across multiple time points and observed a significant main effect of strain, with Taok2−/− mice exhibiting decreased BEC values (F(1, 13) = 4.56, P < 0.05, Fig. 2h).
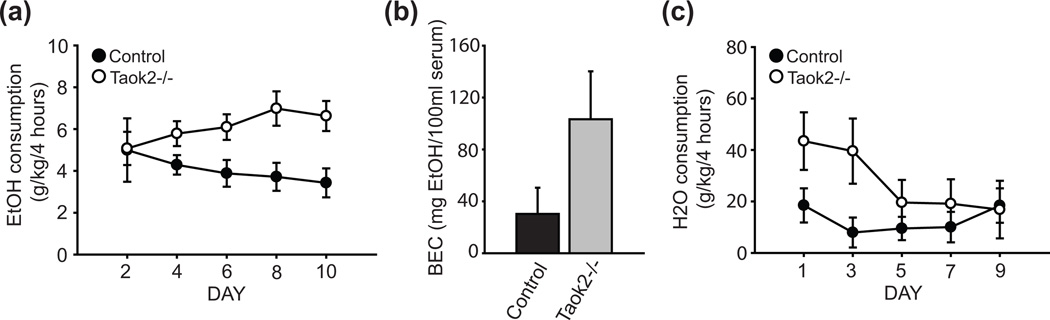
Studies in humans and several mouse genetic models have established a negative correlation between sensitivity to the acute effects of ethanol and ethanol consumption or propensity for abuse (Boyce-Rustay et al. 2006; Crabbe et al. 1996; Fee et al. 2004; Kapfhamer et al. 2008; Naassila et al. 2002, 2004; Newton and Messing 2007; Palmer et al. 2004; Schuckit 1994). To assess ethanol consumption in Taok2−/− and control mice, we tested subjects in a limited-access, “drinking-in-the dark” (DID) assay. This assay involves acclimating subjects to a reverse light-dark schedule and providing access to ethanol for several hours during the middle of the dark period when mice are most active. In contrast to standard, continuous access models, it has been shown that mice in the DID assay typically consume pharmacologically relevant (intoxicating) levels of ethanol, resulting in the blood alcohol levels in excess of 1.0mg/ml (Rhodes et al. 2005). We observed a significant strain effect for ethanol consumption in the DID assay, with Taok2−/− mice consuming increased amounts of ethanol compared to control animals (main effect of genotype: F(1, 14) = 5.96, P < 0.05, genotype X time interaction: F(4, 56) = 2.44, P = 0.06, Fig. 3a). Following the final ethanol session, we measured BEC and found a trend toward increased BEC in Taok2−/− mice (t(14) = 1.73, P = 0.11, Fig. 3b). We measured taste preference for a 0.06% w/v saccharin solution and a 0.03 mM quinine solution and did not observe a significant main effect of genotype for either tastant (t(16) = 0.82, P = 0.43, t(16) = 1.49, P = 0.16, for saccharin and quinine, respectively), indicating the increased ethanol consumption by Taok2−/− mice is likely not due to altered gustation. We also measured water consumption on alternate days during the course of the study and found a trend toward increased consumption (main effect of genotype: F(1, 14) = 2.56, P = 0.13, genotype X time interaction: F(4, 56) = 2.16, P = 0.09, Fig. 3c), however water consumption toward the end of the study (days 5, 7 and 9) was comparable between strains, in contrast to ethanol consumption during this time period (days 6, 8 and 10).
Figure 3. Constitutive loss of Taok2 increases ethanol consumption.
(a) Taok2−/− mice (n = 8) show increased ethanol consumption in the DID assay relative to controls (n = 8). (b) BEC values in Taok2−/− and control mice on the last day of ethanol access. (c) Water consumption on alternate days during the DID assay. Error bars are mean ± SEM.
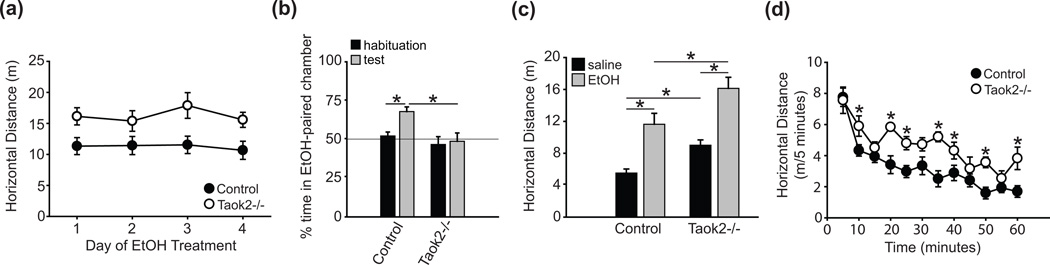
Based on our data showing that Taok2−/− mice consume more ethanol than control mice, we investigated whether disruption of the Taok2 gene results in enhanced ethanol-induced conditioned place preference (CPP), a commonly used assay of the rewarding or motivational effects of drugs of abuse (Chester et al. 1998; Liu et al. 2008). On day 1 of the study (habituation to the chamber), both genotypes exhibited a slight preference for the right side of the apparatus (control: 16.6 ± 0.6 minutes versus Taok2−/−: 15.9 ± 1.0 minutes), however the genotypes did not differ significantly from one another (t(22) = −0.59, P = 0.56). We trained mice using a 2g/kg dose ethanol that did not produce ethanol-induced locomotor sensitization in either genotype in our study (effect of day: F(3, 22) = 1.28, P = 0.29, Fig. 4a) and observed that expression of ethanol-CPP was completely blocked in Taok2−/−?mice (main effect of genotype: F(1, 22) = 8.50, P < 0.01; main effect of day: F(1, 22) = 12.75, P < 0.05; genotype X day interaction: F(1, 22) = 6.91, P < 0.05, Fig. 4b).
Figure 4. Constitutive loss of Taok2 in mice impairs ethanol-induced conditioned place preference without affecting ethanol-induced locomotor hyperactivity.
(a) 2g/kg ethanol dose used in CPP training does not result in locomotor sensitization (b) Ethanol-induced CPP is blocked in Taok2−/− mice (n = 8); controls: (n = 16). (c) Control and Taok2−/− mice show similar ethanol-induced locomotor stimulation in response to 2g/kg ethanol. Note the increased baseline activity in Taok2−/− mice. (d) Taok2−/− mice (n = 10) show increased activity and impaired locomotor habituation to an open field apparatus compared to controls (n = 12). Error bars are mean ± SEM. Asterisks indicate level of significance (* P < 0.05).
In addition to place preference, the CPP assay measures general locomotor activity and ethanol-induced hyperactivity. Both wild-type and Taok2−/−?mice increased activity in response to ethanol compared to saline treatment (F(1, 22) = 45.9, P < 0.001; Fig. 4c). However, Taok2−/−?mice were generally more active than controls (F(1, 22) = 9.54, P < 0.005), such that we failed to detect a significant genotype X treatment interaction (F(1, 22) = 0.41, P = 0.53; Fig. 4c). These data indicate that Taok2−/−?mice exhibit normal ethanol-induced hyperactivity when normalized for an increased baseline activity level, in contrast to dtao mutant flies. We further characterized the locomotor hyperactivity phenotype of Taok2−/− and control mice by measuring locomotor activity in an open field apparatus and observed a significant main effect of genotype (F(1, 20) = 13.2, P < 0.05), time (F(11, 20) = 23.1, P < 0.001), and genotype X time interaction: (F(11, 220) = 2.00, P < 0.05), indicating that Taok2−/− mice do not habituate normally to the open field (Fig. 4d).
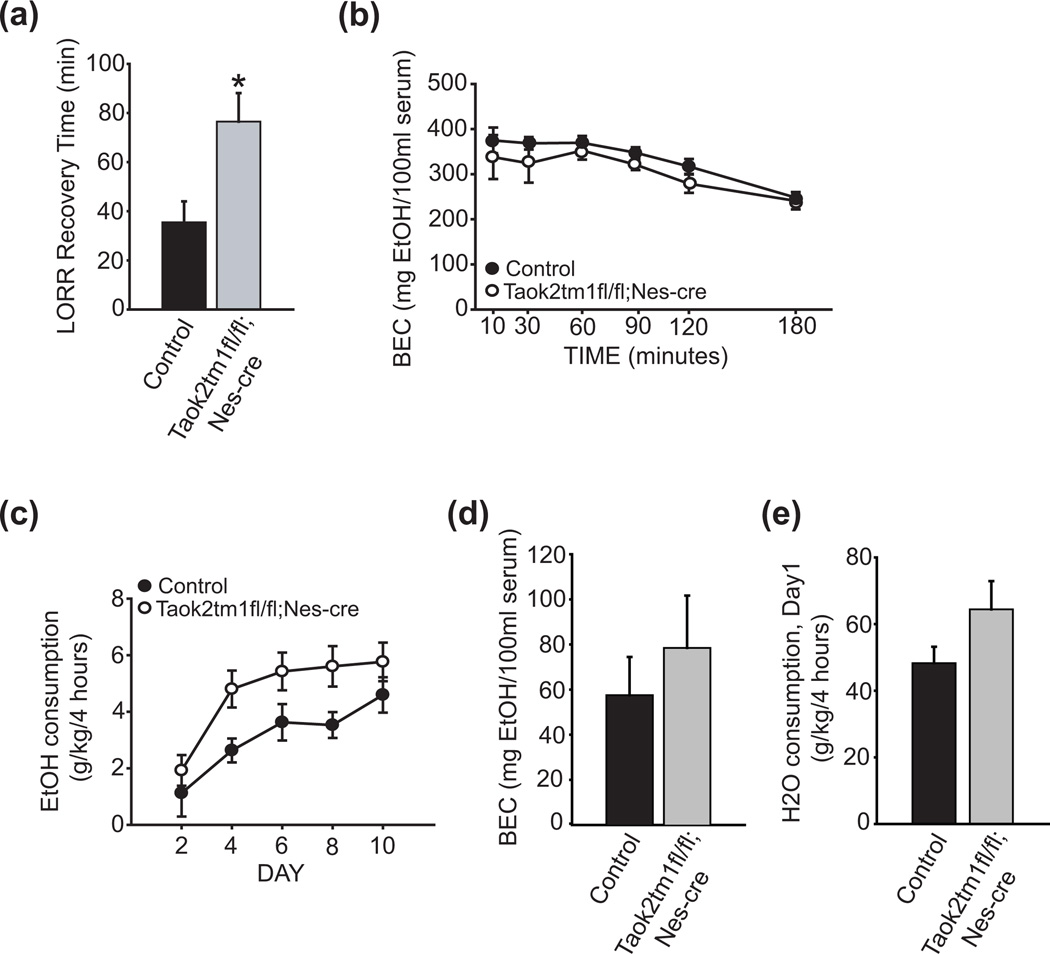
Based on our observations that Taok2−/− mice exhibit multiple ethanol-dependent behavioral phenotypes that may result, wholly or in part, from increased ethanol metabolism, we investigated whether nervous system-specific disruption of the Taok2 gene would result in similar ethanol-dependent phenotypes. To assess sensitivity to the sedating effects of ethanol, we tested Taok2tm1fl/fl;Nes-cre/+ and control mice in the loss-of-righting assay. Compared with control mice, Taok2tm1fl/fl;Nes-cre/+ mice exhibited a significantly longer recovery time from ethanol (t(12) = 4.10, P < 0.001; Fig. 5a), indicating that nervous system-specific loss of TAOK2 in mice confers increased sensitivity to the sedative effects of ethanol. The increased sensitivity to ethanol observed in Taok2tm1fl/fl;Nes-cre/+ mice does not appear to result from altered ethanol absorption and/or clearance, as a separate cohort of subjects did not show a significant strain effect for blood ethanol content across a series of time points following treatment with 4g/kg ethanol (2-way repeated measures ANOVA, effect of genotype: F(1, 9) = 4.33, P > 0.05; genotype X time interaction: F(5, 45) = 0.18, P > 0.05; Fig. 5b).
Figure 5. Nervous system-specific loss of Taok2 in mice alters ethanol-induced sedation and consumption.
(a) Taok2tm1fl/fl;Nes-cre mice (n = 8) show increased recovery time relative to control animals (n = 6) to 4g/kg ethanol in the loss of right reflex (LORR) assay. (b) Normal metabolism of a 4g/kg dose of ethanol is observed in Taok2tm1fl/fl;Nes-cre mice (n = 5) compared to controls (n = 6). (c) Taok2tm1fl/fl;Nes-cre mice (n = 12) consume more ethanol than controls (n = 9) in a limited-access, drinking in the dark (DID) assay. (d) BEC levels are comparable between Taok2tm1fl/fl;Nes-cre and control mice on the final day of ethanol access (e) H20 consumption is normal in Taok2tm1fl/fl;Nes-cre mice. BEC = blood ethanol content. Error bars are mean ± SEM. Asterisks indicate level of significance (* P < 0.05).
To assess ethanol consumption in Taok2tm1fl/fl;Nes-cre/+ and control mice, we tested subjects in the DID assay. We observed a significant strain effect for ethanol consumption in the DID assay, with Taok2tm1fl/fl;Nes-cre/+ mice consuming more ethanol than control mice (effect of genotype: F(1, 20) = 4.65, P < 0.05, Fig. 5c). BEC levels between strains at the end of the study were comparable (t(20) = 0.49, P = 0.63, Fig. 5d), as was expected since both mutants and control animals consumed similar amounts of ethanol on the last day of the study (Fig. 5c). The increased ethanol consumption of Taok2tm1fl/fl;Nes-cre/+ mice was not a result of an overall increase in fluid consumption as control and mutant mice consumed comparable amounts of water during the habituation period (t(12) = 1.77, P = 0.09, Fig. 5e).
Discussion
We have generated a conditionally-disrupted allele of the Taok2 gene in mice using the Cre/loxP system and demonstrate that loss of Taok2 affects multiple behaviors following exposure to ethanol. Specifically, disruption of Taok2 constitutively results in earlier recovery from the ataxic and sedative effects of ethanol, increased ethanol consumption, and reduced ethanol-induced conditioned place preference. By comparison, nervous system-specific disruption of Taok2 increases recovery time from the sedative effects of ethanol and increases ethanol consumption, indicating that global versus nervous system-specific disruption of Taok2 results in specific and overlapping ethanol-dependent phenotypes.
The earlier recovery time from ethanol-induced ataxia and sedation we observe in Taok2−/− mice occurs at lower blood ethanol contents, suggesting that these phenotypes may result from altered ethanol absorption and/or clearance. In fact, we observe altered pharmacokinetics of a 4g/kg dose ethanol in Taok2−/− mice, which may explain the ethanol sensitivity phenotypes in the stationary dowel and loss-of-right-reflex tests. Although TAOK2 is most abundantly expressed in the brain, it is present in other tissues, including liver (http://biogps.org/#goto=genereport&id=9344, and data not shown), the primary site of ethanol metabolism by alcohol dehydrogenase (Lands, 1997). Acute ethanol exposure activates JNK (c-JUN NH2-terminal kinase) and mitogen-activated protein kinase (MAPK) p38 in hepatocytes (Aroor and Shukla 2004; Aroor et al. 2010; Chen et al. 1998), two signaling pathways also engaged by TAOK2 activation (Calderon de Anda et al. 2012; Chen and Cobb 2001; Yasuda et al. 2007). If and how TAOK2-dependent MAPK signaling may affect ethanol metabolism merits further investigation.
Similarly, an increased rate of ethanol metabolism may underlie the increased ethanol consumption and reduced ethanol-induced CPP phenotypes of Taok2−/− mice. We also observed an increase in water consumption of Taok2−/− mice relative to controls during the beginning of the drinking in the dark study. Interestingly, water consumption among mutant mice normalized to control levels later in the study, when ethanol consumption increased. Since water and ethanol consumption were measured on alternate days, a straightforward interaction between both is not clear. It will be important in future studies to investigate water and ethanol consumption concomitantly using a 2-bottle choice paradigm.
Global disruption of Taok2 impairs ethanol-induced conditioned place preference, but fails to affect locomotor hyperactivity, in contrast to disruption of the Drosophila homolog, dtao (King et al. 2011). The ethanol-induced hyperactivity defect of dtao mutants has been shown to be mediated through regulation of the downstream target Par-1, a kinase that has been implicated in the activation of TAU protein and microtubule destabilization. In mammals, PAR1 activity is dependent on TAOK1 (Timm et al. 2003), however the regulation of PAR1 by TAOK2 or TAOK3 has not been investigated. The possible functional redundancy among mammalian TAOK proteins in regard to PAR1 regulation may explain why we fail to observe altered ethanol-induced hyperactivity defects in Taok2 mutant mice.
In addition to the ethanol ataxia, sedation and consumption phenotypes of Taok2-disrupted mice, we observe that Taok2−/− mice exhibit reduced ethanol-induced CPP. It should be noted that several confounding factor may have affected the behavior of Taok2−/− mice in this assay: as previously mentioned, altered ethanol pharmacokinetics may have affected the formation of ethanol-induced CPP in this line. In addition, Taok2−/− mice are generally hyperactive and show impaired habituation to an open field. Hyperactivity has been reported to disrupt expression of ethanol-induced CPP (Gremel and Cunningham, 2007) and we observed a significant negative correlation between CPP score and activity during the test session (r = −0.479, P < 0.05).
The CPP assay measures the rewarding aspects of a drug of abuse through an associative learning process in which the subject learns to associate the subjective effects of a drug with a specific environment (Schechter and Calcagnetti, 1993). Functionally, TAOK2 is involved in the trafficking of cell adhesion molecules NCAM and Arcadlin (Yasuda et al. 2007), which may alter neuroplasticity and, in the case of NCAM disruption, cause spatial learning deficits (Becker et al. 1996; Bukalo et al. 2004; Cremer et al. 1994; Moy et al. 2009; Tang et al. 1998; Venero et al. 2006; Yamagata et al. 1999). Furthermore, ethanol may directly interact with cell adhesion molecules to affect their function (Aravelo et al. 2008; Dou et al. 2011). Taken together, these observations suggest a model whereby altered cell adhesion molecule trafficking and/or function in Taok2 mutant mice may impair ethanol-induced CPP by disrupting normal associative learning processes. Such a model might assume that under normal conditions, the interaction of ethanol with cell adhesion molecules somehow facilitates the associative learning that occurs during CPP training and this process is impaired when TAOK2 is absent, perhaps through its role in cell adhesion molecule trafficking. Additional experiments will be important to determine if Taok2 mutants are deficient in other spatial learning tasks and to what degree trafficking of cell adhesion molecules may be involved. It is interesting to note that mice in which the cell adhesion molecule CD81 has been disrupted are impaired in cocaine-induced CPP (Michna et al. 2001), underscoring the importance of cell adhesion in this behavior.
Perhaps our most interesting finding is that although ethanol metabolism is altered in Taok2−/− mice, ethanol metabolism appears normal in mice in which the Taok2 gene has been disrupted specifically in the nervous system, yet Tao2fl/fl;Nes/cre mice consume increased amounts of ethanol and in contrast to Taok2−/− mice, are sensitive to ethanol in the LORR assay. These data suggest that the nervous system-specific effect of TAOK2 on ethanol sensitivity is masked by peripheral disruption of Taok2 in Taok2−/− mice and that the increased consumption of ethanol in Taok2−/− mice may be independent of the gene’s effect on ethanol pharmacokinetics. To further characterize the function of TAOK2 in the nervous system in regulating behavioral sensitivity to ethanol, it will be important to measure BEC levels in Tao2fl/fl;Nes/cre mice upon LORR recovery as well as assessment of acute functional tolerance to ethanol.
Previous studies in humans and rodent models have suggested a negative correlation between initial sensitivity to ethanol and ethanol consumption (Boyce-Rustay et al., 2006; Crabbe et al. 1996; Fee et al. 2004; Kapfhamer et al. 2008; Naassila et al. 2002, 2004; Newton and Messing 2007; Palmer et al. 2004; Schuckit 1994). Based on our observation that nervous system-specific disruption of Taok2 confers greater sensitivity to ethanol in the LORR assay, we might predict that Taok2 mutants would consume less ethanol. Instead, we observe that Taok2 mutants actually consume more ethanol than control mice. It may be noted that the LORR assay measures sensitivity to the sedative/hypnotic effects of ethanol, which may be mechanistically distinct from the ataxic effects of lower doses of ethanol and a more accurate predictor of ethanol consumption (Shuckit, 1994; Thiele et al. 2002). It will therefore be interesting to examine the ataxic effects of ethanol in Taok2fl/fl;Nes/cre mice.
Cell culture studies have also established a role for TAOK2 in the regulation of the actin cytoskeleton (Moore et al. 2000), a function that may also be critical for neuroplasticity (Huntley et al. 2002; Lisman et al. 2003; Matus 2000). A growing body of literature suggests that ethanol exposure may alter actin organization both pre- and post-synaptically (Funk et al. 2007; Mulholland and Chandler, 2007; Offenhäuser et al. 2006). Mice lacking the Eps8 gene, which encodes an EGFR substrate and regulator of actin dynamics, are resistant to the acute intoxicating effects of ethanol and show increased ethanol consumption (Offenhäuser et al. 2006), supporting the notion that not only are actin dynamics ethanol-sensitive, but alterations in the cytoskeleton may affect behavioral responses to ethanol. Future experiments should address if altered actin cytoskeleton dynamics underlie the abnormal ethanol responsiveness of Taok2 mutant mice.
Our data provide, to our knowledge, the first evidence that a Ste20p family kinase functions to regulate ethanol-dependent behaviors in mammals. Specifically, TAOK2 regulates ethanol-induced ataxia, sedation, consumption and conditioned place preference. Previously, we have shown that the fly homolog, dtao, is required for normal ethanol-induced hyperactivity (King et al. 2011) and mutations in a related Ste20p family kinase gene, happyhour, confers resistance to ethanol-induced sedation in Drosophila (Corl et al. 2009). These data support the conservation of intracellular signaling through Ste20p family kinases in the regulation of behavioral responses to ethanol in flies and mammals and identify novel candidate genes for ethanol use disorders in humans.
Acknowledgments
The authors thank Dr. Y. Wada at Osaka University, Osaka, Japan for plasmids used to generate the Taok2 targeting construct, and Drs. R. Messing and D. Ron for helpful discussions. This work was supported by the National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health and by the State of California for medical research on alcohol and substance abuse through UCSF.
References
- Aoyama M, Agari K, Sun-Wada GH, Futai M, Wada Y. Simple and straightforward construction of a mouse gene targeting vector using in vitro transposition reactions. Nucleic Acids Res. 2005;33:e52. doi: 10.1093/nar/gni055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo E, Shanmugasundararaj S, Wilkemeyer MF, Dou X, Chen S, Charness ME, Miller KW. An alcohol binding site on the neural cell adhesion molecule L1. Proc Natl Acad Sci U S A. 2008;105:371–375. doi: 10.1073/pnas.0707815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arror AR, Shukla SD. MAP kinase signaling in diverse effects of ethanol. Life Sci. 2004;74:2339–2364. doi: 10.1016/j.lfs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Aroor AR, James TT, Jackson DE, Shukla SD. Differential Changes in MAP Kinases, Histone Modifications, and Liver Injury in Rats Acutely Treated With Ethanol. Alcohol Clin Exp Res. 2010;34:1543–1551. doi: 10.1111/j.1530-0277.2010.01239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker CG, Artola A, Gerardy-Schahn R, Becker T, Welzl H, Schachner M. The polysialic acid modification of the neural cell adhesion molecule is involved in spatial learning and hippocampal long-term potentiation. J Neurosci Res. 1996;45:143–152. doi: 10.1002/(SICI)1097-4547(19960715)45:2<143::AID-JNR6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Boyce-Rustay JM, Wiedholz LM, Millstein RA, Carroll J, Murphy DL, Daws LC, Holmes A. Ethanol-related behaviors in serotonin transporter knockout mice. Alcohol Clin Exp Res. 2006;30:1957–1965. doi: 10.1111/j.1530-0277.2006.00241.x. [DOI] [PubMed] [Google Scholar]
- Bradley A. Production and analysis of chimeric mice. In: Robertson EJ, editor. Teratocarcinoma and Embryonic Stem Cells: A Practical Approach. Oxford, England: IRL Press; 1987. [Google Scholar]
- Buck KJ, Harris RA. Neuroadaptive responses to chronic ethanol. Alcohol Clin Exp Res. 1991;15:460–470. doi: 10.1111/j.1530-0277.1991.tb00544.x. [DOI] [PubMed] [Google Scholar]
- Bukalo O, Fentrop N, Lee AY, Salmen B, Law JW, Wotjak CT, Schweizer M, Dityatev A, Schachner M. Conditional ablation of the neural cell adhesion molecule reduces precision of spatial learning, long-term potentiation, and depression in the CA1 subfield of mouse hippocampus. J Neurosci. 2004;24:1565–1577. doi: 10.1523/JNEUROSCI.3298-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon de Anda F, Rosario AL, Durak O, Tran T, Graff J, Meletis K, Rei D, Soda T, Madabhushi R, Ginty DD, Kolodkin AL, Tsai LH. Autism spectrum disorder susceptibility gene TAOK2 affects basal dendrite formation in the neocortex. Nat Neurosci. 2012;15:1022–1031. doi: 10.1038/nn.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler LJ. Ethanol and brain plasticity: receptors and molecular networks of the postsynaptic density as targets of ethanol. Pharmacol Ther. 2003;99:311–326. doi: 10.1016/s0163-7258(03)00096-2. [DOI] [PubMed] [Google Scholar]
- Chen J, Ishac EJ, Dent P, Kunos G, Gao B. Effects of ethanol on mitogen-activated protein kinase cascades in normal and regenerating liver. Biochem J. 1998;334:669–676. doi: 10.1042/bj3340669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Cobb MH. Regulation of stress-responsive mitogen-activated protein (MAP) kinase pathways by TAO2. J Biol Chem. 2001;276:16070–16075. doi: 10.1074/jbc.M100681200. [DOI] [PubMed] [Google Scholar]
- Chester JA, Risinger FO, Cunningham CL. Ethanol reward and aversion in mice bred for sensitivity to ethanol withdrawal. Alcohol Clin Exp Res. 1998;22:468–473. [PubMed] [Google Scholar]
- Corl AB, Berger KH, Ophir-Shohat G, Gesch J, Simms JA, Bartlett SE, Heberlein U. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137:949–960. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Corominas M, Roncero C, Ribases M, Castells X, Casas M. Brain-derived neurotrophic factor and its intracellular signaling pathways in cocaine addiction. Neuropsychobiology. 2007;55:2–13. doi: 10.1159/000103570. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Feller DJ, Hen R, Wenger CD, Lessov CN, Schafer GL. Elevated alcohol consumption in null mutant mice lacking 5-HT1B serotonin receptors. Nat Genet. 1996;14:98–101. doi: 10.1038/ng0996-98. [DOI] [PubMed] [Google Scholar]
- Cremer H, Lange R, Christoph A, Plomann M, Vopper G, Roes J, Brown R, Baldwin S, Kraemer P, Scheff S, Barthels D, Rajewsky K, Wille W. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature. 1994;367:455–459. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- Daniels GM, Buck KJ. Expression profiling identifies strain-specific changes associated with ethanol withdrawal in mice. Genes Brain Behav. 2002;1:35–45. doi: 10.1046/j.1601-1848.2001.00008.x. [DOI] [PubMed] [Google Scholar]
- Davis RJ. MAPKs: new JNK expands the group. Trends Biochem Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Dou X, Menkari CE, Shanmugasundararaj S, Miller KW, Charness ME. Two alcohol binding residues interact across a domain interface of the L1 neural cell adhesion molecule and regulate cell adhesion. J Biol Chem. 2011;286:16131–16139. doi: 10.1074/jbc.M110.209254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fee JR, Sparta DR, Knapp DJ, Breese GR, Picker MJ, Thiele TE. Predictors of high ethanol consumption in RIIbeta knock-out mice: assessment of anxiety and ethanol-induced sedation. Alcohol Clin Exp Res. 2004;28:1459–1468. doi: 10.1097/01.ALC.0000141809.53115.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman WM, Lull ME, Patel KM, Brucklacher RM, Morgan D, Roberts DC, Vrana KE. Gene expression changes in the medial prefrontal cortex and nucleus accumbens following abstinence from cocaine self-administration. BMC Neurosci. 2010;11:29. doi: 10.1186/1471-2202-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk CK, Dohrman DP. Chronic ethanol exposure inhibits dopamine release via effects on the presynaptic actin cytoskeleton in PC12 cells. Brain Res. 2007;1185:86–94. doi: 10.1016/j.brainres.2007.09.069. [DOI] [PubMed] [Google Scholar]
- Gremel CM, Cunningham CL. Role of test activity in ethanol-induced disruption of place preference expression in mice. Psychopharm. 2007;191:195–202. doi: 10.1007/s00213-006-0651-5. [DOI] [PubMed] [Google Scholar]
- Harris RA, Hitzemann RJ. Membrane fluidity and alcohol actions. Curr Alcohol. 1981;8:379–404. [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2007;64:830–842. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- Hassan S, Duong B, Kim KS, Miles MF. Pharmacogenomic analysis of mechanisms mediating ethanol regulation of dopamine beta-hydroxylase. J Biol Chem. 2003;278:38860–38869. doi: 10.1074/jbc.M305040200. [DOI] [PubMed] [Google Scholar]
- Hebert MA, O'Callaghan JP. Protein phosphorylation cascades associated with methamphetamine-induced glial activation. Ann N Y Acad Sci. 2000;914:238–262. doi: 10.1111/j.1749-6632.2000.tb05200.x. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo, A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Huntley GW, Benson DL, Colman DR. Structural remodeling of the synapse in response to physiological activity. Cell. 2002;108:1–4. doi: 10.1016/s0092-8674(01)00631-6. [DOI] [PubMed] [Google Scholar]
- Hutchison M, Berman KS, Cobb MH. Isolation of TAO1, a protein kinase that activates MEKs in stress-activated protein kinase cascades. J Biol Chem. 1998;273:28625–28632. doi: 10.1074/jbc.273.44.28625. [DOI] [PubMed] [Google Scholar]
- Johne C, Matenia D, Li XY, Timm T, Balusamy K, Mandelkow EM. Spred1 and TESK1--two new interaction partners of the kinase MARKK/TAO1 that link the microtubule and actin cytoskeleton. Mol Biol Cell. 2008;19:1391–1403. doi: 10.1091/mbc.E07-07-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung ME, Ju X, Simpkins JW, Metzger DB, Yan LJ, Wen Y. Ethanol withdrawal acts as an age-specific stressor to activate cerebellar P38 kinase. Neurobiol Aging. 2010;32:2266–2278. doi: 10.1016/j.neurobiolaging.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapfhamer D, Bettinger JC, Davies AG, Eastman CL, Smail EA, Heberlein U, McIntire SL. Loss of RAB-3/A in Caenorhabditis elegans and the mouse affects behavioral response to ethanol. Genes Brain Behav. 2008;7:669–676. doi: 10.1111/j.1601-183X.2008.00404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I, Tsai LT, Pflanz R, Voigt A, Lee S, Jackle H, Lu B, Heberlein U. Drosophila tao controls mushroom body development and ethanol-stimulated behavior through par-1. J Neurosci. 2011;31:1139–1148. doi: 10.1523/JNEUROSCI.4416-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konu O, Kane JK, Barrett T, Vawter MP, Chang R, Ma JZ, Donovan DM, Sharp B, Becker KG, Li MD. Region-specific transcriptional response to chronic nicotine in rat brain. Brain Res. 2001;909:194–203. doi: 10.1016/s0006-8993(01)02685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna M, Narang H. The complexity of mitogen-activated protein kinases (MAPKs) made simple. Cell Mol Life Sci. 2008;65:3525–3544. doi: 10.1007/s00018-008-8170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku BM, Lee YK, Jeong JY, Mun J, Han JY, Roh GS, Kim HJ, Cho GJ, Choi WS, Yi GS, Kang SS. Ethanol-induced oxidative stress is mediated by p38 MAPK pathway in mouse hippocampal cells. Neurosci Lett. 2007;419:64–67. doi: 10.1016/j.neulet.2007.03.049. [DOI] [PubMed] [Google Scholar]
- Lands WEM. A review of alcohol clearance in humans. Alcohol. 1998;15:147–160. doi: 10.1016/s0741-8329(97)00110-9. [DOI] [PubMed] [Google Scholar]
- Lisman J. Actin's actions in LTP-induced synapse growth. Neuron. 2003;38(3):361–362. doi: 10.1016/s0896-6273(03)00257-5. [DOI] [PubMed] [Google Scholar]
- Liu Y, Le Foll B, Liu Y, Wang X, Lu L. Conditioned place preference induced by licit drugs: establishment, extinction, and reinstatement. Scientific World Journal. 2008;8:1228–1245. doi: 10.1100/tsw.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290:754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- Michna L, Brenz Verca MS, Widmer DA, Chen S, Lee J, Rogove J, Zhou R, Tsitsikov E, Miescher GC, Dreyer JL, Wagner GC. Altered sensitivity of CD81-deficient mice to neurobehavioral effects of cocaine. Brain Res Mol Brain Res. 2001;90:68–74. doi: 10.1016/s0169-328x(01)00092-4. [DOI] [PubMed] [Google Scholar]
- Mitsopoulos C, Zihni C, Garg R, Ridley AJ, Morris JD. The prostate-derived sterile 20-like kinase (PSK) regulates microtubule organization and stability. J Biol Chem. 2003;278:18085–18091. doi: 10.1074/jbc.M213064200. [DOI] [PubMed] [Google Scholar]
- Moonat S, Starkman BG, Sakharkar A, Pandey SC. Neuroscience of alcoholism: molecular and cellular mechanisms. Cell Mol Life Sci. 2010;67:73–88. doi: 10.1007/s00018-009-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TM, Garg R, Johnson C, Coptcoat MJ, Ridley AJ, Morris JD. PSK, a novel STE20-like kinase derived from prostatic carcinoma that activates the c-Jun N-terminal kinase mitogen-activated protein kinase pathway and regulates actin cytoskeletal organization. J Biol Chem. 2000;275:4311–4322. doi: 10.1074/jbc.275.6.4311. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nonneman RJ, Young NB, Demyanenko GP, Maness PF. Impaired sociability and cognitive function in Nrcam-null mice. Behav Brain Res. 2009;205:123–131. doi: 10.1016/j.bbr.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland PJ, Chandler LJ. The thorny side of addiction: adaptive plasticity and dendritic spines. Scientific World Journal. 2007;7:9–21. doi: 10.1100/tsw.2007.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naassila M, Ledent C, Daoust M. Low ethanol sensitivity and increased ethanol consumption in mice lacking adenosine A2A receptors. J Neurosci. 2002;22:10487–10493. doi: 10.1523/JNEUROSCI.22-23-10487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naassila M, Pierrefiche O, Ledent C, Daoust M. Decreased alcohol self-administration and increased alcohol sensitivity and withdrawal in CB1 receptor knockout mice. Neuropharmacology. 2004;46:243–253. doi: 10.1016/j.neuropharm.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Newton PM, Messing RO. Intracellular signaling pathways that regulate behavioral responses to ethanol. Pharmacol Ther. 2006;109:227–237. doi: 10.1016/j.pharmthera.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Newton PM, Messing RO. Increased sensitivity to the aversive effects of ethanol in PKCepsilon null mice revealed by place conditioning. Behav Neurosci. 2007;121:439–442. doi: 10.1037/0735-7044.121.2.439. [DOI] [PubMed] [Google Scholar]
- Offenhäuser N, Castelletti D, Mapelli L, Soppo BE, Regondi MC, Rossi P, D'Angelo E, Frassoni C, Amadeo A, Tocchetti A, Pozzi B, Disanza A, Guarnieri D, Betsholtz C, Scita G, Heberlein U, Di Fiore PP. Increased ethanol resistance and consumption in Eps8 knockout mice correlates with altered actin dynamics. Cell. 2006;127:213–226. doi: 10.1016/j.cell.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Palmer AA, Sharpe AL, Burkhart-Kasch S, McKinnon CS, Coste SC, Stenzel-Poore MP, Phillips TJ. Corticotropin-releasing factor overexpression decreases ethanol drinking and increases sensitivity to the sedative effects of ethanol. Psychopharmacology (Berl) 2004;176:386–397. doi: 10.1007/s00213-004-1896-5. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Best K, Belknap JK, Finn DA, Crabbe JC. Evaluation of a simple model of ethanol drinking to intoxication in C57BL/6J mice. Physiol Behav. 2005;84:53–63. doi: 10.1016/j.physbeh.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Schechter MD, Calcagnetti DJ. Trends in place preference conditioning with a cross-indexed bibliography; 1957–1991. Neurosci Biobehav. 1993;17:21–41. doi: 10.1016/s0149-7634(05)80228-3. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. Am J Psychiatry. 1994;151:184–189. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- Seger R, Krebs EG. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- Seong E, Saunders TL, Stewart CL, Burmeister M. To knockout in 129 or in C57BL/6: that is the question. Trends Genet. 2004;20:59–62. doi: 10.1016/j.tig.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Sugden PH, Clerk A. Regulation of the ERK subgroup of MAP kinase cascades through G protein-coupled receptors. Cell Signal. 1997;9:337–351. doi: 10.1016/s0898-6568(96)00191-x. [DOI] [PubMed] [Google Scholar]
- Takeda K, Ichijo H. Neuronal p38 MAPK signalling: an emerging regulator of cell fate and function in the nervous system. Genes Cells. 2002;7:1099–1111. doi: 10.1046/j.1365-2443.2002.00591.x. [DOI] [PubMed] [Google Scholar]
- Tang L, Hung CP, Schuman EM. A role for the cadherin family of cell adhesion molecules in hippocampal long-term potentiation. Neuron. 1998;20:1165–1175. doi: 10.1016/s0896-6273(00)80497-3. [DOI] [PubMed] [Google Scholar]
- Tassi E, Biesova Z, Di Fiore PP, Gutkind JS, Wong WT. Human JIK, a novel member of the STE20 kinase family that inhibits JNK and is negatively regulated by epidermal growth factor. J Biol Chem. 1999;274:33287–33295. doi: 10.1074/jbc.274.47.33287. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Koh MT, Pedrazzini T. Voluntary alcohol consumption is controlled via the neuropeptide Y Y1 receptor. J Neurosci. 2002;22:RC208. doi: 10.1523/JNEUROSCI.22-03-j0006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm T, Li XY, Biernat J, Jiao J, Mandelkow E, Vandekerckhove J, Mandelkow EM. MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. EMBO J. 2003;22:5090–5101. doi: 10.1093/emboj/cdg447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schütz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Venero C, Herrero AI, Touyarot K, Cambon K, López-Fernández MA, Berezin V, Bock E, Sandi C. Hippocampal up-regulation of NCAM expression and polysialylation plays a key role on spatial memory. Eur J Neurosci. 2006;23:1585–1595. doi: 10.1111/j.1460-9568.2006.04663.x. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Andreasson KI, Sugiura H, Maru E, Dominique M, Irie Y, Miki N, Hayashi Y, Yoshioka M, Kaneko K, Kato H, Worley PF. Arcadlin is a neural activity-regulated cadherin involved in long term potentiation. J Biol Chem. 1999;274:19473–19479. doi: 10.1074/jbc.274.27.19473. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Tanaka H, Sugiura H, Okamura K, Sakaguchi T, Tran U, Takemiya T, Mizoguchi A, Yagita Y, Sakurai T, De Robertis EM, Yamagata K. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2beta and p38 MAP kinases. Neuron. 2007;56:456–471. doi: 10.1016/j.neuron.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yustein JT, Xia L, Kahlenburg JM, Robinson D, Templeton D, Kung HJ. Comparative studies of a new subfamily of human Ste20-like kinases: homodimerization, subcellular localization, and selective activation of MKK3 and p38. Oncogene. 2003;22:6129–6141. doi: 10.1038/sj.onc.1206605. [DOI] [PubMed] [Google Scholar]
- Zihni C, Mitsopoulos C, Tavares IA, Ridley AJ, Morris JD. Prostate-derived sterile 20-like kinase 2 (PSK2) regulates apoptotic morphology via C-Jun N-terminal kinase and Rho kinase-1. J Biol Chem. 2006;281:7317–7323. doi: 10.1074/jbc.M513769200. [DOI] [PubMed] [Google Scholar]