Abstract
Aim:
To investigate the mechanisms underlying the biphasic redox regulation of hypoxia-inducible factor-1 (HIF-1) transcriptional activity under different levels of oxidative stress caused by reactive oxidative species (ROS).
Methods:
HeLa cells were exposed to different concentrations of H2O2 as a simple model for mild and severe oxidative stress. Luciferase reporter assay and/or quantitative real-time PCR were used to investigate the transcriptional activity. Immunoblot was used to detect protein expression. Chromatin immunoprecipitation assay was used to detect HIF-1/DNA binding. The interaction of p300 with HIF-1α or with SENP3, and the SUMO2/3 conjugation states of p300 were examined by coimmunoprecipitation.
Results:
HIF-1 transcriptional activity in HeLa cells was enhanced by low doses (0.05–0.5 mmol/L) of H2O2, but suppressed by high doses (0.75–8.0 mmol/L) of H2O2. The amount of co-activator p300 bound to HIF-1α in HeLa cells was increased under mild oxidative stress, but decreased under severe oxidative stress. The ROS levels differentially modified cysteines 243 and 532 in the cysteine protease SENP3, regulating the interaction of SENP3 with p300 to cause different SUMOylation of p300, thus shifting HIF-1 transcriptional activity.
Conclusion:
The shift of HIF-1 transactivation by ROS is correlated with and dependent on the biphasic redox sensing of SENP3 that leads to the differential SENP3/p300 interaction and the consequent fluctuation in the p300 SUMOylation status.
Keywords: ROS, redox, SENP3, HIF-1, p300, SUMOylation, HeLa cells
Introduction
Oxidative stress, a common challenge to cellular homeostasis, is caused predominantly through the excessive production of reactive oxygen species (ROS). Many extracellular insults, such as irradiation, drugs and various disturbances in temperature, pH, osmotic pressure, oxygen tension and sugar concentration, may all increase the production of ROS1,2,3,4,5. The extent of the increase in ROS production usually determines the consequences of cellular adaptive responses to oxidative stress. Mild oxidative stress promotes cell survival or proliferation, while severe oxidative stress causes proliferation arrest, senescence or cell death, during which global alterations of gene expression and protein post-translational modifications are differentially regulated according to the differences in ROS levels1,2,6,7,8.
Hypoxia-inducible factor-1 (HIF-1) is a well-known transcription factor that controls gene expression in response to hypoxia. Interestingly, HIF-1 can also be activated under normoxic conditions. For instance, HIF-1 activation occurs during inflammation9,10,11, growth factor activation12,13 and insulin administration14,15,16,17. Notably, ROS generation accompanies these events and is required for normoxic HIF-1 activation. Moreover, it has been demonstrated that ROS production is paradoxically increased during hypoxia and is even required for hypoxic HIF-1 activation18,19. A correlation of increased ROS and over-expressed/activated HIF-1 has also been suggested in cancer cells20,21,22.
While an increasing body of work has reported that ROS can stabilize HIF-1α and enhance HIF-1 transactivation23,24,25, we have previously shown that HIF-1 can be suppressed in the prostate cancer cell line Du145 by the overwhelming production of ROS induced by a combined administration of the ROS producer emodin and the conventional chemotherapeutic agent cisplatin21. The capacity of HIF-1 to be activated by mild oxidative stress but inhibited by severe oxidative stress, ie, the mechanism for the biphasic regulation of HIF-1 trans-activation, has thus become an intriguing question.
We have recently found that the SUMO2/3-specific protease SENP3 and its de-SUMOylating catalytic activity are required for the ROS-induced enhancement of HIF-1 transactivation, based on ROS-induced HIF-1α stabilization26. The SENP3-mediated deconjugation of SUMO2/3 from p300, the co-activator for HIF-1, is beneficial for HIF-1 transcriptional activity under both hypoxic and normoxic conditions. Being different with the other SENPs family members, SENP3 is specifically and sensitively responsive to low oxidative stress26,27. SENP3 is stabilized upon exposure to hydrogen peroxide (H2O2) at very low doses, for instance, 0.05 mmol/L, and redistributes from the nucleolus to the nucleoplasm, which allows it to modulate various nuclear events, including the enhancement of HIF-1 transactivation. However, SENP3, along with all SENP family members, is a cysteine protease28,29,30, meaning that its enzymatic activity relies on cysteine residues in the catalytic site. It has been demonstrated that highly oxidizing conditions (10 mmol/L H2O2) inhibit the deconjugation activity of SENP1 and SENP228,31. We therefore hypothesize that the oxidation of the catalytic cysteine in SENP3 under severe oxidative stress might inhibit its de-SUMOylating activity that is required for the physiological functions of various substrates, including p300, and HIF-1 transactivation might be thus suppressed.
To test this hypothesis, we used HeLa cells exposed to a series of concentrations of H2O2 as a simple model of mild and severe oxidative stress, examined the steps of HIF-1 activation, and analyzed SENP3 activity and the SUMOylation status of p300. Our findings in the present study suggest that the shift of HIF-1 transactivation by ROS is correlated with and dependent on the differential SENP3/p300 interaction and consequent fluctuation in the SUMOylation status of p300. SENP3 protein is induced by mild oxidative stress but its catalytic activity is inactivated by severe oxidative stress, thus leading to opposite SUMOylation status of its substrate p300. This biphasic ROS effect on SENP3 is achieved through a sequential oxidative modification at two cysteine residues.
Materials and methods
Cell culture and treatments
HeLa cells were used for all experiments. Cells were cultured in Dulbecco's modified Eagle's medium (GibcoBRL, Gaithersburg, MD, USA). All media were supplemented with 100 U/mL penicillin, 100 mg/L streptomycin and 10% newborn calf serum (Biochrom AG, Germany). Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2.
To set a cell model of oxidative stress, cells were treated with hydrogen peroxide (H2O2, Sigma-Aldrich, St Louis, MO, USA) at increasing doses. When needed, anti-oxidant N-acetylcysteine (NAC, Sigma-Aldrich, St Louis, MO, USA) was pre-incubated with cells for 4 h and protein reducing agent dithiothreitol (DTT, Sigma-Aldrich, St Louis, MO, USA) was pre-incubated for 2 h.
Cell viability assay
Cells were seeded at 1.5×104/mL cells per well in 96-microculture-well plates. After exposed to the various concentration of H2O2 as indicated for 24 h, cell viability was assayed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) (Sigma-Aldrich, St Louis, MO, USA)32.
ROS detection
2,7-Dichlorodihydrofluorescein diacetate (DCFH-DA; Sigma-Aldrich, St Louis, MO, USA) was used as ROS capturing reagent with the method described previously33,34.
Luciferase reporter assay
The constructs of the luciferase reporter specific for hypoxia response element (HRE) and the renilla control were transfected and the reporter assay were performed as described previously26. Briefly, cells were transfected with the double reporters, and 40 h post transfection, cells were exposed to H2O2 at varied doses once for 6 h before relative luciferase activity was assayed.
Quantitative real-time PCR
Quantitative real-time PCR was carried out on the ABI Prism 7300 system (Applied Biosystems, Foster City, CA, USA) using SYBR Green following the manufacturer's instructions. The primers for the vascular endothelial growth factor (VEGF) gene were 5′-CGGTATAAGTCCTGGAGCGTGT-3′ and 5′-TCACCGCCTCGGCTTGTCA-3′. The primers for carbonic anhydrase 9 (CA9) were 5′-CTGTCACTGCTGCTTCTGAT-3′ and 5′-TCCTCTCCAGGTAGATCCTC-3′. For real-time PCR, up to 1 μL of cDNA were used as template. Thermal cycling conditions were 95 °C for 60 s, followed by 45 cycles of 95 °C for 30 s, 56 °C for 30 s and 72 °C for 35 s. Primer efficiency of >90% was confirmed with a standard curve spanning four orders of magnitude. Following the reactions, the raw data were exported using the 7300 System Software v1.3.0 (Applied Biosystems, Foster City, CA, USA) and analyzed.
Immunoblotting (IB)
Cells were lysed in sample solution. Proteins were separated on 6%, 8%, or 10% SDS-PAGE gels, transferred to nitrocellulose membranes, and bands were detected using various antibodies as indicated. The membranes were incubated with the primary antibodies at 4 °C overnight and horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 2 h at the room temperature (RT) before detection using an enhanced chemiluminescence (ECL) system (Pierce Biotechnology, Rockford, IL, USA).
Immunofluorescence
Cell monolayers were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and were blocked with 5% BSA before incubation with the mouse monoclonal anti-HIF-1α at 4 °C overnight. Subsequently, the cells were incubated with fluorescent isothiocyanate (FITC)-conjugated anti-mouse antibody (SouthenBiotech, Birmingham, AL, USA) for 1.5 h at RT. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were then examined under an Axioplan 2 fluorescent microscope (Zeiss, Germany).
Chromatin immunoprecipitation (ChIP) assay
Cells were incubated with 150 μmol/L CoCl2 for 20 h to ensure the stabilization of HIF-1α, and then exposed to various concentration of H2O2 for 6 h. Protein-DNA were cross-linked in 1% formaldehyde for 10 min at 37 °C before the reaction was quenched by glycine (0.125 mol/L) for 5 min at RT. Cells were then harvested and the ChIP assay was performed using a kit (Upstate Biotechnology Inc, Lake Placid, NY, USA) according to the manufacturer's instructions and as in our previous work26. The sonicated chromatin samples were precipitated with anti-HIF-1α antibody. The primer sequences for HRE were 5′-CCTTTGGGTTTTGCCAGA-3′ and 5′-CCAAGTTTGTGGAGCTGA-3′. The input fractions were used as the internal control.
Co-immunoprecipitation (co-IP)
Cells were washed once with cold PBS and lysed with cold 1×IP buffer (1% Triton X-100, 150 mmol/L NaCl, 10 mmol/L Tris, pH 7.4, 1 mmol/L EDTA, 1 mmol/L EGTA, pH 8.0, 0.2 mmol/L sodium ortho-vanadate, 1 mmol/L PMSF, 0.5% protease inhibitor cocktail, 0.5% IGEPAL CA-630) at 4 °C for 30 min. N-Ethylmaleimide (NEM 20 mmol/L) was included in the above buffer when SUMOylation of proteins needed to be examined. Cells were transiently co-transfected with various tagged constructs. At 48 h post-transfection, cells were exposed to various reagents as indicated before harvesting using the IP buffer. Cell lysates were passed several times through a 26-gauge needle and centrifuged at 14 000×g for 30 min at 4 °C. The supernatants were incubated with various antibodies overnight at 4 °C, followed by incubation with Protein A/ Protein G-coated agarose beads (Santa Cruz Biotechnology Inc, Santa Cruz, CA, USA) for another 4 h at 4 °C. After samples were washed four times with ice-cold IP buffer and supernatants were removed by centrifugation at 2000×g for 1 min, proteins were co-precipitated. The proteins were then separated from the beads using IB loading buffer for 15 min at 95 °C. The supernatants were collected and subjected to SDS-PAGE analysis and detected using various antibodies as indicated.
Intermolecular cross-linkage assay
HeLa cells were transfected with RGS-SENP3. At 48 h post-transfection, cells were treated with H2O2 for 1 h with or without DTT pretreatment for 2 h. Cells were lysed in SDS loading buffers and proteins were separated on 10% SDS-PAGE. IB was performed with anti-RGS antibody to detect the intermolecular SENP3 cross-linkage via disulfide formation28.
Antibodies
The mouse monoclonal antibodies against HIF-1α for IB and immunofluorescence (BD Pharmingen, San Diego, CA, USA), HIF-1α for IP (ABR Affinity Bioreagents, Golden, CO, USA), RGS (Qiagen, Germany), HA, β-actin (Sigma-Aldrich), p300 for IB, p300 for IP (BD Bioscience, Franklin Lakes, NJ, USA) were purchased. The goat polyclonal anti-SUMO2/3 antibody and mouse monoclonal anti-SENP3 antibody were from Santa Cruz Biotechnology Inc (Santa Cruz, CA, USA).
Constructs and the site-mutagenesis
RGS tagged SENP3, HA tagged SUMO3, SENP3 mutants C243S and C532A were previously used in our work35. The mutant C532S was generated by QuikChange lighting Site-Directed Mutagenesis Kit (Agilent Technology, Santa Clara, CA, USA) with previously used method35.
siRNA
siRNA specific for SENP3, SENP3 (3′UTR) and non-specific control (NC) siRNA were synthesized (RIBOBIO, China), and transfected using Lipofectamine 2000. The sequences of siRNA oligonucleotides for endogenous human SENP3 were: 5′-CAAUAAGGAGCUACUGCUAdTdT-3′ and 5′-GUUAUUCCUCGAUGACGAUdTdT-3′. While those for 3′-UTR of SENP3 were: 5′-GATCCTTTGTTGATACGTAdTdT-3′.
Transfection, co-transfection and rescue
The constructs were transiently transfected or co-transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
HeLa cells were first transfected with non-specific siRNA control or siRNA for endogenous SENP3. After 24 h, cells were again transfected with RGS-SENP3 WT, the mutant C243S or the mutant C532S for another 48 h to rescue SENP3 functions.
Statistical analysis
Results were derived from at least three independent experiments and expressed as Mean±SD. The difference between groups were compared using Student's t test.
Results
HIF-1 transcriptional activity is enhanced by low doses of H2O2 but suppressed by high doses of H2O2
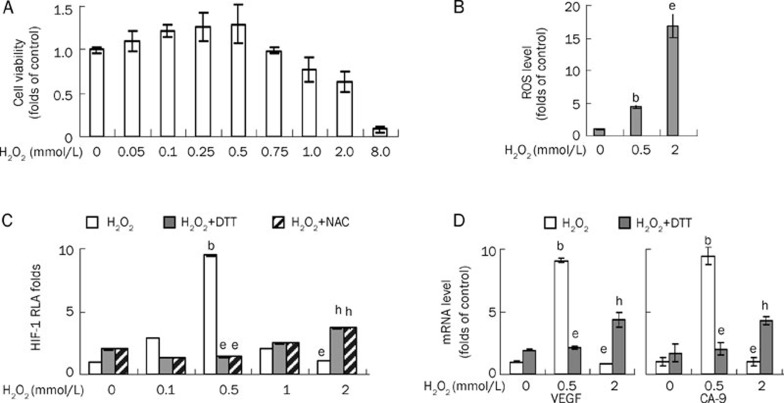
Varied doses of H2O2 were administered to cultured HeLa cells and incubated for 24 h before the cell viability was assayed. The results showed that the cell viability was promoted at doses ranging from 0.05 mmol/L to 0.5 mmol/L with a peak at approximately 0.5 mmol/L, and was suppressed at doses higher than 0.75 mmol/L (Figure 1A). After screening the doses of H2O2 that resulted in opposite cellular effects, 0.5 mmol/L and 2 mmol/L were used as the doses to create the low and high oxidative stress conditions respectively. Truly, ROS increased to modest or dramatic levels following treatments with these doses of H2O2 (Figure 1B).
Figure 1.
HIF-1 transcriptional activity is enhanced by low doses of H2O2, but suppressed by high doses of H2O2. (A) HeLa cells were treated once with the indicated concentrations of H2O2 for 24 h and then cell viabilities were evaluated by MTT. (B) ROS levels were detected by flow cytometry after HeLa cells were exposed to the indicated concentrations of H2O2 for 1 h and incubated with DCFH-DA for 10 min. (C) HeLa cells were co-transfected with the constructs of luciferase reporter for HRE and Renilla. Fourty hours posttransfection, HeLa cells were exposed to H2O2 as indicated for 6 h. When used, 5 mmol/L NAC was pretreated for 4 h or 10 mmol/L DTT pretreated for 2 h before H2O2 exposure. Cells were then lysed, and the relative luciferase activity (RLA) for HIF-1 was assayed as described in “Materials and methods”. (D) HeLa cells were treated with the indicated concentrations of H2O2 for 6 h. HIF-1 target gene VEGF and CA-9 mRNA levels were evaluated by quantitative real-time PCR and shown as folds of control. The results were shown as the Mean±SD of three independent experiments, and samples were duplicated in each experiment. Data were represented as Mean±SD. bP<0.05 vs H2O20 mmol/L. eP<0.05 vs H2O20.5 mmol/L. hP<0.05 vs H2O22 mmol/L.
The transcriptional activity of HIF-1 was assessed using luciferase reporters after cells were exposed to H2O2 for 6 h. The results showed that the transcriptional activity of HIF-1 was enhanced by 0.5 mmol/L of H2O2 but suppressed by 2 mmol/L of H2O2 (Figure 1C). The antioxidant NAC and the reducing agent DTT could reverse the alteration of HIF-1 transactivation caused by ROS regardless of whether transactivation was enhanced or suppressed, indicating that this effect was under redox control.
As 2 mmol/L of H2O2 led to the repression of viability or mitotic senescence at later time points, it was necessary to distinguish a specific inhibition toward HIF-1 transactivation from a general transcriptional suppression. The luciferase reporter activity for the control (renilla) at this level of oxidative stress was compared with the activity at the lower level, and the result excluded a general transcriptional suppression (data not shown). A reporter assay for p53 showed that the transcriptional activity of p53 was induced while the one for HIF-1 was suppressed by 2 mmol/L of H2O2 (Supplementary Figure 1), implying that the inhibition of HIF-1 transactivation by oxidative stress was not a non-specific phenomenon.
The transcriptional activity of HIF-1 was also assessed using real-time PCR for the mRNA levels of the HIF-1 target genes VEGF and CA-9 after the cells were exposed to H2O2 for 6 h. The results showed that the transcriptional levels of the HIF-1 target genes were enhanced by 0.5 mmol/L of H2O2 and suppressed by 2 mmol/L of H2O2 (Figure 1D). The reducing agent DTT could reverse the alterations in the mRNA levels of HIF-1 target genes caused by ROS, regardless of whether they were enhanced or suppressed (Figure 1D).
Differential HIF-1 transcriptional activity under mild or severe oxidative stress results from altered HIF-1α/p300 binding
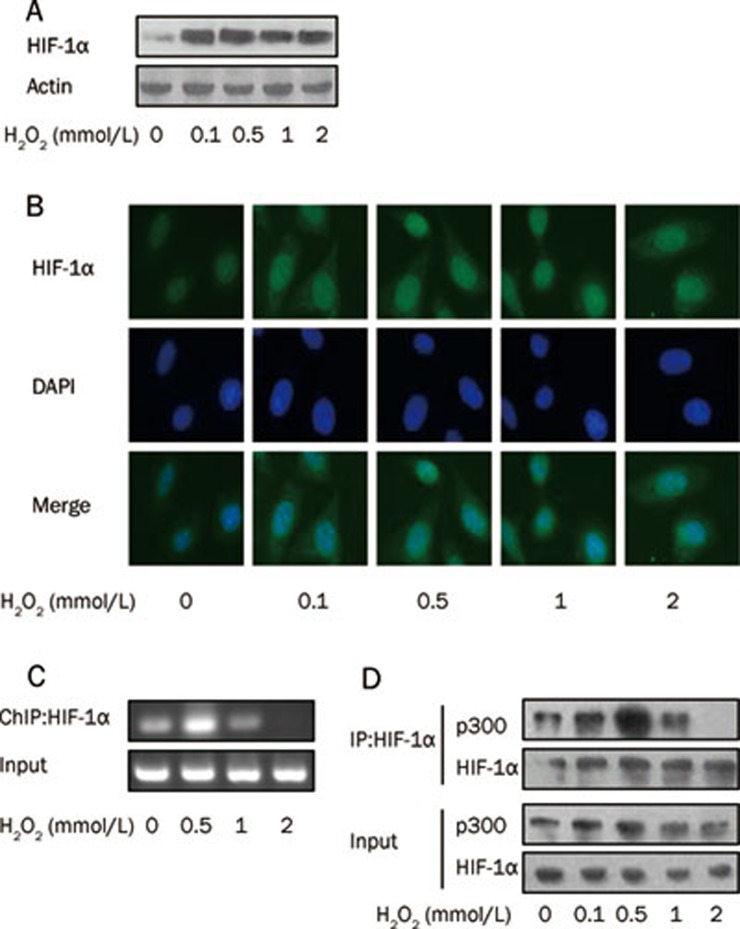
Because HIF-1 activation is a multistep process involving the stabilization of the HIF-1α protein, the dimerization of the HIF-α and -β subunits, translocation to the nucleus, binding to HIF-1 responsive elements HRE, and the formation of active transcriptional complexes with the co-activators p300/CBP36,37,38,39,40, we examined these processes during mild and severe oxidative stress. The result of the immunoblot showed that, although HIF-1α was stabilized by treatment with a very low dose of H2O2, it remained unchanged under increasing doses of H2O2 (Figure 2A). In addition, the nuclear accumulation of HIF-1α remained stable in the presence of increasing doses of H2O2, as shown by immunofluorescence (Figure 2B). However, a ChIP assay showed that the amount of HIF-1α bound to HRE was remarkably increased by 0.5 mmol/L H2O2 but was decreased by 2 mmol/L H2O2 (Figure 2C). Likewise, a co-IP assay showed that the amount of p300 bound to HIF-1α increased at low doses of H2O2 but decreased at high doses (Figure 2D). These data clearly demonstrated that the biphasic regulation of HIF-1 transactivation by different levels of ROS was not attributable to the altered expression or nuclear accumulation of HIF-1α but rather to an alteration in HIF-1/p300 binding and a concomitant alteration in HIF/DNA binding.
Figure 2.
Differential HIF-1 transcriptional activity under mild or severe oxidative stress results from altered HIF-1α/p300 binding (A) HIF-1α protein levels were evaluated by IB after HeLa cells were incubated with cobalt chloride for 20 h and then treated with the indicated H2O2 for additional 1 h. β-Actin was shown as a loading control. (B) HIF-1α translocation to nuclei was shown by immunofluorescence. Cells were treated with the indicated H2O2 for 1 h. Immunofluorescence was performed with anti-HIF-1α antibody and the FITC-second antibody. DAPI was used for counterstaining the nuclei. (C) Ability of HIF-1 binding with HRE was tested by ChIP assay. HeLa cells were incubated with cobalt chloride for 20 h and then treated with the indicated H2O2 for additional 1 h followed by ChIP assay. (D) Interaction of HIF-1 with p300 was detected by co-IP. HeLa cells were incubated with cobalt chloride for 20 h and then treated with the indicated H2O2 for additional 1 h. Cells were lysed and endogenous HIF-1α was pulled down using antibody against HIF-1α. The pulled-down HIF-1α and whole cell lysates as input were analyzed by IB as indicated.
The ROS level regulates the SENP3-p300 interaction to cause differential SUMOylation of p300
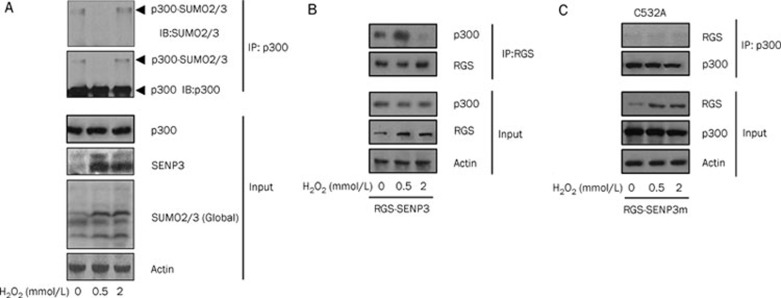
As we had previously found that the SUMO protease SENP3 accumulated in response to a mild increase in ROS and could specifically deconjugate SUMO2/3 from p300 thus promoting p300 binding with HIF-126, the SUMOylation status of p300 at different ROS levels was examined. The results of the co-IP showed that, as predicted, the endogenous SUMO2/3 modification of endogenous p300 obviously decreased after the cells were exposed to a low dose of H2O2, while the SUMO2/3 modification of p300 in the cells treated with a high dose of H2O2 remained similar to that in untreated cells. However, the level of the SENP3 protein remained stable in cells undergoing low or high stress (Figure 3A).
Figure 3.
The ROS level regulates the SENP3-p300 interaction to cause differential SUMOylation of p300. (A) The SUMOylation of endogenous p300 was detected by co-IP. HeLa cells were treated with different concentrations of H2O2 for 1 h as indicated. Co-IP was carried out with p300 antibody and IB was carried out using the antibodies as indicated. The binding of SENP3 with its substrate p300 was detected by co-IP. HeLa cells were transfected with RGS-SENP3 for 48 h, then were incubated with different concentrations of H2O2 for 1 h. (B) Co-IP was carried out with the antibody against RGS tag and IB was carried out using the antibodies against RGS and endogenous p300. The accumulation of SENP3 (RGS) was shown in the input. β-actin was used as a loading control. (C) The binding of the SENP3 mutant with its substrate p300 was detected by co-IP. HeLa cells were transfected with RGS-SENP3-C532A for 40 h. Co-IP and IB procedures were the same to above in B. The accumulation of SENP3 (RGS) was shown in the input. β-Actin was used as a loading control.
We hypothesized that the unchanged SUMOylation status of p300 under high stress might be caused by the inactivation of the enzymatic activity of SENP3, given that the SENP3 protein level was unchanged. To clarify this, RGS-tagged SENP3 was ectopically expressed in cells, and co-IP for RGS-SENP3 was then used to detect the interaction of SENP3 with p300. The results showed that the SENP3 interaction with p300 was enhanced under low stress condition. However, this interaction was markedly blocked under high stress and was even weaker than that under non-stress condition (Figure 3B). This result indicated that the inactivation of SENP3 catalytic capability might occur under severe oxidative stress, and this inactivation could be manifested as the loss of its ability to bind to its substrates.
SENP3 is a cysteine protease, and its enzymatic activity relies on a specific cysteine, C53229. As the mutant that substitutes C532 with alanine, C532A, is typically used as a dominant-negative SENP329, we used this mutant to examine SENP3-p300 binding under low and high stress conditions. The results showed that this mutant could not bind the substrate under any conditions, clearly confirming that C532, which is responsible for enzymatic activity, is required for substrate binding (Figure 3C). Hence, SENP3-p300 binding capacity could be used as a readout for the enzymatic activity of SENP3 in the following experiments.
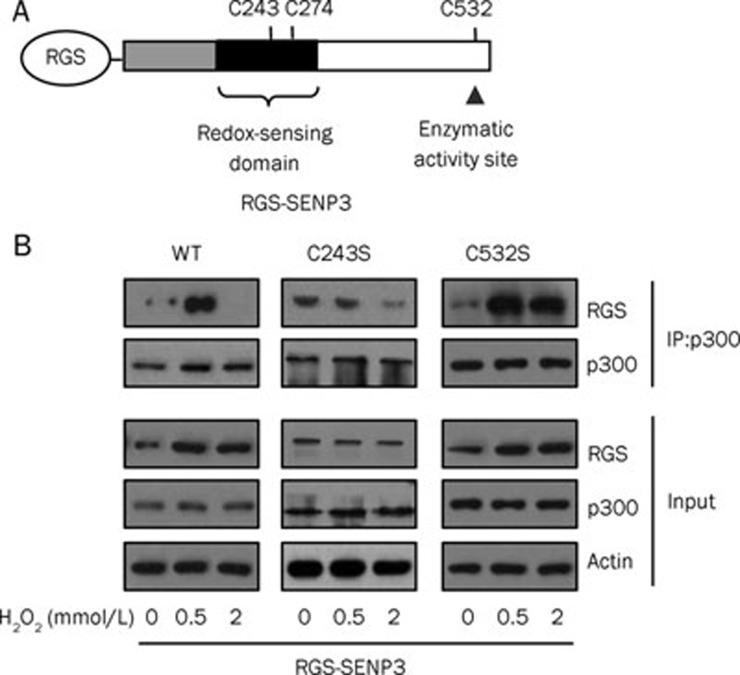
The ROS level regulates the SENP3-p300 interaction by affecting different cysteines
Our previous studies26,27 have shown that the decrease in the SUMO2/3 modification of p300 during mild stress is due to an accumulation of SENP3. We also found that this accumulation following a blockage of ubiquitination can be attributed to the oxidation of cysteines 243 or 274 within the redox sensing domain of SENP3; the mutants (C243S or C274S) in which these cysteines were replaced by serine, an amino acid residue non-responsive to ROS, failed to accumulate under stress35. Given that the SENP3 protein level was unchanged upon high stress as compared with low stress and that the enzymatic activity of SENP3, as represented by its substrate binding capacity, was lost only under high stress, we hypothesized that, whereas mild stress oxidizes cysteines 243 or 274 to induce SENP3 stabilization, severe stress might oxidize another cysteine and inactivate the enzymatic activity of SENP3, ie, its capacity to bind with its substrates. We then generated another mutant in which C532 was replaced with serine. The SENP3-p300 binding was then analyzed by co-IP in cells transfected with the wild-type SENP3 or the mutants C243S or C532S.
The results showed that the C243S mutant could not be stabilized in response to either high or low oxidative stress conditions (Figure 4B, compare the blot of the input RGS of C243S with the wild-type). However, this mutant failed to bind to p300 when exposed to high stress conditions, showing a similar response as the wild-type SENP3. Interestingly, the C532S mutant was sensitively responsive to ROS-induced accumulation, regardless the degree of stress (Figure 4B, compare the blot of the input RGS of C532S with the wild-type). Furthermore, the co-IP results showed that it retained the capacity to bind with p300 under both low and high stress, being not responsive to ROS-induced inactivation (Figure 4B). These data indicate that the oxidation of C243 is responsible for SENP3 accumulation under both low and high stress conditions, while the oxidation of C532 is responsible for the blockage of the SENP3-substrate interaction under high stress condition.
Figure 4.
The ROS level regulates the SENP3-p300 interaction by affecting different cysteines. The identified domains and important cysteine residues of SENP3. (A) The plasmid RGS-SENP3 was constructed and the site mutagenesis was conducted with serines to replace cysteines at 243 and 532 sites. (B) The accumulation of the SENP3 wild-type (WT) or mutants in response to stress conditions and the binding with its substrate p300 was detected by co-IP. HeLa cells were transfected with RGS-SENP3 WT, C243S, or C532S for 40 h before treated with H2O2 for 1 h. The proteins were co-immunoprecipitated using anti-p300 and detected by IB using the indicated antibodies. The accumulation of SENP3 (RGS) was shown in the input. β-Actin was used as a loading control.
Oxidative modification of the individual cysteines in SENP3 regulates p300 SUMOylation and HIF-1 transactivation
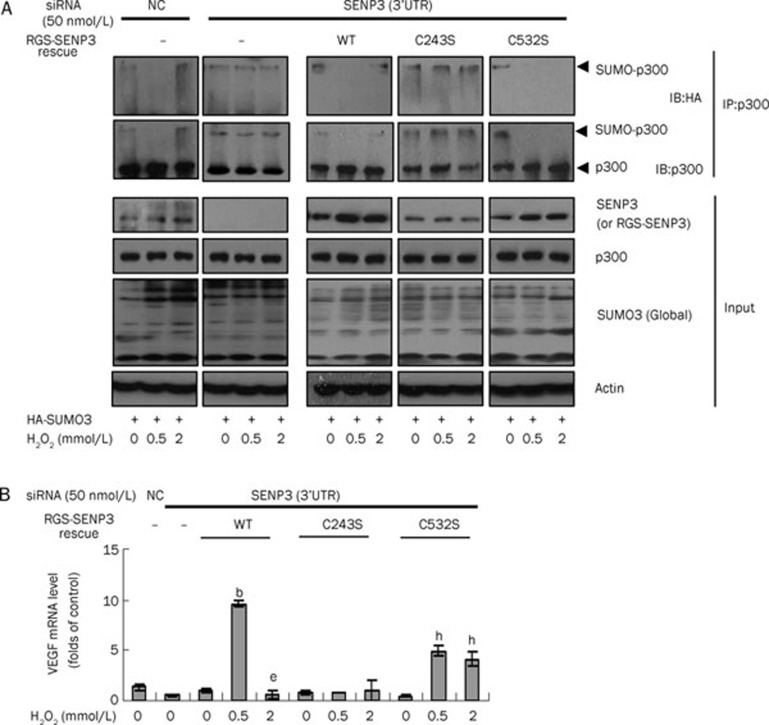
To elucidate the biochemical consequences of SENP3 regulation by ROS in a biphasic manner, we investigated whether C243S and C532S truly affected p300 SUMOylation. Endogenous SENP3 was knocked down by siRNA that did not target exogenous SENP3, and the SENP3 functions were then rescued by transfection with the wild-type or the C243S or C532S mutants before the co-IP experiments were conducted. The results showed that siRNA effectively abolished basal and the ROS-enhanced SENP3 accumulation and that the conjugation of p300 with SUMO2/3 was unchanged upon stress conditions. The wild-type SENP3 could rescue the ROS-enhanced SENP3 accumulation and p300 de-SUMOylation under low stress, but SENP3 accumulation could not mediate p300 de-SUMOylation under high stress. The mutant C243S could not rescue SENP3 accumulation or p300 de-SUMOylation under both stress conditions. Remarkably, the mutant C532S could de-SUMOylate p300 under both mild and severe stress conditions, because SENP3 could accumulate under both stress conditions, and the enzyme remained catalytically active under severe stress (Figure 5A).
Figure 5.
Oxidative modification of the individual cysteines in SENP3 regulates p300 SUMOylation and HIF-1 transactivation. (A) The SUMOylation of p300 was detected by co-IP. HeLa cells were transfected with non-specific siRNA control or siRNA for endogenous SENP3 for 24 h, and then transfected with RGS-SENP3 WT or the mutants C243S or C532S to rescue for another 48 h. HA-SUMO3 was simultaneously co-tranfected. Cells were treated with H2O2 for 1 h. The proteins were co-immunoprecipitated using anti-p300 and detected by IB using the antibodies against p300 and HA. β-Actin as a control. (B) HeLa cells were transfected with non-specific siRNA control or SENP3 siRNA for 24 h, and then transfected with RGS-SENP3 WT, the mutants C243S or C532S to rescue for another 48 h. Cells were treated with H2O2 for 6 h. The VEGF mRNA levels were evaluated by real-time PCR and shown as folds of control. The results were shown as the Mean±SD of three independent experiments, and samples were duplicated in each experiment. Data were represented as Mean±SD. bP<0.05 vs siRNA SENP3 rescue WT H2O20 mmol/L. eP<0.05 vs siRNA SENP3 rescue WT H2O20.5 mmol/L. hP<0.05 vs siRNA SENP3 rescue C532S H2O20 mmol/L.
Taken together, these data suggested that the two redox-sensing cysteines have different functions. C243 is responsible for sensing the increase in ROS production and in turn blocking ubiquitination. After SENP3 becomes stabilized upon mild stress, C532 is responsible for the interaction of SENP3 with its substrates, but this site is inactivated during a further increased ROS generation. The sequential oxidation of these two cysteines leads to changes in SENP3 protein abundance and its catalytic activity during the increase in ROS level, which eventually results in a fluctuation in the SUMOylation status of p300.
We then examined the HIF-1 transcriptional activity by measuring the mRNA level of the HIF-1 specific target gene VEGF in cells with SENP3 knockdown and recue. The results showed that the HIF-1 target gene expression was also differently affected by the two SENP3 mutants, which was consistent with the SUMOylation statuses of p300 (Figure 5B).
To exclude the possibility that 2 mmol/L H2O2 might cause oxidative damage to SENP3, we finally performed an intermolecular cross-linkage assay. This experiment showed that SENP3 cross-linking only occurred after treatment with 10 mmol/L of H2O2 (supplementary Figure 2). This result suggested that SENP3 underwent a selective oxidative modification upon treatment with 2 mmol/L H2O2.
Discussion
The dual or biphasic regulatory effects of ROS have been reported for many years2,6,41,42,43. For instance, ROS can mediate phenotypes ranging from survival and growth to apoptosis in vascular endothelial and smooth muscle cells, which may be considered both physiological and pathophysiological41. It has been concluded that the specific response elicited by ROS is determined by their specific intracellular target(s), which, in turn, depends on the species of oxidant(s), the source and subcellular localization of the oxidant(s), the kinetics of production, and the quantities produced41. For example, in some cell types, low levels of ROS (usually submicromolar concentrations) induce growth, but higher concentrations (usually >10 micromolar) induce apoptosis or necrosis44. These studies, although using the words dual or biphasic, actually intended to highlight the fact that ROS may cause opposing effects at different doses.
It has been observed that cellular signaling pathways are generally subjected to dual redox regulation in which ROS have opposing effects on upstream signaling systems and downstream transcription factors21,26,32,45,46. Our previous research has shown that, when increased by chemical compounds or genetic manipulation, ROS may enhance the transcriptional activities of NF-κB, AP-1 and HIF-1 at lower doses but suppress them at higher doses21,26,32,45,47,48. For instance, when ROS increase modestly, they activate NF-κB by triggering the degradation of IκBα and other events in the cytoplasm, which usually lead to prosurvival signaling events. However, as the ROS levels become further excessive, the nuclear environment is shifted from reductive to oxidative, which inhibits the activation of NF-κB and other transcription factors by preventing them from binding to DNA. As a consequence, the prosurvival transcriptional activity is abolished. This finding shows how ROS can shift the activation/inactivation control of the NF-κB pathway45.
Based on findings in the literature and our own observations, we propose that the opposing effects caused by low vs high levels of ROS may be mediated by the biphasic redox sensing of proteins, ie, the ability of proteins to sense redox changes through the oxidation of their cysteine residues by different levels of ROS. Oxidative modification of protein thiols is now considered as an emerging role in cell signaling. It has been well recognized that the thiol oxidation state of reactive cysteine residues in proteins controls the function of the proteins and the pathways that they are part of49,50,51,52,53,54. We hypothesize that, during the biphasic redox sensing, the different cysteines of a given protein molecule might be sequentially oxidized by different levels of ROS, thus leading to different conformational characteristics and functional statuses of the protein.
HIF-1 is one of the most well known redox-sensitive transcription factors. An increasing body of evidence shows that ROS induce the stabilization of HIF-1α under hypoxic or normoxic conditions14,26,55,56. Our previous study proposed an additional mechanism in which mild oxidative stress induced by low doses of H2O2 can rapidly stabilize SENP3, in turn promoting the transcriptional activity of HIF-1 through the deconjugation of SUMO2/3 from the co-activator p300. This activating mechanism functions independently of HIF-1α stabilization, and is required for ROS-mediated HIF-1 transcriptional activity under both hypoxia and normoxia13. To establish why the transcriptional activity of HIF-1 can be suppressed by a further dramatic increase of ROS, we hypothesized that SENP3 is a biphasic redox sensor that mediates the biphasic redox regulation of HIF-1. We used a variety of SENP3 mutants to study the responsiveness of SENP3 to increasing levels of ROS. The results of the present study demonstrated that the oxidation of C243 is required for ROS-induced SENP3 accumulation and consequent HIF-1 transactivation, while in contrast, the oxidation of C532 is responsible for ROS-induced SENP3 inactivation and the consequent suppression of the transcriptional activity of HIF-1. Therefore, we suggest that the biphasic redox sensing of SENP3 mediates, at least in part, the bidirectional ROS regulation of HIF-1. To our knowledge, the present study has demonstrated for the first time that the different cysteines in a redox-sensitive protein can sense different ROS levels, and thus mediate a shift in protein function and the related signaling activity.
The cell model of the biphasic redox effects in this study is produced by treating HeLa cells with 0.5 and 2 mmol/L of H2O2. This dose range of H2O2 used in cancerous or transformed cells is considered to be appropriate in mimicking oxidative stress conditions28,31,57,58,59. Bossis et al reported that ROS at low concentrations, 1 mmol/L, result in the rapid disappearance of most SUMO conjugates, which is due to direct and reversible inhibition of SUMO conjugating enzymes through the formation of (a) disulfide bond(s) involving the catalytic cysteines of the SUMO E1 subunit Uba2 and the E2-conjugating enzyme Ubc9. And they also observed the same phenomenon in a physiological scenario of endogenous ROS production, the respiratory burst in macrophages31. Xu et al showed a reversible oxidative modification of SUMO protease SENP1 by 10 mmol/L of H2O2 that serves as a protective mechanism for the enzyme28. Thus, the cell model of low vs high stress in this study may represent ROS fluctuation that is correlated with protein oxidation.
The findings in the present study will shed light on biphasic redox effects in cells and increase our understanding of how the generation of ROS may be relevant to both cancer therapy and to cancer genesis and progression. A typical example of this rationale is arsenic, which is classically recognized as a ROS-dependent environmental carcinogen but becomes an anticancer agent by inducing the apoptosis of leukemic cells60,61.
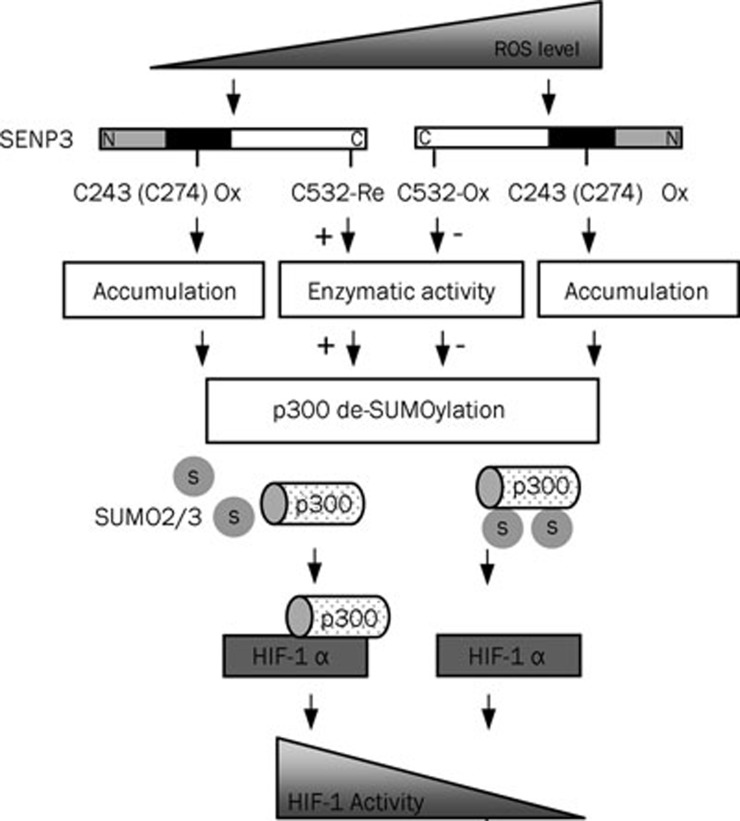
In summary, SENP3 is a biphasic redox sensor. The increase in intracellular ROS generation could induce SENP3 stabilization through the oxidation of the cysteines 243 or 274 that in turn blocks its ubiquitin-mediated degradation. But, when an overwhelming ROS are generated, the cysteine 532 that is required for the substrate interaction undergoes oxidation and consequent inactivation. This biphasic redox sensing of SENP3 leads to a fluctuation in the SUMOylation status of its substrate p300, a co-activator of HIF-1, thus making a shift of HIF-1 transcriptional activity from activation to inactivation (Figure 6).
Figure 6.
The schematic illustration for the biphasic redox sensing of SENP3 and its role in mediating the shift of HIF-1 transcriptional activity by ROS
Notes: “↓”, to lead to a result; “+ ↓”, to enhance; “↓ −”, to suppress;  , from low to high;
, from low to high;  , from high to low.
, from high to low.
Abbreviation
ROS, reactive oxygen species; SUMO, small ubiquitin-like modifier; NAC, N-acetylcysteine; DTT, dithiothreitol; RT-PCR, real-time polymerase chain reaction; DCFH-DA, 2, 7-dichlorodihydrofluorescein diacetate; HRE, hypoxia response element; IB, immunoblotting; co-IP, co-immunoprecipitation; ChIP, chromatin immunoprecipitation; siRNA, small interfering RNA; RT, room temperature.
Author contribution
Ying WANG, Jie YANG, Xin-zhi HUANG, and Jing YI designed the experiments and analyzed the data; Ying WANG, Kai YANG, Hui CANG, and Hui LI performed the experiments; Ying WANG, Jie YANG, and Jing YI prepared the manuscript.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (91013012, 30971437 to Jing YI, 31000613 to Xin-zhi HUANG), Shanghai Municipal Science and Technology Commission (11ZR1419100 to Ying WANG, 11DZ2260200 to Jing YI), and Shanghai Municipal Education Commission (J50201, to Jing YI).
Footnotes
Supplementary figures are available at website of Acta Pharmacologica Sinica on NPG.
Supplementary Information
The procedures were as the same as figue 1C.
HeLa cells were transfected with RGS-SENP3 for 48 h.
References
- Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey MR, Sharpless NE. ROS as a tumour suppressor. Nat Cell Biol. 2006;8:1213–15. doi: 10.1038/ncb1106-1213. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–93. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Gough DR, Cotter TG.Hydrogen peroxide: a Jekyll and Hyde signalling molecule Cell Death Dis 20112e213doi: 10.1038/cddis.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita A, Weinberger M, Silva A, Sampaio-Marques B, Almeida B, Leão C, et al. Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proc Natl Acad Sci U S A. 2010;107:15123–8. doi: 10.1073/pnas.1004432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale JL, Holbrook NJ. Cellular response to oxidative stress: signaling for suicide and survival. J Cell Physio. 2002;192:1–15. doi: 10.1002/jcp.10119. [DOI] [PubMed] [Google Scholar]
- Gao L, Mann GE. Vascular NAD(P)H oxidase activation in diabetes: a double-edged sword in redox signalling. Cardiovasc Res. 2009;82:9–20. doi: 10.1093/cvr/cvp031. [DOI] [PubMed] [Google Scholar]
- Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–90. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer T, Yamnishi Y, Clausen BE, Förster I, Pawlinski R, Mackman N, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–57. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YJ, Isaacs JS, Lee S, Trepel J, Neckers L. IL-1beta-mediated up-regulation of HIF-1alpha via an NFkappaB/COX-2 pathway identifies HIF-1 as a critical link between inflammation and oncogenesis. FASEB J. 2003;17:2115–7. doi: 10.1096/fj.03-0329fje. [DOI] [PubMed] [Google Scholar]
- Walmsley SR, Cadwallader KA, Chilvers ER. The role of HIF-1alpha in myeloid cell inflammation. Trends Immunol. 2005;26:434–9. doi: 10.1016/j.it.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Tacchini L, Dansi P, Matteucci E, Desiderio MA. Hepatocyte growth factor signalling stimulates hypoxia inducible factor-1 (HIF-1) activity in HepG2 hepatoma cells. Carcinogenesis. 2001;22:1363–71. doi: 10.1093/carcin/22.9.1363. [DOI] [PubMed] [Google Scholar]
- Patten DA, Lafleur VN, Robitaille GA, Chan DA, Giaccia AJ, Richard DE. Hypoxia-inducible factor-1 activation in nonhypoxic conditions: the essential role of mitochondrial-derived reactive oxygen species. Mol Biol Cell. 2010;21:3247–57. doi: 10.1091/mbc.E10-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelzer E, Levy Y, Kahana C, Shilo BZ, Rubinstein M, Cohen B. Insulin induces transcription of target genes through the hypoxia-inducible factor HIF-1alpha/ARNT. EMBO J. 1998;17:5085–94. doi: 10.1093/emboj/17.17.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll VA, Ashcroft M. Role of hypoxia-inducible factor (HIF)-1alpha versus HIF-2alpha in the regulation of HIF target genes in response to hypoxia, insulin-like growth factor-I, or loss of von Hippel-Lindau function: implications for targeting the HIF pathway. Cancer Res. 2006;66:6264–70. doi: 10.1158/0008-5472.CAN-05-2519. [DOI] [PubMed] [Google Scholar]
- Roth U, Curth K, Unterman TG, Kietzmann T. The transcription factors HIF-1 and HNF-4 and the coactivator p300 are involved in insulin-regulated glucokinase gene expression via the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2004;279:2623–31. doi: 10.1074/jbc.M308391200. [DOI] [PubMed] [Google Scholar]
- Treins C, Murdaca J, Van Obberghen E, Giorgetti-Peraldi S. AMPK activation inhibits the expression of HIF-1alpha induced by insulin and IGF-1. Biochem Biophys Res Commun. 2006;342:1197–202. doi: 10.1016/j.bbrc.2006.02.088. [DOI] [PubMed] [Google Scholar]
- Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci U S A. 1998;95:11715–20. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS, Budinger G. The cellular basis for diverse responses to oxygen. Free Radic Biol Med. 2007;42:165–74. doi: 10.1016/j.freeradbiomed.2006.10.048. [DOI] [PubMed] [Google Scholar]
- Jung SN, Yang W, Kim J, Kim HS, Kim EJ, Yun H, et al. Reactive oxygen species stabilize hypoxia-inducible factor-1 alpha protein and stimulate transcriptional activity via AMP-activated protein kinase in DU145 human prostate cancer cells. Carcinogenesis. 2008;29:713–21. doi: 10.1093/carcin/bgn032. [DOI] [PubMed] [Google Scholar]
- Huang XZ, Wang J, Huang C, Chen YY, Shi GY, Hu QS, et al. Emodin enhances cytotoxicity of chemotherapeutic drugs in prostate cancer cells: the mechanisms involve ROS-mediated suppression of multidrug resistance and hypoxia inducible factor-1. Cancer Biol Ther. 2008;7:468–75. doi: 10.4161/cbt.7.3.5457. [DOI] [PubMed] [Google Scholar]
- Galanis A, Pappa A, Giannakakis A, Lanitis E, Dangaj D, Sandaltzopoulos R. Reactive oxygen species and HIF-1 signalling in cancer. Cancer Lett. 2008;266:12–20. doi: 10.1016/j.canlet.2008.02.028. [DOI] [PubMed] [Google Scholar]
- Tuttle SW, Maity A, Oprysko PR, Kachur AV, Ayene IS, Biaglow JE, et al. Detection of reactive oxygen species via endogenous oxidative pentose phosphate cycle activity in response to oxygen concentration: implications for the mechanism of HIF-1alpha stabilization under moderate hypoxia. J Biol Chem. 2007;282:36790–6. doi: 10.1074/jbc.M700327200. [DOI] [PubMed] [Google Scholar]
- Comito G, Calvani M, Giannoni E, Bianchini F, Calorini L, Torre E, et al. HIF-1α stabilization by mitochondrial ROS promotes Met-dependent invasive growth and vasculogenic mimicry in melanoma cells. Free Radic Biol Med. 2011;51:893–904. doi: 10.1016/j.freeradbiomed.2011.05.042. [DOI] [PubMed] [Google Scholar]
- Schumacker PT. SIRT3 controls cancer metabolic reprogramming by regulating ROS and HIF. Cancer Cell. 2011;19:299–300. doi: 10.1016/j.ccr.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Han Y, Wang YM, Sun XX, Yan S, Yeh ET, et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. 2009;28:2748–62. doi: 10.1038/emboj.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Huang C, Sun X, Xiang B, Wang M, Yeh ET, et al. SENP3-mediated de-conjugation of SUMO2/3 from promyelocytic leukemia is correlated with accelerated cell proliferation under mild oxidative stress. J Biol Chem. 2010;285:12906–15. doi: 10.1074/jbc.M109.071431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Lam LS, Lam LH, Chau SF, Ng TB, Au SW. Molecular basis of the redox regulation of SUMO proteases. FASEB J. 2008;22:127–37. doi: 10.1096/fj.06-7871com. [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ET. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281:15869–77. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Mukhopadhyay D, Mathew S, Hasebe T, Heimeier RA, Azuma Y, et al. Identification and developmental expression of Xenopus laevis SUMO proteases. PLoS One. 2009;4:e8462. doi: 10.1371/journal.pone.0008462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–57. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Yi J, Yang J, He R, Gao F, Sang H, Tang XM, et al. Emodin enhances arsenic trioxide induced apoptosis via generation of reactive oxygen species and inhibition of survival signaling. Cancer Res. 2004;64:108–16. doi: 10.1158/0008-5472.can-2820-2. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Xiang BG, Yang J, Sun XX, Wang YM, Cang H, et al. oxidative modification of caspase-9 facilitates its activation via disulfide-mediated interaction with Apaf-1. Cell Res. 2009;19:449–57. doi: 10.1038/cr.2009.19. [DOI] [PubMed] [Google Scholar]
- Sang J, Yang K, Sun YP, Han Y, Cang H, Chen YY, et al. SUMO2 and SUMO3 transcription is differentially regulated by oxidative stress in an Sp1-dependent manner. Biochem J. 2011;435:489–98. doi: 10.1042/BJ20101474. [DOI] [PubMed] [Google Scholar]
- Yan S, Sun XX, Xiang BG, Cang H, Kang XL, Chen YY, et al. Redox regulation of the stability of the SUMO protease SENP3 via interactions with CHIP and Hsp90. EMBO J. 2010;29:3773–86. doi: 10.1038/emboj.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D, Peet D, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295:858–61. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- Kallio PJ, Okamoto K, O'Brien S, Carrero P, Makino Y, Tanaka H, et al. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. EMBO J. 1998;17:6573–86. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G, et al. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci U S A. 2002;99:5367–72. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Bunn HF. Regulation of transcription by hypoxia requires a multiprotein complex that includes hypoxia-inducible factor 1, an adjacent transcription factor, and p300/CREB binding protein. Mol Cell Biol. 1998;18:4089–96. doi: 10.1128/mcb.18.7.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996;271:32253–9. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- Irani K. Oxidant signaling in vascular cell growth, death, and survival: a review of the roles of reactive oxygen species in smooth muscle and endothelial cell mitogenic and apoptotic signaling. Circ Res. 2000;87:179–83. doi: 10.1161/01.res.87.3.179. [DOI] [PubMed] [Google Scholar]
- Deshpande SS, Angkeow P, Huang J, Ozaki M, Irani K. Rac1 inhibits TNF-alpha-induced endothelial cell apoptosis: dual regulation by reactive oxygen species. FASEB J. 2000;14:1705–14. doi: 10.1096/fj.99-0910com. [DOI] [PubMed] [Google Scholar]
- Lopes N, Gregg D, Vasudevan S, Hassanain H, Goldschmidt-Clermont P, Kovacic H. Thrombospondin 2 regulates cell proliferation induced by Rac1 redox-dependent signaling. Mol Cell Biol. 2003;23:5401–8. doi: 10.1128/MCB.23.15.5401-5408.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RM, Suzuki YJ. Cell proliferation, reactive oxygen and cellular glutathione. Dose Response. 2006;3:425–42. doi: 10.2203/dose-response.003.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing YW, Yang J, Wang YM, Li H, Chen YY, Hu QS, et al. Alteration of subcellular redox equilibrium and the consequent oxidative modification of nuclear factor kappaB are critical for anticancer cytotoxicity by emodin, a reactive oxygen species-producing agent. Free Radic Biol Med. 2006;40:2183–97. doi: 10.1016/j.freeradbiomed.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Wang YM, Huang XZ, Cang H, Gao F, Yamamoto T, Osaki T, et al. The endogenous reactive oxygen species promote NF-kappaB activation by targeting on activation of NF-kappaB-inducing kinase in oral squamous carcinoma cells. Free Radic Res. 2007;41:963–71. doi: 10.1080/10715760701445045. [DOI] [PubMed] [Google Scholar]
- Yi J, Yang J, He R, Gao F, Sang H, Tang XM, et al. Emodin enhances arsenic trioxide-induced apoptosis via generation of reactive oxygen species and inhibition of survival signaling. Cancer Res. 2004;64:108–16. doi: 10.1158/0008-5472.can-2820-2. [DOI] [PubMed] [Google Scholar]
- Wang J, Yi J. Cancer cell killing via ROS: to increase or decrease, that is the question. Cancer Biol Ther. 2008;7:1875–84. doi: 10.4161/cbt.7.12.7067. [DOI] [PubMed] [Google Scholar]
- Thamsen M, Jakob U. The redoxome: proteomic analysis of cellular redox networks. Curr Opin Chem Biol. 2011;15:113–9. doi: 10.1016/j.cbpa.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17:381–92. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A. 2008;105:8197–202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Chida AS, Rahman I. Redox modifications of protein-thiols: emerging roles in cell signaling. Biochem Pharmacol. 2006;71:551–64. doi: 10.1016/j.bcp.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Eaton P. Protein thiol oxidation in health and disease: techniques for measuring disulfides and related modifications in complex protein mixtures. Free Radic Biol Med. 2006;40:1889–99. doi: 10.1016/j.freeradbiomed.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang J, Yi J. Redox sensing by proteins: oxidative modifications on cysteines and the consequent events. Antioxid Redox Signal. 2012;16:649–57. doi: 10.1089/ars.2011.4313. [DOI] [PubMed] [Google Scholar]
- Liu XH, Kirschenbaum A, Lu M, Yao S, Dosoretz A, Holland JF, et al. Prostaglandin E2 induces hypoxia-inducible factor-1alpha stabilization and nuclear localization in a human prostate cancer cell line. J Biol Chem. 2002;277:50081–6. doi: 10.1074/jbc.M201095200. [DOI] [PubMed] [Google Scholar]
- Sun W, Chang SS, Fu Y, Liu Y, Califano JA. Chronic CSE treatment induces the growth of normal oral keratinocytes via PDK2 upregulation, increased glycolysis and HIF1α stabilization. PLoS One. 2011;6:e16207. doi: 10.1371/journal.pone.0016207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Song G, Cai Q, Liu M, Luo H, Shi M, et al. Hydrogen peroxide-induced apoptosis in human gastric carcinoma MGC803 cells. Cell Biol Int. 2006;30:332–7. doi: 10.1016/j.cellbi.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Hardwick JS, Sefton BM. Activation of the Lck tyrosine protein kinase by hydrogen peroxide requires the phosphorylation of Tyr-394. Proc Natl Acad Sci U S A. 1995;92:4527–31. doi: 10.1073/pnas.92.10.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SG, Chiu DT, Errasfa M, Wang JM, Qi JS, Stern A. Effects of H2O2 on protein tyrosine phosphatase activity in HER14 cells. Free Radic Biol Med. 1994;16:399–403. doi: 10.1016/0891-5849(94)90042-6. [DOI] [PubMed] [Google Scholar]
- Chou WC, Dang CV. Acute promyelocytic leukemia: recent advances in therapy and molecular basis of response to arsenic therapies. Curr Opin Hematol. 2005;12:1–6. doi: 10.1097/01.moh.0000148552.93303.45. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–55. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The procedures were as the same as figue 1C.
HeLa cells were transfected with RGS-SENP3 for 48 h.