Abstract
Purpose
To investigate associations between computed tomography (CT) features of clear-cell renal cell carcinoma (ccRCC) and mutations in VHL, PBRM1, SETD2, KDM5C or BAP1 genes.
Materials and Methods
The institutional review board approved this retrospective, hypotheses-generating study of 233 patients with ccRCC and waived the informed consent requirement. The study was HIPAA compliant. Three radiologists independently reviewed pre-treatment CT images of all ccRCC without knowledge of their genomic profile. One radiologist measured largest diameter and enhancement parameters of each ccRCC. Associations between CT features and mutations in VHL, PBRM1, SETD2, KDM5C and BAP1 genes were tested using Fisher’s exact tests. Associations between mutations and size/enhancement were assessed using independent t-tests. Interreader agreements were calculated using Fleiss’ Kappa.
Results
Mutation frequencies among ccRCC were: VHL, 53.2% (124/233); PBRM1, 28.8% (67/233); SETD2, 7.3% (17/233); KDM5C, 6.9% (16/233); BAP1, 6% (14/233). Mutations of VHL were significantly associated with well-defined tumor margins (p=0.013), nodular tumor enhancement (p=0.021) and gross appearance of intratumoral vascularity (p=0.018). Mutations of KDM5C and BAP1 were significantly associated with evidence of renal vein invasion (p=0.022 and 0.046, respectively). The genotype of solid ccRCC differed significantly from the one of multicystic ccRCC. While mutations of SETD2, KDM5C and BAP1 were absent in multicystic ccRCC, mutations of VHL (p=0.016) and PBRM1 (p=0.017) were significantly more common among solid ccRCC. Interreader agreements for CT feature assessments ranged from substantial to excellent (κ=0.791–0.912).
Conclusion
This preliminary Radiogenomics analysis of ccRCC revealed associations between CT features and underlying mutations which warrant further investigation and validation.
INTRODUCTION
The genomic landscape of clear-cell renal cell carcinoma (ccRCC) was long thought to be dominated by the mutation of the von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase (VHL) gene located on the short arm of chromosome 3 (3p25). Loss of VHL function has up-regulating effects on hypoxia inducible factors, which play a key role in triggering neo-angiogenic activity of ccRCC. Recent advances in whole genome sequencing of ccRCC have led to the identification of the following histone modifying and chromatin remodeling gene mutations: polybromo 1 (PBRM1), SET domain containing 2 (SETD2), lysine (K)-specific demethylase 5C (KDM5C), and BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (BAP1) (1–5). While KDM5C is part of the short arm of chromosome X (Xp11), PBRM1, SETD2, and BAP1 are located on the short arm of chromosome 3 (3p21) - in close proximity to VHL. Mutations of VHL, PBRM1, BAP1, SETD2 and KDM5C were recently found to be associated with advanced stage, advanced grade, and possibly worse cancer specific survival (6, 7).
Diagnostic imaging of RCC is primarily based on tumor detection, cytological subtype characterization, definition of location and extent, and treatment response assessment. Computed tomography (CT), by its potential to fulfill these tasks (8), continues to contribute to clinical decision-making and serves as the primary basis for staging and treatment response assessment (9, 10). However, as the diagnostic standard of reference is rapidly expanding to the genomic level, the role of CT in ccRCC needs to be refined. In the near future, demonstrating the presence, location, and extent of ccRCC may not be sufficient when challenged by critical questions of which molecular drug to apply, which patients to select for active surveillance, or whether early response to treatment is evident or not (11). For an integrated diagnostic approach between Radiology and Genomics, the term “Radiogenomics” has been established (12, 13).
The critical first step of testing the robustness of Radiogenomics embodies the establishment of predictable and systematic associations between imaging features and underlying molecular and genomic alterations of ccRCC. A recent study investigated a similar approach in lung cancer and discovered significant associations between imaging features and mutations (14). Another recent study investigated magnetic resonance imaging (MRI) features of breast cancer in correlation to underlying mutations and identified 21 imaging traits that were globally correlated with multiple, recently discovered breast cancer mutations (15). In another study, cerebral edema and cellular invasion caused by glioblastoma and illustrated by MRI had been linked to a specific up-regulated gene (16). In ccRCC, one study had been published on the investigation of associations between CT features of 58 ccRCC and the underlying karyotype (17). However, so far no study has investigated associations between CT features of ccRCC and the underlying genotype.
The objective of this hypotheses-generating Radiogenomics study was to investigate associations between CT features of ccRCC and mutations of the genes VHL, PBRM1, SETD2, KDM5C, and BAP1.
MATERIALS AND METHODS
Patients
The institutional review board approved this retrospective study and waived the requirement for informed consent regarding the acquisition of CT data. However, all patients provided written informed consent for their ccRCC tissue to be used for genome sequencing. The study was compliant with the health insurance portability and accountability act (HIPAA).
Patients were included in this study upon fulfillment of the following criteria:
Histopathological diagnosis of ccRCC and genome sequencing including information on mutations of the chromatin modifying genes PBRM1, SETD2, BAP1, KDM5C and VHL, which have been considered the most frequent mutations in ccRCC (6).
Availability of a pre-treatment, contrast-enhanced CT study through our institution’s picture archiving and communications system (PACS), either in digital imaging and communications in medicine (DICOM) format or as scanned film prints.
Information regarding the mutations of PBRM1, SETD2, BAP1, KDM5C and VHL were available for 289 ccRCC from two distinct cohorts (i.e. institutional cohort and TCGA cohort). We were able to retrieve pre-treatment contrast-enhanced CT studies for 80.6% (233/289) of patients. 6.4% (15/233) were scanned film prints and 93.6% (218/233) were available in DICOM format. Of the CT studies performed at our institution, 79.6% (121/152) had been acquired using our institutional, tri-phasic kidney protocol consisting of a non-contrast-enhanced data acquisition and contrast-enhanced acquisitions during the nephrographic and excretory phases. Demographic and tumor characteristics of all 233 patients are summarized in Table 1.
Table 1. Demographics and mutations.
Patient demographics and mutational information.
| Gender | Female | 27.5% (64/233) |
| Male | 72.5% (169/233) | |
| Clinical Presentation | Incidental | 75.5% (176/233) |
| Localized | 19.3% (45/233) | |
| Systemic | 4.3% (10/233) | |
| Unknown | 0.9% (2/233) | |
| Type of Surgery | Radical | 41.2% (96/233) |
| Partial | 58.8% (137/233) | |
| Side | Left | 49.4% (115/233) |
| Right | 50.6% (118/233) | |
| Localized clear-cell RCC | no | 46.4% (108/233) |
| yes | 53.6% (125/233) | |
| Fuhrman Grade | 1 | 0.4% (1/233) |
| 2 | 41.6% (97/233) | |
| 3 | 48.5% (113/233) | |
| 4 | 9.4% (22/233) | |
| Mutations | VHL | 53.2% (124/233) |
| PBRM1 | 28.8% (67/233) | |
| SETD2 | 7.3% (17/233) | |
| BAP1 | 6% (14/233) | |
| KDM5C | 6.9% (16/233) |
Select gene sequencing and identification of mutations
Mutation information for the 233 ccRCC, for whom we were able to retrieve contrast-enhanced CT studies, was retrieved from The Cancer Genome Atlas (TCGA) web portal for 34.3% (80/233) of cases, and from a distinct cohort sequenced at our institution for 65.7% (153/233) of ccRCC. Mutation analyses of the entire coding regions of VHL, PBRM1, SETD2, BAP1, and KDM5C for 65.7% (153/233) of ccRCC were performed at our institution using polymerase chain reaction amplification and bidirectional Sanger sequencing, as previously described by Hakimi et al. (6). For the remaining 80 cases, mutation data was acquired from our institution’s contribution to the TCGA ccRCC project. Non-silent, coding mutations were considered for both cohorts, with truncating mutations defined as nonsense, frameshift, or essential splice site (within first 2 base-pairs of coding region).
CT image acquisition and analysis
65.2% (152/233) of pre-treatment, contrast-enhanced CT studies had been performed at our institution and 34.8% (81/233) at outside institutions. Through our PACS, 81.5% (66/81) of outside CT studies were available as complete DICOM data sets and 18.5% (15/81) as scanned film prints. Regarding the CT studies from our institution, 79.6% (121/152) had been performed using a dedicated renal mass protocol consisting of a non-enhanced data acquisition and data acquisitions during the nephrographic (delay, 90 sec) and excretory (delay, 3 minutes) phase after the application of 150 mL of iodinated contrast agent at a constant flow rate of 3.5 mL/sec. At identical scan and contrast material parameters, 21.4% (31/152) of CT studies from our institution had been acquired during the nephrographic phase only.
For the qualitative ccRCC feature analysis, we used all available contrast-enhanced CT studies (n=233). While we performed tumor size measurements only on contrast-enhanced CT images available in DICOM format (n=218), tumor enhancement measurements were performed exclusively on CT studies from our institution that had been acquired using the dedicated renal mass protocol (n=121) in order to avoid inhomogeneity in scan parameters.
Three radiologists with different degrees of experience in interpreting genitourinary CT images independently performed all qualitative image analyses. One radiologist was an assistant attending with 4 years of experience (__), the other two radiologists were research fellows with 5 years (__) and 4 years (__) of experience. All three radiologists analyzed all CT studies without access to genomic data. All three radiologists were aware that each patient had at least one ccRCC. After each Radiologist had finished the analysis, a consensus was established for each qualitative CT feature as follows: If there was disagreement between the readers, the matching results of two readers were chosen for further analysis. This consensus was performed for all features and in all patients.
The following eight qualitative features of ccRCC were analyzed by all readers (Figure 1): (I) necrosis, defined as either the presence or absence of areas within the tumor that did not demonstrate contrast enhancement during the nephrographic and delayed phase; (II) presence or absence of intratumoral calcifications; (III) definition of tumor margin as either ill- or well-defined; (IV) definition of tumor architecture as either solid or multicystic; (V) absence or presence of collecting system invasion, defined as evidence of a filling defect on excretory phase images; (VI) absence or presence of renal vein invasion, defined as evidence of a filling defect within the renal vein or its branches on nephrographic-phase images; (VII) gross evidence of intratumoral vascularity on nephrographic-phase CT images; (VIII) definition of tumor enhancement pattern on nephrographic-phase images as either homogeneous or nodular.
Figure 1.
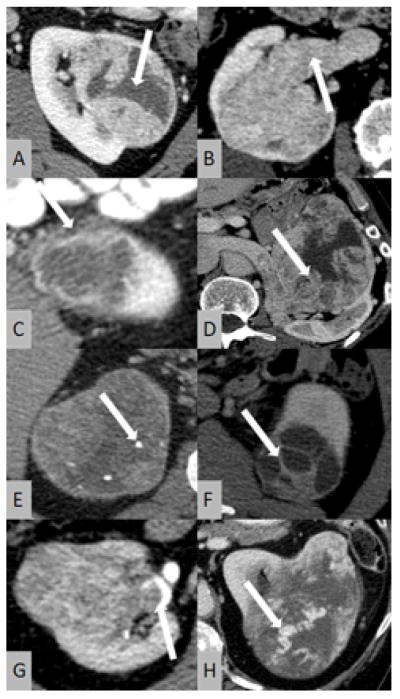
Illustration of CT features of clear-cell renal cell carcinoma (ccRCC) investigated in this study: (A) necrosis; (B) renal vein invasion; (C) ill-defined margin; (D) nodular enhancement; (E) calcifications; (F) multicystic architecture; (G) collecting system invasion; (H) gross appearance of intratumoral vasculature.
In addition to the qualitative feature analysis, one radiologist measured the greatest diameter of each ccRCC on transverse nephrographic-phase images and the enhancement within the most enhancing component of each ccRCC and within the renal cortex on transverse images during the non-enhanced, nephrographic and excretory, contrast-enhanced phases, and noted the average enhancement from region of interest analysis (ROI) in Hounsfield units (HU). This enhancement analysis was performed only for the CT studies performed at our institution using a dedicated renal mass protocol. Nephrographic-phase enhancement was calculated as the percentage increase between non-enhanced and nephrographic-phase, contrast-enhanced HU measurements for the renal cortex and the ccRCC. The percentage difference in enhancement between the ccRCC and the renal cortex were calculated. The percentage difference in enhancement of ccRCC between the nephrographic and excretory, contrast enhanced phases was calculated.
Statistical analysis
To assess interreader agreements regarding qualitative feature analyses between the three readers, Fleiss’ Kappa was calculated separately for each feature and interpreted as follows: < 0.20, poor agreement; 0.20–0.40, fair agreement; 0.40–0.60, moderate agreement; 0.60–0.80, substantial agreement; and 0.80–1.00, excellent agreement. Fisher’s exact tests were performed to assess for significant differences in distribution of each qualitative CT feature among each mutation (i.e. VHL, PBRM1, BAP1, SETD2 and KDM5C). To assess for significant differences in quantitative features (enhancement and size parameters) among each mutation, independent t-tests were performed. Relative risk (including 95% confidence intervals) for the evidence of each CT imaging feature was calculated for each mutation.
Two-sided P values of less than .05 were considered to indicate statistically significant differences. All statistical analyses were performed by using commercially available statistics software (SPSS, version 19; IBM, Armonk, NY).
RESULTS
Demographic and tumor characteristics of all 233 patients are summarized in Table 1.
Frequency of mutations
Mutation of VHL was identified in 53.2% (124/233) of ccRCC, followed by mutations of PBRM1 (28.8% [67/233]), SETD2 (7.3% ([17/233]), KDM5C (6.9% [16/233]), and BAP1 (6% [14/233]) (Figure 2).
Figure 2.
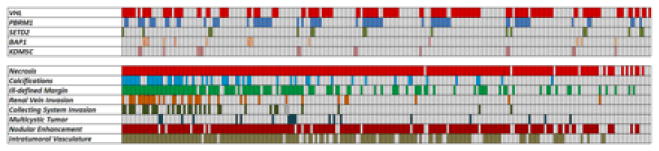
Subject-based color map illustrating mutations and CT features.
Qualitative CT Feature Analysis
Frequencies of all CT features per mutation, as illustrated in Figures 2 and 3, are summarized in Table 2. Interreader agreements for the assessment of qualitative tumor features on CT images ranged from substantial to excellent (κ=0.791–0.912).
Table 2. CT features and mutations.
Summary of CT features per mutation including p-values from fisher’s exact tests and Fleiss’ kappa values from interreader agreement analyses.
| VHL | PBRM1 | SETD2 | BAP1 | KDM5C | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | − | + | p | − | + | p | − | + | p | − | + | p | − | + | p | κ | ||
| Necrosis | − | 17 | 58.8% 10/17 |
41.2% 7/17 |
0.324 | 88.2% 15/17 |
11.8% 2/17 |
0.163 | 82.4% 14/17 |
17.6% 3/17 |
0.116 | 94.1% 16/17 |
5.9% 1/17 |
1.000 | 94.1% 16/17 |
5.9% 1/17 |
1.000 | 0.820 |
| + | 216 | 45.8% 99/216 |
54.2% 117/216 |
69.9% 151/216 |
30.1% 65/216 |
93.5% 202/216 |
6.5% 14/216 |
94% 203/216 |
6% 13/216 |
93.1% 201/216 |
6.9% 15/216 |
|||||||
| Calcifications | − | 181 | 48.6% 88/181 |
51.4% 93/181 |
0.345 | 69.6% 126/181 |
30.4% 55/181 |
0.385 | 92.3% 167/181 |
7.7% 14/181 |
0.770 | 95.6% 173/181 |
4.4% 8/181 |
0.090 | 92.3% 167/181 |
7.7% 14/181 |
0.534 | 0.899 |
| + | 52 | 40.4% 21/52 |
59.6% 31/52 |
76.9% 40/52 |
23.1% 12/52 |
94.2% 49/52 |
5.8% 3/52 |
88.5% 46/52 |
11.5% 6/52 |
96.2% 50/52 |
3.8% 2/52 |
|||||||
| Ill-defined Margin | − | 118 | 38.1% 45/118 |
61.9% 73/118 |
0.009 | 71.2% 84/118 |
28.8% 34/118 |
1.000 | 93.2% 110/118 |
6.8% 8/118 |
0.805 | 96.6% 114/118 |
3.4% 4/118 |
0.104 | 94.9% 112/118 |
5.1% 6/118 |
0.310 | 0.839 |
| + | 115 | 55.7% 64/115 |
44.3% 51/115 |
71.3% 82/115 |
28.7% 33/115 |
92.2% 106/115 |
7.8% 9/115 |
91.3% 105/115 |
8.7% 10/115 |
91.3% 105/115 |
8.7% 10/115 |
|||||||
| Renal Vein Invasion | − | 198 | 47% 93/198 |
53% 105/198 |
1.000 | 71.2% 141/198 |
28.8% 57/198 |
1.000 | 92.9% 184/198 |
7.1% 14/198 |
0.726 | 95.5% 189/198 |
4.5% 9/198 |
0.042 | 94.9% 188/198 |
5.1% 10/198 |
0.019 | 0.866 |
| + | 35 | 45.7% 16/35 |
54.3% 19/35 |
71.4% 25/35 |
28.6% 10/35 |
91.4% 32/35 |
8.6% 3/35 |
85.7% 30/35 |
14.3% 5/35 |
82.9% 29/35 |
17.1% 6/35 |
|||||||
| Collecting System Invasion | − | 201 | 47.8% 96/201 |
52.2% 105/201 |
0.568 | 72.1% 145/201 |
27.9% 56/201 |
0.528 | 93.5% 188/201 |
6.5% 13/201 |
0.264 | 94.5% 190/201 |
5.5% 11/201 |
0.417 | 92.5% 186/201 |
7.5% 15/201 |
0.705 | 0.868 |
| + | 32 | 40.6% 13/32 |
59.4% 19/32 |
65.6% 21/32 |
34.4% 11/32 |
87.5% 28/32 |
12.5% 4/32 |
90.6% 29/32 |
9.4% 3/32 |
96.9% 31/32 |
3.1% 1/32 |
|||||||
| Multicystic Tumor | − | 214 | 44.4% 95/214 |
55.6% 119/214 |
0.017 | 69.2% 148/214 |
30.8% 66/214 |
0.017 | 92.1% 197/214 |
7.9% 17/214 |
0.373 | 93.5% 200/214 |
6.5% 14/214 |
0.612 | 92.5% 198/214 |
7.5% 16/214 |
0.375 | 0.815 |
| + | 19 | 73.7% 14/19 |
26.3% 5/19 |
94.7% 18/19 |
5.3% 1/19 |
100% 19/19 |
0% 0/19 |
100% 19/19 |
0% 0/19 |
100% 19/19 |
0% 0/19 |
|||||||
| Nodular Tumor Enhancement | − | 44 | 63.6% 28/44 |
36.4% 16/44 |
0.018 | 81.8% 36/44 |
18.2% 8/44 |
0.098 | 86.4% 38/44 |
13.6% 6/44 |
0.101 | 93.2% 41/44 |
6.8% 3/44 |
0.723 | 97.7% 43/44 |
2.3% 1/44 |
0.318 | 0.900 |
| + | 189 | 42.9% 81/189 |
57.1% 108/189 |
68.8% 130/189 |
31.2% 59/189 |
94.2% 178/189 |
5.8% 11/189 |
94.2% 178/189 |
5.8% 11/189 |
92.1% 174/189 |
7.9% 15/189 |
|||||||
| Intratumoral Vasculature | − | 78 | 35.9% 28/78 |
64.1% 50/78 |
0.026 | 74.4% 58/78 |
25.6% 20/78 |
0.540 | 89.7% 70/78 |
10.3% 8/78 |
0.285 | 97.4% 76/78 |
2.6% 2/78 |
0.150 | 89.7% 70/78 |
10.3% 8/78 |
0.173 | 0.886 |
| + | 155 | 52.3% 81/155 |
47.7% 74/155 |
69.7% 108/155 |
30.3% 47/155 |
94.2% 146/155 |
5.8% 9/155 |
92.3% 143/155 |
7.7% 12/155 |
94.8% 147/155 |
5.2% 8/155 |
|||||||
Radiogenomic Associations
Mutations of the VHL gene were significantly associated with the following phenotypic characteristics of clear-cell renal cell carcinoma (ccRCC) on contrast-enhanced computed tomography (CT): well-defined tumor margins (p=0.013), nodular tumor enhancement (p=0.021) and gross appearance of intratumoral vascularity (p=0.018).
Mutations of the KDM5C and BAP1 genes were significantly associated with evidence of renal vein invasion on contrast-enhanced CT of ccRCC (p=0.022 and 0.046, respectively) The genotype of solid ccRCC differed significantly from the one of multicystic ccRCC. While mutations of SETD2, KDM5C and BAP1 were completely absent in multicystic ccRCC, mutations of VHL (p=0.016) and PBRM1 (p=0.017) were significantly more common among solid ccRCC. SETD2 mutations were not significantly associated with any qualitative CT feature investigated. Radiogenomic associations are summarized in Table 2 and illustrated in Supplementary Figure 3. Results from all risk analyses are presented in Table 3.
Table 3.
Results from risk analyses (odds ratios, 95% confidence intervals in parentheses);
| VHL | PBRM1 | SETD2 | BAP1 | KDM5C | |
|---|---|---|---|---|---|
| Necrosis | 1.69 (0.62–4.60) | 3.29 (0.72–14.52) | 0.32 (0.08–1.26) | 1.03 (0.13–8.34) | 1.19 (0.15–9.63) |
| Calcifications | 1.40 (0.75–2.61) | 0.69 (0.34–1.41) | 0.73 (0.20–2.65) | 2.82 (0.93–8.54) | 0.48 (0.11–2.17) |
| Ill-defined margin | 0.49 (0.29–0.83) | 0.99 (0.56–1.75) | 1.17 (0.43–3.14) | 2.71 (0.83–8.92) | 1.78 (0.62–5.06) |
| Renal vein invasion | 1.05 (0.51–2.16) | 0.99 (0.45–2.19) | 1.23 (0.34–4.53) | 3.50 (1.09–11.16) | 3.89 (1.31–11.51) |
| Collecting system invasion | 1.34 (0.63–2.85) | 1.36 (0.61–2.99) | 2.07 (0.63–6.78) | 1.79 (0.47–6.79) | 0.40 (0.05–3.14) |
| Multicystic tumor | 0.28 (0.09–0.82) | 0.12 (0.01–0.95) | n/a | n/a | n/a |
| Nodular enhancement | 2.33 (1.18–4.60) | 2.04 (0.89–4.66) | 0.39 (0.14–1.12) | 0.85 (0.23–3.16) | 3.71 (0.48–28.84) |
| Intratumoral vasculature | 0.51 (0.29–0.89) | 1.26 (0.68–2.33) | 0.54 (0.20–1.46) | 3.19 (0.69–14.62) | 0.48 (0.17–1.32) |
Quantitative Feature Analysis
In the presence of a mutation of KDM5C, ccRCC demonstrated significantly lower contrast enhancement during the nephrographic phase when compared to the renal cortex (p=0.030) (Table 4). No other associations between enhancement parameters and mutations were discovered. Tumor size, as measured on contrast-enhanced CT images, was not associated with any of the investigated mutations (Table 4).
Table 4.
Associations between mutations and CT enhancement parameters
| Tumor1 | Renal Cortex2 | %Difference3 | Tumor Washout4 | Tumor Size5 | ||
|---|---|---|---|---|---|---|
| VHL | − | 336 ± 222 (94–1682) | 487 ± 125 (227–791) | 49 ± 33 (−73–124) | −25 ± 11 (−47–9) | 5.7 (1.5–139) |
| + | 339 ± 249 (91–1710) | 498 ± 150 (235–1081) | 48 ± 19 (11–107) | −26 ± 12 (−56–4) | 5.9 (1.4–16.3) | |
| p | 0.632 | 0.815 | 0.825 | 0.772 | 0.707 | |
| PBRM1 | − | 323 ± 213 (91–1682) | 493 ± 137 (235–1081) | 48 ± 28 (−73–124) | −25 ± 11 (−47–9) | 5.8 (1.4–16.3) |
| + | 374 ± 279 (134–1710) | 489 ± 139 (227–757) | 49 ± 26 (−40–107) | −26 ± 12 (−56–4) | 5.9 (2.3–13.6) | |
| p | 0.301 | 0.941 | 0.496 | 0.933 | 0.726 | |
| SETD2 | − | 339 ± 238 (91–1710) | 490 ± 127 (227–791) | 48 ± 28 (−73–124) | −25 ± 12 (−56–9) | 5.8 (1.5–16.3) |
| + | 313 ± 148 (184–605) | 530 ± 281 (333–1081) | 56 ± 13 (43–80) | −29 ± 8 (−38–−17) | 5.8 1.4–12.8) | |
| p | 0.844 | 0.659 | 0.249 | 0.459 | 0.954 | |
| BAP1 | − | 343 ± 237 (94–1710) | 495 ± 138 (227–1081) | 49 ± 28 (−73–124) | −26 ± 12 (−56–9) | 5.7 (1.4–16.3) |
| + | 208 ± 101 (91–352) | 416 ± 98 (326–576) | 40 ± 10 (22–49) | −21 ± 14 (−34–0) | 7.1 (2.0–12.1) | |
| p | 0.086 | 0.176 | 0.405 | 0.499 | 0.093 | |
| KDM5C | − | 339 ± 240 (94–1710) | 493 ± 138 (227–1081) | 50 ± 28 (−73–124) | −25 ± 12 (−56–9) | 5.8 (1.4–16.3) |
| + | 320 ± 150 (91–551) | 483 ± 131 (268–656) | 31 ± 18 (0–57) | −31 ± 8 (−42–−15) | 6.5 (.4–12.1) | |
| p | 0.722 | 0.992 | 0.030 | 0.177 | 0.371 |
tumor and renal cortex enhancement during the nephrographic CT phase in Hounsfield Units (mean ± standard deviation [range]).
percentage difference between tumor and renal cortex enhancement during the nephrographic CT phase (mean ± standard deviation [range]).
percentage difference in tumor enhancement between the nephrographic and delayed CT phase (mean ± standard deviation [range]).
mean tumor size as measured on CT images in cm (range)
DISCUSSION
The investigation of individual associations between diagnostic imaging features and mutations is considered the critical first step of Radiogenomics of ccRCC (11). While the genomic landscape of ccRCC has long been dominated by the loss of VHL function, recent advances in cancer genome sequencing of ccRCC have led to the identification of additional, prognostically significant mutations in ccRCC. This is the first hypotheses-generating study of potential associations between individual CT features of ccRCC and mutations of the genes VHL, PBRM1, BAP1, SETD2, and KDM5C. This study yielded the following results:
Well-defined tumor margins were significantly more common among ccRCC with loss of VHL function. Well-defined tumor margins are considered an indicator of less infiltrative behavior and thus lower aggressiveness of ccRCC when compared to ccRCC with ill-defined margins (18). This observation therefore warrants further investigation and validation, because well-defined margins of ccRCC on CT imaging may indicate a loss of VHL function.
Nodular, heterogeneous enhancement of ccRCC and evidence of intratumoral vasculature on contrast-enhanced CT images were significantly more common among ccRCC with underlying VHL mutations. This may be explained by the fact, that a loss of VHL function is associated with an up-regulation of hypoxia inducible factors and an overexpression of angiogenic factors such as vascular endothelial growth factor. This angiogenic activity may thus be reflected on contrast-enhanced CT images by evidence of nodular, heterogeneous enhancement and visibility of intratumoral blood vessels. This finding should be further investigated and validated using standardized CT imaging in larger cohorts.
Another finding of this study was that ccRCC with mutations of BAP1 and KDM5C were associated with an increased evidence of renal vein invasion. This observation supports previously published reports demonstrating that mutations of PBRM1, SETD2, BAP1, and/or KDM5C in ccRCC were associated with advanced stage, grade, and tumor invasiveness (6, 7).
Solid ccRCC were associated with a substantially different genotype when compared to multicystic ccRCC. While mutations of VHL and PBRM1 were more common among solid ccRCC, mutations of SETD2, KDM5C and BAP1 were absent in multicystic ccRCC. This finding supports a recently published study that multicystic ccRCC in fact represent less aggressive ccRCC of relatively low malignant potential (19). However, due to the small sample size of multicystic ccRCC in our study (n=19), this observation warrants further validation in a larger cohort.
This study had the following limitations: First, it is a discovery-phase study without validation of the findings. However, we discovered associations between mutations and CT imaging features of ccRCC, which may assist in the selection of features for future studies. We did not correct for multiple hypothesis testing, which may be regarded as an additional limitation of this study. However, this study was primarily executed as a preliminary, discovery-phase analysis of Radiogenomic associations in ccRCC. Another limitation of this study was that the CT studies had been performed at different institutions using different CT scanners. However, this was a retrospective analysis in which we aimed to keep the sample size as lage as possible. In addition, we only used CT studies from our institution, which had been performed using a dedicated renal mass protocol, for enhancement analyses. In addition, the amount of contrast material applied to each patient was not weight corrected, therefore CT features associated with enhancement could be affected due to the different weight based doses that were administered. Regarding the genomic findings, our data was recorded as mutations or no mutations. This may be considered a limitation, because genes may loose their function through copy number errors or epigenetic defects as well. However, this is thought to be rather limited to VHL, with a methylation rate of 5–10%. Data from the TCGA project suggest that none of the other genes investigated in this study were methylated or homozygously deleted. Furthermore, ccRCC frequently exhibit varying amounts of intratumoral heterogeneity, which may substantially alter the genomic landscape within ccRCC (20). That is, ccRCC frequently consist of several genetically different components that may be illustrated as a whole by CT, but represent a challenge for tissue sampling for whole genome sequencing. Therefore, imaging may be useful to guide tissue sampling in the future in order to ensure that the most aggressive or prognostically relevant tumor component is being chosen for the acquisition of tissue for genomic sequencing.
In summary, this preliminary Radiogenomics analysis of ccRCC revealed associations between CT features and underlying mutations and therefore warrants further investigation and validation. Moreover, future studies are necessary to combine individual CT features into CT imaging phenotypes of ccRCC and correlate them to underlying genomic profiles rather than individual gene mutations.
Supplementary Material
Advances in knowledge.
Mutations of the VHL gene were significantly associated with the following phenotypic characteristics of clear-cell renal cell carcinoma (ccRCC) on contrast-enhanced computed tomography (CT): well-defined tumor margins (p=0.013), nodular tumor enhancement (p=0.021) and gross appearance of intratumoral vascularity (p=0.018).
Mutations of the KDM5C and BAP1 genes were significantly associated with evidence of renal vein invasion on contrast-enhanced CT of ccRCC (p=0.022 and 0.046, respectively)
The genotype of solid ccRCC differed significantly from the one of multicystic ccRCC. While mutations of SETD2, KDM5C and BAP1 were completely absent in multicystic ccRCC, mutations of VHL (p=0.016) and PBRM1 (p=0.017) were significantly more common among solid ccRCC.
Footnotes
Implications for Patient Care:
n/a
References
- 1.Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–3. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duns G, van den Berg E, van Duivenbode I, et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer research. 2010;70(11):4287–91. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 3.Guo G, Gui Y, Gao S, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nature genetics. 2012;44(1):17–9. doi: 10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- 4.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nature genetics. 2012;44(7):751–9. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–42. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hakimi AA, Chen YB, Wren J, et al. Clinical and Pathologic Impact of Select Chromatin-modulating Tumor Suppressors in Clear Cell Renal Cell Carcinoma. European urology. 2013;63(5):848–54. doi: 10.1016/j.eururo.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapur P, Pena-Llopis S, Christie A, et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. The lancet oncology. 2013;14(2):159–67. doi: 10.1016/S1470-2045(12)70584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reznek RH. CT/MRI in staging renal cell carcinoma. Cancer imaging : the official publication of the International Cancer Imaging Society. 2004;4(Spec No A):S25–32. doi: 10.1102/1470-7330.2004.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powles T, Albers P. Management of favorable-risk patients with metastatic renal cell carcinoma: when to start and when to stop targeted therapy. Clinical genitourinary cancer. 2012;10(4):213–8. doi: 10.1016/j.clgc.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Carles J, Chirivella I, Climent MA, et al. Evaluation of patients with metastatic renal cell carcinoma after failure of first-line treatment. Cancer metastasis reviews. 2012;31 (Suppl 1):S3–9. doi: 10.1007/s10555-012-9353-0. [DOI] [PubMed] [Google Scholar]
- 11.Kuo MD, Yamamoto S. Next generation radiologic-pathologic correlation in oncology: Rad-Path 2.0. AJR American journal of roentgenology. 2011;197(4):990–7. doi: 10.2214/AJR.11.7163. [DOI] [PubMed] [Google Scholar]
- 12.Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. European journal of radiology. 2009;70(2):232–41. doi: 10.1016/j.ejrad.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 13.Jaffe CC. Imaging and genomics: is there a synergy? Radiology. 2012;264(2):329–31. doi: 10.1148/radiol.12120871. [DOI] [PubMed] [Google Scholar]
- 14.Gevaert O, Xu J, Hoang CD, et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology. 2012;264(2):387–96. doi: 10.1148/radiol.12111607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto S, Maki DD, Korn RL, Kuo MD. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. AJR American journal of roentgenology. 2012;199(3):654–63. doi: 10.2214/AJR.11.7824. [DOI] [PubMed] [Google Scholar]
- 16.Zinn PO, Mahajan B, Sathyan P, et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PloS one. 2011;6(10):e25451. doi: 10.1371/journal.pone.0025451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauk SC, Hsu MS, Margolis DJ, et al. Clear cell renal cell carcinoma: multiphasic multidetector CT imaging features help predict genetic karyotypes. Radiology. 2011;261(3):854–62. doi: 10.1148/radiol.11101508. [DOI] [PubMed] [Google Scholar]
- 18.Ro JY, Ayala AG, Sella A, Samuels ML, Swanson DA. Sarcomatoid renal cell carcinoma: clinicopathologic. A study of 42 cases. Cancer. 1987;59(3):516–26. doi: 10.1002/1097-0142(19870201)59:3<516::aid-cncr2820590327>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 19.Hindman NM, Bosniak MA, Rosenkrantz AB, Lee-Felker S, Melamed J. Multilocular cystic renal cell carcinoma: comparison of imaging and pathologic findings. AJR American journal of roentgenology. 2012;198(1):W20–6. doi: 10.2214/AJR.11.6762. [DOI] [PubMed] [Google Scholar]
- 20.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366(10):883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


