Abstract
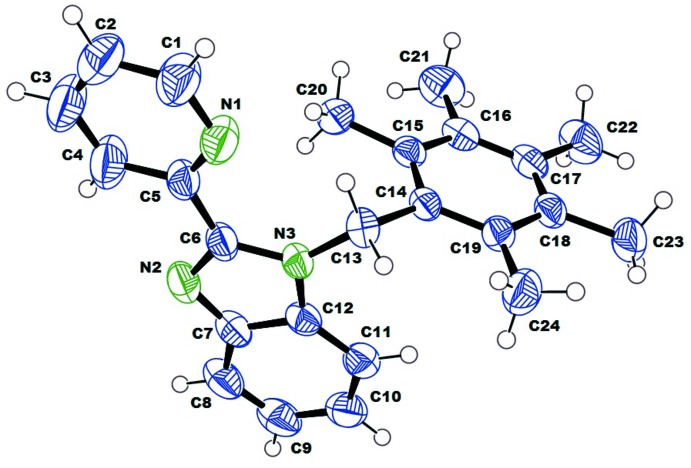
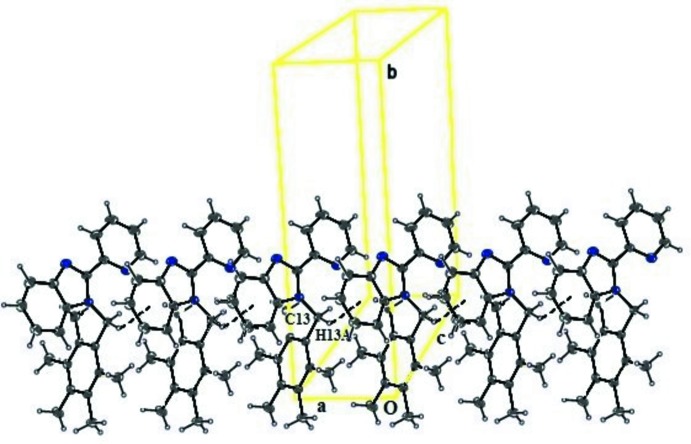
In the title compound, C24H25N3, the benzimidazole ring system is essentially planar, with an r.m.s. deviation of 0.017 Å, and forms dihedral angles of 7.81 (5) and 87.61 (4)° with the pyridine and benzene rings, respectively. An intramolecular C—H⋯N hydrogen bond is observed. In the crystal, molecules are stacked along the a axis by weak C—H⋯π interactions.
Related literature
For the use of 2-(2-pyridyl)benzimidazole in coordination chemistry, see: Sahin et al. (2010 ▶); Harkins et al. (1956 ▶); Chiswell et al. (1964 ▶); De Castro et al. (1991 ▶); Maekawa et al. (1994 ▶); Khalil et al. (2001 ▶); Boca et al. (1997 ▶). For the use of N—N-type ligand systems involving 2,2′-bipyridine, see: Lippert (1999 ▶); Wong & Giandomenico (1999 ▶), Kelland & Farrell (2000 ▶). For related structures, see: Çelik et al. (2007 ▶, 2009 ▶, 2014 ▶).
Experimental
Crystal data
C24H25N3
M r = 355.47
Monoclinic,
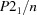
a = 5.3470 (3) Å
b = 21.0622 (12) Å
c = 17.0379 (9) Å
β = 97.699 (3)°
V = 1901.50 (18) Å3
Z = 4
Mo Kα radiation
μ = 0.07 mm−1
T = 296 K
0.25 × 0.20 × 0.15 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (Blessing, 1995 ▶) T min = 0.982, T max = 0.989
25354 measured reflections
3741 independent reflections
3056 reflections with I > 2σ(I)
R int = 0.026
Refinement
R[F 2 > 2σ(F 2)] = 0.052
wR(F 2) = 0.150
S = 1.04
3741 reflections
254 parameters
H-atom parameters constrained
Δρmax = 0.24 e Å−3
Δρmin = −0.17 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL2013 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536814007934/rz5115sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814007934/rz5115Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814007934/rz5115Isup3.cml
CCDC reference: 996309
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg is the centroid of the C7–C12 benzene ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C13—H13A⋯Cg i | 0.97 | 2.91 | 3.6941 (18) | 139 |
| C13—H13B⋯N1 | 0.97 | 2.30 | 3.029 (2) | 131 |
Symmetry code: (i)  .
.
Acknowledgments
The authors are indebted to the X-ray laboratory of Dicle University Science and the Technology Application and Research Center, Diyarbakir, Turkey, for use of the X-ray diffractometer.
supplementary crystallographic information
1. Comment
The N—N type ligand system 2-(2-pyridyl)benzimidazole has a venerable history in coordination chemistry (Sahin et al., 2010; Harkins et al., 1956; Chiswell et al., 1964; De Castro et al., 1991; Maekawa et al., 1994; Khalil et al., 2001; Boca et al., 1997). Lots of platinum chemistry bearing N—N type ligand systems involving 2,2'-bipyridine has been reported aimed at the design of drugs having less serious side-effects than those of cisplatin, or which could extend the scope of Pt based chemotherapy to tumours (Lippert, 1999; Wong & Giandomenico, 1999; Kelland & Farrell, 2000).
The molecular structure of the title compound is shown in Fig. 1. Bond lengths and angles are in good agreement with those reported for related structures (Çelik et al., 2007; Çelik et al., 2009; Çelik et al., 2014). The benzimidazole ring system is substantially planar, the maximum deviation being 0.027 (2) Å for atom C8, and forms dihedral angles of 7.81 (5) and 87.61 (4)° with the mean planes through the pyridine (N1/C1–C5) and phenyl (C14–C19) rings, respectively. An intramolecular C—H···N hydrogen bond is present (Table 1). In the crystal structure (Fig. 2), molecules are stacked along the a axis by weak C—H···π hydrogen interactions (Table 1) involving the C7—C12 benzene ring of the benzimidazole moiety.
2. Experimental
NaH (60%) (398 mg, 11.0 mmol) was washed two times with dry hexane, filtered off via cannula and a solution of the 2-pyridiylbenzimidazole (1.95 g, 10.0 mmol) in dry toluene (10 ml) was added, then the solution was heated at 90°C for 24 h. Evolution of hydrogen was observed at this temperature. 2,3,4,5,6-Pentamethylbenzyl bromide (2.45 g, 10.0 mmol) was added to this mixture at room temperature and then heated again at 90°C for 1 day. Then volatiles were evaporated in vacuum to dryness. The residue was dissolved in CH2Cl2 and filtered via cannula on celite. The desired product was obtained after concentration of CH2Cl2 (15 ml) and then precipitated with hexane (30 ml). The off-white solid obtained in 86% yield. M. p. 416–418 K. 1H NMR (400 MHz, CDCl3, δ p.p.m.): 1.26 (s, 6H, o-(CH3)2); 1.36(s, 6H, m-(CH3)2); 2.13–2.20 (s, 3H, p-CH3); 5.50 (s, 2H, N—CH2); 6.86 (s, 1H, Ar—CH); 7.11 (s, 1H, Ar—CH); 7.26 (s, 1H, Ar—CH); 7.35 (s, 1H, Ar—CH); 7.49 (s, 1H, Ar—CH); 8.46 (s, 1H, Ar—CH); 10.19 (s, 2H, Ar—CH). 13C NMR (100.56 MHz, CDCl3, δ p.p.m.): 17.0; 17.4; 29.6; 31.7; 34.4; 35.2; 48.7; 49.3; 56.2; 117.9; 121.2; 122.4; 125.5; 126.5; 133.8; 134.0; 137.5; 140.7; 158.0; 167.9.
3. Refinement
All H atoms were placed geometrically and refined using a riding model approximation, with C—H = 0.93–0.97 Å, and with Uiso(H) = 1.2 Ueq(C) or 1.5 Ueq(C) for methyl H atoms. A rotating group model was applied to the methyl groups.
Figures
Fig. 1.
ORTEP-3 of the title compound, showing displacement ellipsoids drawn at 50% probability level.
Fig. 2.
The stacking of the title compound along the a axis with C—H···π type hydrogen-bond interactions (dashed lines). Displacement ellipsoids are drawn at the 50% probability level.
Crystal data
| C24H25N3 | F(000) = 760 |
| Mr = 355.47 | Dx = 1.242 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 5.3470 (3) Å | Cell parameters from 1276 reflections |
| b = 21.0622 (12) Å | θ = 2.3–31.5° |
| c = 17.0379 (9) Å | µ = 0.07 mm−1 |
| β = 97.699 (3)° | T = 296 K |
| V = 1901.50 (18) Å3 | Prism, yellow |
| Z = 4 | 0.25 × 0.20 × 0.15 mm |
Data collection
| Bruker APEXII CCD diffractometer | 3056 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.026 |
| φ and ω scans | θmax = 26.0°, θmin = 2.3° |
| Absorption correction: multi-scan (Blessing, 1995) | h = −6→6 |
| Tmin = 0.982, Tmax = 0.989 | k = −25→25 |
| 25354 measured reflections | l = −21→20 |
| 3741 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.052 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.150 | H-atom parameters constrained |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0775P)2 + 0.5927P] where P = (Fo2 + 2Fc2)/3 |
| 3741 reflections | (Δ/σ)max < 0.001 |
| 254 parameters | Δρmax = 0.24 e Å−3 |
| 0 restraints | Δρmin = −0.17 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 1.0873 (5) | 0.86580 (11) | 0.44231 (14) | 0.0724 (6) | |
| H1 | 1.2174 | 0.8428 | 0.4246 | 0.087* | |
| C2 | 1.0060 (5) | 0.91952 (12) | 0.40152 (14) | 0.0737 (6) | |
| H2 | 1.0790 | 0.9327 | 0.3577 | 0.088* | |
| C3 | 0.8151 (5) | 0.95308 (12) | 0.42706 (16) | 0.0842 (7) | |
| H3 | 0.7548 | 0.9900 | 0.4011 | 0.101* | |
| C4 | 0.7133 (5) | 0.93151 (10) | 0.49165 (15) | 0.0771 (7) | |
| H4 | 0.5809 | 0.9535 | 0.5094 | 0.092* | |
| C5 | 0.8068 (3) | 0.87716 (7) | 0.53054 (11) | 0.0465 (4) | |
| C6 | 0.6926 (3) | 0.85653 (7) | 0.60059 (10) | 0.0452 (4) | |
| C7 | 0.4595 (4) | 0.86110 (8) | 0.69313 (11) | 0.0507 (4) | |
| C8 | 0.2891 (4) | 0.87790 (10) | 0.74483 (13) | 0.0658 (6) | |
| H8 | 0.2014 | 0.9161 | 0.7387 | 0.079* | |
| C9 | 0.2541 (4) | 0.83708 (10) | 0.80460 (13) | 0.0667 (6) | |
| H9 | 0.1414 | 0.8477 | 0.8396 | 0.080* | |
| C10 | 0.3850 (4) | 0.77962 (10) | 0.81392 (12) | 0.0598 (5) | |
| H10 | 0.3597 | 0.7531 | 0.8558 | 0.072* | |
| C11 | 0.5507 (4) | 0.76104 (9) | 0.76284 (10) | 0.0505 (4) | |
| H11 | 0.6360 | 0.7225 | 0.7690 | 0.061* | |
| C12 | 0.5841 (3) | 0.80251 (8) | 0.70185 (10) | 0.0432 (4) | |
| C13 | 0.8894 (3) | 0.74472 (7) | 0.62583 (10) | 0.0422 (4) | |
| H13A | 1.0264 | 0.7410 | 0.6691 | 0.051* | |
| H13B | 0.9632 | 0.7525 | 0.5777 | 0.051* | |
| C14 | 0.7459 (3) | 0.68215 (7) | 0.61734 (9) | 0.0370 (4) | |
| C15 | 0.5434 (3) | 0.67529 (7) | 0.55618 (9) | 0.0391 (4) | |
| C16 | 0.4047 (3) | 0.61901 (8) | 0.54888 (10) | 0.0452 (4) | |
| C17 | 0.4753 (3) | 0.56796 (8) | 0.59946 (11) | 0.0471 (4) | |
| C18 | 0.6838 (3) | 0.57318 (7) | 0.65801 (10) | 0.0448 (4) | |
| C19 | 0.8190 (3) | 0.63066 (7) | 0.66771 (10) | 0.0412 (4) | |
| C20 | 0.4783 (4) | 0.72858 (9) | 0.49769 (10) | 0.0511 (4) | |
| H20A | 0.406 (3) | 0.7111 (2) | 0.4472 (7) | 0.077* | |
| H20B | 0.630 (2) | 0.7521 (5) | 0.4912 (6) | 0.077* | |
| H20C | 0.357 (2) | 0.7567 (5) | 0.5172 (5) | 0.077* | |
| C21 | 0.1762 (4) | 0.61342 (11) | 0.48609 (14) | 0.0703 (6) | |
| H21A | 0.2254 (10) | 0.5965 (8) | 0.4399 (8) | 0.105* | |
| H21B | 0.106 (2) | 0.6535 (6) | 0.4754 (8) | 0.105* | |
| H21C | 0.057 (2) | 0.5868 (8) | 0.5043 (5) | 0.105* | |
| C22 | 0.3235 (5) | 0.50692 (10) | 0.59025 (15) | 0.0710 (6) | |
| H22A | 0.314 (3) | 0.4923 (5) | 0.5373 (9) | 0.107* | |
| H22B | 0.158 (3) | 0.5146 (2) | 0.6025 (10) | 0.107* | |
| H22C | 0.403 (2) | 0.4757 (5) | 0.6251 (9) | 0.107* | |
| C23 | 0.7616 (5) | 0.51661 (9) | 0.71064 (13) | 0.0655 (6) | |
| H23A | 0.626 (2) | 0.5047 (5) | 0.7383 (8) | 0.098* | |
| H23B | 0.904 (3) | 0.5276 (3) | 0.7476 (8) | 0.098* | |
| H23C | 0.803 (3) | 0.4820 (6) | 0.6790 (5) | 0.098* | |
| C24 | 1.0435 (4) | 0.63574 (10) | 0.73176 (13) | 0.0630 (5) | |
| H24A | 1.168 (2) | 0.6592 (7) | 0.7134 (4) | 0.094* | |
| H24B | 1.104 (2) | 0.5960 (6) | 0.7452 (7) | 0.094* | |
| H24C | 0.9950 (10) | 0.6551 (7) | 0.7754 (7) | 0.094* | |
| N1 | 0.9926 (3) | 0.84413 (8) | 0.50574 (10) | 0.0616 (4) | |
| N2 | 0.5315 (3) | 0.89400 (7) | 0.63031 (10) | 0.0561 (4) | |
| N3 | 0.7309 (3) | 0.79969 (6) | 0.64092 (8) | 0.0406 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0797 (15) | 0.0651 (13) | 0.0780 (14) | 0.0052 (11) | 0.0312 (12) | 0.0205 (11) |
| C2 | 0.0844 (16) | 0.0737 (14) | 0.0641 (13) | −0.0104 (12) | 0.0138 (12) | 0.0232 (11) |
| C3 | 0.1041 (19) | 0.0620 (14) | 0.0871 (17) | 0.0111 (13) | 0.0152 (15) | 0.0351 (13) |
| C4 | 0.0940 (17) | 0.0516 (12) | 0.0889 (16) | 0.0184 (11) | 0.0244 (14) | 0.0216 (11) |
| C5 | 0.0539 (10) | 0.0329 (8) | 0.0517 (10) | −0.0066 (7) | 0.0031 (8) | 0.0006 (7) |
| C6 | 0.0522 (10) | 0.0305 (8) | 0.0519 (10) | −0.0019 (7) | 0.0034 (8) | −0.0032 (7) |
| C7 | 0.0580 (11) | 0.0396 (9) | 0.0551 (10) | −0.0025 (8) | 0.0099 (9) | −0.0119 (8) |
| C8 | 0.0756 (14) | 0.0522 (11) | 0.0726 (13) | 0.0050 (10) | 0.0212 (11) | −0.0188 (10) |
| C9 | 0.0737 (14) | 0.0657 (13) | 0.0651 (13) | −0.0053 (11) | 0.0261 (11) | −0.0210 (11) |
| C10 | 0.0732 (13) | 0.0590 (11) | 0.0490 (10) | −0.0122 (10) | 0.0152 (9) | −0.0084 (9) |
| C11 | 0.0582 (11) | 0.0457 (9) | 0.0476 (10) | −0.0050 (8) | 0.0070 (8) | −0.0042 (7) |
| C12 | 0.0451 (9) | 0.0397 (8) | 0.0442 (9) | −0.0072 (7) | 0.0038 (7) | −0.0087 (7) |
| C13 | 0.0405 (9) | 0.0345 (8) | 0.0514 (9) | 0.0012 (6) | 0.0059 (7) | 0.0014 (7) |
| C14 | 0.0395 (8) | 0.0319 (7) | 0.0407 (8) | 0.0008 (6) | 0.0096 (6) | −0.0031 (6) |
| C15 | 0.0416 (8) | 0.0382 (8) | 0.0389 (8) | 0.0030 (6) | 0.0108 (7) | −0.0045 (6) |
| C16 | 0.0441 (9) | 0.0449 (9) | 0.0479 (9) | −0.0036 (7) | 0.0108 (7) | −0.0122 (7) |
| C17 | 0.0536 (10) | 0.0381 (9) | 0.0538 (10) | −0.0077 (7) | 0.0227 (8) | −0.0116 (7) |
| C18 | 0.0574 (10) | 0.0339 (8) | 0.0473 (9) | 0.0041 (7) | 0.0230 (8) | −0.0003 (7) |
| C19 | 0.0453 (9) | 0.0367 (8) | 0.0426 (8) | 0.0044 (7) | 0.0094 (7) | 0.0000 (6) |
| C20 | 0.0573 (11) | 0.0504 (10) | 0.0441 (9) | 0.0048 (8) | 0.0010 (8) | 0.0003 (8) |
| C21 | 0.0589 (12) | 0.0747 (14) | 0.0738 (14) | −0.0118 (11) | −0.0033 (11) | −0.0163 (11) |
| C22 | 0.0808 (15) | 0.0505 (11) | 0.0862 (15) | −0.0231 (10) | 0.0274 (12) | −0.0111 (10) |
| C23 | 0.0940 (16) | 0.0398 (10) | 0.0665 (13) | 0.0052 (10) | 0.0247 (11) | 0.0093 (9) |
| C24 | 0.0675 (13) | 0.0540 (11) | 0.0624 (12) | 0.0037 (9) | −0.0094 (10) | 0.0093 (9) |
| N1 | 0.0691 (10) | 0.0521 (9) | 0.0671 (10) | 0.0057 (8) | 0.0216 (8) | 0.0180 (8) |
| N2 | 0.0697 (10) | 0.0346 (7) | 0.0650 (10) | 0.0040 (7) | 0.0129 (8) | −0.0039 (7) |
| N3 | 0.0456 (7) | 0.0305 (6) | 0.0455 (7) | −0.0029 (5) | 0.0056 (6) | −0.0021 (5) |
Geometric parameters (Å, º)
| C1—N1 | 1.334 (3) | C13—H13B | 0.9700 |
| C1—C2 | 1.368 (3) | C14—C19 | 1.405 (2) |
| C1—H1 | 0.9300 | C14—C15 | 1.406 (2) |
| C2—C3 | 1.360 (4) | C15—C16 | 1.395 (2) |
| C2—H2 | 0.9300 | C15—C20 | 1.510 (2) |
| C3—C4 | 1.369 (3) | C16—C17 | 1.398 (3) |
| C3—H3 | 0.9300 | C16—C21 | 1.517 (3) |
| C4—C5 | 1.382 (3) | C17—C18 | 1.397 (3) |
| C4—H4 | 0.9300 | C17—C22 | 1.517 (2) |
| C5—N1 | 1.328 (2) | C18—C19 | 1.408 (2) |
| C5—C6 | 1.477 (3) | C18—C23 | 1.515 (2) |
| C6—N2 | 1.319 (2) | C19—C24 | 1.514 (3) |
| C6—N3 | 1.382 (2) | C20—H20A | 0.968 (12) |
| C7—N2 | 1.373 (2) | C20—H20B | 0.968 (13) |
| C7—C8 | 1.395 (3) | C20—H20C | 0.968 (12) |
| C7—C12 | 1.401 (2) | C21—H21A | 0.933 (14) |
| C8—C9 | 1.365 (3) | C21—H21B | 0.933 (14) |
| C8—H8 | 0.9300 | C21—H21C | 0.933 (14) |
| C9—C10 | 1.396 (3) | C22—H22A | 0.948 (14) |
| C9—H9 | 0.9300 | C22—H22B | 0.948 (14) |
| C10—C11 | 1.379 (3) | C22—H22C | 0.948 (14) |
| C10—H10 | 0.9300 | C23—H23A | 0.951 (14) |
| C11—C12 | 1.387 (2) | C23—H23B | 0.951 (14) |
| C11—H11 | 0.9300 | C23—H23C | 0.951 (13) |
| C12—N3 | 1.384 (2) | C24—H24A | 0.915 (14) |
| C13—N3 | 1.478 (2) | C24—H24B | 0.915 (13) |
| C13—C14 | 1.522 (2) | C24—H24C | 0.915 (13) |
| C13—H13A | 0.9700 | ||
| N1—C1—C2 | 124.4 (2) | C15—C16—C17 | 120.14 (16) |
| N1—C1—H1 | 117.8 | C15—C16—C21 | 119.82 (17) |
| C2—C1—H1 | 117.8 | C17—C16—C21 | 120.05 (16) |
| C3—C2—C1 | 117.9 (2) | C18—C17—C16 | 120.23 (15) |
| C3—C2—H2 | 121.0 | C18—C17—C22 | 120.34 (17) |
| C1—C2—H2 | 121.0 | C16—C17—C22 | 119.42 (18) |
| C2—C3—C4 | 118.7 (2) | C17—C18—C19 | 119.95 (15) |
| C2—C3—H3 | 120.6 | C17—C18—C23 | 119.35 (16) |
| C4—C3—H3 | 120.6 | C19—C18—C23 | 120.69 (17) |
| C3—C4—C5 | 120.2 (2) | C14—C19—C18 | 119.69 (15) |
| C3—C4—H4 | 119.9 | C14—C19—C24 | 120.95 (15) |
| C5—C4—H4 | 119.9 | C18—C19—C24 | 119.35 (15) |
| N1—C5—C4 | 121.30 (19) | C15—C20—H20A | 109.5 |
| N1—C5—C6 | 120.74 (15) | C15—C20—H20B | 109.5 |
| C4—C5—C6 | 117.96 (18) | H20A—C20—H20B | 109.5 |
| N2—C6—N3 | 112.86 (16) | C15—C20—H20C | 109.5 |
| N2—C6—C5 | 119.77 (15) | H20A—C20—H20C | 109.5 |
| N3—C6—C5 | 127.37 (15) | H20B—C20—H20C | 109.5 |
| N2—C7—C8 | 129.74 (18) | C16—C21—H21A | 109.5 |
| N2—C7—C12 | 110.39 (16) | C16—C21—H21B | 109.5 |
| C8—C7—C12 | 119.87 (18) | H21A—C21—H21B | 109.5 |
| C9—C8—C7 | 118.6 (2) | C16—C21—H21C | 109.5 |
| C9—C8—H8 | 120.7 | H21A—C21—H21C | 109.5 |
| C7—C8—H8 | 120.7 | H21B—C21—H21C | 109.5 |
| C8—C9—C10 | 120.93 (19) | C17—C22—H22A | 109.5 |
| C8—C9—H9 | 119.5 | C17—C22—H22B | 109.5 |
| C10—C9—H9 | 119.5 | H22A—C22—H22B | 109.5 |
| C11—C10—C9 | 121.92 (19) | C17—C22—H22C | 109.5 |
| C11—C10—H10 | 119.0 | H22A—C22—H22C | 109.5 |
| C9—C10—H10 | 119.0 | H22B—C22—H22C | 109.5 |
| C10—C11—C12 | 116.88 (18) | C18—C23—H23A | 109.5 |
| C10—C11—H11 | 121.6 | C18—C23—H23B | 109.5 |
| C12—C11—H11 | 121.6 | H23A—C23—H23B | 109.5 |
| N3—C12—C11 | 132.66 (16) | C18—C23—H23C | 109.5 |
| N3—C12—C7 | 105.53 (15) | H23A—C23—H23C | 109.5 |
| C11—C12—C7 | 121.80 (17) | H23B—C23—H23C | 109.5 |
| N3—C13—C14 | 113.67 (13) | C19—C24—H24A | 109.5 |
| N3—C13—H13A | 108.8 | C19—C24—H24B | 109.5 |
| C14—C13—H13A | 108.8 | H24A—C24—H24B | 109.5 |
| N3—C13—H13B | 108.8 | C19—C24—H24C | 109.5 |
| C14—C13—H13B | 108.8 | H24A—C24—H24C | 109.5 |
| H13A—C13—H13B | 107.7 | H24B—C24—H24C | 109.5 |
| C19—C14—C15 | 119.78 (14) | C5—N1—C1 | 117.39 (17) |
| C19—C14—C13 | 120.99 (14) | C6—N2—C7 | 105.26 (15) |
| C15—C14—C13 | 119.19 (14) | C6—N3—C12 | 105.94 (13) |
| C16—C15—C14 | 120.06 (15) | C6—N3—C13 | 129.98 (14) |
| C16—C15—C20 | 119.99 (15) | C12—N3—C13 | 124.08 (13) |
| C14—C15—C20 | 119.95 (14) | ||
| N1—C1—C2—C3 | −0.2 (4) | C15—C16—C17—C22 | −179.52 (16) |
| C1—C2—C3—C4 | −0.3 (4) | C21—C16—C17—C22 | 0.7 (3) |
| C2—C3—C4—C5 | 1.0 (4) | C16—C17—C18—C19 | 1.9 (2) |
| C3—C4—C5—N1 | −1.2 (4) | C22—C17—C18—C19 | −178.06 (16) |
| C3—C4—C5—C6 | 179.1 (2) | C16—C17—C18—C23 | −178.23 (16) |
| N1—C5—C6—N2 | 171.36 (17) | C22—C17—C18—C23 | 1.8 (2) |
| C4—C5—C6—N2 | −9.0 (3) | C15—C14—C19—C18 | −1.9 (2) |
| N1—C5—C6—N3 | −9.7 (3) | C13—C14—C19—C18 | −179.49 (14) |
| C4—C5—C6—N3 | 169.96 (19) | C15—C14—C19—C24 | 177.28 (16) |
| N2—C7—C8—C9 | −178.4 (2) | C13—C14—C19—C24 | −0.3 (2) |
| C12—C7—C8—C9 | 1.9 (3) | C17—C18—C19—C14 | −1.2 (2) |
| C7—C8—C9—C10 | −0.1 (3) | C23—C18—C19—C14 | 178.93 (15) |
| C8—C9—C10—C11 | −1.2 (3) | C17—C18—C19—C24 | 179.62 (16) |
| C9—C10—C11—C12 | 0.7 (3) | C23—C18—C19—C24 | −0.3 (2) |
| C10—C11—C12—N3 | −179.82 (17) | C4—C5—N1—C1 | 0.7 (3) |
| C10—C11—C12—C7 | 1.1 (3) | C6—C5—N1—C1 | −179.67 (18) |
| N2—C7—C12—N3 | −1.48 (19) | C2—C1—N1—C5 | 0.1 (4) |
| C8—C7—C12—N3 | 178.23 (17) | N3—C6—N2—C7 | 0.2 (2) |
| N2—C7—C12—C11 | 177.79 (16) | C5—C6—N2—C7 | 179.26 (15) |
| C8—C7—C12—C11 | −2.5 (3) | C8—C7—N2—C6 | −178.8 (2) |
| N3—C13—C14—C19 | −121.53 (16) | C12—C7—N2—C6 | 0.8 (2) |
| N3—C13—C14—C15 | 60.88 (19) | N2—C6—N3—C12 | −1.10 (19) |
| C19—C14—C15—C16 | 4.3 (2) | C5—C6—N3—C12 | 179.91 (16) |
| C13—C14—C15—C16 | −178.05 (14) | N2—C6—N3—C13 | 178.34 (15) |
| C19—C14—C15—C20 | −174.84 (15) | C5—C6—N3—C13 | −0.7 (3) |
| C13—C14—C15—C20 | 2.8 (2) | C11—C12—N3—C6 | −177.65 (18) |
| C14—C15—C16—C17 | −3.6 (2) | C7—C12—N3—C6 | 1.50 (17) |
| C20—C15—C16—C17 | 175.52 (15) | C11—C12—N3—C13 | 2.9 (3) |
| C14—C15—C16—C21 | 176.10 (16) | C7—C12—N3—C13 | −177.98 (14) |
| C20—C15—C16—C21 | −4.7 (2) | C14—C13—N3—C6 | −125.33 (17) |
| C15—C16—C17—C18 | 0.5 (2) | C14—C13—N3—C12 | 54.0 (2) |
| C21—C16—C17—C18 | −179.21 (16) |
Hydrogen-bond geometry (Å, º)
Cg is the centroid of the C7–C12 benzene ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C13—H13A···Cgi | 0.97 | 2.91 | 3.6941 (18) | 139 |
| C13—H13B···N1 | 0.97 | 2.30 | 3.029 (2) | 131 |
Symmetry code: (i) x−1, y, z.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: RZ5115).
References
- Blessing, R. H. (1995). Acta Cryst. A51, 33–38. [DOI] [PubMed]
- Boca, R., Baran, P., Dlhán, L., Sima, J., Wiesinger, G., Renz, F., El-Ayaan, U. & Linert, W. (1997). Polyhedron, 16, 47–55.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Çelik, Ö., Anĝay, F., Gündoĝan, M. & Ulusoy, M. (2014). Acta Cryst. E70, o485. [DOI] [PMC free article] [PubMed]
- Çelik, Ö., Kasumov, V. T. & Şahin, E. (2009). Acta Cryst. E65, o2786. [DOI] [PMC free article] [PubMed]
- Çelik, Ö., Ulusoy, M., Taş, E. & Íde, S. (2007). Anal. Sci. 23, 185–186.
- Chiswell, B., Lions, F. & Morris, B. S. (1964). Inorg. Chem. 3, 110–114.
- De Castro, B., Freire, C., Domingues, D. & Gomes, J. (1991). Polyhedron, 10, 2541–2549.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Harkins, T. R., Walter, J. L., Harris, O. E. & Freiser, H. (1956). J. Am. Chem. Soc. 78, 260–264.
- Kelland, L. R. & Farrell, N. (2000). In Platinum-based Drugs in Cancer Chemotherapy Tatowa: Humana Press.
- Khalil, M. M. H., Ali, S. A. & Ramadan, R. M. (2001). Spectrochim. Acta Part A, 57, 1017–1024. [DOI] [PubMed]
- Lippert, B. (1999). In Cisplatin: Chemistry and Biochemistry of a Leading Anticancer Drug Weinheim: Wiley–VCH.
- Maekawa, M., Munakata, M., Kuroda-Sowa, T. & Hachiya, K. (1994). Inorg. Chim. Acta, 227, 137–143.
- Sahin, C., Ulusoy, M., Zafer, C., Ozsoy, C., Varlikli, C., Dittrich, T., Cetinkaya, B. & Icli, S. (2010). Dyes Pigments, 84, 88–94.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wong, E. & Giandomenico, C. M. (1999). Chem. Rev. 99, 2451–2466. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536814007934/rz5115sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814007934/rz5115Isup2.hkl
Supporting information file. DOI: 10.1107/S1600536814007934/rz5115Isup3.cml
CCDC reference: 996309
Additional supporting information: crystallographic information; 3D view; checkCIF report