Abstract
In the title compound, C14H20O5, an intermediate in the synthesis of oligosaccharides, the glycosidic [H—C—O—C(H3)] torsion angle ϕH is 52.3° and the exo-cyclic [H—C—O—C(H2)] torsion angle θH is −11.7°. The hexapyranose ring has a chair conformation. In the crystal, molecules are linked by O—H⋯O hydrogen bonds, forming chains propagating along [010]. Enclosed within the chains are R 3 3(12) ring motifs involving three molecules. The chains are linked via C—H⋯π interactions, forming a three-dimensional network.
Related literature
For a description of l-rhamnose as part of polysaccharides, see: Ansaruzzaman et al. (1996 ▶); Marie et al. (1998 ▶); Säwén et al. (2012 ▶). For a description of syntheses in which the title compound has been used, see: Eklund et al. (2005 ▶); Handa et al. (1979 ▶). For the structure of rhamnosyl-containing trisaccharides, see: Eriksson & Widmalm (2012 ▶); Eriksson et al. (1999 ▶); Jonsson et al. (2006 ▶). For further related literature on l-rhamnose, see: Anderson & Ijeh (1994 ▶); Varki et al. (1999 ▶); Haines (1969 ▶); Herget et al. (2008 ▶); Olsson et al. (2005 ▶). For puckering analysis, see: Cremer & Pople (1975 ▶).
Experimental
Crystal data
C14H20O5
M r = 268.30
Orthorhombic,

a = 6.5377 (1) Å
b = 9.1848 (2) Å
c = 23.2699 (5) Å
V = 1397.30 (5) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 293 K
0.25 × 0.12 × 0.05 mm
Data collection
Oxford Diffraction Xcalibur 3 with sapphire 3 CCD diffractometer
Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2004 ▶) T min = 0.921, T max = 1.000
9540 measured reflections
1665 independent reflections
1407 reflections with I > 2σ(I)
R int = 0.040
Refinement
R[F 2 > 2σ(F 2)] = 0.035
wR(F 2) = 0.081
S = 1.00
1665 reflections
177 parameters
H-atom parameters constrained
Δρmax = 0.14 e Å−3
Δρmin = −0.13 e Å−3
Data collection: CrysAlis CCD (Oxford Diffraction, 2004 ▶); cell refinement: CrysAlis CCD; data reduction: CrysAlis RED (Oxford Diffraction, 2004 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: DIAMOND (Brandenburg, 2001 ▶); software used to prepare material for publication: enCIFer (Allen et al., 2004 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, rp1. DOI: 10.1107/S1600536814007922/su2708sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814007922/su2708Isup2.hkl
CCDC reference: 996312
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg is the centroid of the C41–C46 benzyl ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2A⋯O3i | 0.82 | 2.00 | 2.813 (2) | 172 |
| O3—H3A⋯O5ii | 0.82 | 2.05 | 2.799 (2) | 151 |
| C7—H7C⋯Cg iii | 0.96 | 2.89 | 3.652 (3) | 137 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
This work was supported by grants from the Swedish Research Council and the Knut and Alice Wallenberg foundation.
supplementary crystallographic information
1. Comment
Bacteria contain many different sugar residues (Herget et al., 2008) in contrast to man where only a dozen monosaccharides are utilized in the formation of polysaccharides, glycoproteins and glycolipids [see Varki et al. (1999)]. In lipopolysaccharides L-rhamnose (6-deoxy-L-mannose) is often present as a sugar component, ranging from one residue per repeating unit, for example, as the terminal residue in the biosynthesized and polymerized oligosaccharide; consequently, it forms the side-chain residue in the O-antigen (Olsson et al., 2005), which often has 10 – 25 repeating units. Alternatively, L-rhamnose can make up the O-antigen polysaccharide per se, as a homopolymer (Ansaruzzaman et al., 1996).
The title compound, Fig. 1, has been used in the synthesis of a rhamnosyl-containing trisaccharide (Eklund et al., 2005), the crystal structure of which was recently determined (Eriksson & Widmalm, 2012). The title monosaccharide is the methyl glycoside of α-L-rhamnopyranose and carries a benzyl protecting group at O4 in an ether linkage; the remaining two hydroxyl groups are unprotected and available for further synthetic modifications.
The glycosidic torsion angle defined by H1-C1-O1-C7, φH is 52.3° (Fig. 1). The exo-cyclic torsion angle defined by H4—C4—O4—C40, θH = -11.7°, shows an almost eclipsed conformation. The corresponding torsion angle in the crystal structure of 4-O-Benzyl-2,3-O-isopropylidene-α-L-rhamnopyranose was 36.8° (Eriksson et al., 1999). Moreover, in the title compound the C4—O4—C40—C41 torsion is antiperiplanar and the benzyl ring plane deviates significantly from that defined by plane O4/C40/C41, with a dihedral angle of 54.85 (18)°.
The hexapyranose ring O5/C1-C5 has a chair conformation, with puckering parameters (Cremer & Pople, 1975) Q = 0.570 (2) Å, θ = 177.4 (2)° and φ = 11 (4)°. These puckering parameters reveal a 1C4 conformation close to the south pole, in contrast to another protected methyl α-L-rhamnopyranoside derivative carrying an isopropylidene group at O2 and O3 (Jonsson et al., 2006).
In the crystal, molecules are linked via O—H···O hydrogen bonds, involving both hydroxyl groups, forming chains along the a axis (Table 1 and Fig. 2). They enclose 12-membered R33(12) ring motifs. There are also C—H ··· π interactions present, between the C7 methyl group and the centroid of the (C41–C46) benzyl ring (Table 1), that link the chains forming a three-dimensional network.
The conformation of the exo-cyclic torsion angle (H4—C4—O4—C40) was analyzed by NMR measurements (see details in the archived CIF) of the long-range heteronuclear coupling constant between nuclei H4 and C40 using a J-HMBC experiment, which resulted in 3JCH = 6.25 Hz. Interpretation of this coupling constant using the Karplus-type relationship 3JC,H = 7.6 cos2θ - 1.7 cosθ + 1.6 (Anderson & Ijeh, 1994) leads to |θH| = 26° when interpreted as a single conformation, i.e., quite similar to the structure determined in the solid state. The corresponding torsion angle in the crystal structure of 4-O-Benzyl-2,3-O-isopropylidene-α-L-rhamnopyranose was 36.8° (Eriksson et al., 1999).
2. Experimental
The synthesis of the title compound was performed according to a published procedure (Haines, 1969), where the rhamnosyl residue has the L absolute configuration. The title monosaccharide was crystallized at ambient temperature by slow evaporation from chloroform yielding colourless prismatic crystals. Spectroscopic data and details of the NMR measurements are given in the archived CIF.
3. Refinement
The OH and C-bound atoms were positioned geometrically and allowed to ride on their parent atoms: O-H = 0.82 Å, C-H = 0.98, 0.96 and 0.92 Å, for CH, CH3, and CH(aromatic) H atoms, respectively, with Uiso(H) = 1.2Ueq(C) and = 1.5Ueq(O). In the final cycles of refinement, in the absence of significant anomalous scattering effects, Friedel pairs were merged and Δf " set to zero. The absolute configuration was set by the a priori knowledge of the absolute configuration of the starting reagent.
Figures
Fig. 1.
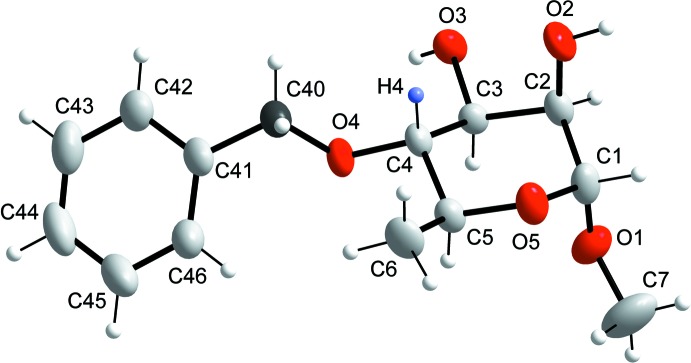
The molecular structure of the title molecule with atom labelling. Displacement ellipsoids are drawn at the 50% probability level. The long-range heteronuclear NMR coupling constant was measured beteen nuclei H4 (blue) and C40 (graphite).
Fig. 2.
A partial view along the b axis of the crystal packing of the title compound. Hydrogen bonds are shown as dashed lines (see Table 1 for details).
Crystal data
| C14H20O5 | F(000) = 576 |
| Mr = 268.30 | Dx = 1.275 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 4263 reflections |
| a = 6.5377 (1) Å | θ = 3.8–32.2° |
| b = 9.1848 (2) Å | µ = 0.10 mm−1 |
| c = 23.2699 (5) Å | T = 293 K |
| V = 1397.30 (5) Å3 | Prism, colourless |
| Z = 4 | 0.25 × 0.12 × 0.05 mm |
Data collection
| Oxford Diffraction Xcalibur 3 with sapphire 3 CCD diffractometer | 1665 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 1407 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.040 |
| Detector resolution: 16.5467 pixels mm-1 | θmax = 26.4°, θmin = 3.8° |
| ω scans at different φ | h = −3→8 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2004) | k = −11→11 |
| Tmin = 0.921, Tmax = 1.000 | l = −28→29 |
| 9540 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.035 | H-atom parameters constrained |
| wR(F2) = 0.081 | w = 1/[σ2(Fo2) + (0.0524P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.00 | (Δ/σ)max < 0.001 |
| 1665 reflections | Δρmax = 0.14 e Å−3 |
| 177 parameters | Δρmin = −0.13 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.013 (2) |
Special details
| Experimental. Spectroscoptic data for the title compound: 1H NMR (CDCl3, ppm, 298K, selected 3JH,H values are given in parenthesis): H1 4.645(1.55); H2 3.903(3.49); H3 3.877(9.12); H4 3.333(9.52); H5 3.700(6.31); H6 1.352; H7 3.345; H40 4.738; H42,H46 7.355; H43,H45 7.360; H44 7.304; HO2 2.574(3.92); HO3 2.470(5.31). 13C NMR (CDCl3, ppm, 298K): C1 100.48; C2 71.20; C3 71.60; C4 81.77; C5 67.14; C6 18.14; C7 54.97; C40 75.11; C41 138.40; C42,C46 128.06; C43,C45 128.75; C44 128.12.NMR experiments were performed on a Bruker Avance III spectrometer operating at a 1H frequency of 700 MHz. The title compound was dissolved in chloroform-d and 1H and 13C resonances were referenced to internal TMS (δ = 0.0) and the solvent resonance (δ = 77.16), respectively. Resonance assignments were performed using standard experiments for oligosaccharides (Widmalm, G. (2007). NMR spectroscopy of carbohydrates and conformational analysis in solution. Comprehensive glycoscience, J. P. Kamerling, Ed., Elsevier, Oxford, Vol. 2, pp. 101–132) and measurement of the heteronuclear coupling constant was carried out by a J-HMBC experiment (Meissner, A. & Sørensen, O. W. (2001). Magn. Reson. Chem.39, 49–52) using two separate experiments with κ values of 59.0 and 99.0, respectively (Jonsson, K. H. M., Pendrill, R. & Widmalm, G. (2011). Magn. Reson. Chem.49, 117–124). |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 1.0867 (3) | 0.5237 (2) | 0.04789 (8) | 0.0370 (5) | |
| H1 | 1.1807 | 0.5419 | 0.0159 | 0.044* | |
| C2 | 0.8931 (3) | 0.6130 (2) | 0.03845 (8) | 0.0317 (5) | |
| H2 | 0.8232 | 0.5790 | 0.0037 | 0.038* | |
| C3 | 0.7515 (3) | 0.5959 (2) | 0.09008 (8) | 0.0273 (4) | |
| H3 | 0.7109 | 0.4935 | 0.0933 | 0.033* | |
| C4 | 0.8644 (3) | 0.6397 (2) | 0.14447 (8) | 0.0282 (4) | |
| H4 | 0.8990 | 0.7435 | 0.1432 | 0.034* | |
| C5 | 1.0597 (3) | 0.5481 (2) | 0.15042 (8) | 0.0314 (4) | |
| H5 | 1.0212 | 0.4456 | 0.1549 | 0.038* | |
| O5 | 1.1851 (2) | 0.56299 (17) | 0.09964 (6) | 0.0382 (4) | |
| C6 | 1.1946 (3) | 0.5915 (3) | 0.20013 (9) | 0.0444 (6) | |
| H6A | 1.3178 | 0.5347 | 0.1994 | 0.067* | |
| H6B | 1.1234 | 0.5747 | 0.2356 | 0.067* | |
| H6C | 1.2284 | 0.6929 | 0.1970 | 0.067* | |
| O1 | 1.0298 (2) | 0.37637 (17) | 0.04700 (6) | 0.0458 (4) | |
| C7 | 1.1990 (5) | 0.2788 (3) | 0.04520 (14) | 0.0819 (10) | |
| H7A | 1.2799 | 0.2985 | 0.0117 | 0.123* | |
| H7B | 1.1498 | 0.1804 | 0.0437 | 0.123* | |
| H7C | 1.2813 | 0.2918 | 0.0790 | 0.123* | |
| O2 | 0.9419 (2) | 0.76226 (15) | 0.03268 (6) | 0.0444 (4) | |
| H2A | 0.9920 | 0.7768 | 0.0009 | 0.067* | |
| O3 | 0.5733 (2) | 0.68189 (16) | 0.08031 (6) | 0.0355 (3) | |
| H3A | 0.4761 | 0.6461 | 0.0975 | 0.053* | |
| O4 | 0.7372 (2) | 0.60980 (15) | 0.19307 (5) | 0.0337 (3) | |
| C40 | 0.7222 (3) | 0.7259 (2) | 0.23403 (8) | 0.0380 (5) | |
| H40A | 0.6411 | 0.8049 | 0.2183 | 0.046* | |
| H40B | 0.8573 | 0.7630 | 0.2430 | 0.046* | |
| C41 | 0.6222 (3) | 0.6670 (2) | 0.28745 (8) | 0.0355 (5) | |
| C42 | 0.4478 (3) | 0.7314 (3) | 0.31025 (9) | 0.0424 (5) | |
| H42 | 0.3947 | 0.8154 | 0.2937 | 0.051* | |
| C43 | 0.3531 (4) | 0.6700 (3) | 0.35777 (10) | 0.0563 (7) | |
| H43 | 0.2358 | 0.7130 | 0.3727 | 0.068* | |
| C44 | 0.4302 (5) | 0.5471 (3) | 0.38299 (10) | 0.0601 (7) | |
| H44 | 0.3630 | 0.5050 | 0.4141 | 0.072* | |
| C45 | 0.6085 (5) | 0.4856 (3) | 0.36203 (10) | 0.0594 (7) | |
| H45 | 0.6650 | 0.4044 | 0.3799 | 0.071* | |
| C46 | 0.7013 (4) | 0.5455 (3) | 0.31468 (9) | 0.0471 (6) | |
| H46 | 0.8205 | 0.5033 | 0.3006 | 0.057* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0251 (10) | 0.0573 (15) | 0.0288 (10) | −0.0006 (10) | 0.0027 (9) | −0.0024 (9) |
| C2 | 0.0274 (9) | 0.0430 (12) | 0.0247 (9) | −0.0035 (10) | −0.0007 (8) | 0.0023 (8) |
| C3 | 0.0220 (9) | 0.0325 (10) | 0.0275 (10) | −0.0004 (8) | −0.0010 (8) | 0.0023 (8) |
| C4 | 0.0274 (9) | 0.0345 (11) | 0.0226 (9) | −0.0045 (8) | 0.0032 (8) | 0.0023 (8) |
| C5 | 0.0263 (9) | 0.0400 (11) | 0.0277 (9) | −0.0027 (9) | −0.0010 (8) | 0.0033 (9) |
| O5 | 0.0216 (6) | 0.0611 (9) | 0.0320 (7) | −0.0031 (7) | −0.0010 (6) | 0.0002 (7) |
| C6 | 0.0342 (11) | 0.0628 (15) | 0.0362 (11) | −0.0018 (11) | −0.0118 (10) | 0.0040 (10) |
| O1 | 0.0394 (8) | 0.0460 (9) | 0.0520 (9) | 0.0094 (8) | −0.0054 (8) | −0.0125 (7) |
| C7 | 0.0693 (19) | 0.0734 (19) | 0.103 (2) | 0.0371 (18) | −0.0094 (18) | −0.0190 (18) |
| O2 | 0.0489 (9) | 0.0499 (10) | 0.0344 (8) | −0.0046 (8) | 0.0109 (7) | 0.0105 (7) |
| O3 | 0.0228 (7) | 0.0481 (8) | 0.0357 (8) | 0.0034 (7) | 0.0002 (6) | 0.0070 (7) |
| O4 | 0.0351 (7) | 0.0410 (8) | 0.0250 (7) | −0.0071 (7) | 0.0058 (6) | −0.0019 (6) |
| C40 | 0.0469 (12) | 0.0375 (11) | 0.0295 (10) | −0.0021 (11) | 0.0033 (10) | −0.0013 (9) |
| C41 | 0.0449 (11) | 0.0374 (12) | 0.0244 (10) | −0.0076 (10) | −0.0002 (9) | −0.0050 (8) |
| C42 | 0.0451 (12) | 0.0494 (13) | 0.0326 (11) | −0.0021 (12) | −0.0008 (10) | −0.0062 (10) |
| C43 | 0.0519 (14) | 0.0757 (18) | 0.0414 (13) | −0.0124 (14) | 0.0130 (12) | −0.0143 (13) |
| C44 | 0.0819 (19) | 0.0642 (16) | 0.0343 (12) | −0.0272 (17) | 0.0159 (13) | −0.0018 (12) |
| C45 | 0.097 (2) | 0.0465 (14) | 0.0349 (12) | −0.0048 (15) | 0.0069 (15) | 0.0030 (10) |
| C46 | 0.0603 (15) | 0.0441 (12) | 0.0369 (11) | 0.0019 (12) | 0.0111 (11) | −0.0007 (10) |
Geometric parameters (Å, º)
| C1—O1 | 1.404 (3) | C7—H7A | 0.9600 |
| C1—O5 | 1.412 (2) | C7—H7B | 0.9600 |
| C1—C2 | 1.524 (3) | C7—H7C | 0.9600 |
| C1—H1 | 0.9800 | O2—H2A | 0.8200 |
| C2—O2 | 1.414 (2) | O3—H3A | 0.8200 |
| C2—C3 | 1.525 (3) | O4—C40 | 1.433 (2) |
| C2—H2 | 0.9800 | C40—C41 | 1.505 (3) |
| C3—O3 | 1.426 (2) | C40—H40A | 0.9700 |
| C3—C4 | 1.519 (2) | C40—H40B | 0.9700 |
| C3—H3 | 0.9800 | C41—C46 | 1.383 (3) |
| C4—O4 | 1.431 (2) | C41—C42 | 1.390 (3) |
| C4—C5 | 1.535 (3) | C42—C43 | 1.387 (3) |
| C4—H4 | 0.9800 | C42—H42 | 0.9300 |
| C5—O5 | 1.445 (2) | C43—C44 | 1.369 (4) |
| C5—C6 | 1.508 (3) | C43—H43 | 0.9300 |
| C5—H5 | 0.9800 | C44—C45 | 1.384 (4) |
| C6—H6A | 0.9600 | C44—H44 | 0.9300 |
| C6—H6B | 0.9600 | C45—C46 | 1.373 (3) |
| C6—H6C | 0.9600 | C45—H45 | 0.9300 |
| O1—C7 | 1.424 (3) | C46—H46 | 0.9300 |
| O1—C1—O5 | 112.30 (17) | H6B—C6—H6C | 109.5 |
| O1—C1—C2 | 107.24 (16) | C1—O1—C7 | 113.7 (2) |
| O5—C1—C2 | 111.33 (16) | O1—C7—H7A | 109.5 |
| O1—C1—H1 | 108.6 | O1—C7—H7B | 109.5 |
| O5—C1—H1 | 108.6 | H7A—C7—H7B | 109.5 |
| C2—C1—H1 | 108.6 | O1—C7—H7C | 109.5 |
| O2—C2—C1 | 110.35 (16) | H7A—C7—H7C | 109.5 |
| O2—C2—C3 | 108.14 (15) | H7B—C7—H7C | 109.5 |
| C1—C2—C3 | 109.60 (15) | C2—O2—H2A | 109.5 |
| O2—C2—H2 | 109.6 | C3—O3—H3A | 109.5 |
| C1—C2—H2 | 109.6 | C4—O4—C40 | 115.00 (14) |
| C3—C2—H2 | 109.6 | O4—C40—C41 | 108.17 (15) |
| O3—C3—C4 | 112.53 (15) | O4—C40—H40A | 110.1 |
| O3—C3—C2 | 108.27 (14) | C41—C40—H40A | 110.1 |
| C4—C3—C2 | 109.53 (15) | O4—C40—H40B | 110.1 |
| O3—C3—H3 | 108.8 | C41—C40—H40B | 110.1 |
| C4—C3—H3 | 108.8 | H40A—C40—H40B | 108.4 |
| C2—C3—H3 | 108.8 | C46—C41—C42 | 118.4 (2) |
| O4—C4—C3 | 108.97 (13) | C46—C41—C40 | 120.40 (19) |
| O4—C4—C5 | 107.88 (14) | C42—C41—C40 | 121.2 (2) |
| C3—C4—C5 | 109.51 (15) | C41—C42—C43 | 119.8 (2) |
| O4—C4—H4 | 110.1 | C41—C42—H42 | 120.1 |
| C3—C4—H4 | 110.1 | C43—C42—H42 | 120.1 |
| C5—C4—H4 | 110.1 | C44—C43—C42 | 120.9 (2) |
| O5—C5—C6 | 105.68 (15) | C44—C43—H43 | 119.6 |
| O5—C5—C4 | 110.25 (15) | C42—C43—H43 | 119.6 |
| C6—C5—C4 | 114.22 (17) | C43—C44—C45 | 119.7 (2) |
| O5—C5—H5 | 108.8 | C43—C44—H44 | 120.2 |
| C6—C5—H5 | 108.8 | C45—C44—H44 | 120.2 |
| C4—C5—H5 | 108.8 | C46—C45—C44 | 119.4 (3) |
| C1—O5—C5 | 114.51 (14) | C46—C45—H45 | 120.3 |
| C5—C6—H6A | 109.5 | C44—C45—H45 | 120.3 |
| C5—C6—H6B | 109.5 | C45—C46—C41 | 121.7 (2) |
| H6A—C6—H6B | 109.5 | C45—C46—H46 | 119.1 |
| C5—C6—H6C | 109.5 | C41—C46—H46 | 119.1 |
| H6A—C6—H6C | 109.5 | ||
| O1—C1—C2—O2 | −173.76 (14) | C4—C5—O5—C1 | −57.5 (2) |
| O5—C1—C2—O2 | 63.0 (2) | O5—C1—O1—C7 | −67.8 (2) |
| O1—C1—C2—C3 | 67.3 (2) | C2—C1—O1—C7 | 169.56 (19) |
| O5—C1—C2—C3 | −55.9 (2) | C3—C4—O4—C40 | −132.63 (17) |
| O2—C2—C3—O3 | 58.98 (19) | C5—C4—O4—C40 | 108.58 (18) |
| C1—C2—C3—O3 | 179.27 (15) | C4—O4—C40—C41 | −168.12 (15) |
| O2—C2—C3—C4 | −64.05 (19) | O4—C40—C41—C46 | 54.2 (2) |
| C1—C2—C3—C4 | 56.2 (2) | O4—C40—C41—C42 | −124.9 (2) |
| O3—C3—C4—O4 | 65.15 (19) | C46—C41—C42—C43 | −2.5 (3) |
| C2—C3—C4—O4 | −174.41 (15) | C40—C41—C42—C43 | 176.6 (2) |
| O3—C3—C4—C5 | −177.09 (14) | C41—C42—C43—C44 | 0.5 (3) |
| C2—C3—C4—C5 | −56.64 (19) | C42—C43—C44—C45 | 2.2 (4) |
| O4—C4—C5—O5 | 174.32 (14) | C43—C44—C45—C46 | −2.8 (4) |
| C3—C4—C5—O5 | 55.90 (18) | C44—C45—C46—C41 | 0.7 (4) |
| O4—C4—C5—C6 | −66.9 (2) | C42—C41—C46—C45 | 1.9 (3) |
| C3—C4—C5—C6 | 174.69 (16) | C40—C41—C46—C45 | −177.2 (2) |
| O1—C1—O5—C5 | −62.6 (2) | C40—O4—C4—H4 | −11.7 |
| C2—C1—O5—C5 | 57.6 (2) | C7—O1—C1—H1 | 52.3 |
| C6—C5—O5—C1 | 178.65 (18) |
Hydrogen-bond geometry (Å, º)
Cg is the centroid of the C41–C46 benzyl ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2A···O3i | 0.82 | 2.00 | 2.813 (2) | 172 |
| O3—H3A···O5ii | 0.82 | 2.05 | 2.799 (2) | 151 |
| C7—H7C···Cgiii | 0.96 | 2.89 | 3.652 (3) | 137 |
Symmetry codes: (i) x+1/2, −y+3/2, −z; (ii) x−1, y, z; (iii) −x+2, y−1/2, −z+1/2.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: SU2708).
References
- Allen, F. H., Johnson, O., Shields, G. P., Smith, B. R. & Towler, M. (2004). J. Appl. Cryst. 37, 335–338.
- Anderson, J. E. & Ijeh, A. I. (1994). J. Chem. Soc. Perkin Trans. 2, pp. 1965–1967.
- Ansaruzzaman, M., Albert, M. J., Holme, T., Jansson, P.-E., Rahman, M. M. & Widmalm, G. (1996). Eur. J. Biochem. 237, 786–791. [DOI] [PubMed]
- Brandenburg, K. (2001). DIAMOND Crystal Impact GbR, Germany.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Eklund, R., Lycknert, K., Söderman, P. & Widmalm, G. (2005). J. Phys. Chem. B, 109, 19936–19945. [DOI] [PubMed]
- Eriksson, L., Söderman, P. & Widmalm, G. (1999). Acta Cryst. C55, 1736–1738.
- Eriksson, L. & Widmalm, G. (2012). Acta Cryst. E68, o2221–o2222. [DOI] [PMC free article] [PubMed]
- Haines, A. H. (1969). Carbohydr. Res. 10, 466–467.
- Handa, V. K., Piskorz, C. F., Barlow, J. J. & Matta, K. L. (1979). Carbohydr. Res. 74, C5–C7.
- Herget, S., Toukach, P. V., Ranzinger, R., Hull, W. E., Knirel, Y. A. & von der Lieth, C.-W. (2008). BMC Struct. Biol. 8, article No. 35 (pp. 1–20). [DOI] [PMC free article] [PubMed]
- Jonsson, K. H. M., Eriksson, L. & Widmalm, G. (2006). Acta Cryst. C62, o447–o449. [DOI] [PubMed]
- Marie, C., Weintraub, A. & Widmalm, G. (1998). Eur. J. Biochem. 254, 378–381. [DOI] [PubMed]
- Olsson, U., Lycknert, K., Stenutz, R., Weintraub, A. & Widmalm, G. (2005). Carbohydr. Res. 340, 167–171. [DOI] [PubMed]
- Oxford Diffraction (2004). CrysAlis CCD and CrysAlis RED Oxford Diffraction Ltd, Abingdon, England.
- Säwén, E., Östervall, J., Landersjö, C., Edblad, M., Weintraub, A., Ansaruzzaman, M. & Widmalm, G. (2012). Carbohydr. Res. 348, 99–103. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Varki, A., Cummings, R., Esko, J., Freeze, H., Hart, G. & Marth, J. (1999). Editors. Essentials of Glycobiology Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, rp1. DOI: 10.1107/S1600536814007922/su2708sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536814007922/su2708Isup2.hkl
CCDC reference: 996312
Additional supporting information: crystallographic information; 3D view; checkCIF report



