Abstract
The way in which the DNA damage response signals the presence of DNA lesions and mediates DNA repair has not been fully elucidated. Now, Gao et al. reveal that diRNA-Ago2 complexes recruit RAD51 to the break sites.
Eukaryotic genomic DNA is bound and protected by a variety of chromatin proteins, but cells in our body receive constant threats from endogenous and environmental agents on their DNA1,2,3,4. Therefore, cells have evolved mechanisms, collectively referred to as the DNA damage response (DDR) that detects DNA lesions, signals their presence, and mediates repair. Double-strand DNA breaks (DSBs) represent the most harmful DNA lesions because if not repaired correctly, they can trigger permanent growth arrest and cell death, and even induce oncogenic translocations of chromosomes. Thus, DSB signaling and repair are crucial to maintain genome integrity.
There are two principal eukaryotic repair mechanisms: non-homologous end-joining (NHEJ) which is error-prone and can operate in any phase of the cell cycle, and homologous recombination (HR) which is restricted to the late S and G2 phases of growing cells because it uses sister chromatid sequence as a template to mediate faithful repair. The basic biochemical requirements of DSB repair directed by HR can be broken down into four steps1,2,3: (1) sensing and resecting of the DSB by nuclease and helicase complexes to produce long stretches of 3′ single-stranded DNA (ssDNA). These ssDNAs are coated by replication protein A (RPA) with concomitant recruitment of protein kinases such as ataxia telangiectasia (ATR), which in turn phosphorylate a histone H2A variant H2AX to generate γH2AX on the chromatin in cis, starting from the region most proximal to the DNA lesion and spreading distally; (2) RPA is then displaced by RAD51 to form a RAD51-ssDNA filament; (3) the RAD51-ssDNA filament then searches for and synapses with a homologous sequence within the undamaged double-stranded DNA (dsDNA) template to form a physical connection between the invading ssDNA and the homologous dsDNA target, generating a displacement D-loop in which DNA synthesis is initiated to replace the DNA surrounding the former break site; (4) finally, resolution of the D-loop occurs either by dissociation of one of the invading strands or through migration of double Holliday junction intermediates.
Each of these steps requires the concerted activity of several factors, including small RNAs. Qi and colleagues recently discovered a class of 21 nt-long small RNAs, which are induced by DSBs and originate from the vicinity of the DSBs by a Dicer-dependent mechanism5. These small RNAs, termed DSB-induced small RNAs (diRNAs), are loaded onto Ago2, the key component of the RNA-induced silencing complex (RISC) in RNAi6. Dicer or Ago2 knockdown reduces DSB repair efficiency, suggesting a model in which diRNAs trigger DSB repair by guiding Ago2 complexes (diRISCs) to target DSB sites by means of base pairing. This model also implies that diRISCs recruit DSB repair factors to the target DSB sites. Similar small RNAs induced by DDR have been reported in other systems7,8,9. However, the specific function of small RNAs has remained largely unknown.
Gao et al.10 now take advantage of the DR-GFP/U2OS HR reporter system to determine whether Ago2 and other RNAi factors contribute to DSB repair. This system uses a modified nonfunctional green fluorescent protein (GFP) gene with an I-SceI endonuclease recognition site to introduce DSBs11. When cleaved, an HR repair event with a linked donor GFP fragment restores functional GFP expression, which is readily detected by flow cytometry. The authors found that depletion of tested RNAi factors impairs HR repair in the reporter system. It was also revealed that the function of diRNAs in HR repair specifically relies on Ago2 among four Argonautes (Ago1-4) expressed in human cells. Ago2 is the only Argonaute that exhibits small RNA-directed endonuclase activity or Slicer activity in mammals5.
To determine whether diRNAs could directly participate in HR repair, the authors incubated Dicer-depleted cells with small RNAs isolated from I-SceI-transfected reporter cells. Small RNAs isolated from DSB-introduced cells could restore HR repair in Dicer-depleted cells. These findings show that RNAi directly mediates HR repair and suggest that both diRNAs and Ago2, and therefore presumably diRISCs, are required for certain steps in the repair.
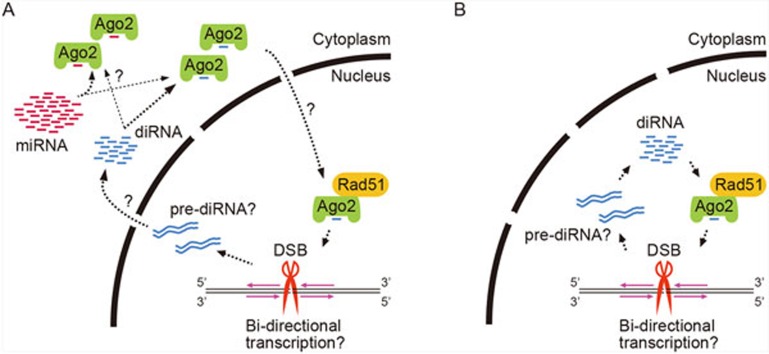
An important step toward understanding how Ago2 mediates HR repair is to determine its precise localization on chromatin in DSB-introduced cells. The authors performed chromatin immunoprecipitation and observed enrichment of Ago2 at I-SceI-induced DSB sites. This prompted them to examine whether Ago2 interacts with DSB repair proteins. In untreated cells, Ago2 interacted specifically with RAD51 among other tested DDR proteins and this interaction was enhanced by ionizing radiation (IR) treatment that generated DSBs. RAD51, a eukaryotic homolog of E. coli RecA, plays a central role in the strand exchange during HR and accumulates at DSB sites (RAD51 foci) upon DNA damage3,4. To examine whether the efficient formation of RAD51 foci depends on diRNAs and/or Ago2, the authors performed immunostaining and found that depletion of Ago2 or Dicer severely impaired RAD51 recruitment at DSBs in IR-treated cells. However, accumulation of other DDR factors including γH2AX and RPA was unaffected. Although this contradicts an earlier report that Dicer and Drosha regulate the recruitment of many DDR proteins8, the current findings suggest that Ago2 directly recruits RAD51 to DSBs probably with the aid of diRNAs that originate from the DSBs (Figure 1).
Figure 1.
Two models of diRNA biogenesis and its Ago2 loading pathways. Both models depend on bi-directional transcription (magenta arrow) around the DSB site (red scissors) and the presence of a double-stranded intermediate transcript of diRNA (pre-diRNA). The main difference between models A and B is the location of Ago2-diRNA complex formation.
Finally, to determine whether small RNA binding and/or Slicer activity of Ago2 is required for the recruitment of RAD51 to DSB sites, the authors used Ago2 mutants deficient for small RNA binding or Slicer. These mutants interacted with RAD51, indicating that both small RNA binding and Slicer activity are dispensable for the Ago2-RAD51 association. However, the Ago2 mutants did not restore recruitment of RAD51 to DSB sites nor rescue HR repair in Ago2-depleted cells. Based on these data, the authors propose a new model for RAD51 recruitment to target DSB sites: namely, transcripts produced from the DSB vicinity form dsRNAs that are converted into diRNA duplexes by Dicer. These duplexes are loaded onto Ago2 and are unwound by Ago2 Slicer activity, where one strand is cleaved by Slicer and displaced by Ago2. diRISCs with RAD51 then target DSBs using base pairing with DNA sequences surrounding the break sites or RNA transcripts generated from around the break sites.
The work by Gao and colleagues opens the door to studies of how DSB responses signal the presence of DNA lesions and mediate their repair. However, there are many future challenges. For example, how is the transcription of diRNA precursors initiated at DSB sites? Where are diRNAs produced in the cell (Figure 1)? It is conceivable that diRNAs are produced and loaded onto Ago2 to form diRISCs in the cytoplasm, similar to mammalian miRNAs which are produced from precursors by Dicer. If so, how are diRISCs imported into the nucleus while miRNA-primed Ago2-RISCs remain in the cytoplasm? Once imported into the nucleus, how is the Ago2-RAD51 interaction enhanced upon IR treatment? Moreover, how are diRISC-RAD51 repair intermediates stabilized and retained at DSB sites? By analogy with heterochromatin assembly in the fission yeast S. pombe in which the RNAi pathway targets both transcripts of silenced loci and histone H3 lysine 9 methylation12, diRISCs may also target both transcripts at DSBs and specific histone modifications such as γH2AX.
References
- Jackson SP, Bartek J. Nature. 2009. pp. 1071–1078. [DOI] [PMC free article] [PubMed]
- d'Adda di Fagana F. Trends Cell Biol. 2014. pp. 171–178. [DOI] [PubMed]
- Krejci L, Altmannova V, Spirek M, et al. Nucleic Acids Res. 2012. pp. 5795–5818. [DOI] [PMC free article] [PubMed]
- Forget al, Kowalczykowski SC. Trends Cell Biol. 2010. pp. 269–276. [DOI] [PMC free article] [PubMed]
- Wei W, Ba Z, Gao M, et al. Cell. 2012. pp. 101–112. [DOI] [PubMed]
- Siomi H, Siomi MC. Nature. 2009. pp. 396–404. [DOI] [PubMed]
- Lee HC, Chang SS, Choudhary S, et al. Nature. 2009. pp. 274–277. [DOI] [PMC free article] [PubMed]
- Francia S, Michelini F, Saxena A, et al. Nature. 2012. pp. 231–235. [DOI] [PMC free article] [PubMed]
- Michalik KM, Bottcher R, Forstemann K. Nucleic Acids Res. 2012. pp. 9596–9603. [DOI] [PMC free article] [PubMed]
- Gao M, Wei W, Li MM, et al. Cell Res. 2014. pp. 532–541. [DOI] [PMC free article] [PubMed]
- Pierce AJ, Johnson RD, Thompson LH, et al. Genes Dev. 1999. pp. 2633–2638. [DOI] [PMC free article] [PubMed]
- Reyes-Turcu FE, Grewal SI. Curr Opin Genet Dev. 2012. pp. 156–163. [DOI] [PMC free article] [PubMed]