Abstract
Hedgehog (Hh) signaling plays vital roles in animal development and tissue homeostasis, and its misregulation causes congenital diseases and several types of cancer. Suppressor of Fused (Su(fu)) is a conserved inhibitory component of the Hh signaling pathway, but how it is regulated remains poorly understood. Here we demonstrate that in Drosophila Hh signaling promotes downregulation of Su(fu) through its target protein HIB (Hh-induced BTB protein). Interestingly, although HIB-mediated downregulation of Su(fu) depends on the E3 ubiquitin ligase Cul3, HIB does not directly regulate Su(fu) protein stability. Through an RNAi-based candidate gene screen, we identify the spliceosome factor Crooked neck (Crn) as a regulator of Su(fu) level. Epistasis analysis indicates that HIB downregulates Su(fu) through Crn. Furthermore, we provide evidence that HIB retains Crn in the nucleus, leading to reduced Su(fu) protein level. Finally, we show that SPOP, the mammalian homologue of HIB, can substitute HIB to downregulate Su(fu) level in Drosophila. Our study suggests that Hh regulates both Ci and Su(fu) levels through its target HIB, thus uncovering a novel feedback mechanism that regulates Hh signal transduction. The dual function of HIB may provide a buffering mechanism to fine-tune Hh pathway activity.
Keywords: Hh, Su(fu), HIB, Crn, SPOP, Cul3
Introduction
Hedgehog (Hh) signaling plays essential roles in embryonic development and adult tissue homeostasis in species ranging from insects to human1,2,3,4. Aberrant Hh signaling activity is associated with many human disorders including birth defects and cancers4,5,6,7.
Hh transduces signal through binding to its receptor, a 12-transmembrane protein Patched (Ptc), which alleviates Ptc-mediated suppression of Smoothened (Smo), a GPCR-like seven-transmembrane protein8,9,10,11,12,13,14. Activated Smo functions as the Hh signal transducer to recruit an intracellular protein complex containing the kinesin-like protein Cos2 and the kinase Fused (Fu), leading to Fu phosphorylation and activation15,16,17,18,19,20,21,22,23. Activated Fu then inhibits Ci processing to prevent the production of a Ci repressor form (CiR) and antagonizes Su(fu) to convert the latent full-length Ci into an activator form (CiA)21,22,24, leading to the expression of target genes including decapentaplegic (dpp), ptc and engrailed (en)1,25,26.
In Drosophila wing discs, posterior (P) compartment cells produce Hh that moves into anterior (A) compartment to form a concentration gradient and induces the A-compartment cells of the A/P boundary to express dpp. Dpp functions as a long-range morphogen and moves into both the A and P compartments to control the growth and patterning of cells in the whole wing1,27,28. Hh also functions as a local morphogen to specify cell patterning near the A/P boundary with low, intermediate and peak levels of Hh to induce dpp, ptc and en expression, respectively29,30.
In A-compartment cells distant from the A/P boundary where Hh is very low or absent, Ci is processed into a truncated form Ci75 by the SCF (Skp1-Cullin-F-box protein complex) E3 ligase containing the F-box protein Slimb31. Ci75 (also called CiR) functions as a transcription repressor to block the expression of a subset of Hh target genes, including dpp31,32,33,34,35,36,37, whereas in A-compartment cells near the A/P boundary Hh signaling antagonizes Ci processing and converts the accumulated full-length Ci (Ci155) into an active but unstable form (CiA), which activates Hh target gene expression24. The BTB protein HIB is upregulated by Hh signaling in A-compartment cells abutting the A/P boundary, where it forms a complex with Cul3 (Cul3-HIB) that functions as an ubiquitin E3 ligase. HIB recognizes the active form of Ci and targets it for ubiquitin/proteasome-mediated degradation to attenuate Hh signaling activity in a negative feedback loop38,39,40,41,42,43.
In Drosophila, Su(fu) is a genetic suppressor of the Fu kinase and is antagonized by Fu function44,45,46,47,48,49,50. Su(fu) and the kinesin-like protein Cos2 retain Ci155 in the cytoplasm to oppose its nuclear import51,52,53,54,55,56. Studies in mammals show that SUFU can both tether Gli in the cytoplasm and enter the nucleus with Gli1 to repress transcription from Gli-dependent promoters through specific interaction with SAP18, a component of the Sin3-HDAC co-repressor complex46,57,58. In mice, abrogation of sufu leads to ligand-independent Hh signaling activation, and homozygous sufu mutant embryos die at mid-gestation with a ventralized spinal cord59,60. Also, sufu mutations are present in 9% of medulloblastomain human patients, suggesting that sufu is a tumor suppressor gene61.
Although Su(fu)/SUFU plays a conserved role in regulating Ci/Gli activity, its regulation remains poorly understood. One study has identified that Hh signaling promotes SUFU degradation through the ubiquitin-proteasome pathway in mammalian NCI-H322M lung cell line, but the E3 ligase involved remains unknown62.
In this study, we demonstrate that Hh signaling can downregulate Su(fu) through its target protein HIB. Interestingly, regulation of Su(fu) by HIB depends on Cul3 but occurs at a level distinct from Su(fu) degradation. Through a candidate gene screen, we find that the spliceosome factor Crn can modulate Su(fu) level. Epistasis analysis suggests that HIB downregulates Su(fu) through Crn. Furthermore, we provide evidence that HIB and Crn may inhibit the formation of functional Su(fu) mRNA but not its nuclear export. Finally, we show that the mammalian homologue of HIB, SPOP, can substitute HIB to downregulate Su(fu) in flies.
Results
Hh signaling downregulates Su(fu)
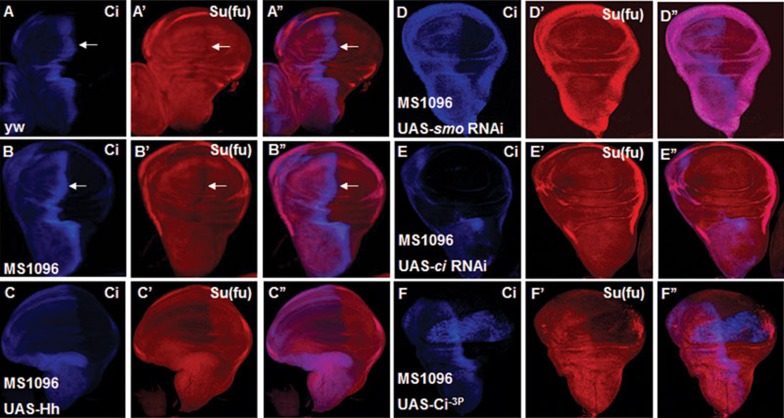
In Drosophila, Su(fu) forms a complex with Ci and inhibits its transcriptional activity through tethering Ci in the cytoplasm and inhibits Ci activity even after Ci enters the nucleus55,63; however, how Su(fu) is regulated by Hh signaling is still poorly understood. In wild-type wing discs either from yw or from MS1096 Gal4-expressing flies, Su(fu) level is lower in A-compartment cells near the A/P boundary where Hh signaling is activated (arrows in Figure 1A-1A'', 1B-1B''). This observation prompted us to test whether Hh signaling is involved in Su(fu) downregulation. We overexpressed UAS-Hh through the MS1096 Gal4 line, which drives target gene expression in the pouch region of the wing disc, and found that Hh severely reduced Su(fu) level in wing pouch (Figure 1C-1C''). Of note, overexpression of Hh could only dramatically downregulate Su(fu) in A-compartment cells but not P-compartment cells where Ci is not expressed (Figure 1C'), suggesting that Hh signaling but not Hh itself is essential for Su(fu) downregulation. To verify this hypothesis, we knocked down smo and ci by expressing UAS-smo-RNAi and UAS-ci-RNAi, respectively, with MS1096 and found that the level of Su(fu) near A/P boundary increased to the level similar to that outside the A/P boundary (Figure 1D-1E''). Furthermore, MS1096-driven overexpression of UAS-Ci−3P, which is one active form of Ci with three mutated PKA kinase sites39, downregulated Su(fu) in both A and P compartments (Figure 1F-1F''). Taken together, these results support the notion that Hh signal transduction is both necessary and sufficient for downregulating Su(fu).
Figure 1.
Hh signaling is sufficient for downregulating Su(fu). All the wing discs shown in this study are oriented with anterior to the left and ventral to the top. UAS transgene expression with MS1096 gal4 are usually stronger in the dorsal region than in the ventral of wing pouch. (A-A'') A wing disc from yw flies was immunostained to show full-length Ci (CiFL) (blue) and Su(fu) (red) expression by immunostaining with corresponding antibodies. Of note, Su(fu) level is lower in A-compartment cells abutting the A/P boundary. (B-B'') A wing disc of MS1096 gal4 line was immunostained to show full-length Ci (CiFL) (blue) and Su(fu) (red) expression. Su(fu) level is also lower in A/P boundary. (C-C'') Overexpression of UAS-Hh with MS1096 decreased Su(fu) level in A-compartment cells but not in P-compartment cells where Ci is not expressed. (D-E'') MS1096-driven knockdown of smo (D-D'') or ci (E-E'') resulted in Ci reduction in the pouch of wing discs and Su(fu) (red) level at the A/P boundary was upregulated to an equal level outside the A/P boundary. (F-F'') UAS-Ci-3P, which is an active form of Ci with three PKA kinase sites mutated, was expressed with MS1096 in wing discs. Su(fu) (red) level was decreased in both anterior and posterior compartments of the wing pouch.
Hh signaling downregulates Su(fu) through its target HIB
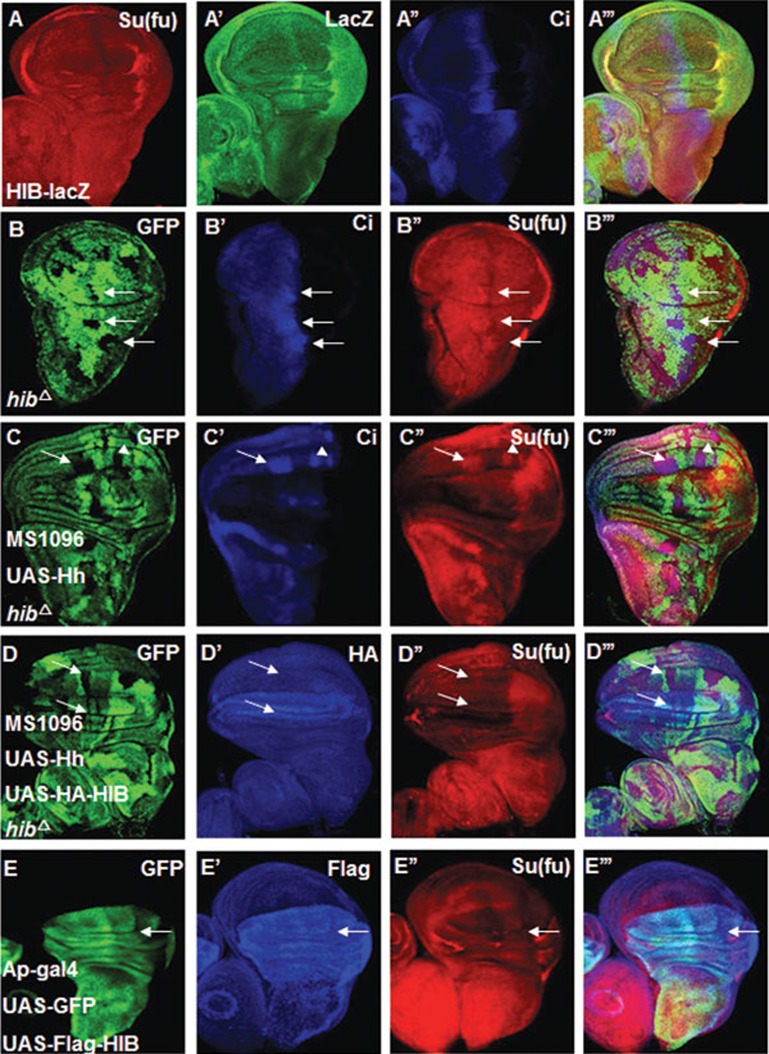
We next asked how Hh signaling downregulates Su(fu). hib is a target gene of Hh signaling pathway and its upregulation overlaps with low Su(fu) domain, as indicated by LacZ staining from the hib-lacZ enhancer trap line, which mimics endogenous hib expression pattern (Figure 2A-2A''')39. HIB could function as a substrate recognition component of Cul3-based E3 ubiquitin ligase to promote ubiquitination and proteasome-mediated degradation of its target proteins39. These observations prompted us to test whether HIB mediates the reduction of Su(fu) induced by Hh signaling. hibΔ mutant clones were generated using the FRT/FLP-mediated mitotic recombination in wing discs and were marked by the lack of GFP expression. As shown in Figure 2B-2B''', Su(fu) level was upregulated in hibΔ mutant clones in A-compartment cells near the A/P border. In addition, overexpression of Hh by MS1096 failed to downregulate Su(fu) in anteriorly situated hibΔ mutant clones (arrows in Figure 2C-2C'''). Co-expression of UAS-HIB with UAS-Hh restored the ability of Hh to downregulate Su(fu) in hibΔ mutant clones (arrows in Figure 2D-2D'''). These results suggest that HIB is responsible for downregulating Su(fu) in response to Hh. Furthermore, overexpression of HIB in the dorsal (D) compartment of wing discs using the Ap-Gal4 driver downregulated Su(fu) in both A- and P-compartment cells of the dorsal regions (arrows in Figure 2E-2E'''). Taken together, these results support the notion that Hh signaling induces HIB, which in turns downregulates Su(fu).
Figure 2.
Hh signaling downregulates Su(fu) through HIB. (A-A''') LacZ expression in wing discs from a hib enhancer trap line, l(3)03477 (hib-Z). High LacZ staining (green) overlapped with low Su(fu) (red) region along the A/P border. (B-B''') Su(fu) (red) was upregulated in the hibΔ clones near the A/P boundary. hibΔ clones were marked by the lack of GFP expression and were indicated by arrows. (C-C''') A wing disc carrying hibΔ clones and expressing UAS-Hh with MS1096 was immunostained with GFP (green), CiFL (blue) and Su(fu) (red) antibodies. Su(fu) and CiFL accumulated in hibΔ clones when Hh was overexpressed. (D-D''') Co-expression of UAS-Hh and UAS-HA-HIB (blue) with MS1096 could erase the accumulated Su(fu) (red) inside hibΔ clones of wing discs. (E-E''') Wing discs expressing UAS-Flag-HIB plus UAS-GFP with Ap-Gal4 were immunostained with Flag (blue), GFP (green) and Su(fu) (red) antibodies. GFP marks the cells that express UAS-Flag-HIB. HIB overexpression led to diminishing Su(fu) level in both A- and P-compartment cells in the dorsal region of wing pouch (indicated by arrows).
Regulation of Su(fu) level by HIB depends on E3 ligase Cul3
HIB contains an N-terminal MATH domain and a C-terminal BTB domain39. Usually the BTB domain interacts with the Cul family member Cul3 and the MATH domain recognizes the substrates for the 26S proteasome degradation64. Our previous study showed that HIB interacts with Cul3 to form an E3 ligase complex that mediates Ci degradation39,43. We therefore tested whether HIB acts in conjunction with Cul3 to regulate Su(fu).
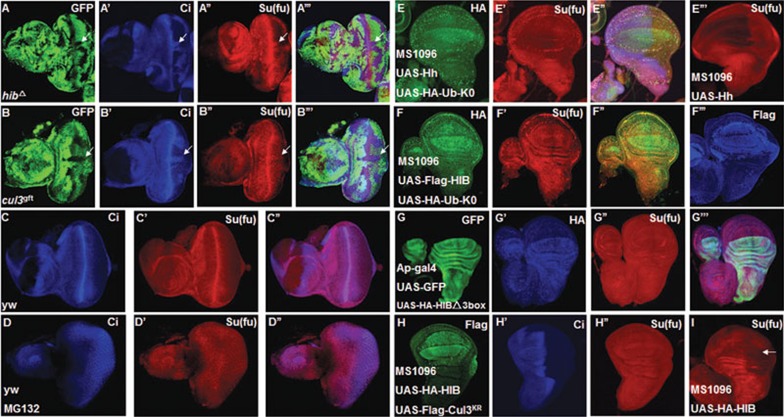
We made hibΔ mutant clones or mutant clones for a strong loss-of-function allele of cul3, cul3gft2, in eye discs since hib is expressed specifically in the posterior region of eye discs. We found that both hibΔ and cul3gft2 clones in the posterior region of eye discs accumulated Su(fu) (Figure 3A-3B'''), suggesting that both HIB and Cul3 are necessary for the downregulation of Su(fu).
Figure 3.
HIB downregulates Su(fu) depending on Cul3 and the ubiquitin/proteasome pathway. (A-A''') hibΔ clones were generated in eye discs, followed by immunostaining with GFP (green), CiFL (blue) and Su(fu) (red) antibodies. CiFL and Su(fu) were accumulated in posterior hibΔ clones. Clones were marked by the lack of GFP expression and indicated by arrows. (B-B''') An eye disc carrying cul3gft2 clones was immunostained to show the accumulation of CiFL (blue) and Su(fu) (red) in the posterior compartments of eye discs. (C-C'') Endogenous Su(fu) expression pattern was shown in eye discs. (D-D'') Proteasome inhibitor MG132 blocked Su(fu) reduction in the posterior region of eye discs. (E-E''') Co-expression of Ub-K0 and UAS-Hh with MS1096 attenuated the Su(fu) reduction in wing discs. (F-F''') Co-expression of Ub-K0 and UAS-HIB with MS1096 blocked the Su(fu) reduction in wing discs. (G-G''') HIB-Δ3box did not downregulate Su(fu) in wing discs. HIB-Δ3box has lost Cul3-HIB E3 ligase activity due to lack of its C-terminal 299 aa-330 aa that mediates formation of a functional Cul3-HIB complex. (H-I) UAS-HIB was expressed alone (I) or together with UAS-Cul3KR (H-H''), a dominant-negative form of Cul3, in the wing discs using MS1096. Co-expression of UAS-Cul3KR with UAS-HIB prevented HIB-mediated reduction of Su(fu).
To explore whether Cul3-HIB downregulates Su(fu) through the ubiquitin/proteasome pathway, we firstly employed a proteasome inhibitor MG132 to block 26S proteasome-mediated degradation in eye discs and found that MG132 blocked Su(fu) downregulation in the posterior region of eye discs (Figure 3D-3D''), suggesting that 26S proteasome is involved in Su(fu) regulation. Furthermore, we applied the Ub-K0, which blocks polyubiquitination. To test whether polyubiquitination is necessary for Su(fu) regulation, we co-expressed Ub-K0 with UAS-Hh (Figure 3E-3E'') or UAS-HIB (Figure 3F-3F''') in wing discs, and found that Ub-K0 attenuated both Hh- and HIB-mediated Su(fu) reduction in wing discs, suggesting that polyubiquitination is indispensable for Su(fu) reduction. Moreover, we overexpressed HIB-Δ3box by AP-Gal4 in wing discs to investigate the change of Su(fu) level. Compared with wild-type HIB, HIB-Δ3box lacks amino acids from 299 aa to 330 aa that are essential for conjugating with Cul3 and forming a functional Cul3-HIB E3 ligase (our unpublished data). We found that HIB-Δ3box no longer downregulated Su(fu) level (Figure 3G-3G'''). We also applied a dominant-negative form of Cul3, designated Cul3KR, to address the effect of Cul3 on Su(fu) level in wing discs. Cul3KR impairs its Cul3 E3 ligase activity due to the lysine (K) to arginine (R) replacement on the neddylation site39,65. We found that co-expression of Cul3KR with HIB blocked Su(fu) downregulation mediated by HIB (Figure 3H-3I). Taken together, these results suggest that Cul3-HIB E3 ligase activity and the ubiquitin/proteasome pathway are required for Su(fu) downregulation.
HIB does not directly regulate Su(fu)
In the Cul3-HIB E3 ligase complex, HIB specifically recognizes substrates for ubiquitination through direct binding to them39,64. Therefore, we test whether HIB forms a complex with Su(fu) by immunoprecipitation (IP) experiments. Intriguingly, the result showed that HIB did not bind Su(fu) in S2 cells (Supplementary information, Figure S1A). Previous studies indicate that HIB/SPOP binds S/T-rich degrons that are found in multiple copies in a large number of HIB-interacting proteins43,66. Su(fu) does not contain such motifs. Taken together, these observations suggest that Su(fu) is not a direct substrate of the Cul3-HIB E3 ligase and its level is indirectly affected by Cul3-HIB.
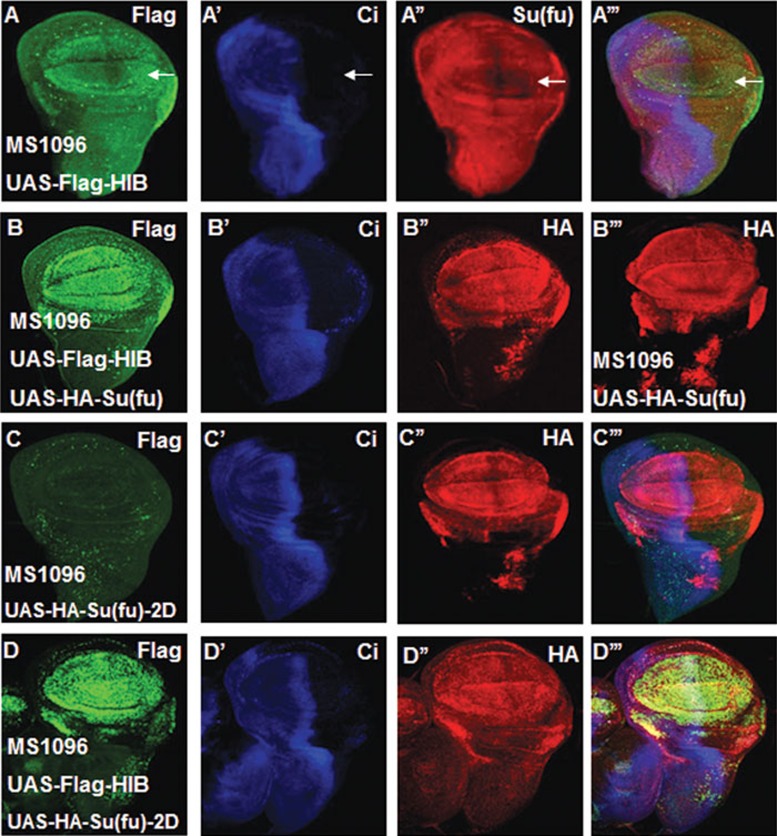
In Drosophila wing discs, overexpression of Flag-HIB downregulated endogenous Su(fu), as indicated by anti-Su(fu) antibody staining (Figure 4A-4A'''); however, co-expression of Flag-HIB with HA-Su(fu) did not affect the exogenously expressed Su(fu) recognized by anti-HA antibody staining (Figure 4B-4B'''). To further determine whether HIB regulates Su(fu) protein stability, we transfected S2 cells with Flag-HIB alone or together with HA-Su(fu) and treated the cells with cycloheximide (CHX) to block protein synthesis. We found that HIB did not affect protein stability of the endogenous Su(fu) or exogenous HA-Su(fu) (Supplementary information, Figure S1B and S1C); similarly, Hh did not affect the stability of endogenous Su(fu) in S2 cells (Supplementary information, Figure S4B), suggesting that Su(fu) is unlikely to be affected by Cul3-HIB at post-translational stage.
Figure 4.
HIB does not affect Su(fu) stability. (A-A''') A wing disc overexpressing UAS-Flag-HIB with MS1096 was immunostained with Flag (green), CiFL (blue) and Su(fu) (red) antibodies. Overexpression of HIB downregulated the endogenous Su(fu) level (arrow in A'') in the dorsal region of wing pouch. (B-B''') HA-Su(fu) was expressed alone (B''') or together with Flag-HIB (B-B'') with MS1096. Overexpression of HIB did not change the exogenous HA-Su(fu) (red) level shown by HA antibody staining (compare B'' with B'''). (C-D''') Wing discs overexpressing HA-Su(fu)-2D alone (C-C''') or together with Flag-HIB (D-D''') with MS1096 were immunostained to show the expression of HA-Su(fu)-2D, Flag-HIB and Ci. Co-expression of Flag-HIB with HA-Su(fu)-2D did not change HA-Su(fu)-2D level (compare D'' with C'').
CKI and Fu kinases trigger Su(fu) phosphorylation at S321 and S324 upon Hh stimulation21. To test whether phosphorylated Su(fu) at S321 and S324 stimulated by Hh facilitates its reduction by Cul3-HIB E3 ligase, we made a construct HA-Su(fu)-2D in which amino acids S321 and S324 were changed to aspartic acids (D) to mimic phosphorylation. We overexpressed HA-Su(fu)-2D together with HIB in wing discs and found that HA-Su(fu)-2D level did not change in the presence of HIB overexpression (Figure 4D-4D'''). In addition, we co-transfected HA-Su(fu)-2D and Flag-HIB into S2 cells and treated them with CHX for different periods of time, followed by western blot analysis. We found that HA-Su(fu)-2D stability was not affected by HIB (Supplementary information, Figure S1D). These results rule out the possibility that phosphorylation of Su(fu) at S321 and S324 might render its regulation by Cul3-HIB.
HIB affects Su(fu) level through Crn
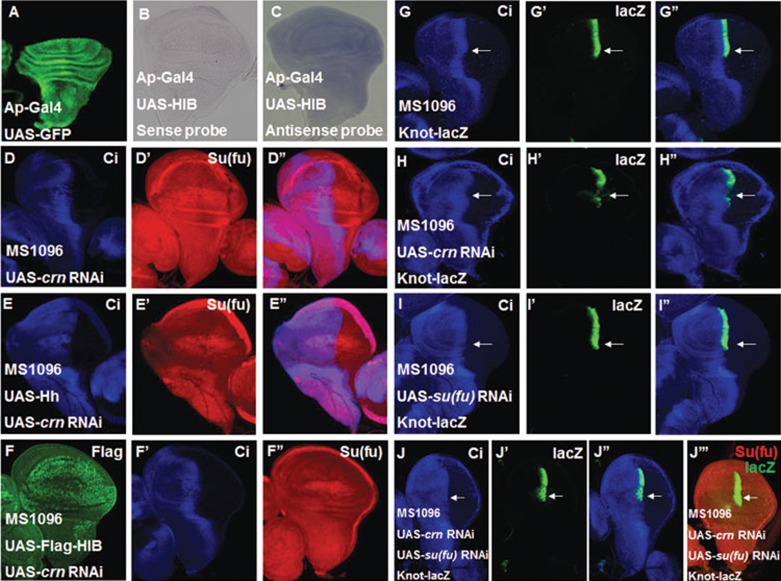
According to the above results, Su(fu) level is not affected by HIB at post-translation stage, raising the possibility that Su(fu) could be regulated at the level of mRNA. To determine whether HIB regulates Su(fu) mRNA expression, we overexpressed HIB with AP-Gal4 in the dorsal region of the wing disc and used the ventral region of the same disc as internal control. We checked the su(fu) mRNA level by in situ hybridization and found that overexpression of HIB driven by AP-Gal4 did not affect su(fu) mRNA level in dorsal region of the wing discs (Figure 5A-5C). In addition, knocking down hib in S2 cells did not change endogenous su(fu) mRNA level (Supplementary information, Figure S6B). However, it is still possible that HIB affects Su(fu) mRNA nuclear export or inhibits the formation of functional su(fu) mRNA to finally result in downregulating Su(fu).
Figure 5.
HIB inhibits Su(fu) level through splicesome factor Crooked neck (Crn). (A) UAS-GFP (green) marked the Ap-Gal4-mediated gene expression pattern. AP-Gal4 drives UAS transgenes to be specifically expressed in the dorsal region of wing discs. (B, C) Overexpression of HIB with AP-Gal4 did not change su(fu) mRNA level in the dorsal region of wing discs as determined by in situ hybridization assay. Su(fu) sense probe was used as control (B), and Su(fu) antisense probe detected su(fu) mRNA level (C). (D-D'') Knockdown of crn with MS1096 upregulated Su(fu) protein level in the wing pouch region. (E-F'') Wing discs expressing UAS-crn-RNAi together with UAS-Hh (E-E'') or UAS-Flag-HIB (F-F'') were immunostained with CiFL (blue) and Su(fu) (red) antibodies. Simultaneous overexpression of UAS-Hh or UAS-Flag-HIB and UAS-crn-RNAi phenocopied crn knockdown. (G-J'') crn knockdown through MS1096 significantly reduced the expression of Knot-lacZ (indicated by arrows) near the A/P border (compare H' with G'), and co-overexpression of su(fu) RNAi prevented the reduction in Knot-lacZ expression (indicated by arrows) in wing discs (I-J''). (J''') Co-overexpression of su(fu) RNAi downregulated endogenous Su(fu) level (red) revealed by immunostaining with an anti-Su(fu) antibody. Of note, MS1096 exhibited higher expression in the dorsal region than in the ventral region of wing discs.
Considering that HIB E3 ligase activity is necessary for Su(fu) downregulation, we reasoned that HIB inhibits Su(fu) through an intermediate substrate. To identify the HIB substrate that regulates Su(fu), we knocked down over 30 genes that encode putative HIB-interacting proteins annotated by the flyBase through their transgenic RNAi expression in wing discs and examined Su(fu) level via immunostaining with the anti-Su(fu) antibody (Supplementary information, Table S1). We found that knockdown of the spliceosome factor Crn using MS1096 resulted in an upregulation of Su(fu) level (Figure 5D-5D''). Of note, the more dramatic effect on Su(fu) expression in the dorsal region is because MS1096 drives higher levels of UAS-crn-RNAi in this region. Furthermore, we found that knockdown of crn blocked the downregulation of Su(fu) induced by overexpression of either Hh or HIB (Figure 5E-5F''). Similarly, we found that knockdown of crn in S2 cells upregulated the level of endogenous Su(fu) in the absence or presence of HIB or Hh overexpression (Supplementary information, Figure S6A, S6C and S6E). Taken together, these results suggest that Crn may act downstream of Hh and HIB to downregulate Su(fu). Multiple crn RNAi lines showed the same effect on Su(fu) level and a UAS-Crn transgene could rescue the phenotype of crn RNAi, suggesting that Su(fu) upregulation is caused by crn inactivation but not by an off-target effect (data not shown). Finally, we found that the read-out of high levels of Hh signaling activity, knot-lacZ, was significantly downregulated by crn RNAi (Figure 5G-5H''), and this reduction was alleviated when Su(fu) level was downregulated by co-overexpression of su(fu) RNAi in wing discs (Figure 5I-5J'''), suggesting that inactivation of Crn attenuates Hh signaling activity through Su(fu).
HIB reduces Su(fu) level by modulating Crn subcellular localization
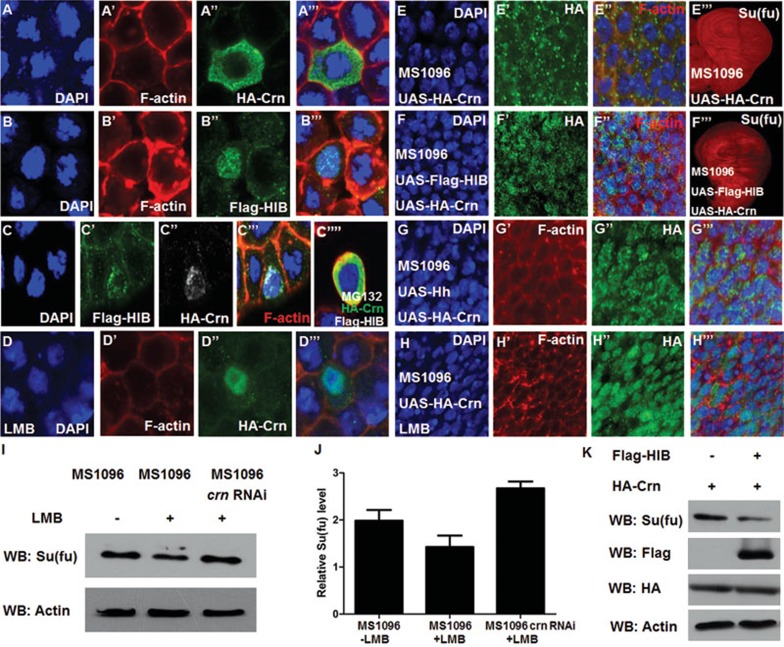
Based on the information shown in flyBase, HIB binds Crn in a yeast two-hybrid assay. However, HIB failed to bind Crn through IP assay when they were co-transfected in S2 cells (Supplementary information, Figure S2), indicating that Crn might not be a direct substrate of HIB-Cul3 E3 ligase. Strikingly, when transfected into S2 cells the majority of HA-tagged Crn (HA-Crn) was localized in the cytoplasm (Figure 6A-6A'''), whereas when co-transfected with HIB, HA-Crn was largely retained in the nucleus and co-localized with HIB (Figure 6C-6C'''). Similarly, when expressed in wing discs, HA-Crn was localized mainly in the cytoplasm in A-compartment cells away from the A/P boundary (Figure 6E-6E''); however, when co-expressed with either HIB (Figure 6F-6F'') or Hh (Figure 6G-6G''', and Supplementary information, Figure S5E-S5E'''), Crn was enriched in the nucleus, suggesting that Hh signaling and HIB can modulate Crn cellular localization. Furthermore, upon treatment with Leptomycin B (LMB), which is a specific nuclear export inhibitor, we found that the majority of Crn was localized in the nucleus both in S2 cells and in the wing discs in the absence of HIB co-overexpression (Figure 6D-6D''', 6H-6H'''). As shown in Figure 6F''', nuclear localization of Crn correlated with lower Su(fu) level in the cells of the wing disc. Meanwhile, in the wing discs treated with LMB, the endogenous Su(fu) level was reduced as shown by western blot analysis (Figure 6I-6J, lane 2 compared with lane 1). Knockdown of crn blocked LMB-induced downregulation of Su(fu) and accumulated Su(fu) to a level higher than that of control discs (Figure 6I-6J, compare lane 3 with lanes 1 and 2). In addition, when co-transfected with HIB in S2 cells, HA-Crn could downregulate endogenous Su(fu) level (Figure 6K). Taken together, these results suggest that HIB may reduce Su(fu) level by modulating Crn cellular localization.
Figure 6.
HIB affects Su(fu) level by modulating Crn cellular localization. (A-C'''') S2 cells transfected with UAS-HA-Crn, UAS-Flag-HIB, or both were immunostained to show the subcellular localization of HA-Crn and Flag-HIB. The nuclei were stained with DAPI (blue) and the membrane was stained with TRITC-labeled phalloidin that preferentially labels filamentous actin (F-actin) (red). When expressed alone, HA-Crn was located mainly in the cytoplasm and barely in the nucleus (A-A'''), whereas Flag-HIB was located in the nucleus (B-B'''). When co-expressed with Flag-HIB, HA-Crn was co-localized with Flag-HIB at speckle-like sites in the nucleus (C-C'''); however, treating cells with MG132 resulted in cytoplasmic localization of HA-Crn (C''''). (D-D''') LMB treatment resulted in nuclear localization of HA-Crn in the absence of Flag-HIB co-expression in S2 cells. (E-G''') Wing discs expressing UAS-HA-Crn alone (E-E''') or together with UAS-Flag-HIB (F-F''') or UAS-Hh (G-G''') with MS1096 were immunostained to show HA (green), Su(fu) (red in E''', F'''), DAPI (blue) and F-actin (red in E'', F''). High-magnification views of anterior compartment cells were shown in (E-E''), (F-F''), and (G-G'''). When expressed alone, HA-Crn was located mainly in cytoplasm (E-E'') and did not affect endogenous Su(fu) level (E'''); however, when co-overexpressed with Flag-HIB, Crn was localized in the nucleus (F-F'') and downregulated endogenous Su(fu) level (F'''). Similarly, when co-overexpressed with UAS-Hh, Crn was also localized in the nucleus (G-G'''). (H-H''') LMB treatment led to nuclear localization of HA-Crn in the wing discs in the absence of HIB or Hh co-expression. (I) Endogenous Su(fu) level was reduced as shown by western blot in LMB-treated wing discs; however, the endogenous Su(fu) level was restored or even upregulated when crn was knocked down at the same time. (J) Quantification of the western blot results of endogenous Su(fu) level in (I) analyzed by ImageJ. (K) Co-expression of Flag-HIB with HA-Crn decreased Su(fu) protein level in S2 cells as determined by western blot analysis. Actin was used as a loading control.
We further checked whether HIB regulates su(fu) mRNA nuclear export through Crn and found that it did not. Cytoplasmic and nuclear mRNA for the Su(fu) coding region were similar among S2 cells transfected with either HIB or Crn alone, or their combination (Supplementary information, Figure S8), suggesting that HIB and Crn downregulate Su(fu), most possibly by inhibiting the formation of functional su(fu) mRNA.
SPOP can regulate Su(fu) in Drosophila
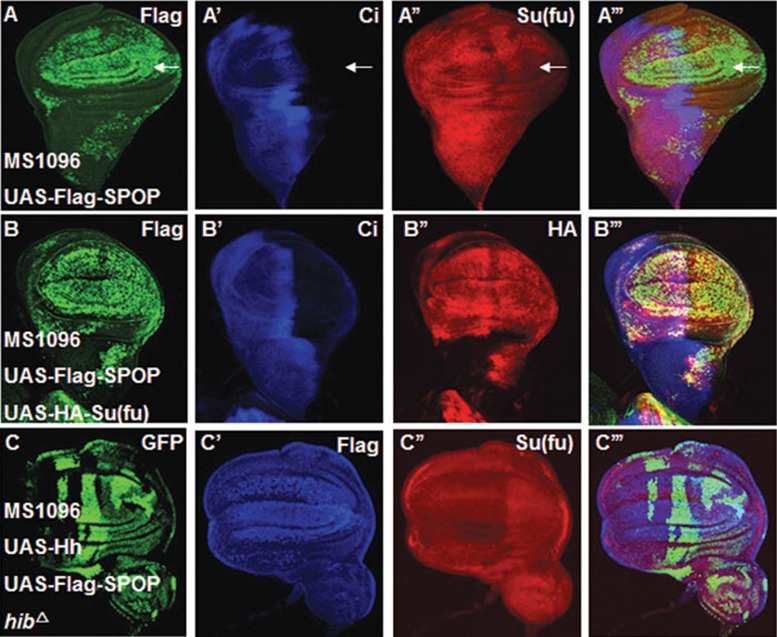
SPOP, the mammalian homologue of HIB, plays a conserved role in regulation of Ci/Gli degradation39,47,67. To investigate whether SPOP can substitute HIB to suppress Su(fu) in flies, first we checked whether SPOP has the ability to regulate Su(fu). The results showed that overexpression of SPOP in wing discs driven by MS1096 or AP-gal4 could downregulate endogenous Su(fu) (Figure 7A-7A''', and Supplementary information, Figure S3A-S3B'''); however, like HIB, overexpression of SPOP together with HA-Su(fu) did not affect exogenous HA-tagged Su(fu) level (Figure 7B-7B'''). Similarly, in S2 cells, exogenous HA-Su(fu) stability was not affected by SPOP (Supplementary information, Figure S3C). Second, when generating hibΔ mutant clones and overexpressing Hh and SPOP with MS1096, we found that SPOP could erase the accumulated Su(fu) inside hibΔ clones (Figure 7C-7C'''). All these results suggest that SPOP can substitute HIB to affect Su(fu) level in the same manner in Drosophila.
Figure 7.
SPOP can substitute HIB to regulate Su(fu). (A-A''') Overexpression of UAS-Flag-SPOP with MS1096 affected endogenous Su(fu) as revealed by staining with anti-Su(fu) antibody. (B-B''') Overexpression of Flag-SPOP together with HA-Su(fu) did not decrease exogenous HA-Su(fu) level as revealed by staining with anti-HA antibody (red). (C-C''') Su(fu) (red) was not accumulated inside hibΔ clones co-expressing UAS-Hh and Flag-SPOP (blue) with MS1096.
Discussion
Su(fu) is a conserved negative regulator of the Hh pathway, but until now how Su(fu) is regulated is largely unknown. Our work demonstrates that in Drosophila Hh signaling can regulate Su(fu) level and that this regulation is mediated by the Hh target HIB that functions in conjunction with Cul3 to downregulate Su(fu). Furthermore, we identify the splicing factor Crn as a mediator of HIB function and put HIB and Crn in the same pathway to achieve the suppression on Su(fu). Finally, we show that SPOP, like HIB, can downregulate Su(fu) in flies.
As a major signaling pathway involved in many key aspects of animal development, the Hh pathway activity needs to be precisely regulated in vivo, which is likely to rely on multiple layers of regulatory mechanisms. Our previous studies demonstrated that the BTB protein HIB is induced by Hh and forms a negative feedback to attenuate Hh signaling activity by targeting the active form of Ci for degradation39,43. Intriguingly, here we found that HIB also downregulates Su(fu) level in vivo, suggesting that HIB plays a dual role in Hh signaling.
In Drosophila wing imaginal discs we found that Su(fu) level is downregulated by overexpression of Hh, and this regulation relies on Hh signal transduction. We further demonstrate that the Hh pathway target gene hib mediates the regulation of Su(fu) through the HIB/SPOP-Crn axis in Drosophila. Of note, since overexpression of Hh is somehow a little bit more efficient to downregulate Su(fu) than overexpression of HIB, we cannot rule out the possibility that Hh acts through additional factors together to downregulate Su(fu).
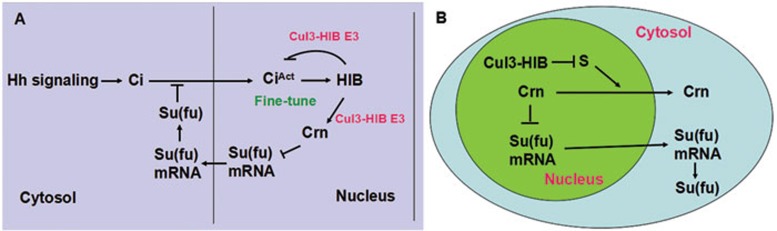
As a target of Hh signaling, hib is upregulated by high levels of Ci activity in A-compartment cells abutting the A/P boundary. HIB-mediated downregulation of Su(fu) in this region may boost the levels of active Ci to counterbalance HIB-mediated degradation of CiA, allowing Ci activity to stay above critical thresholds required for appropriate expression of Hh target genes (Figure 8A). Hence, the dual role of HIB may serve to buffer the steep drop of Ci activity by modulating both Ci and Su(fu), and finally accomplish fine-tuning of Ci activity. The dual role of HIB may explain an earlier observation that Ci accumulated inside hib clones only mildly upregulated dpp-lacZ but not ptc-lacZ level because upregulated Su(fu) might prevent a surge in Ci activity39. Therefore, our study unveils a novel mechanism for fine-tuning Hh pathway activity.
Figure 8.
A model of Su(fu) regulation by HIB. (A) Hh induces hib expression through the transcription factor Ci. HIB degrades activated Ci (CiA) by forming Cul3-HIB E3 ligase complex. Cul3-HIB also downregulates Su(fu) level through Crn that likely interferes with normal translation from su(fu) mRNA. Therefore, Hh modulates both Ci and Su(fu) through its target HIB to fine-tune the pathway activity. (B) Cul3-HIB degrades an unknown substrate “S”, which is essential for Crn nuclear export. In the absence of HIB, “S” promotes cytoplasmic localization of Crn; in the presence of HIB, Cul3-HIB degrades “S” to promote nuclear accumulation of Crn, which inhibits the formation of functional su(fu) mRNA, leading to reduced Su(fu) protein synthesis.
Although in wild-type flies loss of su(fu) does not show overt phenotype due to the presence of another negative regulator Cos2, loss of Su(fu) can impact Hh signaling outcome in a genetically sensitized background, i.e., when the pathway is compromised by expressing a dominant-negative Smo, Smo-PKA. As shown in Supplementary information, Figure S7, expression of Smo-PKA resulted in fusion between veins 3 and 4 in the proximal region of Drosophila adult wings, indicative of reduced Hh signaling. This phenotype was suppressed by removing Su(fu), indicating that Su(fu) plays a negative role in Hh signaling. On the other hand, excessive Su(fu) as shown either by previous overexpression studies or by Crn loss of function can attenuate Hh signaling activity. Hence, altering Su(fu) levels can influence Hh signaling output. Overall, our study demonstrates that Hh regulates both Ci and Su(fu) levels through its target HIB, thus uncovering a novel feedback mechanism that regulates Hh signal transduction. We think that the dual function of HIB may provide a buffer to fine-tune Hh pathway activity to ensure that the appropriate level of pathway activity is achieved for a given level of Hh.
It is interesting that HIB regulates Su(fu) at the mRNA layer instead of protein turnover. We find that HIB does not affect Su(fu) stability, su(fu) mRNA level, or su(fu) mRNA nuclear export (Supplementary information, Figure S8), implying that HIB downregulates Su(fu) most likely by inhibiting the formation of functional su(fu) mRNA. Interestingly, we find that HIB reduces Su(fu) level by modulating Crn subcellular localization. Based on our results that downregulation of Su(fu) by HIB depends on Cul3 and Crn, we propose a working model as shown in Figure 8B. HIB together with Cul3 functions as an E3 ligase to degrade an unknown substrate “S” that is necessary for exporting Crn from nucleus to cytoplasm. Without HIB, Crn is mainly located in the cytoplasm because “S” can promote Crn export, whereas in the presence of HIB, Cul3-HIB E3 ligase degrades “S” to block Crn export. This model is further supported by the observation that MG132 blocked Crn nuclear localization induced by HIB (Figure 6C'''').
Since HIB did not affect su(fu) mRNA nuclear export, Su(fu) gene has no introns and the 5' or 3'UTR structure of su(fu) mRNA plays a very important role in su(fu) translation (Supplementary information, Figure S9A), we speculate that Crn accumulated in the nucleus may affect the 5' or 3'UTR structure of su(fu) mRNA or mediate interaction between 5' and 3'UTR of su(fu) mRNA with some unidentified factors. These changes do not seem to affect su(fu) mRNA export from the nucleus to the cytoplasm, but may affect su(fu) mRNA translation. This idea was supported by the su(fu) mRNA translation assay as shown in Supplementary information, Figure S9B. Compared with control S2 cells, we found more Su(fu) proteins translated from the same amount of su(fu) mRNA upon knockdown of hib during 4 h after blocking the su(fu) transcription with Act D (Supplementary information, Figure S9B and S9C). The exact mechanism by which Crn regulates Su(fu) expression awaits further investigation.
Su(fu)/SUFU plays a conserved role in regulating Ci/Gli activity; however, the regulation of mammalian SUFU remains poorly understood. We found that Hh signaling downregulates Su(fu) through its target HIB/SPOP, which forms a complex with Cul3 to function as an E3 ubiquitin ligase, and that HIB-Cul3 downregulates Su(fu) via regulating the nuclear/cytoplasmic shuttling of a putative spliceosome factor Crn, thus uncovering a novel mechanism for regulating Hh signal transduction. We demonstrated that the mammalian homologue of HIB, SPOP, also downregulates Su(fu) in the same fashion as HIB does in flies. Moreover, the mouse homologue of crn, crnkl1 siRNAs reduced ShhN signaling by 50%68, indicating that both SPOP and Crnkl1 may have a conserved role in vertebrate Hh signaling. Therefore, it will be interesting to investigate whether SPOP and Crnkl1 can regulate SUFU through a similar mechanism in mammalian systems.
Materials and Methods
Mutations and transgenes
yw, hib-lacZ, Ptc-lacZ, Knot-lacZ, MS1096, UAS-ci-RNAi, hibΔ, Ap-Gal4, UAS-Hh, UAS-HA-Su(fu), UAS-Flag-Su(fu), UAS-Myc-Ci, UAS-HA-Ci−3P, UAS-Flag-HIB, UAS-HA-HIB, UAS-HA-CiN, UAS-hib-RNAi, UAS-Flag-SPOP, UAS-Flag-Cul3KR, UAS-Smo−PKA and UAS-GFP have been described39,42,55,63,68,69. The crn RNAi lines, su(fu) RNAi lines and other RNAi lines used in small-scale genetic screen were obtained from the National Institute of Genetics Stock Center, Japan and the Vienna Drosophila RNAi Center, Austria. eyflp, hsflp, cul3gft2 and FRT82 flies were obtained from the Bloomington Stock Center. To make UAS-HA-Ub-K0, ubiquitin (ub) cDNA was amplified by PCR with substitution of amino acids at all seven lysine residues (K6, K11, K27, K29, K33, K48 and K63) to Arg, then inserted into NotI and XhaI sites of the pUAST-HA vector. For UAS-HA-HIB-Δ3box, the sequence encoding hib lack of amino acids from 299 aa to 330 aa was cloned into pUAST-HA digested with BglII and XhoI. To construct UAS-HA-Su(fu) 2D, su(fu) cDNA was amplified by PCR with substitution of amino acids at both S321 and S324 to Asp, then cloned into BglII and XhoI sites of pUAST-HA vectors. The coding sequence for crn was amplified and inserted into BglII and XhoI sites of the pUAST-HA vector to make construct UAS-HA-Crn. The dsDNA sequences for 5′UTR and 3′UTR of su(fu) were synthesized in Genscript, then cloned into BglII or KpnI or between these two sites of the pUAST-Flag-Su(fu) vector to make the constructs UAS-5′UTR-Flag-Su(fu), UAS-Flag-Su(fu)-3′UTR and UAS-5′UTR-Flag-Su(fu)-3′UTR, respectively.
Immunostaining and in situ hybridization
Immunostaining and in situ hybridization of imaginal discs were performed with standard protocols70. Antibodies were used in this study: rat anti-Ci (2A) (1:50; DSHB); mouse anti-Flag (M2) (1:200; Sigma); mouse anti-HA (F7) (1:200; Santa Cruz); rabbit anti-β Gal (1:500; Cappel); mouse anti-Su(fu) (1:100; DSHB); rabbit anti-GFP (1:500; Santa Cruz); TRITC-labeled phalloidin (1:250; Sigma) and 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) (1:1000; Santa Cruz). For in situ hybridization assay, the primers for su(fu) and crn sub-cloning are as follows: su(fu)-F, 5′-ATAAGAATGCGGCCGCATGGCCGAGGCGAATT-TGGACA-3′, su(fu)-R, 5′-GGGGTACCTCACATTTTCA-GAGGAGC-3′ crn-F, 5′-GGGGTACCGCCGGCGGAGGTGCAAATTAC-3′, crn-R, 5′-CCCAAGCTTCGCGCCCTGTCGT-GCTCCTT-3′. su(fu) and crn RNA probes were synthesized from the above corresponding regions.
Generating clones of mutant cells
Clones of mutant cells were generated by FLP/FRT-mediated mitotic recombination as described39,70. Genotypes for generating clones are as follows: hib clones in wing discs with or without expressing UAS transgenes: MS1096 hs-flp; (UAS-Hh or/and UAS transgenes); FRT82 hibΔ/FRT82 hs-Myc-GFP. hib clones in eye discs with or without expressing UAS transgenes: ey-flp; (GMR-Gal4/UAS transgenes); FRT82 hibΔ/FRT82 hs-Myc-GFP. cul3 clones in eye discs: ey-flp; cul3gft2FRT40/hs-Myc-GFP FRT40.
Cell culture, transfection, RNA interference, immunoprecipitation, western blot assays
S2 cells were cultured in the Schneider's Drosophila Medium (Invitrogen) with 10% fetal bovine serum, 100 U/ml of penicillin and 100 μg/ml of streptomycin. Transfection was carried out using the calcium phosphate transfection method. Usually S2 cells are transfected in 10-cm plates with no more than 20 μg of total DNA for an ubiquitin-Gal4 construct and other co-transfected pUAST expression vectors. 36-48 h after transfection, cells are harvested for immunoprecipitation and western blot analysis with standard protocols as previously described71,72. The following antibodies were used for immunoprecipitation and immunoblotting: mouse anti-Myc (9E10) (1:5 000; Santa Cruz); mouse anti-HA (1:5 000; Santa Cruz); mouse anti-Su(fu) (1:500; DSHB); mouse anti-β-actin (1:10 000; Genscript); and goat anti-mouse HRP (1:10 000; Jackson ImmunoResearch). For RNA interference experiment, dsRNA was generated through in vitro transcription by using the MEGAscript T7 kit (Ambion). After cells were transfected for 24 h, the culture medium was changed to serum-free medium with 15-50 μg dsRNA/106 cells for 8-12 h starvation. Then fresh medium with serum was added and cells were cultured for 48-36 h. The primer sequences of the target genes are as follows: hib RNAi-F, 5′-GAATTAATACGACTCACTATAGGGAGAATGGCGGTCAGCCGTGTACCAT-3′, hib RNAi-R, 5′-GAATTAATACGACTCACTATAGGGAGACCATCTCATGCTCGAACATG-3′ crn RNAi-F, 5′-GAATTAATACGACTCACTATAGGGAGAATGGAGCGGCCACAGAAGATG-3′, crn RNAi-R, 5′-GAATTAATACGACTCACTATAGGGAGACAAATACCCGCCGAGAGCCG-3′ hh RNAi-F, 5′-GAATTAATACGACTCACTATAGGGAGAAGAAGTTCATCCGACGAGAC-3′, hh RNAi-R, 5′-GAATTAATACGACTCACTATAGGGAGAAGCTCATTTTCACGCGTTTC-3′.
MG132, LMB, CHX and Act D treatment
For MG132 (Calbiochem) treatment assay, eye discs were cultured in supplemented M3 medium containing 40 μM MG132 for 4 h, followed by immunostaining. S2 cells were treated with 20 μg/ml MG132 for 4 h before cells were harvested. For LMB (Sigma) treatment, S2 cells were treated with LMB at a final concentration of 5 nM for 2 h before cells were harvested for Crn localization assay. Wing discs from mid-third instar larvae were cultured in Cl-8 cell medium without or with 50 ng/ml LMB treatment for 4 h, endogenous Su(fu) was shown by western blotting and then the results were analyzed by ImageJ. For Cycloheximide (CHX, Calbiochem) treatment, S2 cells were plated in 10-cm dishes and transfected with the indicated plasmids after 18-24 h. After another 24 h, the cells were transferred into six-well cell culture plates at equivalent densities. Cells were treated with 20 μg/ml CHX for the indicated times before harvesting. For Actinomycin D (Act D, Beyotime) treatment, S2 cells were treated with 1 μg/ml Act D for the indicated times before harvesting.
RNA isolation from cell extracts, reverse transcription PCR (RT-PCR) and real-time quantitative PCR (RTQ-PCR)
S2 cells were transfected with UAS-HA-Crn or UAS-Flag-HIB or both of them, respectively. After two days, the cells were harvested, 3 ml of them was prepared for the total RNA isolation, and the rest 7 ml was washed with PBS and incubated in 400 μl of hypotonic lysis buffer (10 mM Tris-HCl, pH 7.4/10 mM NaCl/3 mM MgCl2/0.1% (vol/vol) Nonidet P-400) containing protease inhibitors (Roche) for 20 min on ice. After centrifuged at 2 500× g at 4 °C for 5 min, supernatant was removed in a new tube as cytosol fraction, and the rest (nuclei pellet) was washed for 10 min with hypotonic lysis buffer for three times. Nuclei pellet was resuspended in 100 μl RIPA buffer (50 mM Tris-HCl, pH 7.4/150 mM NaCl/1% (vol/vol) Nonidet P-400/0.5% (wt/vol) sodium desoxycholate/0.1% (wt/vol) SDS) containing protease inhibitors and frozen at −80 °C for 1 h. All of the total cells, cytosol fraction and nuclei pellet RNA were extracted and the reverse transcription was carried out using PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa). RT-PCR primers for Drosophila su(fu) coding region (upstream: 5′-GACAAAAAACCTGAGGTGAAGCC-3′ and downstream: 5′-AGTTTGGTGCCTGCGCTT-3′) were synthesized by Invitrogen (Shanghai, China). For detecting the expression of hh, ci, hib, crn and su(fu) in S2 cells, the PCR primers are as follows: hh (upstream: 5′-ATGGATAACCACAGCTCAGTGCCT-3′ and downstream: 5′-TCAATCGTGGCGCCAGCTCT-3′), ci (upstream: 5′-GCAGTATATGCTTGTTGTGC-3′ and downstream: 5′-TCTTTGACTGAATGAACCCC-3′), hib (upstream: 5′-ATGGCGGTCAGCCGTGTACC-3′ and downstream: 5′-TCAGCTCATTTTCACGCGTT-3′), crn (upstream: 5′-ATGGAGCGGCCACAGAAGAT-3′ and downstream: 5′-TCAGTCACCGCTATCCGTCG-3′), su(fu) (upstream: 5′-ATGGCCGAGGCGAATTTGGA-3′ and downstream: 5′-TCACATTTTCAGAGGAGCAG-3′). For Q-PCR, S2 cell and wing disc RNA were isolated by using Trizol reagent (Invitrogen) and then reverse-transcribed by PrimeScript RT reagent kit (TaKaRa). Finally, the real-time PCR was done using the SYBR Premix Ex Taq (TaKaRa) according to the instrument of StepOnePlus (Applied Biosystem). Standard Q-PCR primers for Drosophila su(fu) (upstream: 5′-AAGCGCAGGCACCAAACT-3′ and downstream: 5′-CCAAAGTGAGCGCCAGAT-3′), hib (upstream: 5′-GCTACACGCAGGTCAAAGTG-3′ and downstream: 5′-CCGTAAACACCATTTTAGTT-3′), ci (upstream: 5′-CCTCTTGCGTATTCTGAATT-3′ and downstream: 5′-GAATCTGATGTTCCACCCGT-3′) and actin (upstream: 5′-CGAAGAAGTTGCTGCTCTGGTTGTCG-3′ and downstream: 5′-GGACGTCCCACAATCGATGGGAAG-3′) were synthesized by Genscript. Actin was used as a control.
Statistical analysis
Imaging data were analyzed using Image J. Statistical tests were performed in GraphPad Prism 5. All measures are reported as mean ± SEM.
Acknowledgments
We thank Dr Yun Zhao (Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China), Fly Stocks of National Institute of Genetics of Japan (NIG-Fly), Vienna Drosophila RNAi Center (VDRC), the Bloomington Stock Center and Developmental Studies Hybridoma Bank at the University of Iowa for providing fly stocks and reagents. This work was supported by grants from the National Natural Science Foundation of China (30971679, 31071264 and 31271531) and the National Key Scientific Program of China (2011CB943902, 2010CB945102) to Qing Zhang. Jin Jiang is supported by grants from NIH (GM061269 and GM06745), Welch foundation (I-1603) and NSFC (31328017).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
HIB neither binds Su(fu) nor affects the stability of Su(fu) protein in S2 cells.
HIB doesn't bind Crn in S2 cells. HA-Crn and Myc-HIB were cotransfected in S2 cells.
SPOP downregulates endogenous Su(fu) in wing discs but does not change the stabililty of exogenous HA-Su(fu) in S2 cells.
Hh doesn't affect su(fu) mRNA level or the stability of Su(fu) in the S2 cells.
Hh affects Su(fu) level in S2 cells.
HIB-Crn affects Su(fu) level in the S2 cells.
Loss of Su(fu) alters Hh signaling in a genetic sensitized background.
HIB doesn't affect su(fu) mRNA nuclear export.
HIB may affect su(fu) translation.
Endogenous crn is ubiquitously expressed in wing (A, B) and eye (C, D) discs detected by in situ hybridization.
The expression of Su(fu) affected by knockdown of the candidate genes in wing discs.
References
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Lum L, Beachy PA. The Hedgehog response network: sensors, switches, and routers. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- Jia J, Jiang J. Decoding the Hedgehog signal in animal development. Cell Mol Life Sci: CMLS. 2006;63:1249–1265. doi: 10.1007/s00018-005-5519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- Hooper JE, Scott MP. The Drosophila patched gene encodes a putative membrane protein required for segmental patterning. Cell. 1989;59:751–765. doi: 10.1016/0092-8674(89)90021-4. [DOI] [PubMed] [Google Scholar]
- Casali A, Struhl G. Reading the Hedgehog morphogen gradient by measuring the ratio of bound to unbound Patched protein. Nature. 2004;431:76–80. doi: 10.1038/nature02835. [DOI] [PubMed] [Google Scholar]
- Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- Stone DM, Hynes M, Armanini M, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450:252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Ingham PW. smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 1996;382:547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper JE. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell. 1996;86:221–232. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- Lum L, Zhang C, Oh S, et al. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol Cell. 2003;12:1261–1274. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- Jia J, Tong C, Jiang J. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes Dev. 2003;17:2709–2720. doi: 10.1101/gad.1136603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mao F, Lu Y, Wu W, Zhang L, Zhao Y. Transduction of the Hedgehog signal through the dimerization of Fused and the nuclear translocation of Cubitus interruptus. Cell Res. 2011;21:1436–1451. doi: 10.1038/cr.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Kalderon D. Hedgehog activates fused through phosphorylation to elicit a full spectrum of pathway responses. Dev Cell. 2011;20:802–814. doi: 10.1016/j.devcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Li S, Jia J, Jiang J. The Hedgehog-induced Smoothened conformational switch assembles a signaling complex that activates Fused by promoting its dimerization and phosphorylation. Development. 2011;138:4219–4231. doi: 10.1242/dev.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, Therond PP. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat Cell Biol. 2003;5:907–913. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- Ohlmeyer JT, Kalderon D. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature. 1998;396:749–753. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- Pan D, Rubin GM. cAMP-dependent protein kinase and hedgehog act antagonistically in regulating decapentaplegic transcription in Drosophila imaginal discs. Cell. 1995;80:543–552. doi: 10.1016/0092-8674(95)90508-1. [DOI] [PubMed] [Google Scholar]
- Methot N, Basler K. An absolute requirement for Cubitus interruptus in Hedgehog signaling. Development. 2001;128:733–742. doi: 10.1242/dev.128.5.733. [DOI] [PubMed] [Google Scholar]
- Tabata T, Kornberg TB. Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell. 1994;76:89–102. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–214. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- Strigini M, Cohen SM. A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development. 1997;124:4697–4705. doi: 10.1242/dev.124.22.4697. [DOI] [PubMed] [Google Scholar]
- Tanimoto H, Itoh S, ten Dijke P, Tabata T. Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Mol Cell. 2000;5:59–71. doi: 10.1016/s1097-2765(00)80403-7. [DOI] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- Jia J, Amanai K, Wang G, Tang J, Wang B, Jiang J. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature. 2002;416:548–552. doi: 10.1038/nature733. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang L, Zhang Q, et al. Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for Slimb/beta-TRCP-mediated proteolytic processing. Dev Cell. 2005;9:819–830. doi: 10.1016/j.devcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Smelkinson MG, Kalderon D. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Curr Biol: CB. 2006;16:110–116. doi: 10.1016/j.cub.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Smelkinson MG, Zhou Q, Kalderon D. Regulation of Ci-SCFSlimb binding, Ci proteolysis, and hedgehog pathway activity by Ci phosphorylation. Dev Cell. 2007;13:481–495. doi: 10.1016/j.devcel.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent D, Bush EW, Hooper JE. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development. 2006;133:2001–2010. doi: 10.1242/dev.02370. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10:719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Jiang J. Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell Cycle. 2006;5:2457–2463. doi: 10.4161/cc.5.21.3406. [DOI] [PubMed] [Google Scholar]
- Ou CY, Wang CH, Jiang J, Chien CT. Suppression of Hedgehog signaling by Cul3 ligases in proliferation control of retinal precursors. Dev Biol. 2007;308:106–119. doi: 10.1016/j.ydbio.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Ou CY, Lin YF, Chen YJ, Chien CT. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 2002;16:2403–2414. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shi Q, Chen Y, et al. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2009;106:21191–21196. doi: 10.1073/pnas.0912008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaeva M, Michelson P, Kogerman P, Toftgard R. Characterization of the physical interaction of Gli proteins with SUFU proteins. J Biol Chem. 2003;278:5116–5122. doi: 10.1074/jbc.M209492200. [DOI] [PubMed] [Google Scholar]
- Sisson BE, Ziegenhorn SL, Holmgren RA. Regulation of Ci and Su(fu) nuclear import in Drosophila. Dev Biol. 2006;294:258–270. doi: 10.1016/j.ydbio.2006.02.050. [DOI] [PubMed] [Google Scholar]
- Barnfield PC, Zhang X, Thanabalasingham V, Yoshida M, Hui CC. Negative regulation of Gli1 and Gli2 activator function by Suppressor of fused through multiple mechanisms. Differentiation. 2005;73:397–405. doi: 10.1111/j.1432-0436.2005.00042.x. [DOI] [PubMed] [Google Scholar]
- Chen MH, Wilson CW, Li YJ, et al. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S. Costal-2: a scaffold for kinases mediates Hedgehog signaling. Dev Cell. 2005;8:140–141. doi: 10.1016/j.devcel.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Monnier V, Dussillol F, Alves G, Lamour-Isnard C, Plessis A. Suppressor of fused links fused and Cubitus interruptus on the hedgehog signalling pathway. Curr Biol. 1998;8:583–586. doi: 10.1016/s0960-9822(98)70227-1. [DOI] [PubMed] [Google Scholar]
- Preat T, Therond P, Limbourg-Bouchon B, et al. Segmental polarity in Drosophila melanogaster: genetic dissection of fused in a Suppressor of fused background reveals interaction with costal-2. Genetics. 1993;135:1047–1062. doi: 10.1093/genetics/135.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- Chen CH, von Kessler DP, Park W, Wang B, Ma Y, Beachy PA. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell. 1999;98:305–316. doi: 10.1016/s0092-8674(00)81960-1. [DOI] [PubMed] [Google Scholar]
- Stegman MA, Vallance JE, Elangovan G, Sosinski J, Cheng Y, Robbins DJ. Identification of a tetrameric hedgehog signaling complex. J Biol Chem. 2000;275:21809–21812. doi: 10.1074/jbc.C000043200. [DOI] [PubMed] [Google Scholar]
- Wang G, Amanai K, Wang B, Jiang J. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev. 2000;14:2893–2905. doi: 10.1101/gad.843900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier V, Ho KS, Sanial M, Scott MP, Plessis A. Hedgehog signal transduction proteins: contacts of the Fused kinase and Ci transcription factor with the kinesin-related protein Costal2. BMC Dev Biol. 2002;2:4. doi: 10.1186/1471-213X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SY, Bishop JM. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc Natl Acad Sci USA. 2002;99:5442–5447. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogerman P, Grimm T, Kogerman L, et al. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- Svard J, Heby-Henricson K, Persson-Lek M, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Cooper AF, Yu KP, Brueckner M, et al. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development. 2005;132:4407–4417. doi: 10.1242/dev.02021. [DOI] [PubMed] [Google Scholar]
- Taylor MD, Liu L, Raffel C, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- Yue S, Chen Y, Cheng SY. Hedgehog signaling promotes the degradation of tumor suppressor Sufu through the ubiquitin-proteasome pathway. Oncogene. 2009;28:492–499. doi: 10.1038/onc.2008.403. [DOI] [PubMed] [Google Scholar]
- Methot N, Basler K. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development. 2000;127:4001–4010. doi: 10.1242/dev.127.18.4001. [DOI] [PubMed] [Google Scholar]
- Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 2004;23:1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JT, Lin HC, Hu YC, Chien CT. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol. 2005;7:1014–1020. doi: 10.1038/ncb1301. [DOI] [PubMed] [Google Scholar]
- Zhuang M, Calabrese MF, Liu J, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Pan Y, Wang B. Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development. 2010;137:2001–2009. doi: 10.1242/dev.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken K, Vokes SA, Lin TY, McMahon AP, Perrimon N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat Genet. 2005;37:1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc P, Lin HY, Ruiz i Altaba A, Kornberg TB. Expression of the vertebrate Gli proteins in Drosophila reveals a distribution of activator and repressor activities. Development. 2000;127:4293–4301. doi: 10.1242/dev.127.19.4293. [DOI] [PubMed] [Google Scholar]
- Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev. 1999;13:2828–2837. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Protein kinase A and hedgehog signaling in Drosophila limb development. Cell. 1995;80:563–572. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhao Y, Tong C, et al. Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of Cubitus interruptus. Dev Cell. 2005;8:267–278. doi: 10.1016/j.devcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HIB neither binds Su(fu) nor affects the stability of Su(fu) protein in S2 cells.
HIB doesn't bind Crn in S2 cells. HA-Crn and Myc-HIB were cotransfected in S2 cells.
SPOP downregulates endogenous Su(fu) in wing discs but does not change the stabililty of exogenous HA-Su(fu) in S2 cells.
Hh doesn't affect su(fu) mRNA level or the stability of Su(fu) in the S2 cells.
Hh affects Su(fu) level in S2 cells.
HIB-Crn affects Su(fu) level in the S2 cells.
Loss of Su(fu) alters Hh signaling in a genetic sensitized background.
HIB doesn't affect su(fu) mRNA nuclear export.
HIB may affect su(fu) translation.
Endogenous crn is ubiquitously expressed in wing (A, B) and eye (C, D) discs detected by in situ hybridization.
The expression of Su(fu) affected by knockdown of the candidate genes in wing discs.