Abstract
Epigenetic changes underlie developmental and age related biology. Promising epidemiologic research implicates epigenetics in disease risk and progression, and suggests epigenetic status depends on environmental risks as well as genetic predisposition. Epigenetics may represent a mechanistic link between environmental exposures, or genetics, and many common diseases, or may simply provide a quantitative biomarker for exposure or disease for areas of epidemiology currently lacking such measures. This great promise is balanced by issues related to study design, measurement tools, statistical methods, and biological interpretation that must be given careful consideration in an epidemiologic setting. This article describes the promises and challenges for epigenetic epidemiology, and suggests directions to advance this emerging area of molecular epidemiology.
Keywords: epigenetics, epidemiology, environment, disease, DNA methylation
Overview
Epidemiology is the study of the distribution of disease in populations and the causes, consequences, prevention, and treatment strategies for those diseases. Epidemiologic studies complement mechanistic toxicologic studies, where associations observed in populations can be tested under controlled conditions in the laboratory, and conversely toxicology findings can inform epidemiology study designs. Collaborative studies between epidemiologists and toxicologists are one of the strongest strategies to efficiently produce science relevant to the public’s health.
Epigenetics is formally defined as heritable changes in gene expression that occur without changes to the underlying DNA sequence. Types of epigenetic regulators include DNA methylation, histone modifications, microRNA, and prions. The majority of epigenetic epidemiology studies have focused on DNA methylation due to its relative stability with storage and the multitude of technical platforms available for analysis. Epidemiologists have observed changes in late-life health status associated with early life environmental exposures, termed the Developmental Origins of Hypothesis of Adult Disease (DOHAD) [Dolinoy et al. 2007] or the Barker hypothesis [Barker 2004]. Some of these lasting effects of exposures long since flushed from the body may be due to persistent epigenetic modifications. Prenatal conditions, epigenetic change, and adverse later life health outcomes have been observed following such extreme cases as the Dutch Hunger Winter during 1944–1945 [Heijmans et al. 2008]. This research has inspired a new wave of epigenetic epidemiology studies.
Epigenetics has been hailed as a missing mechanistic link between environmental exposures or genetics and many common diseases [Cortessis et al. 2012]. The evidence for these associations has largely been provided by promising epidemiologic research [Foley et al. 2009]. These epigenetic epidemiology studies often struggle with common issues related to design, methods, and biology that impair their ability to make causal inferences regarding the role of epigenetics in disease. This article focuses on the promises and challenges for epigenetic epidemiology, and suggests future approaches to move forward this nascent branch of molecular epidemiology.
Promises
Epigenetic mechanisms shape development, aging, and disease
Epigenetic reprogramming is a key element of normal biological development and aging as well as many common diseases. Uncovering the locations and timing of epigenetic changes in development and disease holds the promise to identify time points sensitive to change and potential disease interventions.
During mammalian development, two major waves of epigenetic reprogramming take place [Reik et al. 2001]. The first is during gametogenesis and the second occurs in the preimplantation embryos. After fertilization, the paternal genome is actively demethylated and the maternal genome is passively demethylated with DNA replication. Remethylation occurs several days later during implantation, and is subsequently maintained through cellular replication.
Similar to its role in normal development, epigenetics may be a feature of the aging process. Twin studies show greater epigenetic differences across the lifespan, potentially as a result of accumulated environmental exposures and disease, or as part of aging biology [Fraga et al. 2005; Javierre et al. 2010; Poulsen et al. 2007; Ribel-Madsen et al. 2012]. Further, across epigenomic sites that vary between individuals in a population, particular regions show change in methylation within the same person over time [Bjornsson et al. 2008; Feinberg et al. 2010]. Disease related epigenetic mechanisms may be behind middle to late life functional declines [Feinberg et al. 2010; Heyn et al. 2013]. Indeed, epigenetic modifications play a key role in many forms of cancer [Feinberg and Tycko 2004], as well as Alzheimer’s disease [Bakulski et al. 2012] and other neurodegenerative diseases [Coppede 2013].
Epigenetic marks are influenced by environment and genes
Given the critical roles of epigenetics in development and disease, it follows that identifying and characterizing modulators of epigenetics could lead to methods for disease prevention and intervention. Animal toxicological models are crucial, proof-of-principal studies showing epigenetic susceptibility to environmental conditions. Researchers exploit visible phenotypes that are driven by gene expression and epigenetic state, including the kinky tail AxinFU mouse model. Another highly studied murine epigenetic biosensor is the viable yellow agouti (Avy) allele, which features a retrotransposable element that regulates gene expression when it is inverted and inserted prior to the gene [Dolinoy and Jirtle 2008]. DNA methylation levels at these alleles are stochastic, but depend on fetal in utero and early life developmental environmental and nutritional conditions. Agouti dams fed a high methyl-donor diet produced offspring with a shift in coat color distribution toward brown, driven by increased DNA methylation at the Avy allele [Waterland and Jirtle 2003]. Coat color and DNA methylation shifts were also observed with diet (genistein) [Dolinoy et al. 2006] and environmental exposures, such as bisphenol-A [Anderson et al. 2012] and ethanol [Kaminen-Ahola et al. 2010]. Notably, rat maternal licking and grooming behavior influence offspring stress response and hippocampal DNA methylation at the glucocorticoid receptor promoter [Weaver et al. 2004]. Controlled animal and cell line experiments may be useful to identify dose-response causal relationships and demonstrate that many chemicals and behavioral conditions may be broad epigenetic regulators, potentially in humans.
Toxicology studies inspired emerging environmental epigenetic epidemiology association studies [Bollati and Baccarelli 2010], which have the ability to test these associations in human populations. Paired toxicology and epidemiology research is needed to address shortcomings of either individually, such as generalizability (species or population specificity), dosage range, and causality. In most environmental cases, human exposure levels are orders of magnitude below laboratory toxicologic dosing and epidemiologic research is needed to determine real world risk. Examples of population-based and experimental research on overlapping exposures are listed in Table 1. Specifically, global and gene-specific DNA methylation associations have been observed with diet [Fenech 2001a, b; Fenech and Ferguson 2001], lifestyle and demographic characteristics like maternal smoking [Joubert et al. 2012], and environmental toxicants [Baccarelli and Bollati 2009; Sutherland and Costa 2003]. In addition to DNA methylation, environmental factors also influence histone modifications including metals [Arita et al. 2012; Cantone et al. 2011; Chervona et al. 2012]. Broad epigenetic change does not appear to be specific to a particular class of chemical exposures. Future, replicated genome-wide environmental epigenetic studies will show whether particular chemicals map to corresponding sensitive genomic regions.
Table 1.
Broad environmental DNA methylation regulators and references, by higher order classifications of toxicants.
Inherited genes are another regulator of epigenetics, with several reports describing gene-DNA methylation associations [Bell et al. 2010; Bell et al. 2011; Bjornsson et al. 2004; Liu et al. 2013]. However, the particular locations of gene-epigenotype correspondence have not been well–informed nor mechanisms understood. A recent study observed that single nucleotide polymorphisms (SNPs) influence surrounding DNA methylation, but the size of the methylation region of impact, as well as the size of the genetic signal is inconsistent [Liu et al. 2013]. Combined analysis of genetic and epigenetic data can illuminate relationships (passive and active) between DNA methylation and gene expression[Gutierrez-Arcelus et al. 2013]. Study design options such as Mendelian randomization [Relton and Davey Smith 2012] and potential statistical strategies including mediation analysis [Liu et al. 2013] that may facilitate research in this area will be discussed in the subsequent sections. Further study is needed to understand the gene-epigene spatial relationships; the relative impact of inherited genes versus environment, age, and random noise on epigenetic marks throughout the genome remains unclear.
Roles for epigenetics in epidemiology
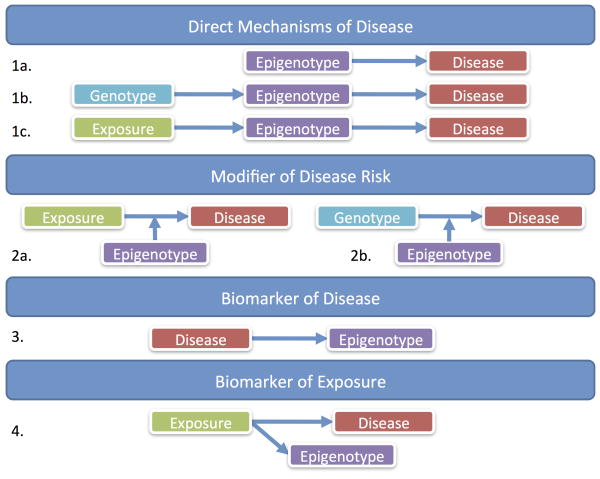
One of the great allures of epigenetics is the potential as a biological mechanism between genetics or environment and disease. We discuss this potential in detail below. It is critical to not only consider the option of epigenetics as a direct mechanism to disease, but also an indirect mechanism - a biomarker of exposure or disease that can be useful to epidemiology even if not mechanistically relevant. These various potential pathways for epigenetic epidemiology are presented in Figure 1.
Figure 1.
Theoretical relationships between epigenetics, disease, exposure and genotype.
Direct effects on disease risk (mediation)
Epigenetic modifications can directly cause disease (Figure 1, 1a). Imprinting disorders, such as Beckwith Weidemann syndrome (BWS), are classic examples where disease is caused by aberrant DNA methylation. Most offspring genes are expressed from both the maternal and paternal copies of chromosomes, but imprinting genes are expressed in only one copy, from a particular type of parent. Thus, changes to the parental balance in epigenetic control of expression at these imprinted sites can interrupt normal biology. BWS is caused by changes in methylation at the imprinting control regions for the genes H19, IGF2, CDKN1C, and KCNQ1, which lead to changes in their gene expression and pediatric overgrowth associated with BWS [Weksberg et al. 2010].
There are also several examples where diseases are caused by genetic mutations that act through epigenetic mechanisms, such as Rett Syndrome (Figure 1, 1b). This predominantly female neurodevelopmental disorder is caused by spontaneous mutations in the methyl-CpG-binding protein 2 (MECP2) gene on the X-chromosome [Amir et al. 1999]. MECP2 is important for recognizing epigenetic modifications controlling gene expression, and mutated MECP2 alters the expression of other genes that are normally regulated by epigenetics. In another example, a particular serotonin receptor (5HT2A) genotype at the T102C position adds 2 CpG sites to the DNA sequence. This variant increases methylation and decreases expression of 5HT2A [Polesskaya et al. 2006], which is involved in psychiatric phenotypes such as schizophrenia [Polesskaya and Sokolov 2002; Serretti et al. 2007]. Similarly in colorectal cancer, the genetic loss of a CpG site through a C-T mutation in the methyltransferase MGMT gene disrupts gene regulation and leads to disease [Ogino et al. 2007]. In a final example, a SNP in the COMT gene introduces a new CpG site and the methylation level of the variant allele is correlated with lifetime stress level and working memory [Ursini et al. 2011]. In these cases, the locations of epigenetic change are restricted to the area of genetic change. Here, the negative effects of a particular genotype are mediated through epigenetics.
In the same way, epigenetics can be the mechanism linking environmental exposures and disease (Figure 1, 1c). Many toxicants are known to create oxidative stress in cells. Oxidative DNA damage from the enivironment interferes with DNA methyltransferase action on DNA [Valinluck et al. 2004], causing aberrant DNA methylation [Turker and Bestor 1997] that can be associated with disease. Toxicants can also directly interact with DNA methylatransferases or one-carbon metabolism enzymes, influencing the global epigenetic state of cells. In the same vein, dietary methyl group and co-factor availability influence global epigenetics. Often, environmental effects on the DNA methylome are global and diffuse, differing from the sequence specificity in the genetic example. Future genome-wide studies may reveal that certain chromatin structural arrangements are more sensitive to environmental influence, so there may be more regional specificity to environmental epigenetic change. Patterns in environmentally induced epigenetic change may also arise from stochastic change followed by cellular proliferation and local selection. Perhaps for some exposures there is a natural regulation of response that yields sequence specific epigenetic sensitivity, but exposure related epigenetic signatures have not yet been defined. Epigenetics as a mediator of the environmental-disease mechanism in particular is appealing for scientists who have noticed exposures in early life can lead to late life disease [Barker 2004]. The lingering toxic effects of non-persistent environmental factors could be maintained in epigenetic marks that are relevant as the body ages.
Modification of disease risk
Epigenetics may also influence the relationships between exposure and disease (Figure 1, 2a), or genotype and disease (Figure 1, 2b) via effect modification. For example, some toxicants impact disease risk via DNA damage and the location of DNA damage (in genes, etc.) dictates disease severity. The surrounding epigenetic state can control the accessibility of DNA, and thus modify the impact of a toxicant on disease risk by titrating the susceptibility of genomic locations to exposure-induced damage [Ha et al. 2002; Jones et al. 2002; Meng et al. 2005; Smith et al. 1998]. Exposure to air pollution increases risk of cardiovascular disease and preliminary epidemiologic research shows susceptibility to elevated intermediate cardiovascular biomarkers following exposure to particles and nitrogen dioxide may in part be due to differences in methylation at the TLR2 gene [Bind et al. 2012]. Effect modification may also occur when a genetic mutation leads to disease, but disease severity and expression of the mutated protein depends on the epigenetic status of the gene. For example, Angelman syndrome is a developmental disorder caused by a genetic mutation in the ubiquitin ligase gene, UBE3A, an imprinted gene where the maternal allele is unmethylated and expressed in the brain. Because the paternal allele is methylated and unexpressed, mutations in the paternal UBE3A gene do not lead to disease while alterations in the maternally expressed sequence do result in Angelman syndrome. Thus, epigenetic context modifies the impact of the mutation. This is a challenging scenario for traditional genetic epidemiology research because the genetic effects would be masked when they are averaged over different epigenetic contexts. Genetic mutations that similarly depend on the parent-of-origin for disease risk have been observed in diseases such as autism [Arking et al. 2008; Fradin et al. 2010], bipolar disorder [Stine et al. 1995], and multiple sclerosis [Ebers et al. 2004], although specific epigenetic mechanisms to explain these parental origin effects have not yet been described.
Biomarkers of disease and environment
Alternatively, there may be specific disease and exposure examples where epigenetic factors are not involved mechanistically, but can serve as useful biomarkers of disease (Figure 1, 3) or of a particular exposure (Figure 1, 4). For example, epigenetic biomarkers are effective tests of disease status or of disease subtype when targeting treatment. In the context of cancer, epigenetic tests are involved in disease prediction, diagnosis, and prognosis [Esteller 2008]. The original example of the utility of epigenetics in personalized cancer treatment was associated with glioblastoma tumors. The DNA repair enzyme O6-methylguanine-DNA methyltransferase (MGMT) was hypermethylated in tumors that responded to alkylating agents, resulting in extended survival [Esteller et al. 2000]. Thus, patients with hypomethylated MGMT tumors would not be targeted for alkylating agent treatment. In addition, hypermethylation of the detoxification enzyme, glutathione s-transferase P (GSTP1), is consistent in prostate cancer. Prostate cancer is a disease where we have a blood-based screening test (prostate-specific antigen, PSA) with high sensitivity. Combining the PSA test with an epigenetic biomarker test could increase diagnosis specificity, by reducing the number of false positives [Sunami et al. 2009].
Secondly, epigenetic marks may represent “tombstones” of previous environmental exposures that have been flushed from the body. For example, in two separate birth cohorts, researchers found matching DNA methylation sites in newborn cord blood that correlated to maternal smoking behavior in pregnancy [Joubert et al. 2012]. Methylation status at these sites may be a biomarker of in utero smoke exposure and appear to persist in children 2–5 years of age (Ladd-Acosta 2013 unpublished). For other exposures that are more challenging to retrospectively assess than maternal smoking, epidemiology studies may greatly benefit from a validated epigenetic biomarker, particularly one that can be measured several years after the exposure.
Finally, epigenetic marks may be used to identify disease- or exposure-susceptible populations or individuals. In cases where epigenetics is not a driver of disease, it may still be useful as a biomarker of disease, exposure, or of susceptible populations.
Epigenetics as potential mechanism for transgenerational effects
Preliminary results suggest epigenetic changes may be transgenerational, in which case the promises of epigenetic epidemiology are even more far-reaching. In principal, transgenerational epigenetic inheritance has been demonstrated in Arabidopsis thaliana [Schmitz et al. 2011], Caenorhabditis elegans [Greer et al. 2011] and the AxinFu mouse model [Rakyan et al. 2003]. Environmentally induced changes in epigenetics may be passed through the germline. Rat in utero exposure to the fungicide vinclozolin produced male offspring with impaired spermatogenesis that persisted to the F4 generations and were correlated with germ line DNA methylation change [Anway et al. 2005]. In humans, early life exposures as known to influence later life health outcomes; for example, low birth weight predicts risk of obesity, type 2 diabetes, and cardiovascular disease [Hales and Barker 1992; McMillen and Robinson 2005]. Women pregnant during the Dutch hunger winter during World War 2 had female offspring with altered lipid profiles in adulthood [Lumey et al. 2009], which was accompanied by persistent epigenetic change [Heijmans et al. 2008; Tobi et al. 2009]. Careful distinction must be made in humans to separate the effects of in utero or early life exposures influencing later life health outcomes, potentially due to epigenetic programming, for which the evidence is mounting and epigenetic inheritance over multiple generations, with much more preliminary evidence [Gluckman et al. 2007; Schmidt 2013]. Rather than in DNA and chromatin, transgenerational inheritance may be linked to RNA factors such as piRNA and microRNA in gametes [Daxinger and Whitelaw 2012]. There is support for transgenerational epigenetic effects in many eukaryote models, but the molecular basis of the inheritance is currently poorly characterized.
Challenges and directions
The outlook surrounding the field of epigenetic epidemiology is very hopeful for the promises detailed above. Practicing researchers, however, have repeatedly described several challenges facing epigenetic epidemiology on the ground [Heijmans and Mill 2012]. In this section, we describe the universal research issues and propose potential approaches for addressing them and moving the field forward.
Catalogue of information
There is a fundamental need to address foundational questions of epigenetic epidemiology prior to more nuanced research questions related to exposures or disease. First, we need to understand which level of the epigenome to examine for changes (DNA methylation, histones, microRNA, etc.). This may depend on the stability of the epigenetic mark over time or in storage, or the sensitivity of the mark to changes in the external or internal environment. Second, there are millions of epigenetic marks in any given cell. We need to know where in the epigenome to look for differences. For example, in DNA methylation studies, there has been an evolution of focus from CpG islands to CpG island shores and a focus on parts of the epigenome with intra- and inter-individual variability [Irizarry et al. 2009]. Epidemiology studies patterns in groups of people and this depends on an understanding of inter-individual differences in the epigenome. We need to characterize normal variability within and between people, in order to comment on what is altered [Bock et al. 2008]. This requires cross-tissue and cross-population measurements to establish variable methylation and catalogue the locations, somewhat analogous to the Haplotype Mapping project for common genetic polymorphisms.
Similarly, we need to understand the spatial relationships in the epigenome. Nearby DNA methylation CpG sites are often correlated in their methylation state. It will be important to understand which sites are individually and which are coordinately methylated. Better knowledge of the spatial structure of the epigenome will help target our discovery searches. Next, we need to understand the temporal changes in the epigenome. These may be related to long-term change with aging, or it may be on a much shorter time scale. Histone acetylation at clock-controlled genes is responsive to circadian rhythm [Bellet and Sassone-Corsi 2010]. Epigenetic state is dynamic and the timing of our sample collection is a critical element.
Epigenome-scale public databases are a potential approach to share foundational epigenetic information. The NIH Roadmap Epigenomics Mapping Consortium (http://www.roadmapepigenomics.org/) was launched to provide a normal reference for ex vivo DNA methylation, chromatin assembly, histone modifications, and small RNA epigenomes from various tissues. This database is already providing comprehensive information on different epigenomic levels, variation across the epigenome, and tissue variation. This ambitious undertaking will inform development of future epigenome-wide technologies and the design of new studies. Unfortunately, documenting inter-individual “normal” variation is outside the scope of this project and additional epidemiologic studies will inform on this point.
Study design
Epigenetic epidemiology requires unique study design considerations. The study timing, sample types, and scale of epigenetic epidemiology are very different than for genetic epidemiology [Foley et al. 2009]. For example, genome-wide association studies (GWAS) often bank cells from participants in culture, which represents a virtually unlimited source of DNA for follow-up studies. However, it is well documented that cell culture conditions impact epigenetic state, so epigenetic epidemiology studies require sufficient primary samples, often with specific collection protocols to preserve the epigenetic mark of interest. In addition, genetics represent a fixed exposure throughout the lifetime; we can assume genetic effects temporally occur prior to disease onset regardless of when the biological sample was taken relative to exposure or disease. Epigenetics, in contrast, are a dynamic exposure that can vary over short intervals. If samples are collected at disease diagnosis, we cannot make assumptions about the role of epigenetics as a driver of disease or a late-stage passenger of the condition. Studies must be very cautious of reverse causation in epigenetic research.
Many of the epigenetic epidemiology study design challenges can be overcome with planning, longitudinal designs, and clearly specified a priori hypotheses and models. Prospective data collection and repeated longitudinal epigenetic measures will help to address questions about temporality, particularly those that begin at conception or in early in utero development. At risk studies, which examine earlier in the disease process during subclinical or disease risk states, will be particularly important. By working with different age-windows, we will be better able to understand the directions of the exposure, epigenetic, and disease relationships. Further, statistical approaches that apply causal inference concepts, such as Mendelian randomization [Relton and Davey Smith 2012] can protect against over-interpretation of results in the face of potential reverse causality, particularly in cross-sectional and case-control studies. In general, careful a priori attention to which role of epigenetic marks is being investigated - causal, a modifier, or a biomarker – should be taken at the design stage.
Sample availability
Related to the study design issues is the challenge of sample availability. The first point related to sampling is that collection timing matters. Etiologic studies require samples collected prior to disease. New prospective collections of large numbers of samples necessitate time and considerable funds. It is prudent, though not always possible, to identify appropriate archived samples. Second, the type of sample, whether fresh or frozen, influences the types of epigenetic analyses that are possible. DNA methylation is considered stable in frozen samples, but collection for histone modifications and microRNA requires additional considerations. Immortalized cell culture derived samples are not appropriate for epigenetic profiling because immortalization processes inherently alter the epigenome [Farwell et al. 2000; Grafodatskaya et al. 2010; Kulaeva et al. 2003], though primary cell culture samples have been used effectively in epigenetic toxicology studies [Rager et al. 2013]. In addition, genetic, RNA, protein, metabolite, and epigenetic data are needed on common samples to answer complex biological questions. Each of these analytes has specific necessary storage conditions [Deng et al. 2004; Masson et al. 2010; Shechter et al. 2007]. Third, most tissue samples are heterogeneous cell populations. To examine individual cell type signals, most physical cell sorting methods require fresh samples [Hawley and Hawley 2011]. As discussed further in the biological interpretation and statistical approaches sections, cell type composition of a mixed sample is an important consideration for interpretation and analytic approach.
The fourth issue with respect to sampling is the paradigm of target tissue and surrogate tissues. There is still debate about the utility of surrogate tissues, such as peripheral blood compared to the primary tissue for a specific disease. However, such primary tissue - for example brain - is simply unavailable for in life sampling. Thus, we may be limited to mixed or indirect signals of “true” effects. There is a growing body of literature to suggest blood or lymphocyte epigenetic profiles may have relevance to tissues of interest. Inter-individual differences in gene expression [Sullivan et al. 2006] and, in a limited set, DNA methylation [Davies et al. 2012] were consistent across blood and brain tissues, however the utility of blood DNA methylation for non-blood related disorders is still an open question. Gene expression and epigenetic findings from blood are evidenced in colorectal neoplasia [Cui et al. 2003], schizophrenia and bipolar disorder [Kuratomi et al. 2008; Tsuang et al. 2005], Alzheimer’s disease [Wang et al. 2008], fragile X syndrome [Darnell et al. 2001; Nishimura et al. 2007], and autism [Gregg et al. 2008; Hu et al. 2006; Nakamura et al. 2008; Nguyen et al. 2010; Wong et al. 2013], among others. Thus, some biological disease differences may span non-target tissues and be detectable via epidemiologic studies. Complementary study designs, where epigenetic associations are examined in both post mortem disease tissues as well as in vivo peripheral tissues will be very powerful.
It is in our best fiscal and time management interest to work with established studies, yet existing cohorts often have limited availability of appropriate biosamples at appropriate collection windows.
Measurement tools
Thus far, epigenetic epidemiology has largely been synonymous with the molecular epidemiology of DNA methylation. These studies measure the percentage of methylated cytosine residues within CpG dinucleotides. It is possible to measure the methylation levels at individual CpG sites, but as a first-pass, studies often measure a proxy of the global methylation level of a sample across the epigenome. Global or total methylation methods include high-performance liquid chromatography (HPLC) [Ehrlich et al. 1982], methylation-specific in situ antibody fluorescence [Miller et al. 1974], repetitive element (such as long interspersed nucleotide element-1, LINE-1) bisulfite sequencing [Yang et al. 2004], and LUminometric Methylation Assay (LUMA) of CGCG sequences [Karimi et al. 2006]. Gene or region-specific DNA methylation is measured by methylation sensitive restriction enzymes [Singer-Sam et al. 1990], bisulfite methylation specific PCR [Herman et al. 1996], restriction digestion (COBRA) [Xiong and Laird 1997], MethyLite fluorescence-based real time PCR [Eads et al. 2000], base-specific cleavage with mass-spectrometry [Ehrich et al. 2005], and bisulfite Pyrosequencing [Tost and Gut 2007], among other methods. Assays have progressed to determine genome-scale DNA methylation coverage including antibody-mediated methylation dependent immunoprecipitation (MeDIP) with oligonucleotide array [Weber et al. 2005], restriction enzyme (mcrBC) digestion with tiling array (CHARM) [Irizarry et al. 2008], whole-genome bisulfite sequencing [Cokus et al. 2008], array capture bisulfite sequencing [Hodges et al. 2009], Illumina Infinium 450k array of bisulfite converted DNA [Bibikova et al. 2011], and reduced-representation bisulfite sequencing (RRBS) [Gu et al. 2011]. Each of these methods features a trade-off between feasibility, cost, input requirements, tissue collection, genome-coverage, and measurement precision. Appropriate epigenomic measurement tools will vary by study. Comparisons across platforms deal with differences in genomic coverage and quantitation, which is sometimes difficult for intermediate levels of methylation.
The technical approaches available to epigenetic researchers are already diverse and they are consistently producing higher quality data at lower costs. As a result, researchers are using multiple platforms with thousands or millions of observations. The data generated across platforms are often not easily comparable and use different genome builds, restricting our ability to make measurements over time or perform pooled or meta-analyses, while keeping up with technological developments. Greater efforts will be needed to bring cohesion to our epigenomics findings, perhaps using informatics to make measurements backward and forward compatible with new technologies.
Statistical analysis and integration
The field of statistical epigenomics is only now developing. Considerations span measurement error issues and signal-to-noise enhancement, estimation and testing of particular association and causal models, and integration of epigenetic measures with other “omics” level data.
First, high-throughput laboratory epigenetic measurements, such as array-based and sequencing protocols are subject to batch effects when signals are variable as a function of environmental conditions, personnel, and reagent group [Leek et al. 2010]. Epigenetic technology development sometimes outpaces analytics development and many of the platforms do not yet have standard pipelines to adjust for batch effects. However, some of these batch concerns can be solved in design by randomizing sample placement across batches. Also, there is not yet uniformity on data quality assurance/quality control measures or normalization procedures [Marabita et al. 2013; Wang et al. 2012], although lessons from expression array analyses and other fields are being applied.
DNA methylation and histone modifications are major drivers of tissue and cell differentiation, and thus particular cell types have specific epigenomic profiles [Irizarry et al. 2009; Jones and Taylor 1980]. As mentioned earlier, epidemiologic samples are often complex mixtures of different cell types, and thus associations with disease may be confounded by cell type distribution, if those distributions are a surrogate of the disease cause. This can be handled via familiar approaches to confounding, such as regression adjustment or stratification, if cell composition is known, or cell types can be isolated. For example, whole blood DNA represents fractions of DNA from granulocytes, monocytes, T-cells, B-cells, etc. Conducting laboratory analyses stratified by cell type can solve this, (and may be most relevant for some questions where effects of interest are isolated to particular cell types –see the Biologically interpreting the results section below) although this is not feasible or cost-effective for most epidemiological studies. Recently, investigators have addressed this by using cell type-specific methylation signatures to predict cell type proportions from mixed tissue samples such as blood and using these predictions for adjustment in disease association analyses [Houseman et al. 2012].
Beyond these technical issues of batch, normalization, and protection from confounding, the field is still settling on the most appropriate analytic approach to detect disease associations within the epigenome. Approaches include single-CpG statistical tests and region-based tests such as “bump hunting” that borrows information from neighboring sites to smooth over findings and exploit correlation in methylation signals [Jaffe et al. 2012] and Aclust that detects clusters between adjacent sites [Sofer et al. 2013]. Gold-standard statistical tests at the epigenome level have not been fully defined and it is currently unclear what reasonable expectations of effect size will be in epidemiologic settings. Calculations of sample size and study power in epigenetic epidemiology are difficult, but requirements may be less than the numbers needed for GWAS given the reduced number of features for many epigenome platforms compared to GWAS and the quantitative nature of epigenetic signals compared to categorical genotypes. Ultimately, replication across studies should drive long-term interpretation.
Concerns that associations may reflect a consequence of the disease rather than an indicator of etiology can be addressed by carefully considering the purpose of the epigenetic analyses (see the Study design section above), and clear causal modeling such as Mendelian Randomization in situations where genotype may be an appropriate instrumental variable [Liu et al. 2013; Smith and Ebrahim 2004].
Measurement and integration across the epigenome with the genome and environment, as key regulators of the epigenome, are needed, and common measures to ensure that our tests are “statistically and biologically sound” are important in this regard [Heijmans and Mill 2012].
Biologically interpreting the results
While epigenetic modifications may be simply biomarkers of exposure or disease, the great hope is that epigenetic research may provide direct mechanistic insight regarding exposure impacts on the body, and potentially on disease. Since epigenetic mechanisms contribute to gene regulation, one functional assessment of epigenetic modification is the correlation with gene expression. However, this correlation is not always apparent when, for example, DNA methylation occurs at non-promoter, or non-genic sites, such that the targeted DNA for regulation/expression is not clear. Examination of correlation between methylation and expression based on single CpGs – single genes versus regional examination at either level may provide different insights. We need to better understand the functional influence of altered marks individually and collectively. Further, most analyses of DNA methylation and RNA gene expression consider both as linear predictors/outcomes. Non-linear relationships may more accurately describe the functional implications of DNA methylation.
Mechanistic work must rely on data from specific target tissues, and at specific developmental or lifespan windows, which, as mentioned earlier, is a particular challenge in large epidemiologic settings. Our partnerships with basic scientists will be critical to characterizing biological relationships and determining biological plausibility of our population-based observations. Mechanistic studies are needed to better understand the implications of epidemiologic associations, leading to therapeutic interventions and treatments.
Public relations and communications
High expectations of the human genome project were deflated by the complexity of the relationships between genetics and most diseases. The public experienced disappointment in human genetics when few clearly disease-causing genetic variants were identified. Epigenetics has experienced considerable hype within the scientific community, which may extend to the public, and could suffer the same fate if expectations are set unreasonably high. Thus far, the observed effect sizes, outside of foundational tissue and cancer studies, have been relatively small. It is important to recognize the promise, but also the limitations of epigenetic studies in humans, including the limitations on available tissue types, cell heterogeneity, and developmental windows. We will need to use appropriately cautious interpretations of results and be clear and deliberate in our communication with the media.
Summary
Recent exciting advances in measurement technology and analytic techniques offer greater epigenetic epidemiology opportunities than ever before. The field has the potential to address questions about mechanisms of basic biology, development, disease, environmental exposures, nutrition, and aging, and may open up new measurement tools for biomarker-based analyses of exposures or disease in epidemiology. Careful designs of new studies, clever leveraging of existing studies, and rigorous statistical and causal modeling will be necessary to overcome challenges inherent to the field related to temporality and biological interpretation. Like other areas of molecular epidemiology, epigenetic epidemiology can provide great insight into the biological changes associated with exposures and disease and may indeed bridge the genetic and environmental epidemiology fields.
Acknowledgments
Funding
NIH R01 ES017646 (PI Fallin/Feinberg). Drs. Bakulski and Fallin together researched, designed, and wrote this review article.
Footnotes
Author Contributions
The authors do not report any conflicts of interest.
References
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in x-linked mecp2, encoding methyl-cpg-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Anderson OS, Nahar MS, Faulk C, Jones TR, Liao C, Kannan K, et al. Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol a. Environ Mol Mutagen. 2012;53:334–342. doi: 10.1002/em.21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita A, Niu J, Qu Q, Zhao N, Ruan Y, Nadas A, et al. Global levels of histone modifications in peripheral blood mononuclear cells of subjects with exposure to nickel. Environ Health Perspect. 2012;120:198–203. doi: 10.1289/ehp.1104140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, et al. A common genetic variant in the neurexin superfamily member cntnap2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba Y, Yu H, Liu F, Geng X, Zhu C, Zhu Q, et al. Relationship of folate, vitamin b12 and methylation of insulin-like growth factor-ii in maternal and cord blood. Eur J Clin Nutr. 2011;65:480–485. doi: 10.1038/ejcn.2010.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21:243–251. doi: 10.1097/mop.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakulski KM, Dolinoy DC, Sartor MA, Paulson HL, Konen JR, Lieberman AP, et al. Genome-wide DNA methylation differences between late-onset alzheimer’s disease and cognitively normal controls in human frontal cortex. J Alzheimers Dis. 2012;29:571–588. doi: 10.3233/JAD-2012-111223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ. The developmental origins of adult disease. J Am Coll Nutr. 2004;23:588S–595S. doi: 10.1080/07315724.2004.10719428. [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Klinge DM, Stidley CA, Issa JP, Herman JG, March TH, et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res. 2003;63:7089–7093. [PubMed] [Google Scholar]
- Bell CG, Finer S, Lindgren CM, Wilson GA, Rakyan VK, Teschendorff AE, et al. Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the fto type 2 diabetes and obesity susceptibility locus. PLoS One. 2010;5:e14040. doi: 10.1371/journal.pone.0014040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. DNA methylation patterns associate with genetic and gene expression variation in hapmap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellet MM, Sassone-Corsi P. Mammalian circadian clock and metabolism - the epigenetic link. J Cell Sci. 2010;123:3837–3848. doi: 10.1242/jcs.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, et al. High density DNA methylation array with single cpg site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Bihaqi SW, Huang H, Wu J, Zawia NH. Infant exposure to lead (pb) and epigenetic modifications in the aging primate brain: Implications for alzheimer’s disease. J Alzheimers Dis. 2011;27:819–833. doi: 10.3233/JAD-2011-111013. [DOI] [PubMed] [Google Scholar]
- Bind MA, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, et al. Air pollution and markers of coagulation, inflammation, and endothelial function: Associations and epigene-environment interactions in an elderly cohort. Epidemiology. 2012;23:332–340. doi: 10.1097/EDE.0b013e31824523f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Walter J, Paulsen M, Lengauer T. Inter-individual variation of DNA methylation and its implications for large-scale epigenome mapping. Nucleic Acids Res. 2008;36:e55. doi: 10.1093/nar/gkn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A. Environmental epigenetics. Heredity (Edinb) 2010;105:105–112. doi: 10.1038/hdy.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghol N, Suderman M, McArdle W, Racine A, Hallett M, Pembrey M, et al. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol. 2012;41:62–74. doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27k discovery and replication. Am J Hum Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-a exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010;24:2273–2280. doi: 10.1096/fj.09-140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantone L, Nordio F, Hou L, Apostoli P, Bonzini M, Tarantini L, et al. Inhalable metal-rich air particles and histone h3k4 dimethylation and h3k9 acetylation in a cross-sectional study of steel workers. Environ Health Perspect. 2011;119:964–969. doi: 10.1289/ehp.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA, Chretien P, Stevenson CW, Zhang TY, Gratton A, Meaney MJ. Variations in nucleus accumbens dopamine associated with individual differences in maternal behavior in the rat. J Neurosci. 2004;24:4113–4123. doi: 10.1523/JNEUROSCI.5322-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervona Y, Arita A, Costa M. Carcinogenic metals and the epigenome: Understanding the effect of nickel, arsenic, and chromium. Metallomics. 2012;4:619–627. doi: 10.1039/c2mt20033c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, et al. Shotgun bisulphite sequencing of the arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppede F. Advances in the genetics and epigenetics of neurodegenerative diseases. Epigenetics of Neurodegenerative Diseases. 2013;1:3–31. [Google Scholar]
- Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, et al. Environmental epigenetics: Prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet. 2012;131:1565–1589. doi: 10.1007/s00439-012-1189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, et al. Loss of igf2 imprinting: A potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile x mental retardation protein targets g quartet mrnas important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CD, Uthus EO. Dietary folate and selenium affect dimethylhydrazine-induced aberrant crypt formation, global DNA methylation and one-carbon metabolism in rats. J Nutr. 2003;133:2907–2914. doi: 10.1093/jn/133.9.2907. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet. 2012;13:153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- Deng J, Davies DR, Wisedchaisri G, Wu M, Hol WG, Mehlin C. An improved protocol for rapid freezing of protein samples for long-term storage. Acta Crystallogr D Biol Crystallogr. 2004;60:203–204. doi: 10.1107/s0907444903024491. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: Linking early developmental environment to adult disease. Reprod Toxicol. 2007;23:297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Jirtle RL. Environmental epigenomics in human health and disease. Environ Mol Mutagen. 2008;49:4–8. doi: 10.1002/em.20366. [DOI] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, et al. Methylight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebers GC, Sadovnick AD, Dyment DA, Yee IM, Willer CJ, Risch N. Parent-of-origin effect in multiple sclerosis: Observations in half-siblings. Lancet. 2004;363:1773–1774. doi: 10.1016/S0140-6736(04)16304-6. [DOI] [PubMed] [Google Scholar]
- Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex MJ, Thomas Boyce W, Hertzman C, Lam LL, Armstrong JM, Neumann SM, et al. Epigenetic vestiges of early developmental adversity: Childhood stress exposure and DNA methylation in adolescence. Child Dev. 2013;84:58–75. doi: 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene mgmt and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, et al. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- Farwell DG, Shera KA, Koop JI, Bonnet GA, Matthews CP, Reuther GW, et al. Genetic and epigenetic changes in human epithelial cells immortalized by telomerase. Am J Pathol. 2000;156:1537–1547. doi: 10.1016/S0002-9440(10)65025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Irizarry RA, Fradin D, Aryee MJ, Murakami P, Aspelund T, et al. Personalized epigenomic signatures that are stable over time and covary with body mass index. Sci Transl Med. 2010;2:49ra67. doi: 10.1126/scitranslmed.3001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech M. The role of folic acid and vitamin b12 in genomic stability of human cells. Mutat Res. 2001a;475:57–67. doi: 10.1016/s0027-5107(01)00079-3. [DOI] [PubMed] [Google Scholar]
- Fenech M. Recommended dietary allowances (rdas) for genomic stability. Mutat Res. 2001b;480–481:51–54. doi: 10.1016/s0027-5107(01)00168-3. [DOI] [PubMed] [Google Scholar]
- Fenech M, Ferguson LR. Vitamins/minerals and genomic stability in humans. Mutat Res. 2001;475:1–6. doi: 10.1016/s0027-5107(01)00069-0. [DOI] [PubMed] [Google Scholar]
- Foley DL, Craig JM, Morley R, Olsson CA, Dwyer T, Smith K, et al. Prospects for epigenetic epidemiology. Am J Epidemiol. 2009;169:389–400. doi: 10.1093/aje/kwn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin D, Cheslack-Postava K, Ladd-Acosta C, Newschaffer C, Chakravarti A, Arking DE, et al. Parent-of-origin effects in autism identified through genome-wide linkage analysis of 16, 000 snps. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Beedle AS. Non-genomic transgenerational inheritance of disease risk. Bioessays. 2007;29:145–154. doi: 10.1002/bies.20522. [DOI] [PubMed] [Google Scholar]
- Grafodatskaya D, Choufani S, Ferreira JC, Butcher DT, Lou Y, Zhao C, et al. Ebv transformation and cell culturing destabilizes DNA methylation in human lymphoblastoid cell lines. Genomics. 2010;95:73–83. doi: 10.1016/j.ygeno.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, et al. Transgenerational epigenetic inheritance of longevity in caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg JP, Lit L, Baron CA, Hertz-Picciotto I, Walker W, Davis RA, et al. Gene expression changes in children with autism. Genomics. 2008;91:22–29. doi: 10.1016/j.ygeno.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- Guerrero-Bosagna CM, Sabat P, Valdovinos FS, Valladares LE, Clark SJ. Epigenetic and phenotypic changes result from a continuous pre and post natal dietary exposure to phytoestrogens in an experimental population of mice. BMC Physiol. 2008;8:17. doi: 10.1186/1472-6793-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Yoo KY, Cho SH. Glycophorin a mutant frequency in radiation workers at the nuclear power plants and a hospital. Mutat Res. 2002;501:45–56. doi: 10.1016/s0027-5107(02)00009-x. [DOI] [PubMed] [Google Scholar]
- Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: The thrifty phenotype hypothesis. Diabetologia. 1992;35:595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- Hass BS, Hart RW, Lu MH, Lyn-Cook BD. Effects of caloric restriction in animals on cellular function, oncogene expression, and DNA methylation in vitro. Mutat Res. 1993;295:281–289. doi: 10.1016/0921-8734(93)90026-y. [DOI] [PubMed] [Google Scholar]
- Hawley TS, Hawley RG. Flow cytometry protocols. 3 2011. [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Mill J. Commentary: The seven plagues of epigenetic epidemiology. Int J Epidemiol. 2012;41:74–78. doi: 10.1093/ije/dyr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific pcr: A novel pcr assay for methylation status of cpg islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Moran S, Esteller M. Aberrant DNA methylation profiles in the premature aging disorders hutchinson-gilford progeria and werner syndrome. Epigenetics. 2013;8:28–33. doi: 10.4161/epi.23366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Ronco AM, Guerrero-Bosagna C, de la Maza MP, Leiva L, Barrera G, et al. Methylation status in healthy subjects with normal and high serum folate concentration. Nutrition. 2008;24:1103–1109. doi: 10.1016/j.nut.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, et al. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–1605. doi: 10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyo C, Murtha AP, Schildkraut JM, Jirtle RL, Demark-Wahnefried W, Forman MR, et al. Methylation variation at igf2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics. 2011;6:928–936. doi: 10.4161/epi.6.7.16263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu VW, Frank BC, Heine S, Lee NH, Quackenbush J. Gene expression profiling of lymphoblastoid cell lines from monozygotic twins discordant in severity of autism reveals differential regulation of neurologically relevant genes. BMC Genomics. 2006;7:118. doi: 10.1186/1471-2164-7-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, et al. Comprehensive high-throughput arrays for relative methylation (charm) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific cpg island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41:200–209. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Zhang L, Peng V, Ren X, McHale CM, Smith MT. A comparison of the cytogenetic alterations and global DNA hypomethylation induced by the benzene metabolite, hydroquinone, with those induced by melphalan and etoposide. Leukemia. 2010;24:986–991. doi: 10.1038/leu.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones IM, Galick H, Kato P, Langlois RG, Mendelsohn ML, Murphy GA, et al. Three somatic genetic biomarkers and covariates in radiation-exposed russian cleanup workers of the chernobyl nuclear reactor 6–13 years after exposure. Radiat Res. 2002;158:424–442. doi: 10.1667/0033-7587(2002)158[0424:tsgbac]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, et al. 450k epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120:1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminen-Ahola N, Ahola A, Maga M, Mallitt KA, Fahey P, Cox TC, et al. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet. 2010;6:e1000811. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Johansson S, Stach D, Corcoran M, Grander D, Schalling M, et al. Luma (luminometric methylation assay)--a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006;312:1989–1995. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Kim KY, Kim DS, Lee SK, Lee IK, Kang JH, Chang YS, et al. Association of low-dose exposure to persistent organic pollutants with global DNA hypomethylation in healthy koreans. Environ Health Perspect. 2010;118:370–374. doi: 10.1289/ehp.0901131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Draghici S, Tang L, Kraniak JM, Land SJ, Tainsky MA. Epigenetic silencing of multiple interferon pathway genes after cellular immortalization. Oncogene. 2003;22:4118–4127. doi: 10.1038/sj.onc.1206594. [DOI] [PubMed] [Google Scholar]
- Kuratomi G, Iwamoto K, Bundo M, Kusumi I, Kato N, Iwata N, et al. Aberrant DNA methylation associated with bipolar disorder identified from discordant monozygotic twins. Mol Psychiatry. 2008;13:429–441. doi: 10.1038/sj.mp.4002001. [DOI] [PubMed] [Google Scholar]
- Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31:142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumey LH, Stein AD, Kahn HS, Romijn JA. Lipid profiles in middle-aged men and women after famine exposure during gestation: The dutch hunger winter families study. Am J Clin Nutr. 2009;89:1737–1743. doi: 10.3945/ajcn.2008.27038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigano J, Baccarelli A, Mittleman MA, Wright RO, Sparrow D, Vokonas PS, et al. Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ Health Perspect. 2011;119:977–982. doi: 10.1289/ehp.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabita F, Almgren M, Lindholm ME, Ruhrmann S, Fagerstrom-Billai F, Jagodic M, et al. An evaluation of analysis pipelines for DNA methylation profiling using the illumina humanmethylation 450 beadchip platform. Epigenetics. 2013;8:333–346. doi: 10.4161/epi.24008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit CJ, Houseman EA, Christensen BC, Eddy K, Bueno R, Sugarbaker DJ, et al. Examination of a cpg island methylator phenotype and implications of methylation profiles in solid tumors. Cancer Res. 2006;66:10621–10629. doi: 10.1158/0008-5472.CAN-06-1687. [DOI] [PubMed] [Google Scholar]
- Masson P, Alves AC, Ebbels TM, Nicholson JK, Want EJ. Optimization and evaluation of metabolite extraction protocols for untargeted metabolic profiling of liver samples by uplc-ms. Anal Chem. 2010;82:7779–7786. doi: 10.1021/ac101722e. [DOI] [PubMed] [Google Scholar]
- McGuinness D, McGlynn LM, Johnson PC, MacIntyre A, Batty GD, Burns H, et al. Socio-economic status is associated with epigenetic differences in the psobid cohort. Int J Epidemiol. 2012;41:151–160. doi: 10.1093/ije/dyr215. [DOI] [PubMed] [Google Scholar]
- McKay JA, Waltham KJ, Williams EA, Mathers JC. Folate depletion during pregnancy and lactation reduces genomic DNA methylation in murine adult offspring. Genes Nutr. 2011;6:189–196. doi: 10.1007/s12263-010-0199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: Prediction, plasticity, and programming. Physiol Rev. 2005;85:571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- Mehedint MG, Niculescu MD, Craciunescu CN, Zeisel SH. Choline deficiency alters global histone methylation and epigenetic marking at the re1 site of the calbindin 1 gene. FASEB J. 2010;24:184–195. doi: 10.1096/fj.09-140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Qin G, Zhang B. DNA damage in mice treated with sulfur dioxide by inhalation. Environ Mol Mutagen. 2005;46:150–155. doi: 10.1002/em.20142. [DOI] [PubMed] [Google Scholar]
- Miller OJ, Schnedl W, Allen J, Erlanger BF. 5-methylcytosine localised in mammalian constitutive heterochromatin. Nature. 1974;251:636–637. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Anitha A, Yamada K, Tsujii M, Iwayama Y, Hattori E, et al. Genetic and expression analyses reveal elevated expression of syntaxin 1a (stx1a) in high functioning autism. Int J Neuropsychopharmacol. 2008;11:1073–1084. doi: 10.1017/S1461145708009036. [DOI] [PubMed] [Google Scholar]
- Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, rora, whose protein product is reduced in autistic brain. FASEB J. 2010;24:3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Martin CL, Vazquez-Lopez A, Spence SJ, Alvarez-Retuerto AI, Sigman M, et al. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum Mol Genet. 2007;16:1682–1698. doi: 10.1093/hmg/ddm116. [DOI] [PubMed] [Google Scholar]
- Ogino S, Hazra A, Tranah GJ, Kirkner GJ, Kawasaki T, Nosho K, et al. Mgmt germline polymorphism is associated with somatic mgmt promoter methylation and gene silencing in colorectal cancer. Carcinogenesis. 2007;28:1985–1990. doi: 10.1093/carcin/bgm160. [DOI] [PubMed] [Google Scholar]
- Pilsner JR, Hu H, Ettinger A, Sanchez BN, Wright RO, Cantonwine D, et al. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect. 2009;117:1466–1471. doi: 10.1289/ehp.0800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya OO, Sokolov BP. Differential expression of the “c” and “t” alleles of the 5-ht2a receptor gene in the temporal cortex of normal individuals and schizophrenics. J Neurosci Res. 2002;67:812–822. doi: 10.1002/jnr.10173. [DOI] [PubMed] [Google Scholar]
- Polesskaya OO, Aston C, Sokolov BP. Allele c-specific methylation of the 5-ht2a receptor gene: Evidence for correlation with its expression and expression of DNA methylase dnmt1. J Neurosci Res. 2006;83:362–373. doi: 10.1002/jnr.20732. [DOI] [PubMed] [Google Scholar]
- Poulsen P, Esteller M, Vaag A, Fraga MF. The epigenetic basis of twin discordance in age-related diseases. Pediatr Res. 2007;61:38R–42R. doi: 10.1203/pdr.0b013e31803c7b98. [DOI] [PubMed] [Google Scholar]
- Rager JE, Bauer RN, Muller LL, Smeester L, Carson JL, Brighton LE, et al. DNA methylation in nasal epithelial cells from smokers: Identification of ulbp3-related effects. Am J Physiol Lung Cell Mol Physiol. 2013;305:L432–438. doi: 10.1152/ajplung.00116.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, et al. Transgenerational inheritance of epigenetic states at the murine axin(fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Relton CL, Davey Smith G. Two-step epigenetic mendelian randomization: A strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol. 2012;41:161–176. doi: 10.1093/ije/dyr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribel-Madsen R, Fraga MF, Jacobsen S, Bork-Jensen J, Lara E, Calvanese V, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS One. 2012;7:e51302. doi: 10.1371/journal.pone.0051302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowling MJ, McMullen MH, Schalinske KL. Vitamin a and its derivatives induce hepatic glycine n-methyltransferase and hypomethylation of DNA in rats. J Nutr. 2002;132:365–369. doi: 10.1093/jn/132.3.365. [DOI] [PubMed] [Google Scholar]
- Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in greenlandic inuit. Environ Health Perspect. 2008;116:1547–1552. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CW. Uncertain inheritance: Transgenerational effects of environmental exposures. Environ Health Perspect. 2013:121. doi: 10.1289/ehp.121-A298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger O, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow WJ, Pesatori AC, Dimont E, Farmer PB, Albetti B, Ettinger AS, et al. Urinary benzene biomarkers and DNA methylation in bulgarian petrochemical workers: Study findings and comparison of linear and beta regression models. PLoS One. 2012;7:e50471. doi: 10.1371/journal.pone.0050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serretti A, Drago A, De Ronchi D. Htr2a gene variants and psychiatric disorders: A review of current literature and selection of snps for future studies. Curr Med Chem. 2007;14:2053–2069. doi: 10.2174/092986707781368450. [DOI] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- Shi ST, Wang ZY, Smith TJ, Hong JY, Chen WF, Ho CT, et al. Effects of green tea and black tea on 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone bioactivation, DNA methylation, and lung tumorigenesis in a/j mice. Cancer Res. 1994;54:4641–4647. [PubMed] [Google Scholar]
- Singer-Sam J, LeBon JM, Tanguay RL, Riggs AD. A quantitative hpaii-pcr assay to measure methylation of DNA from a small number of cells. Nucleic Acids Res. 1990;18:687. doi: 10.1093/nar/18.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GD, Ebrahim S. Mendelian randomization: Prospects, potentials, and limitations. Int J Epidemiol. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- Smith SJ, Li Y, Whitley R, Marion MJ, Partilo S, Carney WP, et al. Molecular epidemiology of p53 protein mutations in workers exposed to vinyl chloride. Am J Epidemiol. 1998;147:302–308. doi: 10.1093/oxfordjournals.aje.a009450. [DOI] [PubMed] [Google Scholar]
- Sofer T, Schifano ED, Hoppin JA, Hou L, Baccarelli AA. A-clustering: A novel method for the detection of co-regulated methylation regions, and regions associated with exposure. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine OC, Xu J, Koskela R, McMahon FJ, Gschwend M, Friddle C, et al. Evidence for linkage of bipolar disorder to chromosome 18 with a parent-of-origin effect. Am J Hum Genet. 1995;57:1384–1394. [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- Sunami E, Shinozaki M, Higano CS, Wollman R, Dorff TB, Tucker SJ, et al. Multimarker circulating DNA assay for assessing blood of prostate cancer patients. Clin Chem. 2009;55:559–567. doi: 10.1373/clinchem.2008.108498. [DOI] [PubMed] [Google Scholar]
- Sutherland JE, Costa M. Epigenetics and the environment. Ann N Y Acad Sci. 2003;983:151–160. doi: 10.1111/j.1749-6632.2003.tb05970.x. [DOI] [PubMed] [Google Scholar]
- Tabish AM, Poels K, Hoet P, Godderis L. Epigenetic factors in cancer risk: Effect of chemical carcinogens on global DNA methylation pattern in human tk6 cells. PLoS One. 2012;7:e34674. doi: 10.1371/journal.pone.0034674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, et al. Effects of particulate matter on genomic DNA methylation content and inos promoter methylation. Environ Health Perspect. 2009;117:217–222. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- Tryndyak VP, Muskhelishvili L, Kovalchuk O, Rodriguez-Juarez R, Montgomery B, Churchwell MI, et al. Effect of long-term tamoxifen exposure on genotoxic and epigenetic changes in rat liver: Implications for tamoxifen-induced hepatocarcinogenesis. Carcinogenesis. 2006;27:1713–1720. doi: 10.1093/carcin/bgl050. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Nossova N, Yager T, Tsuang MM, Guo SC, Shyu KG, et al. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: A preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;133B:1–5. doi: 10.1002/ajmg.b.30161. [DOI] [PubMed] [Google Scholar]
- Turker MS, Bestor TH. Formation of methylation patterns in the mammalian genome. Mutat Res. 1997;386:119–130. doi: 10.1016/s1383-5742(96)00048-8. [DOI] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2010;107:9470–9475. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini G, Bollati V, Fazio L, Porcelli A, Iacovelli L, Catalani A, et al. Stress-related methylation of the catechol-o-methyltransferase val 158 allele predicts human prefrontal cognition and activity. J Neurosci. 2011;31:6692–6698. doi: 10.1523/JNEUROSCI.6631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-cpg sequences inhibits the binding of the methyl-cpg binding domain (mbd) of methyl-cpg binding protein 2 (mecp2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Yan L, Hu Q, Sucheston LE, Higgins MJ, Ambrosone CB, et al. Ima: An r package for high-throughput analysis of illumina’s 450k infinium methylation data. Bioinformatics. 2012;28:729–730. doi: 10.1093/bioinformatics/bts013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset alzheimer’s disease. PLoS One. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Weksberg R, Shuman C, Beckwith JB. Beckwith-wiedemann syndrome. Eur J Hum Genet. 2010;18:8–14. doi: 10.1038/ejhg.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CC, Meaburn EL, Ronald A, Price TS, Jeffries AR, Schalkwyk LC, et al. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, Park SK, et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010;118:790–795. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Laird PW. Cobra: A sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite pcr of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–5503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- Yauk C, Polyzos A, Rowan-Carroll A, Somers CM, Godschalk RW, Van Schooten FJ, et al. Germ-line mutations, DNA damage, and global hypermethylation in mice exposed to particulate air pollution in an urban/industrial location. Proc Natl Acad Sci U S A. 2008;105:605–610. doi: 10.1073/pnas.0705896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa Y, Nagasaki H, Akiyama Y, Hashimoto Y, Takizawa T, Kojima K, et al. DNA methylation status is inversely correlated with green tea intake and physical activity in gastric cancer patients. Int J Cancer. 2009;124:2677–2682. doi: 10.1002/ijc.24231. [DOI] [PubMed] [Google Scholar]
- Zama AM, Uzumcu M. Fetal and neonatal exposure to the endocrine disruptor methoxychlor causes epigenetic alterations in adult ovarian genes. Endocrinology. 2009;150:4681–4691. doi: 10.1210/en.2009-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]