Summary
Muscle-invasive bladder cancers (MIBCs) are biologically heterogeneous and have widely variable clinical outcomes and responses to conventional chemotherapy. We discovered 3 molecular subtypes of MIBC that resembled established molecular subtypes of breast cancer. Basal MIBCs shared biomarkers with basal breast cancers and were characterized by p63 activation, squamous differentiation, and more aggressive disease at presentation. Luminal MIBCs contained features of active PPARγ and estrogen receptor (ER) transcription and were enriched with activating FGFR3 mutations and potentially FGFR inhibitor sensitivity. p53-like MIBCs were consistently resistant to neoadjuvant MVAC chemotherapy, and all chemoresistant tumors adopted a p53-like phenotype after therapy. Our observations have important implications for prognostication, the future clinical development of targeted agents, and disease management with conventional chemotherapy.
INTRODUCTION
Bladder cancer progresses along two distinct pathways that pose distinct challenges for clinical management (Dinney et al., 2004). Low-grade non-muscle invasive (“superficial”) cancers, which account for 70% of tumor incidence, are not immediately life threatening, but they have a propensity for recurrence which necessitates costly life-long surveillance (Botteman et al., 2003). In contrast, high-grade muscle-invasive bladder cancers (MIBCs) progress rapidly to become metastatic and generate the bulk of patient mortality (Shah et al., 2011). Radical cystectomy with perioperative cisplatin-based combination chemotherapy is the current standard of care for high-risk MIBC. Treatment selection depends heavily on clinico-pathologic features, but current staging systems are woefully inaccurate and result in an unacceptably high rate of clinical understaging and consequently inadequate treatment (Svatek et al., 2011). Furthermore, cisplatin-based chemotherapy is only effective in 30–40% of cases, and it is not yet possible to prospectively identify the patients who are likely to obtain benefit (Shah et al., 2011). To add to the quandary, no effective alternative to cisplatin-based chemotherapy has been identified for resistant tumors. Therefore, there is an urgent need to develop a more precise, biology-based approach to the classification of bladder cancer to inform clinical management.
Gene expression profiling has been used widely to identify molecular heterogeneity in other human cancers. For example, Perou and coworkers (Perou et al., 2000) used gene expression profiling to identify molecular subtypes of breast cancer (basal/triple negative, HER2+, luminal A, and luminal B) that behave clinically as though they are distinct disease entities - luminal breast cancers respond to estrogen receptor (ER)-targeted therapy, HER2+ tumors to Herceptin and other ErbB2-blocking agents, and basal tumors to chemotherapy only (Rouzier et al., 2005). Previous studies in bladder cancer identified signatures associated with stage and outcomes (Blaveri et al., 2005; Dyrskjot et al., 2003; Sanchez-Carbayo et al., 2006; Sjodahl et al., 2012) and progression (Kim et al., 2010; Lee et al., 2010), but the biological and clinical significance of these signatures remain unclear. Here we also used gene expression profiling and unsupervised analyses to identify molecular subtypes of MIBC with the goal of defining the biological basis for the molecular heterogeneity that is observed in them.
RESULTS
Muscle-invasive bladder cancers can be grouped into basal and luminal subtypes
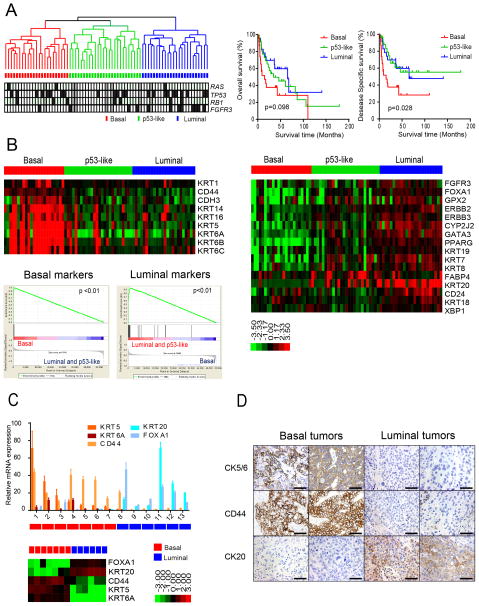
We performed whole genome mRNA expression profiling and unsupervised hierarchical cluster analyses on a cohort of 73 primary fresh frozen MIBCs obtained by transurethral resection at our institution. We identified three distinct molecular subtypes (Fig. 1A, Table 1). The upregulated genes (fold changes) that determined subtype assignments contained signature biomarkers for basal (CD44, KRT5, KRT6, KRT14, CDH3) and luminal (CD24, FOXA1, GATA3, ERBB2, ERBB3, XBP1, and KRT20) breast cancers, respectively (Fig. 1B, heat maps; Fig. S1A) (Perou et al., 2000), and formal gene set enrichment analyses (GSEA) confirmed that the subtypes were enriched with basal and luminal markers (Fig. 1B, below). In control experiments we confirmed that the array-based measurements of basal and luminal marker expression correlated well with the results obtained by quantitative RT-PCR (Fig. 1C) or immunohistochemistry (Fig. 1D) in some of the same tumors. We therefore propose the names “basal” and “luminal” for two of the MIBC subtypes. Although the tumors in the third subtype also expressed luminal biomarkers (Fig. 1B, Fig. S1A), we have termed this MIBC subtype “p53-like” because its distinguishing feature was an activated wild-type p53 gene expression signature that we will discuss further below.
Figure 1. Basal and luminal subtypes of bladder cancer.
A. (Left) Whole genome mRNA expression profiling and hierarchical cluster analysis of a cohort of 73 MIBCs. RNA from fresh frozen tumors was analyzed using Illumina arrays. RAS, TP53, RB1, and FGFR3 mutations were detected by sequencing and are indicated in the color bars below the dendogram. Black, mutation; white, wild type; grey, mutation data were unavailable. (Right) Kaplan-Meier plots of overall survival (p = 0.098) and disease-specific survival (p = 0.028) in the 3 tumor subtypes. B. Expression of basal and luminal markers in the 3 subtypes. The heat maps depict relative expression of basal (left) and luminal (right) biomarkers. GSEA analyses (below, left) were used to determine whether basal and luminal markers were enriched in the subtypes. C. Quantitative RT-PCR was used to evaluate the accuracy of the gene expression profiling results. Relative levels of the indicated basal (red shades) and luminal (blue shades) biomarkers measured by RT-PCR were compared to the levels of the same markers measured by gene expression profiling on RNA isolated from macrodissected FFPE sections of the same tumors. Results are presented as relative quantitation (RQ) and the error bars indicate the range of RQ values as defined by 95% confidence level. RT-PCR results are shown on top, DASL gene expression profiling results are shown below. D. Analysis of basal and luminal marker expression by immunohistochemistry. Results from two representative basal (left) and luminal (right) tumors as defined by gene expression profiling are displayed. The scale bars correspond to 100 microns. See also Figure S1.
Table 1.
Clinicopathologic Characteristics of the MDACC Discovery Cohort (n=73)
| Total | Basal | p53-like | Luminal | p-value | |
|---|---|---|---|---|---|
| Cohort Size | 73 | 23 (32%) | 26 (36%) | 24 (33%) | |
| Mean Age (y) ± SD | 68.8 ± 10.2 | 70.1 ± 9.4 | 69.8 ± 8.9 | 66.4 ± 12.1 | 0.371 |
| Gender | |||||
| Female | 19 (26%) | 10 (44%) | 6 (23%) | 3 (13%) | 0.133 |
| Male | 54 (74%) | 13 (57%) | 20 (77%) | 21 (88%) | |
| Race | |||||
| Caucasian | 54 (74%) | 14 (61%) | 21 (81%) | 19 (79%) | |
| African American | 12 (16%) | 6 (26%) | 2 (7%) | 4 (17%) | 0.352 |
| Hispanic | 7 (10%) | 3 (13%) | 3 (12%) | 1 (4%) | |
| Clinical Stage at TUR (N0, M0) | |||||
| ≤ cT1 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| cT2 | 37 (51%) | 9 (39%) | 16 (62%) | 12 (50%) | 0.990 |
| cT3 | 16 (22%) | 4 (17%) | 7 (27%) | 5 (21%) | |
| cT4 | 6 (8%) | 2 (9%) | 2 (8%) | 2 (8%) | |
| Positive Clinical Lymph Nodes, cN+ | 11 (15%) | 5 (22%) | 1 (4%) | 5 (21%) | 0.137 |
| Positive Clinical Metastasis, cM+ | 7 (10%) | 5 (22%) | 0 (0%) | 2 (8%) | 0.035 |
| Primary Treatment | |||||
| Cystectomy | 57 (78%) | 15 (65%) | 25 (96%) | 17 (71%) | 0.019 |
| Other* | 16 (22%) | 8 (35%) | 1 (4%) | 7 (29%) | |
| Neoadjuvant Chemotherapy, NAC | 18 (25%) | 5 (22%) | 7 (27%) | 6 (25%) | 0.910 |
| Response to NAC^ | |||||
| Yes | 6 (33%) | 2 (40%) | 0 (0%) | 4 (67%) | 0.018 |
| No | 12 (67%) | 3 (60%) | 7 (100%) | 2 (33%) | |
| Pathologic T stage (n=57) | |||||
| pT0 | 4 (7%) | 2 (13%) | 0 (0%) | 2 (12%) | |
| pTa, pT1, pTis | 6 (11%) | 2 (13%) | 1 (4%) | 3 (18%) | |
| pT2 | 10 (18%) | 1 (7%) | 4 (16%) | 5 (29%) | 0.001 |
| pT3 | 25 (44%) | 4 (27%) | 18 (72%) | 3 (18%) | |
| pT4 | 12 (21%) | 6 (40%) | 2 (8%) | 4 (22%) | |
| Positive Pathologic Lymph Nodes | 23 (40%) | 3 (13%) | 14 (54%) | 6 (25%) | 0.010 |
| Variant histology in specimen | |||||
| Squamous Differentiation | 18 (32%) | 13 (57%) | 4 (15%) | 1 (4%) | |
| Sarcomatoid Differentiation | 3 (5%) | 3 (13%) | 0 (0%) | 0 (0%) | <0.001 |
| Other (Micropapillary, Glandular, Adenocarcinoma) | 3 (5%) | 0 (0%) | 3 (12%) | 0 (0%) | |
| Median Overall Survival (m) | 37.2 | 14.9 | 34.6 | 65.6 | 0.098 |
| Median Disease Specific Survival (m) | 46.3 | 14.9 | Not Reached | 65.6 | 0.028 |
Other= Chemotherapy was recommended for all 16 patients, based on available medical records 9 patients had documentation of completion
Response to NAC= Decrease in stage to pT0 or pT1 (for patients with high risk features at TUR: lymphovascular invasion, variant histology, hydronephrosis, or abnormal exam under anesthesia) at cystectomy.
The Kruskal-Wallis test was used to compare differences in mean age between groups. Log-rank test was used to compare differences in survival (overall and disease specific) between groups. For the remainder of categorical variables, Fisher’s exact test was used to determine differences between subtypes. p-values <0.05 were considered significant.
Table 1 depicts the clinical and pathologic characteristics of the discovery cohort by molecular subtype. Basal tumors were enriched with sarcomatoid features and metastatic disease at presentation (Table 1) and were associated with shorter overall survival (14.9 months, p= 0.098), and disease specific survival (median 14.9 months, p= 0.028) (Fig. 1A, right). Although they expressed epithelial cytokeratins, basal tumors also contained “mesenchymal” biomarkers (i.e., TWIST1/2, SNAI2, ZEB2, and VIM)(McConkey et al., 2010; Peinado et al., 2007)(Fig. S1B), as do basal breast cancers (Chaffer et al., 2013). In addition, basal tumors expressed high levels of the epidermal growth factor receptor (EGFR) and several of its ligands (Fig. S1B), similar to basal breast and head and neck squamous cell carcinomas (Perou et al., 2000; Romano et al., 2009; Sorlie et al., 2001). On the other hand, luminal tumors were enriched with “epithelial” biomarkers (E-cadherin/CDH1 and members of the miR-200 family)(Gregory et al., 2008)(Fig. S1B), high levels of fibroblast growth factor receptor-3 (FGFR3), and activating FGFR3 mutations (Fig. 1A,B; Fig. S1C). TP53 mutation frequencies were similar in all of the subtypes (Fig. S1C). To examine cluster stability, we calculated silhouette scores for each subtype. All of the basal and luminal tumors were stable, whereas 9/26 of the p53-like tumors were not (Fig. S1D); five of these unstable tumors were most similar to the luminal subtype (data not shown).
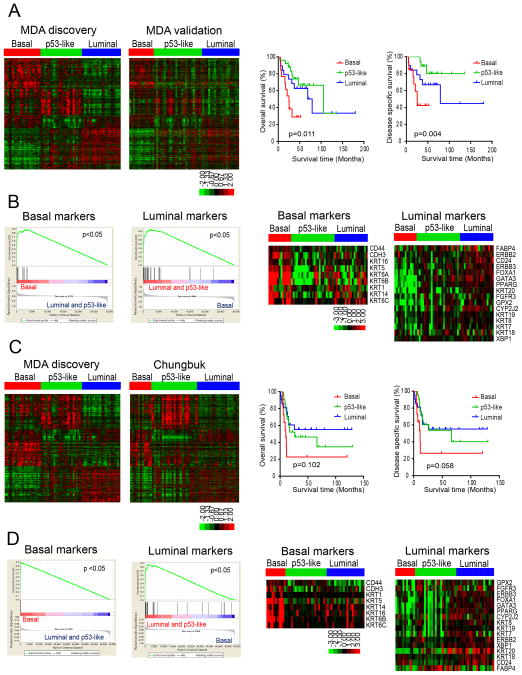
We developed a classifier using the differentially expressed genes associated with subtype membership in the discovery cohort and applied it to whole genome mRNA expression data from an independent cohort of formalin-fixed paraffin-embedded MIBCs (n = 57, MD Anderson validation cohort, Fig. 2A, Table S1). Like the discovery cohort, basal tumors in the validation cohort were associated with shorter overall survival (median 25.0 months, p= 0.011) and disease-specific survival (median 25.3 months, p= 0.004) (Fig. 2A, right side) and were enriched with basal biomarkers compared to tumors in the other subtypes (Fig. 2B). We then used the classifier to make additional predictions in the MIBCs (n = 55) from a third, publicly available gene expression profiling dataset (“Chungbuk cohort”)(Kim et al., 2010)(Fig. 2C, Table S2). The Chungbuk basal tumors were also associated with shorter median disease specific survival (11.2 months, p= 0.102) and overall survival (10.4 months, p= 0.058)(Fig. 2C, right side) and were enriched with basal biomarkers (Fig. 2D). In addition, GSEA confirmed that luminal biomarkers were enriched in luminal subtypes in both of the validation cohorts (Fig. 2B, D).
Figure 2. Characterization of basal and luminal subtypes in other MIBC cohorts.
A. Subtype classification of the MD Anderson validation cohort (n = 57). RNA was isolated from macrodissected FFPE tumor sections and whole genome mRNA expression was measured using Illumina’s DASL platform. Kaplan-Meier plots of overall survival (p = 0.011) and disease-specific survival (p=0.004) associated with the 3 subtypes are presented on the right. B. Expression of basal and luminal markers in the molecular subtypes in the MD Anderson validation cohort. The results of GSEA analyses of basal and luminal marker expression in the subtypes are displayed on the left, and heat maps depicting relative basal and luminal marker levels in the subtypes are displayed on the right. C. Subtype classification of the Chungbuk cohort (n = 55). Whole genome mRNA expression profiling (Illumina platform) and clinical data were downloaded from GEO (GSE13507), and the oneNN classifier was used to assign tumors to subtypes. Tumors were assigned to subtypes using the oneNN prediction model (left). Kaplan-Meier plots of overall survival (p = 0.102) and disease-specific survival (p = 0.058) as a function of tumor subtype (right). D. Expression of basal and luminal markers in the molecular subtypes in the Chungbuk cohort. The results of GSEA analyses of basal and luminal marker expression in the subtypes are displayed on the left, and heat maps depicting basal and luminal marker expression are displayed on the right. See also Tables S1 and S2.
Basal tumors are characterized by squamous differentiation
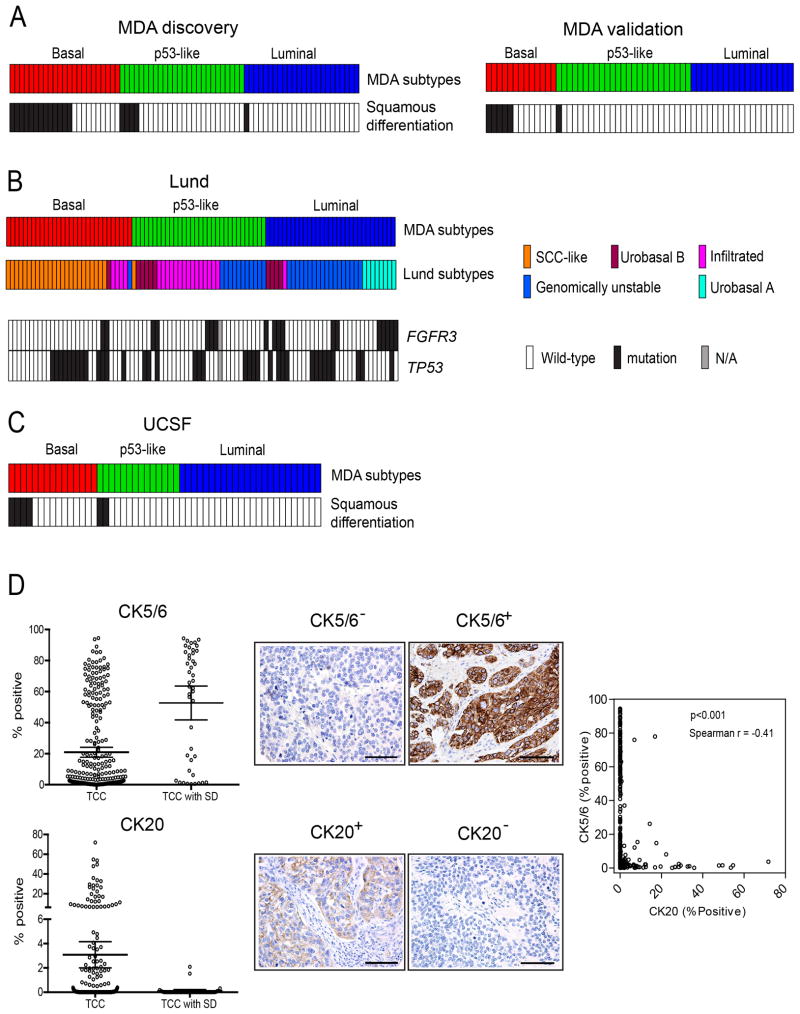
Bladder cancers with squamous histological features are generally considered distinct from conventional urothelial cancers. However, the basal MIBCs in the discovery and validation cohorts were significantly enriched with squamous features (Fig. 3A, Table 1, Table S1), and the basal tumors with squamous features also expressed higher basal biomarker mRNA levels than did basal tumors without squamous features (data not shown). The high molecular weight keratins (KRT5, KRT6, KRT14) that characterized basal MIBCs were also enriched in a lethal “squamous cell carcinoma” MIBC subtype that was identified recently by another group (Sjodahl et al., 2012). We applied our subtype classifier to the other group’s dataset (“Lund cohort”)(Sjodahl et al., 2012) and confirmed that the Lund squamous cell carcinoma subtype (Sjodahl et al., 2012) corresponded to the basal subtype identified here (“Lund” tumors, Fig. 3B, Table S3). Furthermore, like the MD Anderson discovery and validation cohorts, the Lund basal/SCC-like tumors were enriched with squamous differentiation (Sjodahl et al., 2012). Other Lund features also correlated with the subtypes described here - the MD Anderson p53-like subtype and the Lund “infiltrated” (MS2b.1) tumors were enriched with extracellular matrix biomarkers (Fig. 3B; also see Fig. S2)(Sjodahl et al., 2012), and all of the Lund “urobasal A” tumors were confined to the MD Anderson luminal subtype (Fig. 3B). In addition, as was the case in the MD Anderson discovery cohort, the Lund luminal tumors were enriched with activating FGFR3 mutations (p<0.05, Fig. 3B, Fig. S1C). High molecular weight keratins (KRT5 and KRT14) also characterized a bladder cancer “squamous cluster” (cluster D) identified by a group at the University of California-San Francisco (“UCSF cohort”) (Blaveri et al., 2005). We applied our classifier to the UCSF dataset and confirmed that the UCSF basal tumors were also enriched with squamous features (Fig. 3C, “UCSF”, Table S4). Finally, we stained a tissue microarray containing 332 pT3 MIBCs (Table S5) with clinical-grade antibodies specific for basal (CK5/6) or luminal (CK20) cytokeratins, quantified antigen expression across the tissue microarray by image analysis, and correlated cytokeratin levels with the presence of squamous features (Fig. 3D). Mean CK5/6 levels were significantly higher in tumors with squamous differentiation, whereas CK20 was expressed at higher levels in conventional MIBCs, and expression of CK5/6 correlated inversely with CK20 across the cohort (Fig. 3D). Expression of CK5/6 did not correlate with adverse outcomes (data not shown) as careful stage matching had been performed in the tumor cohort. Together, the results demonstrate that squamous differentiation is a common feature of basal MIBCs and that the subtypes described here are similar to those identified independently by other groups.
Figure 3. Presence of squamous features in the subtypes.
A. Tumor squamous feature content in the MD Anderson discovery and validation cohorts. Subtype designations are indicated by the top color bars, and the presence of squamous features (in black) is indicated in the color bars below. B. Relationship between the MD Anderson subtypes and the molecular taxonomy developed by Sjödahl et al (Sjodahl et al., 2012). Whole genome mRNA expression (Illumina platform) and clinical data were downloaded from GEO (GSE32894), and the oneNN classifier was used to assign the Lund tumors to subtypes. Subtype membership is indicated by the top color bars, and FGFR3 and TP53 mutations in the Lund tumors are indicated in color bars below. Black: mutant; white: wild-type; grey (N/A): mutation data were not available. C. Presence of squamous features in the UCSF dataset. Whole genome mRNA expression profiling (in-house platform) and clinical data were downloaded from GEO (GSE1827), and the oneNN classifier was used to assign the UCSF tumors to the subtypes. Subtype memberships for each tumor are indicated in the top color bars, and the presence of squamous features (in black) is indicated in the color bar below. D. Tissue microarray analysis of CK5/6 (basal) and CK20 (luminal) cytokeratin expression. Cytokeratin protein expression was measured by immunohistochemistry and optical image analysis in the MD Anderson Pathology Core on a tissue microarray containing 332 high-grade pT3 tumors. The percentages of positive tumor cells as determined by image analysis are shown. Left panels: mean levels of CK5/6 (top) and CK20 (bottom) in tumors without (TCC) or with (TCC with SD) squamous features. Bars indicate mean values with 95% confidence intervals. Middle panels: representative images of stained cores from tumors that expressed high or low levels of CK5/6 or CK20. The scale bars correspond to 100 microns. Right panel: relationship between CK5/6 and CK20 expression across the cohort. See also Tables S3–5 and Fig. S2.
p63 and PPARγ control basal and luminal biomarker expression
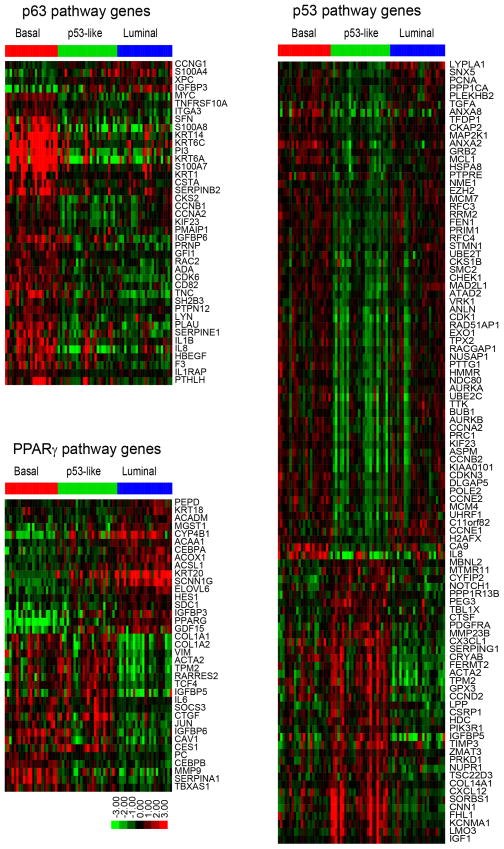
To more clearly define the transcription factors that controlled basal and luminal gene expression, we used the “upstream regulators” function in Ingenuity Pathway Analysis (IPA, Ingenuity® Systems (www.ingenuity.com)) and the gene expression profiling data from the MD Anderson discovery cohort to identify the transcription factors that were responsible for the gene expression signatures observed in the MIBC subtypes (Fig. 4, Table S6). Because the silhouette analyses revealed that 9 of the p53-like tumors were unstable, we compared the IPA results obtained with (n = 73) and without (n = 64) the unstable tumors (Table S7). Transcription factors that have been implicated in the biology of the basal/stem cell compartment of the normal urothelium (Stat-3, NFκB, Hif-1, and p63)(Ho et al., 2012) were predicted to be significantly “activated” in basal MIBCs (Fig. 4, Table S6, Fig. S3A). TP63 has been identified as a biomarker for lethal MIBCs (Choi et al., 2012; Karni-Schmidt et al., 2011), and we used quantitative RT-PCR to confirm that TP63 levels were elevated in the basal MIBCs in the MD Anderson discovery cohort (Fig. S3B). Six of the top 10 upregulated basal MIBC biomarkers (KRT5, KRT6A, KRT6C, PI3, KRT14, and S100A7) based on fold changes are known direct ΔNp63α transcriptional targets in other tissues (Boldrup et al., 2007; Celis et al., 1996; Romano et al., 2009) (Fig. 4, also Fig. 1B). Basal tumors were also enriched for MYC expression, which is controlled by p63 in human bladder cancer cells (Marquis et al., 2012).
Figure 4. Subtype-associated gene expression signatures.
Signatures were identified using the whole genome mRNA expression profiling data from the MD Anderson discovery cohort and the upstream regulators tool in Ingenuity Pathway Analysis (IPA, Ingenuity® Systems (www.ingenuity.com)). Each heat map displays the expression of the corresponding IPA gene signature as a function of tumor subtype membership; note that genes can be either up- or down-regulated by an active transcription factor. Top left: p63-associated gene expression. Below left: PPARγ-associated gene expression. Right: p53-associated gene expression. See also Tables S6 and S7 and Fig. S3.
Luminal MIBCs exhibited strong peroxisome proliferator activator receptor (PPAR) pathway activation as well as high-level expression of PPARG and its direct transcriptional target and coactivator, FABP4 (Fig. 1B, Fig. 4, Table S6) (Ayers et al., 2007). In addition, the estrogen receptor (ER) and its coactivator Trim-24 (Hatakeyama, 2011; Tsai et al., 2010) were among the top “activated” upstream regulators in the luminal MIBCs, whereas the basal Stat-3 and NFκB transcriptional networks were downregulated in them (Table S6, Fig. S3C, D). Conversely, breast luminal transcriptional pathways (ER, Gata-3 and Trim-24) were all downregulated in the basal MIBCs (Table S6). The p53-like luminal MIBCs could be distinguished from the luminal tumors by their expression of an active p53-associated gene expression signature that was not associated with the presence of wild-type TP53 (Fig. 4, also Figs. 1 and 3B, Tables S6 and S7). The p53-like tumors also contained an active p16 (CDKN2A) gene signature (Fig. S3E).
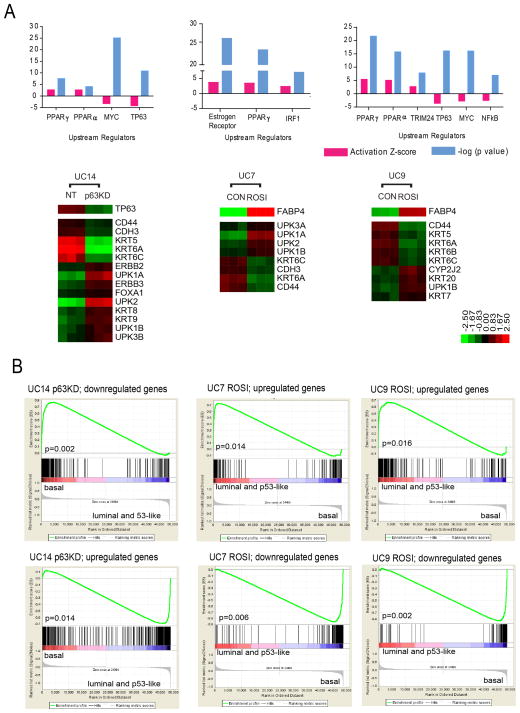
To more directly define p63’s role in controlling basal gene expression, we stably transduced human UM-UC14 bladder cancer cells with non-targeting or TP63-specific shRNAs and used whole genome mRNA expression profiling to create a bladder cancer p63 pathway gene expression signature. IPA analyses indicated that TP63 knockdown decreased basal (p63 and Myc) pathway gene expression, and interestingly, it also increased PPAR pathway gene expression (Fig. 5A, Fig. S4A). GSEA analyses in the discovery cohort confirmed that the p63 gene signature was significantly enriched in primary basal MIBCs (Fig. 5B).
Figure 5. Transcriptional control of the basal and luminal gene expression.
Whole genome mRNA expression profiling was used to analyze the effects of stable p63 knockdown or rosiglitazone-induced PPARγ activation in human bladder cancer cell lines, and the data were used to generate gene expression signatures characteristic of p63 and PPARγ activation. GSEA was then used to determine whether these signatures were present in the MD Anderson discovery cohort tumor subtypes. A. Effects of p63 or PPARγ modulation on basal and luminal transcriptional signatures. Top panels: significantly activated/inhibited transcriptional pathways after p63 knockdown in UM-UC14 cells (top left), PPARγ activation in UM-UC7 (top middle) or PPARγ activation in UM-UC9 (top right) based on IPA analyses. The heat maps below each graph indicate significant changes in basal and luminal marker expression. B. p63 and PPARγ gene expression signatures in the subtypes of primary MIBCs. Separate results and p values are shown for the signatures derived from the up- and down-regulated genes in each condition. ROSI: rosiglitazone. See also Fig. S4.
To determine PPARγ’s in controlling luminal gene expression, we generated PPARγ gene expression signatures using whole genome mRNA expression profiling data collected from two human bladder cancer cell lines (UM-UC7 and UM-UC9) that had been exposed to the PPARγ-selective agonist rosiglitazone. IPA analyses confirmed that rosiglitazone activated PPAR and other luminal transcriptional pathways in both cell lines (Fig. 5A, Figs. S4B, C). GSEA revealed that the UM-UC7 and UM-UC9 PPARγ gene signatures were significantly enriched in primary luminal MIBCs in the discovery cohort (Fig. 5B). Interestingly, rosiglitazone also decreased basal transcription factor activation (Fig. 5A, Fig. S4C). Therefore, PPARγ activation plays an important role in regulating the luminal MIBC gene expression signature, and p63 and PPARγ antagonize each other.
p53-like MIBCs are resistant to neoadjuvant chemotherapy
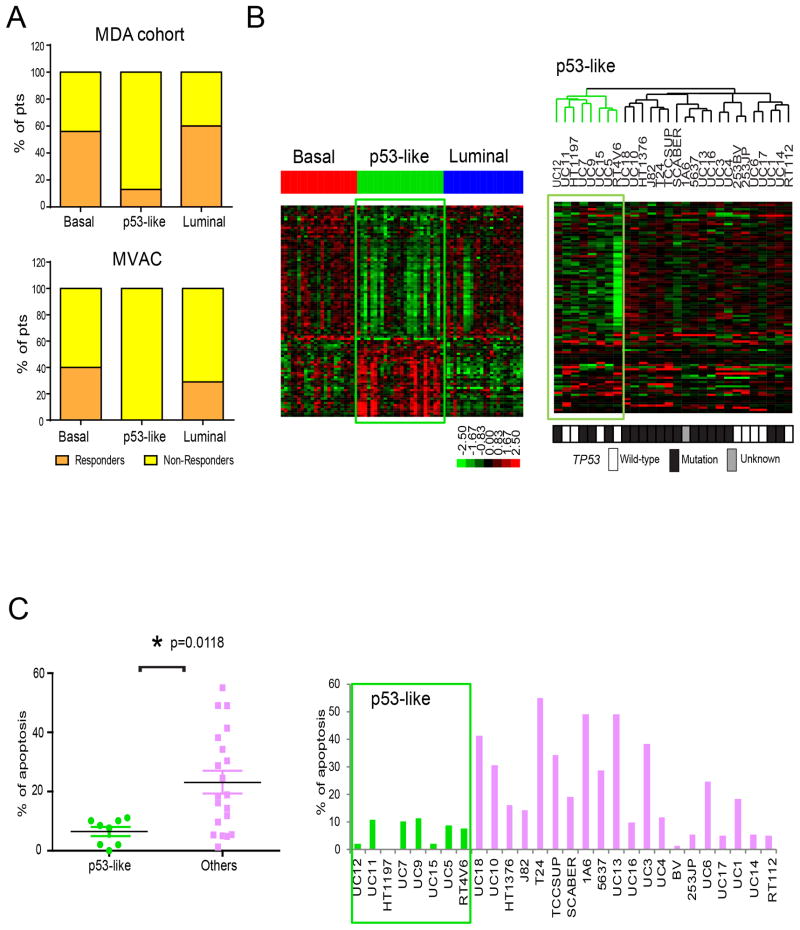
Presurgical (neoadjuvant) cisplatin-based chemotherapy (NAC) is the current standard-of-care for high-risk MIBC (Shah et al., 2011), and pathological response to NAC (downstaging to ≤pT1 at cystectomy) is a strong predictor of disease-specific survival (Grossman et al., 2003), as it is in breast cancer (Esserman et al., 2012b). We noticed that all of the p53-like MIBCs from patients treated with NAC in the discovery cohort (n = 7) were resistant to chemotherapy (Table 1). To examine this relationship further, we explored the chemoresistance of p53-like MIBCs in an expanded NAC cohort (n = 34) and in an additional cohort of 23 archival tumors treated uniformly with MVAC within the context of a Phase III clinical trial (Millikan et al., 2001). The p53-like MIBCs in both cohorts were resistant to NAC (Fig. 6A, Tables S8 and S9). We applied the primary tumor p53 signature to a panel of human bladder cancer cell lines and identified a subset of them that expressed the signature, not all of which retained wild-type TP53 (Fig. 6B). The p53-like cell lines were also resistant to cisplatin-induced apoptosis in vitro (Fig. 6C). In addition, 4/5 of the TP53 wild-type cell lines that did not contain the “p53-like” signature at baseline were cisplatin-resistant (Fig. 6B, C).
Figure 6. Relationship between subtype membership and chemotherapy sensitivity.
A. Responses to neoadjuvant chemotherapy in the MD Anderson NAC (n = 34) and MVAC (n = 23) cohorts. Subtype membership was determined using whole genome mRNA expression profiling data obtained from untreated (TURBT) tumors and the oneNN classifier. Pathological response was defined as downstaging to ≤ pT1. B. The IPA-defined p53 gene expression signature from the p53-like primary MIBCs was used to perform unsupervised hierarchical cluster analysis on whole genome expression data from a panel of human bladder cancer cell lines (n = 28). The green boxes on the heat maps indicate expression of the signature in the MD Anderson discovery cohort (left) or the cell lines (right). TP53 mutational status was determined by sequencing and is indicated by the color bar below the heat map (black = mutant, white = wild-type, grey = data were not available). C. Cells were incubated with or without 10 μM cisplatin for 48 h and apoptosis-associated DNA fragmentation was quantified by propidium iodide staining and FACS analysis in 3 independent experiments. The left panel displays a scatter gram comparing the levels within the two subsets of cell lines (mean ± SEM). The right panel displays the mean value of induced apoptosis in each cell line across the entire cohort. See also Tables S8 and S9.
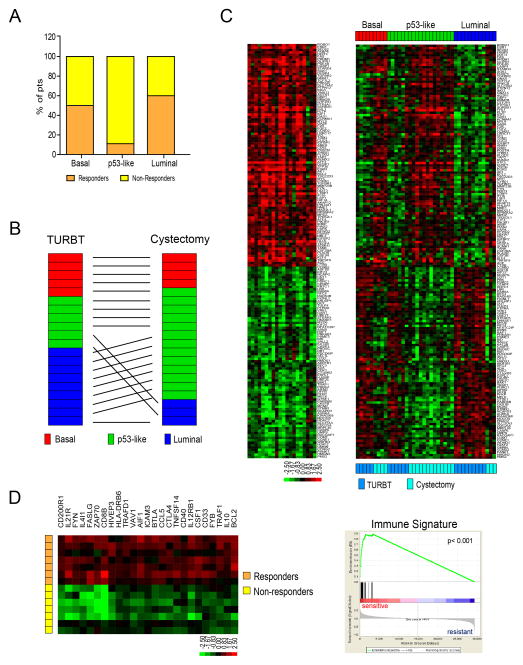
To further examine whether chemoresistance was a consistent feature of the p53-like subtype, we used gene expression profiling and our classifier to perform molecular subtype assignments on matched pre- and post-treatment MIBCs collected within the context of a prospective Phase II clinical trial of neoadjuvant dose-dense MVAC (DDMVAC), conducted at Fox Chase Cancer Center and Thomas Jefferson University Hospital (Table S10). All of the pretreatment tumors with squamous features in this “Philadelphia” cohort were confined to the basal cluster (Table S10, p = 0.012). In addition, and consistent with what we had observed in the MD Anderson cohorts, many of the Philadelphia basal (7/14) and luminal (12/20) tumors responded to NAC, whereas the response rate in the p53-like tumors was significantly lower at 11% (1/9)(Fig. 7A). Furthermore, chemoresistant tumors were enriched with the p53-like subtype after NAC (Fig. 7B).
Figure 7. Wild-type p53 gene signatures in tumors before and after treatment with NAC.
A. Relationship between subtype membership and response to NAC in the Philadelphia DDMVAC cohort. Subtype membership was determined using pretreatment (TURBT) specimens. Pathological response was defined as downstaging to ≤ pT1. B. Comparison of subtype membership in the chemoresistant Philadelphia tumors before and after NAC. Whole genome mRNA expression profiling was performed on matched tumors before and after NAC, and the oneNN classifier was used to assign tumors to subtypes. “TURBT” refers to the pretreatment tumors and “cystectomy” to the post-treatment tumors. C. Expression of a wild-type p53 gene signature in matched pre- and post-treatment tumors. Left: heat map displaying expression of an active p53 gene signature after NAC (log ratio cystectomy/TURBT of matched tumors). Right: relative expression of the p53 signature in matched pre- and post-treatment tumors arranged according to subtype membership. D. Analysis of an immune infiltration signature in basal tumors. A supervised analysis was performed to compare the differences in gene expression between basal tumors that were either sensitive or resistant to neoadjuvant DDMVAC in the Philadelphia cohort. Left: heat map depicting the relative expression of immune signature genes in basal responders and non-responders. Right: GSEA analyses of immune biomarkers in the basal tumors. See also Tables S10 and S11 and Fig. S5.
To further characterize the molecular mechanisms underlying chemoresistance, we compared the matched pre- and post-treatment gene expression profiles of the chemoresistant Philadelphia tumors using the “upstream regulators” function of IPA (data not shown). The results indicated that chemotherapy caused all of the tumors to express an active p53 pathway gene signature after NAC (Figure 7C, Table S11). Importantly, this chemotherapy-induced p53 signature was not very similar to the one that dictated tumor membership within the p53-like subtype (13 overlapping probes, Table S11).
Finally, we searched for pretreatment gene signatures within the Philadelphia basal and luminal MIBC subtypes that might predict chemosensitivity. We were unable to detect such a signature in the luminal tumors (data not shown), but the chemosensitive Philadelphia basal tumors were enriched for biomarkers reflective of immune infiltration (Fig. 7D). Similarly, all of the chemosensitive basal tumors from the MD Anderson MVAC cohort were also enriched with these immune biomarkers (Fig. S5).
DISCUSSION
We conclude that MIBCs can be grouped into basal and luminal subtypes reminiscent of those observed in human breast cancers (Perou et al., 2000). Basal MIBCs were associated with shorter disease-specific and overall survival, presumably because patients with these cancers tended to have more invasive and metastatic disease at presentation. The invasive/metastatic phenotype was associated with expression of “mesenchymal” and bladder cancer stem cell (Chan et al., 2009) biomarkers, and the tumors were enriched with sarcomatoid and squamous features (Blaveri et al., 2005; Sjodahl et al., 2012). The link between squamous features and aggressive behavior is consistent with other recent observations (Kim et al., 2012; Mitra et al., 2013), and the presence of EMT and bladder cancer stem cell biomarkers in basal tumors provides a biological explanation for their aggressive behaviors. p63 plays a central role in controlling the basal gene signature, and our preliminary data suggest that the EGFR, Stat-3, NFκB, and Hif-1α are also involved. Importantly, immune-infiltrated basal MIBCs responded to NAC, as do some basal breast cancers (Esserman et al., 2012a; Esserman et al., 2012b). Because NAC pathological complete response is associated with excellent long-term survival (Grossman et al., 2003), aggressive early management of basal MIBCs with NAC offers the best chance for improved survival for patients with this potentially deadly form of this disease. It also seems likely that T cell modulators (i.e., anti-CTLA4) and EGFR-, NFκB, Hif-1α/VEGF, and/or Stat-3-targeted agents will also be active within this subtype.
Like luminal breast cancers (Perou et al., 2000; Sorlie et al., 2001), luminal MIBCs displayed active ER/TRIM24 pathway gene expression and were enriched with FOXA1, GATA3, ERBB2 and ERBB3. Agents that target the ER (George et al., 2013; Hoffman et al., 2013; Shen et al., 2006) and/or ErbB2 and -3 may therefore be clinically active in luminal MIBCs. In addition, luminal MIBCs contained active PPAR gene expression and activating FGFR3 mutations, so PPARγ- and FGFR-3-targeted agents may be active in this subtype. Because many luminal MIBCs responded to NAC, targeted therapies should probably be combined with conventional chemotherapy for maximum efficacy.
The idea that wild-type p53 is required for DNA damage-induced apoptosis is a central tenet in cancer biology (Lowe et al., 1994; Lowe et al., 1993). Therefore, it was surprising to us that de novo and induced chemoresistance in MIBCs was associated with wild-type p53 gene expression signatures. Nevertheless, this link between “p53-ness” and chemoresistance is another shared property of MIBCs and luminal breast cancers. The recently completed I-SPY 1 TRIAL (“Investigation of Serial Studies to Predict Your Therapeutic Response With Imaging and Molecular Analysis,” CALGB150007/150012) examined the correlation between pathological complete response (pCR) rates and recurrence-free and overall survival in women treated with NAC. One of its main conclusions was that pCR rates varied markedly within the different breast cancer subtypes such that tumors with luminal A and/or wild-type p53-like gene expression signatures responded very poorly to NAC (Esserman et al., 2012a; Esserman et al., 2012b). Wild-type p53-induced reversible senescence has also recently been implicated in chemoresistance in a mouse model of breast cancer (Jackson et al., 2012), and more generally, quiescence is considered an important mechanism of chemoresistance. Importantly, TP53 mutation frequencies were similar in all 3 MIBC subtypes, indicating that wild-type p53 was not responsible for the baseline and chemotherapy-induced p53-like gene expression signatures reported here. We therefore propose that “p53-ness” as measured by mRNA expression would be a more accurate predictor of de novo and induced MIBC chemoresistance than would analyses of TP53 mutational status. It will be important to determine the molecular basis of these p53-like signatures in future studies so that therapeutic approaches can be developed to overcome de novo and/or prevent acquired chemoresistance. We also plan to prospectively test the relationship between the p53-like phenotype and chemoresistance within the context of a SWOG-sponsored multicenter clinical trial (S1314) that is very similar to I-SPY and was designed to prospectively evaluate another gene expression profiling-based algorithm (“CoXEN”)(Lee et al., 2007).
EXPERIMENTAL PROCEDURES
Technical details are provided in the Supplement
Human specimens
Clinical data were obtained from the MD Anderson Genitourinary Cancers Research Database, from Gene Expression Omnibus (GEO), or from patient charts (MVAC and Philadelphia cohorts). All MD Anderson patients signed a “front door” informed consent allowing collection of their tissue and of their clinical data that was approved by the MD Anderson Institutional Review Board (IRB). An additional MD Anderson IRB-approved protocol was obtained specifically for genomics analyses. The Philadelphia tissues were collected and analyzed as part of a Phase II clinical trial that was IRB-approved at the Fox-Chase Cancer Center and Thomas Jefferson University. Unstained tissue sections (10 micron, 5 slides/tumor) and matched H&Es from the DDMVAC clinical trial were transferred to MD Anderson under an executed Materials Transfer Agreement between MD Anderson and Fox-Chase. A genitourinary pathologist (Bogdan Czerniak) reviewed all of the tissue samples.
Tumor cohorts
The discovery cohort consisted of 73 tumors from transurethral resections (TURs) that had been snap frozen in liquid nitrogen within 5 min of isolation and transferred to the MD Anderson Bladder SPORE Tissue Core. The MD Anderson validation cohort consisted of 57 randomly selected, formalin-fixed, paraffin-embedded (FFPE) tumors that were obtained from the main MD Anderson Cancer Center CCSG-supported Pathology Tissue Bank. The MD Anderson NAC cohort (n = 34) contained a mixture of 18 tumors from the discovery cohort plus 16 additional FFPE tumors from patients treated with neoadjuvant chemotherapy on- and off-protocol. The MD Anderson MVAC cohort (n = 23) consisted of all available FFPE pretreatment tumors (TURs) from a previously reported Phase III clinical trial (Millikan et al., 2001). The Philadelphia NAC cohort (n = 43 TURs and 20 cystectomies) consisted of all available pre- and post-treatment FFPE tumors from patients enrolled in a Phase II clinical trial of neodjuvant dose-dense MVAC (DDMVAC) that was conducted at Fox-Chase Cancer Center and The Thomas Jefferson University and will be reported elsewhere. NAC response in all of the cohorts was defined as downstaging to no muscle-invasive disease at cystectomy (≤ pT1)(Millikan et al., 2001).
Gene expression profiling platforms
The MD Anderson discovery cohort and human bladder cancer cell lines were analyzed by direct hybridization on Illumina HT12 v3 and HT12v4 chips, respectively (Illumina, Inc. San Diego, CA). Data from all of the FFPE cohorts were generated using Illumina’s DASL platform (WG-DASL HT12 v4 chips).
Tumor cluster assignments
MIBC clusters (subtypes) were determined in the discovery cohort using unsupervised hierarchical cluster analysis (Eisen et al., 1998). The gene signatures associated with each cluster were then used to generate a one nearest neighbor (oneNN) (Dudoit et al., 2002) prediction model that was used in all subsequent analyses to assign tumors to specific subtypes.
Micro RNA expression
Levels of miR-200b and miR-200c were measured in the discovery cohort by quantitative RT-PCR as described in the Supplement.
Analysis of cytokeratin protein expression
Basal (CK5/6) and luminal (CK20) cytokeratin protein expression was analyzed on a tissue microarray (TMA) consisting of stage-matched (pT3, n = 332) MIBCs collected within the context of the International Bladder Cancer Network’s Bladder Cancer Bank initiative (Goebell et al., 2005). Immunohistochemical staining was performed using clinical-grade (CLIA) antibodies and protocols in the MD Anderson Pathology Core, and staining intensities were quantified by image analysis.
Generation of p63 and PPARγ gene signatures
UM-UC14 human MIBC cells were stably transduced with TP63-specific or non-targeting lentiviral shRNA contructs in the MD Anderson Vector Core. UM-UC7 and UM-UC9 cells were incubated with or without 1 μM rosiglitazone for 48 h. Triplicate RNA isolates were prepared on different days for each condition, and global changes in gene expression were determined by whole genome expression profiling. The statistically significant changes in gene expression were used to create signatures that were subsequently used in the IPA and GSEA analyses presented in Figure 5.
Statistical Analyses
Clinicodemographic characteristics were compared using Fisher’s exact tests and Kruskal-Wallis tests to assess differences between groups in categorical and continuous variables, respectively. Kaplan-Meier analysis with log-rank statistics was used to characterize survival distributions and associations between subtypes and survival outcomes. Statistical analysis was performed using SPSS (version 19) and a p-value of <0.05 was considered significant.
Supplementary Material
Significance.
Using whole genome mRNA expression profiling we identified 3 molecular subtypes of muscle-invasive bladder cancer (MIBC) that shared molecular features with basal and luminal breast cancers. Tumors in one of them expressed an active p53 gene signature, and these “p53-like” MIBCs were consistently resistant to frontline neoadjuvant cisplatin-based combination chemotherapy. Furthermore, comparison of matched gene expression profiles before and after chemotherapy revealed that all resistant tumors expressed wild-type p53 gene expression signatures. Together, the data indicate that “p53-ness” plays a central role in chemoresistance in bladder cancer and suggest that it should be possible to prospectively identify the patients who most likely will not benefit from neoadjuvant chemotherapy.
HIGHLIGHTS.
Chemoresistant bladder cancers express active p53-like gene expression signatures
Bladder cancers form basal and luminal subtypes like those found in breast cancer
Squamous features are characteristic of basal bladder cancers
p63 and PPARγ play opposing roles in controlling basal and luminal biology
Acknowledgments
The authors would like to acknowledge Lyndsay Harris, MD (Southwest Oncology Group, Breast Committee, Case Cancer Center) for her help with the DASL assay and Neema Navai and Matthew Wszolek for sharing their miR-200b and miR-200c PCR data. Grant support: Supported by a grant from the Baker Foundation, the MD Anderson Bladder SPORE (P50 CA91846), R01 CA151489 (to BC), and the MD Anderson CCSG (P30 016672). ERP is supported by grant #IRG-92-027-17 from the American Cancer Society. This paper is dedicated to the memory of Dexter Baker.
Footnotes
Accession numbers: The Chungbuk (n = 55) (Kim et al., 2010), Lund (n = 93) (Sjodahl et al., 2012), and UCSF (n = 53)(Blaveri et al., 2005) cohort data were downloaded from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/, GSE13507, GSE32894 and GSE1827). The gene expression profiling data generated in this study were uploaded to Gene Expression Omnibus with accession numbers GSE48277 and GSE47993.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayers SD, Nedrow KL, Gillilan RE, Noy N. Continuous nucleocytoplasmic shuttling underlies transcriptional activation of PPARgamma by FABP4. Biochemistry. 2007;46:6744–6752. doi: 10.1021/bi700047a. [DOI] [PubMed] [Google Scholar]
- Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, Devries S, Koppie T, Pejavar S, Carroll P, Waldman FM. Bladder cancer outcome and subtype classification by gene expression. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:4044–4055. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- Boldrup L, Coates PJ, Gu X, Nylander K. DeltaNp63 isoforms regulate CD44 and keratins 4, 6, 14 and 19 in squamous cell carcinoma of head and neck. The Journal of pathology. 2007;213:384–391. doi: 10.1002/path.2237. [DOI] [PubMed] [Google Scholar]
- Botteman MF, Pashos CL, Redaelli A, Laskin B, Hauser R. The health economics of bladder cancer: a comprehensive review of the published literature. PharmacoEconomics. 2003;21:1315–1330. doi: 10.1007/BF03262330. [DOI] [PubMed] [Google Scholar]
- Celis JE, Rasmussen HH, Vorum H, Madsen P, Honore B, Wolf H, Orntoft TF. Bladder squamous cell carcinomas express psoriasin and externalize it to the urine. The Journal of urology. 1996;155:2105–2112. [PubMed] [Google Scholar]
- Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D’Alessio AC, Young RA, Weinberg RA. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KS, Espinosa I, Chao M, Wong D, Ailles L, Diehn M, Gill H, Presti J, Jr, Chang HY, van de Rijn M, et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14016–14021. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W, Shah JB, Tran M, Svatek R, Marquis L, Lee IL, Yu D, Adam L, Wen S, Shen Y, et al. p63 expression defines a lethal subset of muscle-invasive bladder cancers. PloS one. 2012;7:e30206. doi: 10.1371/journal.pone.0030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinney CP, McConkey DJ, Millikan RE, Wu X, Bar-Eli M, Adam L, Kamat AM, Siefker-Radtke AO, Tuziak T, Sabichi AL, et al. Focus on bladder cancer. Cancer cell. 2004;6:111–116. doi: 10.1016/j.ccr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Dudoit S, FF, Speed TP. Comparison of discrimination methods for classification of tumors using DNA microarrays. Journal of the American Statistical Association. 2002;97:77–87. [Google Scholar]
- Dyrskjot L, Thykjaer T, Kruhoffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, Wolf H, Orntoft TF. Identifying distinct classes of bladder carcinoma using microarrays. Nature genetics. 2003;33:90–96. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esserman LJ, Berry DA, Cheang MC, Yau C, Perou CM, Carey L, DeMichele A, Gray JW, Conway-Dorsey K, Lenburg ME, et al. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) Breast cancer research and treatment. 2012a;132:1049–1062. doi: 10.1007/s10549-011-1895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esserman LJ, Berry DA, DeMichele A, Carey L, Davis SE, Buxton M, Hudis C, Gray JW, Perou C, Yau C, et al. Pathologic complete response predicts recurrence-free survival more effectively by cancer subset: results from the I-SPY 1 TRIAL--CALGB 150007/150012, ACRIN 6657. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012b;30:3242–3249. doi: 10.1200/JCO.2011.39.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George SK, Tovar-Sepulveda V, Shen SS, Jian W, Zhang Y, Hilsenbeck SG, Lerner SP, Smith CL. Chemoprevention of BBN-Induced Bladder Carcinogenesis by the Selective Estrogen Receptor Modulator Tamoxifen. Translational oncology. 2013;6:244–255. doi: 10.1593/tlo.13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebell PJ, Groshen S, Schmitz-Drager BJ, Sylvester R, Kogevinas M, Malats N, Sauter G, Grossman HB, Dinney CP, Waldman F, Cote RJ. Concepts for banking tissue in urologic oncology--the International Bladder Cancer Bank. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:413–415. [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Grossman HB, Natale RB, Tangen CM, Speights VO, Vogelzang NJ, Trump DL, deVere White RW, Sarosdy MF, Wood DP, Jr, Raghavan D, Crawford ED. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. The New England journal of medicine. 2003;349:859–866. doi: 10.1056/NEJMoa022148. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S. TRIM proteins and cancer. Nature reviews Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- Ho PL, Kurtova A, Chan KS. Normal and neoplastic urothelial stem cells: getting to the root of the problem. Nature reviews Urology. 2012;9:583–594. doi: 10.1038/nrurol.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman KL, Lerner SP, Smith CL. Raloxifene inhibits growth of RT4 urothelial carcinoma cells via estrogen receptor-dependent induction of apoptosis and inhibition of proliferation. Hormones & cancer. 2013;4:24–35. doi: 10.1007/s12672-012-0123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JG, Pant V, Li Q, Chang LL, Quintas-Cardama A, Garza D, Tavana O, Yang P, Manshouri T, Li Y, et al. p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer cell. 2012;21:793–806. doi: 10.1016/j.ccr.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni-Schmidt O, Castillo-Martin M, Shen TH, Gladoun N, Domingo-Domenech J, Sanchez-Carbayo M, Li Y, Lowe S, Prives C, Cordon-Cardo C. Distinct expression profiles of p63 variants during urothelial development and bladder cancer progression. The American journal of pathology. 2011;178:1350–1360. doi: 10.1016/j.ajpath.2010.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SP, Frank I, Cheville JC, Thompson RH, Weight CJ, Thapa P, Boorjian SA. The impact of squamous and glandular differentiation on survival after radical cystectomy for urothelial carcinoma. The Journal of urology. 2012;188:405–409. doi: 10.1016/j.juro.2012.04.020. [DOI] [PubMed] [Google Scholar]
- Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS, Jeong P, Kim MJ, Yun SJ, Lee KM, Moon SK, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Molecular cancer. 2010;9:3. doi: 10.1186/1476-4598-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J, Grimshaw A, Theodorescu D. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13086–13091. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Leem SH, Lee SY, Kim SC, Park ES, Kim SB, Kim SK, Kim YJ, Kim WJ, Chu IS. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:2660–2667. doi: 10.1200/JCO.2009.25.0977. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Bodis S, McClatchey A, Remington L, Ruley HE, Fisher DE, Housman DE, Jacks T. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- Marquis L, Tran M, Choi W, Lee IL, Huszar D, Siefker-Radtke A, Dinney C, McConkey DJ. p63 expression correlates with sensitivity to the Eg5 inhibitor ZD4877 in bladder cancer cells. Cancer biology & therapy. 2012;13:477–486. doi: 10.4161/cbt.19590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey DJ, Lee S, Choi W, Tran M, Majewski T, Lee S, Siefker-Radtke A, Dinney C, Czerniak B. Molecular genetics of bladder cancer: Emerging mechanisms of tumor initiation and progression. Urologic oncology. 2010;28:429–440. doi: 10.1016/j.urolonc.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millikan R, Dinney C, Swanson D, Sweeney P, Ro JY, Smith TL, Williams D, Logothetis C. Integrated therapy for locally advanced bladder cancer: final report of a randomized trial of cystectomy plus adjuvant M-VAC versus cystectomy with both preoperative and postoperative M-VAC. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2001;19:4005–4013. doi: 10.1200/JCO.2001.19.20.4005. [DOI] [PubMed] [Google Scholar]
- Mitra AP, Bartsch CC, Bartsch G, Jr, Miranda G, Skinner EC, Daneshmand S. Does presence of squamous and glandular differentiation in urothelial carcinoma of the bladder at cystectomy portend poor prognosis? An intensive case-control analysis. Urologic oncology. 2013;12:S1078–1439. doi: 10.1016/j.urolonc.2012.08.017. [DOI] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nature reviews Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Romano RA, Ortt K, Birkaya B, Smalley K, Sinha S. An active role of the DeltaN isoform of p63 in regulating basal keratin genes K5 and K14 and directing epidermal cell fate. PloS one. 2009;4:e5623. doi: 10.1371/journal.pone.0005623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- Shah JB, McConkey DJ, Dinney CP. New strategies in muscle-invasive bladder cancer: on the road to personalized medicine. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:2608–2612. doi: 10.1158/1078-0432.CCR-10-2770. [DOI] [PubMed] [Google Scholar]
- Shen SS, Smith CL, Hsieh JT, Yu J, Kim IY, Jian W, Sonpavde G, Ayala GE, Younes M, Lerner SP. Expression of estrogen receptors-alpha and -beta in bladder cancer cell lines and human bladder tumor tissue. Cancer. 2006;106:2610–2616. doi: 10.1002/cncr.21945. [DOI] [PubMed] [Google Scholar]
- Sjodahl G, Lauss M, Lovgren K, Chebil G, Gudjonsson S, Veerla S, Patschan O, Aine M, Ferno M, Ringner M, et al. A molecular taxonomy for urothelial carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:3377–3386. doi: 10.1158/1078-0432.CCR-12-0077-T. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svatek RS, Shariat SF, Novara G, Skinner EC, Fradet Y, Bastian PJ, Kamat AM, Kassouf W, Karakiewicz PI, Fritsche HM, et al. Discrepancy between clinical and pathological stage: external validation of the impact on prognosis in an international radical cystectomy cohort. BJU international. 2011;107:898–904. doi: 10.1111/j.1464-410X.2010.09628.x. [DOI] [PubMed] [Google Scholar]
- Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.