Abstract
The development of, and results from an image analysis system are presented for automated detection and scoring of micronuclei in human peripheral blood lymphocytes. The system is part of the Rapid Automated Biodosimetry Tool, which was developed at the Center for High-Throughput Minimally Invasive Radiation Biodosimetry for rapid radiation dose assessment of many individuals based on single fingerstick samples of blood. Blood lymphocytes were subjected to the cytokinesis-block micronucleus assay and the images of cell cytoplasm and nuclei are analyzed to estimate the frequency of micronuclei in binucleated cells. We describe an algorithm that is based on dual fluorescent labeling of lymphocytes with separate analysis of images of cytoplasm and nuclei. To evaluate the performance of the system, blood samples of seven healthy donors were irradiated in vitro with doses from 0–10 Gy and dose-response curves of micronuclei frequencies were generated. To establish the applicability of the system to the detection of high doses, the ratios of mononucleated cells to binucleated cells were determined for three of the donors. All of the dose-response curves generated automatically showed clear dose dependence and good correlation (R2 from 0.914–0.998) with the results of manual scoring.
INTRODUCTION
In the unfortunate case that a large-scale radiological event occurs in an urban area, there would be a need for screening of hundreds of thousands of people in a short period of time to estimate the doses received by individuals (1). At the Center for High-Throughput Minimally Invasive Radiation Biodosimetry we have developed the Rapid Automated Biodosimetry Tool (RABiT) – a completely automated robotic-based ultra-high-throughput biodosimetry workstation (2–5). The estimation of the dose received by each individual is based on the analysis of peripheral blood lymphocytes obtained from a single fingerstick (6). This approach is minimally invasive and the most compatible with high-throughput automation.
The RABiT utilizes two well-characterized biodosimetry assays: γ-H2AX (7, 8) and cytokinesis-block micronucleus (CBMN) (9). By design of the RABiT both assays will be entirely automated; once the samples are manually loaded into the system, there will be no human intervention through the point when the samples are stored and the dose estimation is presented to the operator (10). The RABiT is currently capable of processing 6,000 samples a day with development in place to achieve an ultimate throughput of 30,000 samples a day (4). This high throughput was achieved by incorporation of the following innovations: (a) using only a fingerstick of blood (30 µl); (b) complete robotic-based assay processing and imaging in multi-well plates; and (c) novel, high-speed imaging techniques (3, 4).
In recent years, much attention has been drawn to systems for high-throughput, high-content cell imaging (11, 12). Despite a wide selection of available commercial high-throughput imaging systems, to our knowledge no system could provide the throughput necessary for the RABiT system, while retaining adequate spatial resolution, for example, for detection of micronuclei (MNi). We have therefore developed such a system ourselves (3–5, 10). The imaging module of the RABiT incorporates three novel techniques for accelerated image acquisition: (1) use of light steering by fast galvanometric mirrors instead of sample motion; (2) single-step auto-focusing; and (3) simultaneous use of multiple cameras for dual-labeled samples. In the current study, which focuses on the CBMN assay, we have developed image analysis software that will rapidly analyze the images produced by the imaging module of the RABiT and quantitate the micronucleus (MN) yield in irradiated blood samples.
Micronuclei are small, generally round objects in the cytoplasm of the cells outside of the main nucleus. They represent chromosome fragments or whole chromosomes that are not incorporated into the daughter cell after nuclear division (9, 13). The use of micronuclei as a biomarker of chromosomal damage was first suggested independently by Heddle (14) and Schmid (15) in 1973. In 1985 Fenech and Morley introduced using cytochalasin B to inhibit cytokinesis upon completion of nuclear cytoplasmic division during the first division cycle (13). Restricting the analysis to binucleated (BN) cells allows selection of lymphocytes that have divided once, thus eliminating confounding parameters associated with nondividing cells or cells that have divided more than once. The CBMN test is easy and reproducible (9, 16) and has become one of the standard tests for genotoxicity assessment (17, 18). Its use in biomonitoring studies has greatly increased in the last 15 years, and recent international efforts such as the HUMN (human micronucleus) project (http://www.humn.org) have greatly contributed to improving the reliability of this assay, providing technical guidelines and analyzing major sources of variability (19–21).
It is natural that there has been demand for automation of the CBMN assay, not only for increasing throughput, but also for better statistics, more objective counting and higher reproducibility (22, 23). Various approaches have been suggested to identify MNi in BN cells. One of the main characteristics that define an algorithm is whether the analysis stems from the detection of nuclei or from the detection of cells. The former approach is used when only the nuclei are stained and no information about cell localizations is then available. It implies that to localize the cell boundaries and to estimate the number of nuclei in each cell, additional analysis of the locations of the nuclei with respect to each other and their relative sizes is required. The disadvantage of this method is that the nuclei may be falsely assigned to a cell. In 1988 Fenech et al. (22) reported difficulty in using computerized image analysis because the cytoplasm was not stained and it was not possible to identify the cytoplasmic boundaries to differentiate BN cells. Varga et al. (24) described a method of localizing the cells based on the occurrence of two similar nuclei, implemented in Metapher slide scanning platform by Metasystems. However, they report a large difference between visual counting and automated scoring with their method.
The approach to use cells as detection units can be achieved either by applying different threshold levels to an image stained with a single dye that labels both the nuclei and the whole cells such as Giemsa as realized in the MIAZ platform (23, 25), Magiscan (26), Pathfinder CellScan (27) and BNCTEST (28) or by counterstaining the cytoplasm as realized in the Matrox platform (29) or Cellomics (30). The advantage of visualizing both nuclei and cytoplasm is that it enables matching the nuclei with their corresponding cells, allowing for a more precise identification of BN cells.
The RABiT system utilizes dual staining of the nuclei and cytoplasm and images them simultaneously with multiple cameras, allowing for high throughput. The image analysis algorithm is based on separate analyses of the images of cytoplasm and nuclei with the cell cytoplasm as the starting point of the analysis. When the nuclei and cell images are analyzed separately, artifacts arising from cytoplasmic staining do not result in false-positive detection of MNi.
This article focuses on the development and validation of the automated image analysis module of the RABiT that analyzes the results of CBMN assay. We present the developed algorithm for detecting and scoring the frequency of MNi in BN cells and validate the performance of the system by producing dose-response curves. Rather than studying the biology of radiation damage, our goal was to confirm the reliable operation of the designed software by comparing the manual and automatic yields of micronuclei under uniform irradiation scenarios. In addition we investigated the possibility of using the software for biodosimetry with a full range of doses, for which a decision about an individual’s hospitalization and treatment has to be made during triage. Low doses, below 2 Gy, cause only transient health effects (31) and outpatient care is recommended (32). Exposures above 2 Gy result in hematopoietic syndrome (33, 34), when the bone marrow function is suppressed. Treatment in this case requires hospitalization, as about half of the exposed people could die after receiving a dose of 3.5 Gy when medical care is not provided, while with appropriate care mortality is low (32, 34). The dose above 10 Gy is almost always lethal due to gastrointestinal syndrome, multisystem organ failure, bleeding and sepsis and only symptomatic care is possible (31–33). However, there is a window of doses from 8–10 Gy within which bone marrow transplantation can be considered as a treatment option (32, 33). Below 8 Gy spontaneous bone marrow recovery is still possible and the risk of graft vs. host disease is high (32, 35). Therefore, it is important to identify the dose range correctly at the time of triage.
The average number of MNi in BN cells is known to be a reliable indicator of the dose for low (1–2 Gy) and intermediate (3–5 Gy) doses of radiation. However, this parameter plateaus or even declines for doses higher than 6 Gy due to cell cycle delay (36). A recent study by Romm et al. (37) demonstrated limitations of the assay for the doses above 4.2 Gy. In situations when the dose cannot be determined based on micronuclei frequency, the use of other cytogenetic endpoints that can be obtained using CBMN assay has previously been suggested. Müller and Rode (36) demonstrated that the number of mononucleated cells, the ratio of tri- to tetranucleated cells and the frequency of MNi in MN-positive BN cells can be used for doses in excess of 5–7 Gy. In 2004 and 2008 the doses received by the victims of the Shandong Jining and Shanxi Taiyauan (China) radiation accidents, respectively, were evaluated using CBMN, along with other assays (38, 39). In both cases, the Nuclear Division Index (NDI), indicative of the cell proliferation activity (40, 41), was used to estimate the dose of those victims who were exposed to more than 10 Gy of γ rays, however it was possible to determine the doses received by other victims using only the frequencies of MNi in BN cells. In the study by Wu et al. (42) the ratio of mononucleated to binucleated cells was calculated across the Bragg curve of energetic particles traversing shielding material and used as a biomarker indicating the inhibition of cell cycle progression. In that study, the ratio of mononucleated to binucleated cells was reported to increase at the Bragg peak, unlike MNi per BN cells. Because the design of our current software allows for detection of cells with different number of nuclei, we have explored the possibility of including one of these parameters, the ratio of mononucleated to binucleated cells, in our analysis at higher doses of irradiation when micronuclei frequencies are not sufficient for dose estimation.
DESIGN
Software Platform
The RABiT image analysis software for scoring MNi was written in “C” language, using the Matrox Imaging Library (MIL) (Release 8.0; Matrox Imaging Systems Ltd, Dorval, QC, Canada). The Matrox Imaging Library consists of modules: image processing, blob analysis, buffer module, etc., implementing different families of image analysis tasks. We used the image processing module for arithmetic operations with images, distance transformation and watershed transformation. In the blob analysis module all cells and nuclei are identified as “blobs” or connected regions of pixels within an image with intensity values different from the background intensity. This module allows measuring various properties of blobs, such as area, perimeter, center of gravity, pixel intensity, etc. These properties can be stored in arrays and, based on their property values, individual blobs can be excluded from or included in the analysis.
Image Preprocessing
The two images corresponding to nuclei and cytoplasm are first subject to background subtraction. An image of a region from the same sample, taken far enough out of focus so that no features are visible, is subtracted from the in-focus image of each field to remove the systematic noise and variations of the background illumination across the field of view. The background subtraction is followed by application of a median filter to remove “salt-and-pepper” noise (pixels with black or white intensity values not related to the neighboring pixels).
Automatic Thresholding of the Nuclei Image
The next step is segmentation of the nuclei and cells by binarization. In the Matrox Imaging Library, the automatic threshold level is the minimum value between the two principal peaks on the image intensity histogram, obtained with 2-means clustering. When applied to the images of lymphocytes stained with DAPI, the automatic thresholding procedure causes some MNi to disappear in the binarized image because the staining intensity of MNi is lower than that of the main nuclei (Fig. 1A). Moreover, the staining intensity of MNi varies between individuals, from dose to dose and from cell to cell, hence there is no constant value for the ratio of the threshold for MNi to the threshold for the main nuclei.
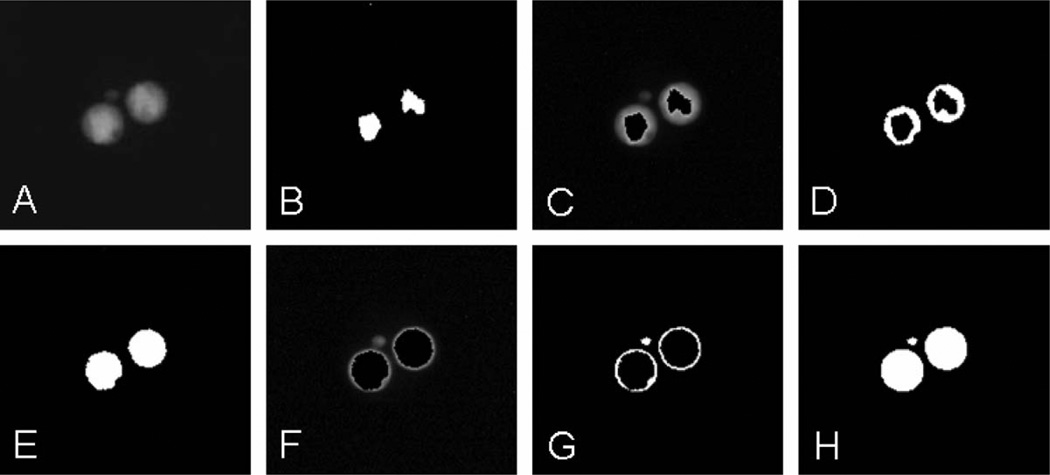
FIG. 1.
Iterative thresholding of the nuclei of a binucleated cell with a micronucleus. Panel A: Original image of the DAPI-stained nuclei, the micronucleus has lower staining intensity. Panel B: Automatic thresholding using the image histogram, no micronuclei visible. Panel C: Masking iteration 1, original image, masked with the binary version. Panel D: Binarized version of panel C. Panel E: Combination of panels B and D, still no micronuclei. Panel F: Masking iteration 2, original image masked with panel E. Panel G: Binarized version of panel F. Panel H: Combination of panels E and G.
To make the detection of MNi less dependent on variations in staining intensity, we developed the following iterative procedure. First, the standard MIL automatic thresholding is applied to the image of the nuclei (Fig. 1B). The nuclear objects from the subsequent binarized image are used as a mask to cover the brightest regions of nuclei in the original image (Fig. 1C). This is equivalent to removing the principle peak with the highest pixel intensity values from the histogram. Then the resulting image is again binarized (Fig. 1D), allowing selection of the dimmer areas. The result of this binarization is combined with the previous binary image to form a new mask and the entire procedure is repeated (Fig. 1E–H).
Sometimes, when the nuclei are binarized at a threshold level sufficient to visualize MNi, their contours become foreground and they appear larger than their apparent size on the grayscale image and have noisier edges. This not only complicates separation of the main nuclei, but also leads to “merging” of the main nuclei with the closest MNi (Fig. 2, top row). The following modification to the iterative thresholding routine eliminates this drawback. First a sufficient number of iterations are performed to detect the nuclei (this number can be determined experimentally, see below). After that at each subsequent iteration all objects with areas greater than a specified nuclear area are excluded. This way the operation is only applied to the smaller objects that could be MNi. When this iterative process ends, the last “micronuclei” buffer is combined with the “nuclei” buffer saved previously. Effectively, with this technique the nuclei and the MNi are binarized at different threshold levels. For the current study, 2 iterations were sufficient to identify nuclei and 3 more (5 total) – to visualize MNi while keeping the area of each binarized nucleus constant. In addition to keeping the binarized nuclear area constant, this technique increases MNi count because of the improved separation of the MNi and nuclei (Fig. 2, right column).
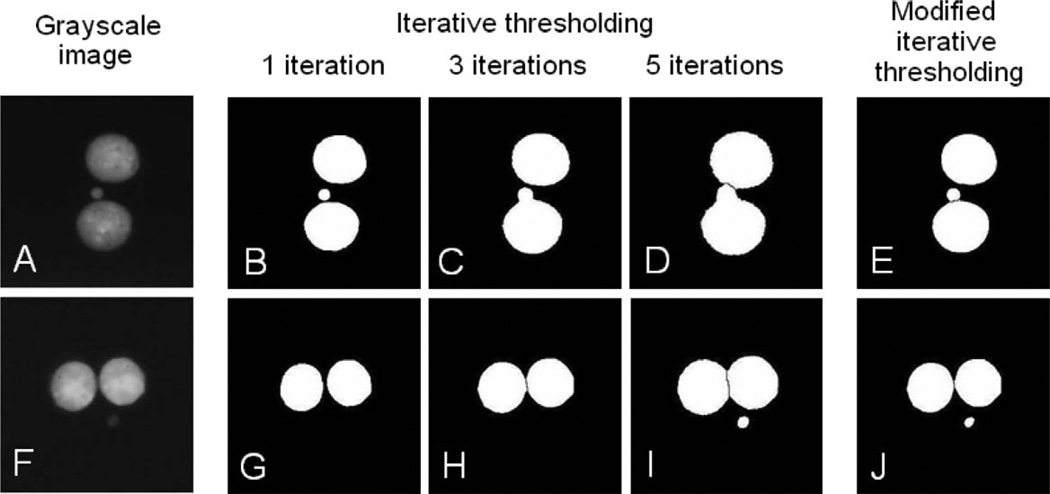
FIG. 2.
Binarized images formed by applying the same number of iterations (“Iterative thresholding”) and different number of iterations (“Modified iterative thresholding”) to the main nuclei and micronuclei. Panels A and F: grayscale images of BN cells with bright (panel A) and dim (panel F) micronuclei. Panels B–D: The micronucleus appears on the binarized image after a single thresholding, while 3 and more iterations produce enlarged nuclei merged with the micronucleus. Panels G–I: The micronucleus is dimmer than the main nuclei; 5 iterations are required for it to appear. Panels E and J: Application of the modified thresholding restricts the size of the main nuclei and allows using lower threshold for the micronuclei. Only the final results after 5 modified iterations are shown; the time required for a single iteration is 0.02 s.
It should be noted that the apparent areas of MNi increase more than that of the main nuclei as a result of this procedure. We performed an experiment to determine the range of possible nuclear areas for all available samples using our software. Then we calculated, knowing the number of iterations, how much the relative MN area would change after the thresholding. For nuclear areas ranging from 3,000–9,000 pixels, which correspond to diameters of 8–18 µm, the maximum relative size of an acceptable MN would be 20% of the nuclear area. Adjusting this parameter ensures selection of MNi whose actual relative area is between 1/256 and 1/9.
Selecting a Thresholding Algorithm
To select the most suitable thresholding method for individual iterations, we compared the standard thresholding operation provided in the Matrox Imaging Library to two other, widely used, thresholding techniques: Otsu’s method (43) and the method proposed by Ridler and Calvard [isodata (44)].
According to the Otsu thresholding technique the optimal threshold value is determined by maximizing a criterion value of separability of the background and object classes (constructed using between-class and within-class variances). In the iterative procedure of Ridler and Calvard, the background and object pixels are integrated separately and the average of the integral values gives an improved estimate of the threshold with each iteration.
Both Otsu’s and Ridler and Calvard’s algorithms were implemented in the C language and tested in comparison to the Matrox Imaging Library thresholding routine. The full set of the acquired data was analyzed with the designed software independently with each of the three thresholding techniques described. For each algorithm, the processing time for thresholding of a single image was recorded and averaged. The iterative Ridler and Calvard algorithm is considerably slower than the other two methods (the average execution time 31 ms vs. 5 ms for MIL and 4 min for Otsu’s algorithm, using a laptop computer with an Intel Core 2 Duo 1.83 GHz processor and 2 GB SDRAM). Since high throughput is one of the objectives of the project, the lower speed of the Ridler and Calvard algorithm is a considerable limitation.
The two fastest methods (Otsu and MIL) were then evaluated by their performance when applied iteratively to the images acquired for this study. The Otsu technique generally produced slightly lower threshold values than did the MIL procedure, which lead to a higher MNi yield (about 36% of the manual yield for MIL and 43% for Otsu). However, when the Otsu routine was applied iteratively, the threshold level became too low after two iterations, reducing the number of analyzable binary images. In contrast, it was possible to apply the MIL thresholding repetitively, in which case the yield of MNi approached the manual count (above 88% of the manual yield for 0–4 Gy and above 64% for 6–10 Gy). After this evaluation, the MIL thresholding routine was selected for this study as optimal in terms of speed and applicability.
Separation of Touching Nuclei
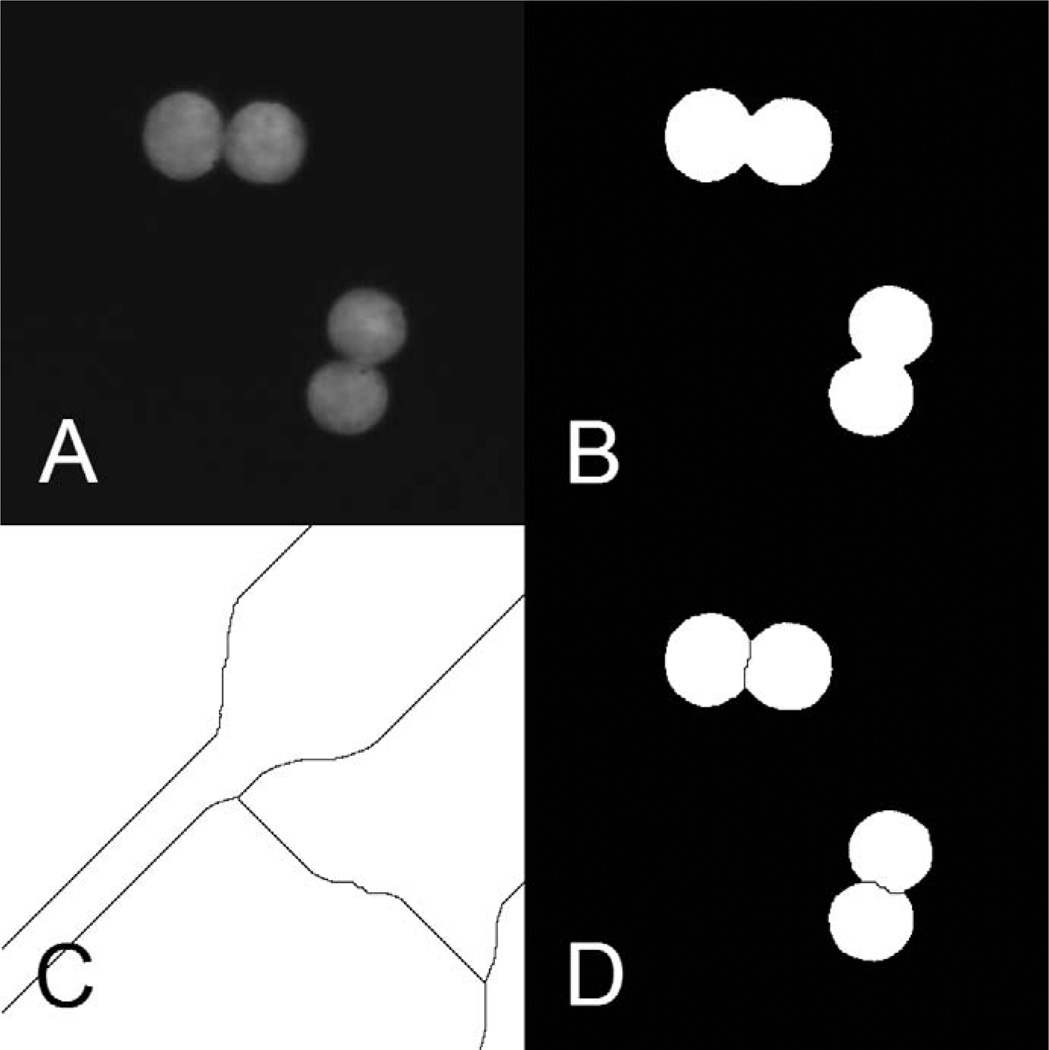
The last step of the processing of the nuclear images is separation of touching nuclei (Fig. 3A). This is done by successive application of a distance transformation and a watershed transformation to the binarized image, shown in Fig. 3B. The distance transformation assigns a value to each pixel of an object that is equal to the geometric distance to the nearest background pixel. This operation creates a topographical map with the “highest” points being the central pixels of the objects and the “lowest” points the pixels in the background. The watershed transformation builds dividing lines between the maxima’s zones of influence (Fig. 3C). The map with the dividing lines is combined with the binarized image to form an image with the separated objects (Fig. 3D).
FIG. 3.
Example of the separation of touching nuclei. Panel A: Original nonbinarized image of nuclei. Panel B: Binarized image before the separation of nuclei. Panel C: Boundaries produced by the “watershed” procedure. Panel D: Binarized image with separated nuclei.
The operation is applied only to separate touching nuclei but not touching cells to restrict the analysis only to isolated cells. A similar approach was used by Szirmai et al. (28).
Automatic Thresholding of the Cell Image
In many of the observed images from the cytoplasm channel the nuclei have higher intensity than the surrounding cytoplasm. In these cases the standard histogram method of binarization may result in segmentation of the nuclei from the cells instead of segmentation of the cells from the background. The performance of the standard binarization technique is also limited when there is residual, nonspecific, staining of the background.
To calculate the correct threshold intensity for cell segmentation, we developed the following procedure (Fig. 4). The binarized image of the nuclei is used to estimate the location of the cytoplasm. First, ring-like shaped areas with pre-defined width are drawn around the nuclei. The width can be determined experimentally and is defined by the average size of cytoplasm. Next, a mask image is created, allowing only the outside vicinities of the nuclei to be visible. This mask image is overlaid over the cell image. The intensities of the pixels of the cell image, located in the areas around the nuclei, are averaged. The average value approximates the intensity of the cell boundaries and is used as a threshold for binarization of the cell image.
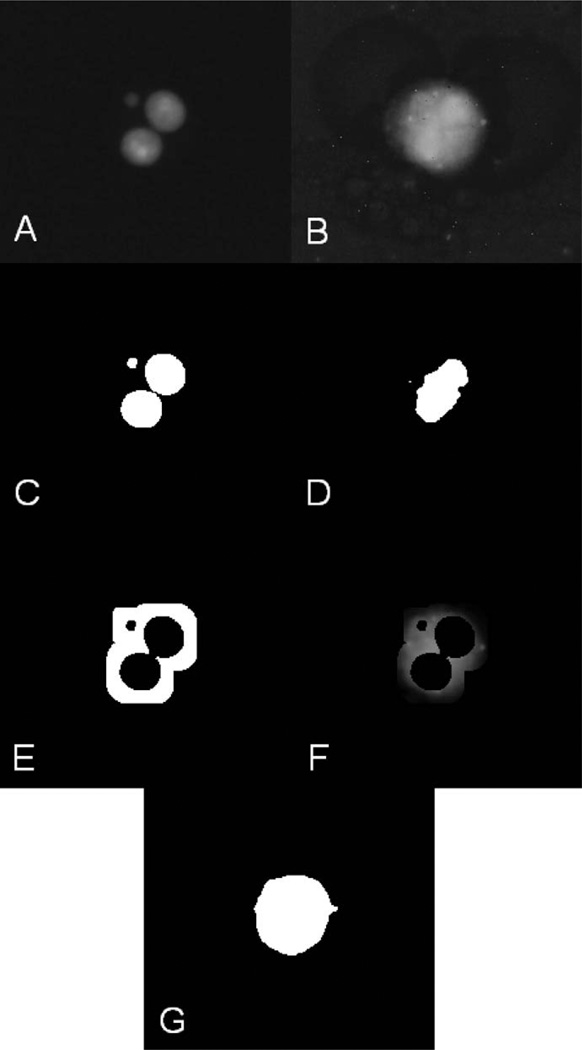
FIG. 4.
Thresholding of the cytoplasm image. Panel A: Original image of the nuclei and a micronucleus. Panel B: Original image of the corresponding cytoplasm. Panel C: Binarized image of the nuclei. Panel D: Binarized image of the cytoplasm when the standard thresholding method (using the image histogram) is applied. Panel E: Mask image, allowing only the vicinities of the nuclei to be seen. Panel F: Mask image from panel E overlaid on the cytoplasm image from panel B. Panel G: Image of the cytoplasm binarized using the threshold obtained by averaging of the nonzero pixels of the image from panel F. This approach provides better localization of cytoplasmic boundaries than the standard thresholding technique, shown in panel D.
Selecting Viable Cells
Both the algorithm development and manual scoring were based on the standard criteria published by the International Collaborative Project on Micronucleus Frequency in Human Populations (19). The criteria require that only viable binucleated cells are scored. Such cells should have intact cytoplasm and so should be approximately circular in shape. The nuclei should also be circular or oval in shape and of similar sizes. To select cells according to these criteria, we used blob parameters describing morphology of cells and nuclei: area, compactness, roughness, elongation and Feret elongation.
Objects that are too small or too large to be counted as cells are excluded from the analysis. The minimum acceptable cell area was set to 2,000 pixels, which corresponds to about 60 µm2; the maximum cell size is 120,000 pixels (350 µm2).
The compactness parameter is equal to p2/(4πA), where p is the perimeter and A is the area of an object. Compactness describes how close the shape of an object is to a circle; it is equal to 1 for a circle and is greater for more convoluted shapes. To process the images used for this study, the maximum compactness of the cells was set to 7.0. Objects that are not sufficiently circular (e.g., cell clusters or aggregations of platelets) are removed from further consideration. The same operation is applied to the nuclei at a later stage after matching cells with their nuclei, with the maximum compactness set to 1.5.
Elongation is a measure of the shape of an object, equal to l/b, where l and b are length and breadth of the object, both derived from its perimeter and area. Elongation is complemented by Feret elongation, which is calculated as a ratio of the maximal Feret diameter to the minimal Feret diameter — the maximal and minimal distances between parallel tangents to the object, respectively. Elongation and Feret elongation are accurate for different types of objects — elongation for longer blobs and Feret elongation for more compact ones. The maximum elongation and Feret elongation were chosen to be 6.0 and 1.35 for the cells and 2.25 and 1.5 for the nuclei.
Compactness, elongation and Feret elongation applied to nuclei are useful in eliminating cells with incorrectly separated nuclei, because of the differences in the shape of nuclei and nuclear fragments. The cells, in which such nuclear fragments have been detected, are not counted.
Roughness describes how rough the blob is; it is equal to the ratio of perimeter and convex perimeter. A smooth convex object has roughness as low as 1.0. The limit on this parameter was set to 2.2 for the cells and 1.2 for the nuclei in our study.
Restricting the acceptable morphological parameters of a cell allows for elimination of cellular clusters and debris from the analysis. To avoid situations when a cluster is round enough to be falsely identified as a cell, the software calculates the ratio of the “cell” area to the area of its largest nucleus. If the nuclear objects are too small compared to the corresponding cytoplasm object, it is likely a cluster. Based on the statistics generated using the collected samples, the maximum value of this ratio was set to 16.0. Using the cell-to-nuclear area ratio also helps to eliminate cells with nuclei not fully reconstructed during binarization due to weak staining.
This method of selecting viable cells based on the parameters described above was visually confirmed and proved to be sufficiently reliable for eliminating clusters, debris, cellular and nuclear fragments, and remnant aggregations of platelets, as well as cells with incorrectly separated nuclei. For this work, the software parameters were set so that all objects in these categories are rejected. The strict criteria also resulted in some loss of viable cells, but micronuclei yields were maintained. All chosen values depend on various characteristics of the imaging system, such as magnification, resolution, signal-to-noise ratio, etc. All the values are stored in a separate file and can be easily modified by the user.
Automatic Identifying and Scoring Micronuclei
If a cell appears to be viable, the analysis continues to identify its nuclei and to score them as well as micronuclei. The area of the micronuclei in binucleated human lymphocytes varies between 1/256 and 1/9 of the area of the main nuclei (9, 19). The analysis of the binucleated cells obtained by the CBMN assay showed that the cell and nuclear sizes may differ not only between samples (i.e., between different individuals), but also within a single field of view. This variability is large enough that the size criterion described above can only be applied if each nucleus is compared to other nuclei of the same cell, which requires each cell to be analyzed individually. Therefore, after cells have been selected for analysis based on their size and compactness, the cells are analyzed one-by-one.
To determine which nuclei in the nuclei image belong to a particular cell in the cytoplasm image, a rectangular area is circumscribed around the cell. Next, an area with the same coordinates is selected on the image containing nuclei. The coordinates of the centers of gravity of the nuclei from that area are determined and matched with the outlines of the cell under consideration. If the center of gravity of a nucleus is not located within the cell boundary, the nucleus is excluded from the analysis.
The number of nuclei in a single cell and their areas are calculated and the nuclei are sorted by size. After that the areas of all nuclei are calculated as fractions of the area of the biggest nucleus and all nuclei are divided into two categories: the main nuclei and the micronuclei. The numbers of the main nuclei and the MNi are recorded. If the cell has one, two or more than two nuclei, the number of mononucleated, binucleated or multinucleated cells is incremented. Similarly, if the cell has MNi, then the number of micronucleated mononucleated cells, micronucleated BN cells or micronucleated cells with multiple nuclei is incremented and the number of MNi is added to the total number of MNi in the corresponding category of cells. All information is accumulated as the analysis continues from frame to frame. The analysis of the sample terminates either when a preset number of BN cells are scored or when a certain set of images is analyzed. Increasing the number of cells scored increases the precision of the dose determination at the expense of the throughput. In triage situations, the RABiT system will analyze 250 cells per dose per individual (3, 45).
Staining Optimization
As discussed in the Introduction section, the RABiT workstation utilizes dual labeling of the nuclei and cytoplasm. To facilitate high-throughput imaging and scoring, the cell staining needs to be bright enough for the cells to be visualized with short exposure times and stable enough to withstand sample storage. In addition, the excitation and emission spectra of the nuclear and the cytoplasmic probes have to be sufficiently separated to allow simultaneous imaging of the stained lymphocytes by two different cameras with no crosstalk. We tested a number of stains for cytoplasm, plasma membrane and cytoskeleton, as well as whole-cell stains [CellTracker Orange, CellMask Orange, CellMask Red, CellMask Deep Red, Rhodamine Phalloidin, Propidium Iodide, CellTracker CM-DiI (Invitrogen) and Red Counterstain C (Trevigen Inc., Gaithersburg, MD)]. CellMask Orange plasma membrane stain satisfies all of the above conditions and provides more uniform staining across the cell and sharper imaging of cell boundaries compared to other stains. DAPI was used as the nuclear stain.
MATERIALS AND METHODS
Hardware Platform
Software development and image analysis were performed on a Dell Latitude laptop with an Intel Core 2 Duo 1.83 GHz processor and 2 GB SDRAM. Image acquisition and timing of the software components were performed on a desktop computer with an Intel Core 2 Quad 2.40 GHz processor and 3.21 GB of RAM.
Blood Collection and Irradiation
With informed consent, peripheral blood from seven healthy volunteers was collected by venipuncture into vacutainer tubes containing sodium heparin (Becton Dickinson, Franklin Lakes, NJ). The total volume of blood collected was 10–12 ml per donor. Samples of whole blood were individually irradiated from 2–10 Gy using a Gammacell 40 137Cs irradiator (Atomic Energy of Canada, Ltd.) at a dose rate of 0.84 Gy/min. The total doses were 2, 4, 6 and 8 Gy for donors 1 through 4 and 2, 4, 6, 8 and 10 Gy for donors 5 through 7. Two milliliters of blood from each donor were maintained as control.
The study was reviewed by Columbia University’s IRB and the use of human materials was approved under the IRB protocols AAAB5846 and AAAF2671, Center for High-Throughput Minimally Invasive Radiation Biodosimetry.
Lymphocyte Culture and Slide Preparation
Whole blood samples (1 ml) were diluted in 10 ml of pre-warmed RPMI medium 1640 with l-glutamine (300 mg/l), containing phytohaemagglutinin (PHA, final concentration 20 µg/ml; Invitrogen, Carlsbad, CA) supplemented with 1% penicillin-streptomycin solution (Invitrogen, 10,000 units/ml penicillin and 10,000 µg/ml streptomycin) and 10% heat-inactivated fetal calf serum (Invitrogen). The cells were cultured and maintained in a humidified atmosphere with 5% CO2. After 44 h of incubation, cytochalasin B (Cyt-B; Sigma, St. Louis, MO) was added to the culture medium at a final concentration 6 µg/ml. The culture was maintained for an additional 26 h to arrest cytokinesis and induce the formation of once-divided BN cells. To harvest lymphocytes, the samples were transferred to a centrifuge tube and spun at 1,000 rpm for 10 min and the supernatant was discarded. The pelleted cells in the tube were treated for 10 min with hypotonic solution of KCL (0.075 M) (Sigma-Aldrich, St. Louis, MO) pre-warmed to 37°C. The reaction was stopped by the addition of ~2 ml of ice-cold methanol:glacial acetic acid solution (ratio 3:1). The cells were further centrifuged at 1,000 rpm for 10 min, the supernatant aspirated and fresh fixative added. The cells were washed with fixative two additional times. After the final centrifugation, the cells were resuspended in ~0.5 ml of fixative and a small drop of lymphocyte suspension was transferred to a clean glass slide. The slides were dried at room temperature, and stained with CellMask Orange (Invitrogen) at a concentration of 5 µg/ml for 20 min to label the cell cytoplasm. After the slides were washed two times with phosphate-buffered saline (Invitrogen), the nuclei were counterstained with DAPI [4′,6-diamidino-2-phenylindole, dihydrochloride (Invitrogen)], and the cover slips were mounted using Vectashield mounting medium (Vector Laboratories, Inc., Burlingame, CA).
Imaging
The samples were imaged using a Nikon Eclipse 50i epifluorescence microscope with a Nikon CFI Plan Fluor 60× objective (Morrell Instrument Company Inc., Melville, NY). The microscope was equipped with a Photonfocus MV-1024 CMOS camera (Upstate Technical Equipment Inc, East Syracuse, NY) and a Solios Camera Link Dual Base frame grabber (Matrox Electronic Systems Ltd, Dorval, QC, Canada). The samples were illuminated using an X-Cite 120 Series mercury vapor short arc lamp (EXFO, Life Sciences & Industrial division, Mississauga, Ontario, Canada).
A pair of images was consecutively acquired for each field of view, one through a Nikon UV-2E/C fluorescent filter set (Morrell) to visualize DAPI-stained nuclei and another through a Nikon TRITC HYQ filter set (Morrell) to visualize the outline of the cytoplasm stained with CellMask Orange. This imaging procedure is consistent with the image grabbing procedure in the RABiT.
Data Collection and Analysis
The designed software has the capability of detecting cells with any number of nuclei. Thus, in addition to determining the ratio of micronuclei per binucleated cell, other parameters discussed in the Introduction section can be calculated. In the samples collected for this study we observed that the nuclei in multinucleated cells are closer to each other than the nuclei in BN cells, and their boundaries touch or overlap more often. When such nuclei are separated by the software, their morphological parameters, described in the Design section, could be different from that of the nuclei in mononucleated or BN cells. These nuclei might not be classified as viable and some multinucleated cells could be misinterpreted as clusters. Thus, the ratio of the mononucleated to BN cells was chosen for this study as the most reliable parameter to complement the analysis at high doses.
Two sets of multiple fields of view were acquired: one for analysis of MN frequencies and the other for analysis of the ratios of the mononucleated to BN cells. The fields of view were acquired sequentially by step-wise scanning of the slide. For the first set, only the fields that contained at least one BN cell were captured. This set contains data from all seven donors. The second set was taken from the samples of donors 5–7 and consists of random fields of view containing mononucleated or BN cells. Fields of view containing no cells were excluded. Both sets were analyzed manually and automatically using the developed software described above.
Manual scoring was done by inspecting all of the fields of view in both sets, following the standard scoring criteria (19). The number of BN cells, the number of BN cells with MNi and the number of MNi in BN cells were scored in the first set; mononucleated and BN cells were scored in the second set.
The same fields of view that were presented for manual scoring were then processed and scored automatically. The information presented to the user after the analysis of each dose-point included the numbers of mononucleated cells and BN cells, the numbers of mononucleated cells and BN cells with MNi, and the total numbers of MNi in both groups of cells. This information was used to compute micronuclei frequencies in BN cells for donors 1–7 and the ratio of mononucleated to BN cells for donors 5–7. The obtained dose-response curves were created using Excel (Microsoft Corp., Redmond, WA) and the curve fittings were performed in OriginPro 8.1 (OriginLab Corp, Northampton, MA).
RESULTS
The algorithm and implementation of the designed software are described in the Design section. The operation of the software was validated by establishing automatic and manual dose-response curves of micronuclei frequencies in BN cells and ratios of mononucleated to BN cells.
Validation of the Software by Calculation of Dose Response
The data scored automatically and manually and calculated frequencies of MNi and ratios of mononucleated to BN cells are presented in Tables 1 and 2, respectively.
TABLE 1.
Numbers of Binucleated Cells (BN), Binucleated Cells with Micronuclei (BNMN), Micronuclei (MNi) and the Number of Micronuclei per Binucleated Cell (MNi/BN)
| Donors | Dose (Gy) |
Automated scoring | Manual scoring | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BN | BNMN | MNi | MNi/BN | BN | BNMN | MNi | MNi/BN | ||
| Donor 1 | 0 | 236 | 4 | 5 | 0.021 ± 0.009 | 232 | 5 | 5 | 0.022 ± 0.009 |
| 2 | 166 | 11 | 13 | 0.08 ± 0.02 | 163 | 16 | 18 | 0.11 ± 0.03 | |
| 4 | 135 | 38 | 47 | 0.35 ± 0.05 | 139 | 44 | 54 | 0.39 ± 0.05 | |
| 6 | 58 | 17 | 25 | 0.43 ± 0.09 | 68 | 31 | 42 | 0.62 ± 0.10 | |
| 8 | 20 | 12 | 18 | 0.9 ± 0.2 | 28 | 18 | 30 | 1.1 ± 0.2 | |
| Donor 2 | 0 | 295 | 3 | 3 | 0.010 ± 0.006 | 302 | 2 | 2 | 0.007 ± 0.005 |
| 2 | 288 | 35 | 41 | 0.14 ± 0.02 | 292 | 40 | 43 | 0.15 ± 0.02 | |
| 4 | 190 | 59 | 81 | 0.42 ± 0.05 | 178 | 60 | 78 | 0.44 ± 0.05 | |
| 6 | 145 | 56 | 74 | 0.51 ± 0.06 | 136 | 56 | 80 | 0.59 ± 0.07 | |
| 8 | 65 | 31 | 59 | 0.91 ± 0.12 | 55 | 26 | 46 | 0.84 ± 0.12 | |
| Donor 3 | 0 | 97 | 8 | 9 | 0.09 ± 0.03 | 161 | 2 | 2 | 0.012 ± 0.009 |
| 2 | 278 | 72 | 113 | 0.41 ± 0.04 | 333 | 70 | 88 | 0.26 ± 0.03 | |
| 4 | 404 | 200 | 322 | 0.80 ± 0.04 | 396 | 194 | 275 | 0.69 ± 0.04 | |
| 6 | 132 | 99 | 251 | 1.90 ± 0.12 | 133 | 104 | 255 | 1.92 ± 0.12 | |
| 8 | 68 | 45 | 129 | 1.90 ± 0.17 | 62 | 45 | 123 | 1.98 ± 0.18 | |
| Donor 4 | 0 | 389 | 8 | 8 | 0.021 ± 0.007 | 370 | 3 | 3 | 0.008 ± 0.005 |
| 2 | 358 | 34 | 40 | 0.112 ± 0.018 | 357 | 34 | 40 | 0.112 ± 0.018 | |
| 4 | 254 | 93 | 123 | 0.48 ± 0.04 | 237 | 80 | 103 | 0.44 ± 0.04 | |
| 6 | 168 | 85 | 131 | 0.78 ± 0.07 | 165 | 92 | 141 | 0.86 ± 0.07 | |
| 8 | 55 | 27 | 54 | 0.98 ± 0.13 | 49 | 31 | 57 | 1.16 ± 0.15 | |
| Donor 5 | 0 | 134 | 3 | 3 | 0.022 ± 0.013 | 163 | 3 | 5 | 0.031 ± 0.014 |
| 2 | 88 | 25 | 38 | 0.43 ± 0.07 | 109 | 32 | 39 | 0.36 ± 0.06 | |
| 4 | 53 | 39 | 81 | 1.53 ± 0.17 | 59 | 45 | 75 | 1.27 ± 0.15 | |
| 6 | 51 | 39 | 97 | 1.90 ± 0.19 | 58 | 41 | 98 | 1.69 ± 0.17 | |
| 8 | 102 | 73 | 198 | 1.94 ± 0.14 | 146 | 108 | 264 | 1.81 ± 0.11 | |
| 10 | 49 | 23 | 72 | 1.47 ± 0.17 | 95 | 51 | 126 | 1.33 ± 0.12 | |
| Donor 6 | 0 | 212 | 11 | 14 | 0.066 ± 0.018 | 240 | 9 | 10 | 0.042 ± 0.013 |
| 2 | 418 | 95 | 139 | 0.33 ± 0.03 | 490 | 111 | 144 | 0.29 ± 0.02 | |
| 4 | 410 | 192 | 295 | 0.72 ± 0.04 | 533 | 266 | 417 | 0.78 ± 0.04 | |
| 6 | 215 | 105 | 188 | 0.87 ± 0.06 | 269 | 154 | 253 | 0.94 ± 0.06 | |
| 8 | 229 | 96 | 167 | 0.73 ± 0.06 | 257 | 144 | 254 | 0.99 ± 0.06 | |
| 10 | 132 | 42 | 77 | 0.58 ± 0.07 | 120 | 62 | 102 | 0.85 ± 0.08 | |
| Donor 7 | 0 | 267 | 14 | 18 | 0.067 ± 0.016 | 298 | 6 | 6 | 0.020 ± 0.008 |
| 2 | 206 | 75 | 106 | 0.52 ± 0.05 | 247 | 84 | 112 | 0.45 ± 0.04 | |
| 4 | 122 | 76 | 126 | 1.03 ± 0.09 | 138 | 80 | 128 | 0.93 ± 0.08 | |
| 6 | 198 | 166 | 373 | 1.88 ± 0.10 | 196 | 161 | 358 | 1.83 ± 0.10 | |
| 8 | 56 | 38 | 118 | 2.12 ± 0.19 | 58 | 47 | 140 | 2.41 ± 0.2 | |
| 10 | 38 | 20 | 42 | 1.11 ± 0.17 | 37 | 22 | 39 | 1.05 ± 0.17 | |
TABLE 2.
Numbers of Binucleated (BN) and Mononucleated (Mono) Cells and Calculated Ratios of Mononucleated to Binucleated Cells (Mono/BN)
| Donors | Dose (Gy) | Automated scoring | Manual scoring | ||||
|---|---|---|---|---|---|---|---|
| BN | Mono | Mono/BN | BN | Mono | Mono/BN | ||
| Donor 5 | 0 | 62 | 48 | 0.77 ± 0.15 | 67 | 71 | 1.06 ± 0.18 |
| 2 | 48 | 53 | 1.1 ± 0.2 | 51 | 48 | 0.94 ± 0.19 | |
| 4 | 35 | 54 | 1.5 ± 0.3 | 34 | 69 | 2.0 ± 0.4 | |
| 6 | 48 | 131 | 2.7 ± 0.5 | 41 | 151 | 3.7 ± 0.6 | |
| 8 | 34 | 165 | 4.9 ± 0.9 | 37 | 182 | 4.9 ± 0.9 | |
| 10 | 25 | 315 | 13 ± 3 | 38 | 387 | 10 ± 2 | |
| Donor 6 | 0 | 46 | 31 | 0.67 ± 0.16 | 53 | 48 | 0.91 ± 0.18 |
| 2 | 77 | 56 | 0.73 ± 0.13 | 83 | 63 | 0.76 ± 0.13 | |
| 4 | 37 | 56 | 1.5 ± 0.3 | 38 | 80 | 2.1 ± 0.4 | |
| 6 | 25 | 89 | 3.6 ± 0.8 | 36 | 131 | 3.6 ± 0.7 | |
| 8 | 88 | 691 | 7.9 ± 0.9 | 126 | 777 | 6.2 ± 0.6 | |
| 10 | 125 | 972 | 7.8 ± 0.7 | 158 | 1126 | 7.1 ± 0.6 | |
| Donor 7 | 0 | 101 | 142 | 1.41 ± 0.18 | 110 | 144 | 1.31 ± 0.17 |
| 2 | 66 | 146 | 2.2 ± 0.3 | 81 | 153 | 1.9 ± 0.3 | |
| 4 | 32 | 100 | 3.1 ± 0.6 | 36 | 109 | 3.0 ± 0.6 | |
| 6 | 39 | 194 | 5.0 ± 0.9 | 37 | 212 | 5.7 ± 1.0 | |
| 8 | 39 | 384 | 10 ± 2 | 44 | 433 | 10 ± 2 | |
| 10 | 21 | 294 | 14 ± 3 | 27 | 419 | 16 ± 2 | |
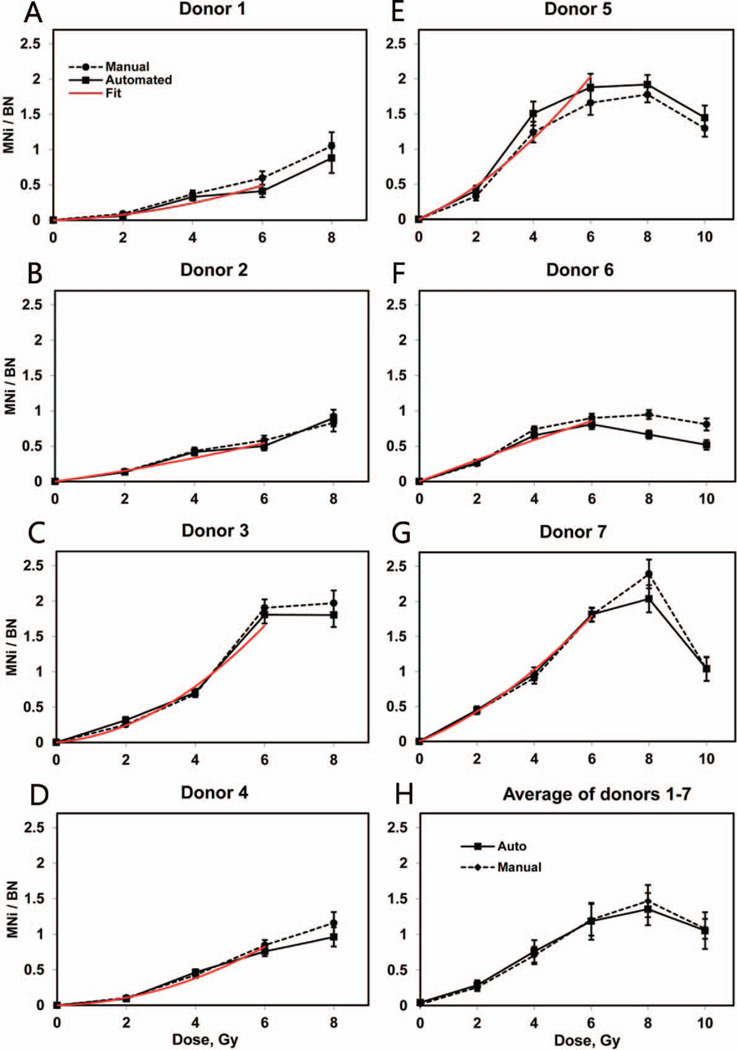
Figure 5 shows the dose-response curves of the frequencies of MNi in BN cells observed for each of the seven individuals (Fig 5A–G) and the average across the seven donors (Fig. 5H). The background micronuclei frequency of each donor was subtracted from the data at each point to demonstrate the frequencies induced in vitro. The automatically and manually obtained individual radiation-induced responses in the range from 0–6 Gy were fitted to second-order polynomials with zero intercepts as represented by the red lines on the panels A–G. The fit was weighted by the reciprocal value of the variance because of the larger errors at high doses. The baseline frequencies, linear and quadratic coefficients of the fit functions as well as the correlation coefficient between the fits and the data are presented in Table 3. All of the manually and automatically scored micronuclei frequencies show an increase with dose: up to 6 Gy for donors 3, 5 and 6 and up to 8 Gy for donors 1, 2, 4 and 7. The dependence on dose is linear-quadratic between 0 and 4 Gy; at higher doses the curvatures change and the functions level-off or decline. The average micronuclei frequencies for 0–8 Gy were calculated across the entire group of seven donors, and for 10 Gy, across donors 5, 6 and 7.
FIG. 5.
Individual (panels A–G) and averaged (panel H) induced frequencies of MNi (number of MNi per binucleated cell). Solid lines: automatically scored, dashed lines: manually scored. The error bars represent Poisson errors for the individual curves and standard error of the mean for the average dose response. The individual errors include uncertainties of the subtracted baseline values. The red lines are error-weighted linear-quadratic fits for the automatically obtained data from 0–6 Gy.
TABLE 3.
Parameters of Linear-Quadratic Fitting of Radiation-Induced Micronuclei Frequencies
| Donors | Correlation between automated and manual scoring, R2 |
Baseline MNi frequencies |
B1 | B2 | Correlation between automated scoring and the fits, R2 |
|---|---|---|---|---|---|
| 1 | 0.986 | 0.021 ± 0.009 | 0.016 ± 0.028 | 0.011 ± 0.011 | 0.872 |
| 2 | 0.978 | 0.010 ± 0.006 | 0.068 ± 0.025 | 0.004 ± 0.005 | 0.952 |
| 3 | 0.998 | 0.093 ± 0.031 | 0.043 ± 0.062 | 0.039 ± 0.014 | 0.970 |
| 4 | 0.987 | 0.021 ± 0.007 | 0.011 ± 0.024 | 0.021 ± 0.006 | 0.961 |
| 5 | 0.994 | 0.022 ± 0.013 | 0.184 ± 0.092 | 0.026 ± 0.020 | 0.944 |
| 6 | 0.914 | 0.066 ± 0.018 | 0.155 ± 0.031 | −0.002 ± 0.006 | 0.982 |
| 7 | 0.981 | 0.067 ± 0.016 | 0.172 ± 0.020 | 0.021 ± 0.004 | 0.998 |
Note. B1 and B2 are the linear and quadratic coefficients of the fit.
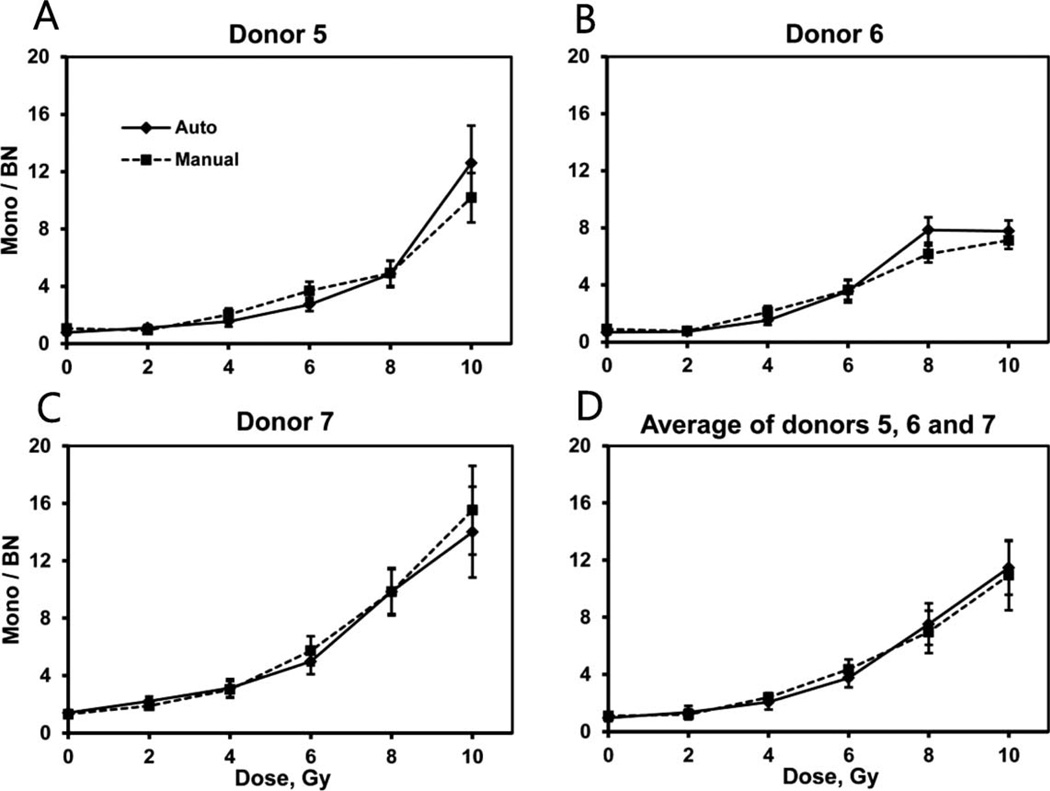
The dose-response dependencies of the ratio of mononucleated to BN cells are shown in Fig. 6. The y-intercepts and linear and quadratic coefficients of error-weighted linear-quadratic fits are given in Table 4. All three individual dose responses (Fig. 6A–C) have positive curvatures (coefficients B2). The ratios of mononucleated cells per BN cells show increasing dose response for the entire range of 0–10 Gy with the exception of the 10 Gy point of donor 6. The average of the three donors is shown in Fig. 6D.
FIG. 6.
Individual (panels A–C) and averaged (panel D) ratios of the number of mononucleated (Mono) cells to binucleated cells. The error bars represent Poisson error for the individual curves and standard error of the mean for the average dose response.
TABLE 4.
Parameters of Linear-Quadratic Fitting of Ratios of Mononucleated to Binucleated Cells
| Donors | Correlation between automated and manual scoring, R2 |
Intercept | B1 | B2 | Correlation between automated scoring and the fits, R2 |
|---|---|---|---|---|---|
| 5 | 0.976 | 0.82 ± 0.19 | −0.11 ± 0.18 | 0.08 ± 0.03 | 0.852 |
| 6 | 0.975 | 0.64 ± 0.25 | −0.13 ± 0.18 | 0.09 ± 0.02 | 0.915 |
| 7 | 0.993 | 1.44 ± 0.14 | 0.06 ± 0.17 | 0.11 ± 0.03 | 0.950 |
Note. B1 and B2 are the linear and quadratic coefficients of the fit.
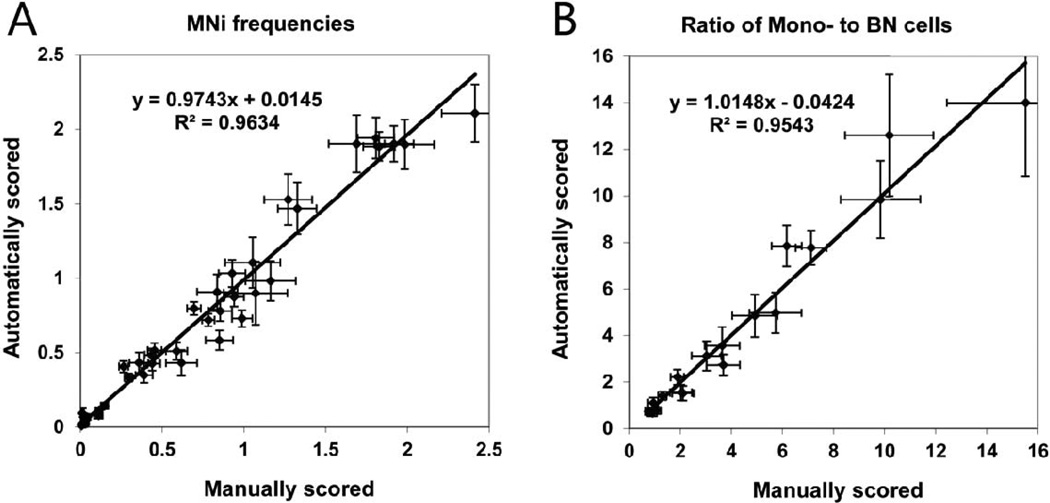
The pooled data from all donors comparing automated and manual calculation of micronuclei frequencies in BN cells (Fig. 7A) and ratio of mononucleated to BN cells (Fig. 7B) are shown in Fig. 7.
FIG. 7.
Regression plots comparing (panel A) number of MNi per BN cell and (panel B) ratios of mononucleated (Mono) cells to binucleated cells obtained by manual scoring (x-axis) and automated scoring (y-axis). The error bars represent Poisson error on both graphs.
The average total time of processing of a single field of view and the times required to execute individual components of the algorithm were measured and are presented in Table 5.
TABLE 5.
| Operation | Time (ms) |
|---|---|
| Nuclear thresholdinga | 100 |
| Separation of nuclei | 82 |
| Cell thresholding | 8.7 |
| Identifying and scoring MNi | 22 |
| Totalb | 260 |
Includes 5 modified iterations.
Includes additional operations.
DISCUSSION
The goal of this work was to design a system for automatic analysis of the results of CBMN assay. The designed software will operate as part of the RABiT biodosimetry workstation. We demonstrated here the suitability of the system for dose assessment by producing the dose-response curves presented in the Results section.
The results show that individual dose responses vary in sensitivity. The individual in vitro radio-sensitivities can be approximately described by the linear coefficients B1 in Table 3. This variability could reasonably be anticipated. It is well know that there exists a considerable variation in the baseline micronuclei frequency and acute radiosensitivity (46–48). In part this is due to variations in demographics, such as age, gender (46), smoking status, occupational exposure (49), as well as health status (50). Additional demographic studies, currently under way, will determine the effects of the confounding factors on the radiation-induced micronuclei formation; thus allowing demographic-specific calibration curves to be produced. It should be noted that in our current study we utilized blood samples of anonymous healthy donors, with information about age, gender or lifestyle parameters unavailable. In part, however, this variation is related to person-to-person variability in sensitivity due to inter-individual genetic or epigenetic variations. This latter variability is, however, potentially a significant advantage for most biodosimetric endpoints (51): an individual who, for example, is particularly sensitive for genetic reasons in terms of response to a biodosimetric DNA damage endpoint, might well be expected to be correspondingly sensitive to acute radiation health effects.
For the purpose of triage, the estimated number of 250 scored BN cells per dose per individual (3, 45) will provide the precision in dose assessment of better than 20% at a 95% confidence level at 1 Gy. Larger number of scored cells will allow higher precision required for epidemiological studies.
The average values of MNi per BN cell (Fig. 5H) increased with the dose between 0 and 6 Gy, leveled off above 6 Gy and decline above 8 Gy. This behavior agrees with previous findings (36). By contrast, the average values of mononucleated to binucleated cells (Fig. 6D) increased and was sufficiently separated at 6, 8 and 10 Gy. This increase, which is also in agreement with the results available in the literature (36, 42, 48), could indicate the delay in cell cycle progression in the presence of DNA damage, which influences the production of BN cells. The monotonically increasing dose dependence of the ratio of mononucleated to binucleated lymphocytes confirms that this endpoint could be taken into account as an additional parameter for dose-assessment purposes at higher doses. For example, the value of the ratio of mononucleated to binucleated cells above a certain cut-off value would indicate that the dose is in the range where the micronuclei frequency is no longer monotonic. In this case the dose will be determined using the mononucleated to binucleated cells ratio. If this parameter is below the cut-off, then micronuclei per binucleated cell would be used for dose evaluation.
The results also demonstrated a significant correlation between manually and automatically acquired data. The individual correlation coefficients R2 ranged from 0.914–0.998 for the frequencies of MNi (Table 3) and from 0.975–0.993 for the mononucleated to binucleated cells ratio. The regression analysis performed on the combined micronuclei frequency data from all donors shows the correlation between automated and manual scoring R2 = 0.963 (Fig. 7A). The detection efficiency of micronuclei varied between 69–99% for individual donors and between 92–98% for the average. The correlation between automated and manual scoring for the combined mononucleated to binucleated cells ratio data was R2 = 0.954 (Fig. 7B).
The ability of the presented automated image analysis system to detect MNi is comparable or superior to that of other available systems (Table 6). For example, Varga et al. (24) reported on using a computerized image analysis system (Metasystems Metafer 4 version 2.12) for scoring MNi to quantify the different radiosensitivities of cancer patients and controls. The algorithm is based on the detection of the DAPI-stained nuclei only and the locations of the cytoplasm are estimated from the distances between nuclei and their relative size. The linear correlation coefficient between manual and automated scoring was R2 =0.83 for 0–2 Gy. The absolute micronuclei frequencies, as detected by the automated analysis, range from 33–38% of the manually found values for the means of 20–27 individuals. Willems et al. (52) describe a study, also employing the MetaSystems software, to establish a method of fast population triage. They reached a higher MNi detection rate, compared to the study by Varga et al., from 71–89% at 0.5–3 Gy for the average of 10 donors. The linear regression coefficient between automated and manual scoring for 0–3 Gy range was R2 = 0.917.
TABLE 6.
Detection Efficiencies and Correlations between Automated and Manual Scoring for Various Studies
| RABiT software | Varga et al. | Willems et al. | Decordier et al. | |
|---|---|---|---|---|
| Blood donors | 7 | 114a | 10 | 1 |
| Investigated doses (Gy) | 0, 2, 4, 6, 8, 10 | 0, 2 | 0, 0.2, 0.5, 1, 2, 3 | 0, 1, 2, 3 |
| Detection efficiency (individual) | 69–99% | n.a. | n.a. | 46–62% |
| Correlation between automated and manual scoring (individual) | 0.914–0.998 | n.a. | n.a. | 0.97 |
| Detection efficiency (for mean frequencies) | 92–98% | 33–38% | 71–89% | n.a. |
| Correlation between automated and manual scoring (for mean micronuclei frequencies) | 0.994 | 0.83 | 0.917 | n.a. |
Seventy-three donors in the test sample (27 patients and 46 controls) and 41 donors in the validation sample (21 control and 20 cases).
The system described by Decordier et al. [utilizing Pathfinder CellScan, IMSTAR (27)] utilized one stain (Giemsa), but the algorithm is similar to our methodology in that the analysis starts from the detection of whole cells. Furthermore, the dose response data are presented for lymphocytes treated with different agents (clastogens, including ionizing radiation, aneugens and apoptogens) and the data for automated scoring were collected with and without visual validation. The dose response for ionizing radiation was established between 0–3 Gy. For micronuclei frequencies scored automatically without visual validation, the percentage of detected MNi was 46–62% of those counted manually and the background was slightly elevated. The correlation coefficient for the frequency of MNi in BN cells between manual and automated scoring was R2 = 0.970. As in our study, the advantage of the described methodology is that the results are not restricted to BN cells only and the detection of mononucleated and multinucleated cells allows for the calculation of the proliferation index.
All of the described systems as well as the developed RABiT software produce slightly elevated baseline micronuclei frequencies relative to manual scoring. The behavior can be described by the formula fauto = DE × fmanual + FP, where fauto and fmanual are the frequencies of micronuclei (MNi/BN) detected by the automated and manual scoring respectively, DE is the detection efficiency of the automated procedure and FP is the false positive yield (29). The higher background frequency can be explained in part by the misinterpretation of released DNA material or staining artifacts as MNi. Such artifacts may be rejected by the human eye during manual processing, but may become indistinguishable from MNi after the binarization stage during automated processing. Lower detection efficiency (DE < 1) of an automated procedure may be caused, for example, by a lower staining intensity of the MNi than that of the main nuclei. The detection efficiency of our system approaches that of manual scoring.
Among the drawbacks of the above studies was the low objective magnification (10×) used for analysis. In the beginning of our study we tried 10× and 50× objectives and discovered that fewer MNi can be seen at a magnification of 10×. However, higher magnification requires more images to be acquired and analyzed for the same statistics. Furthermore, another factor that will be taken into account while selecting the magnification is the surface area of the substrate and the density of the mononucleated cells and binucleated cells to be analyzed; for example, in the RABiT imaging module the cells are imaged on the membranes of the multi-well plates.
To summarize, the developed software provided an estimate of the micronuclei frequency close to the one determined by manual scoring. One reason that high MNi detection efficiency was achieved could be the modified thresholding mechanism, which allowed the rate of the detected MNi to ranges from 88–100%. Another reason for this efficiency is possibly the inclusion of the information about the cytoplasm. Furthermore, our investigated dose range was larger than in the other studies involving automated systems. To our knowledge, this is the first time that automated scoring successfully quantifies MNi at doses above 6.4 Gy. Finally, all of the above referenced studies involved using commercial platforms for cell detection and analysis, and our software does not require a dedicated imaging system. It can be installed on a stand-alone desktop or laptop computer and used to analyze images from a variety of image acquisition systems.
The processing time of 0.26 s per field of view corresponds to an analysis rate of about 13,846 fields of view per hour. Depending on the cell density and magnification, this could be equivalent to 10,000–20,000 or more BN cells scored per hour, which corresponds to 40– 80 or more samples per hour. The system performance will be further optimized by integration of the imaging system of the RABiT with a high-end workstation to achieve faster processing speed.
The work described here was conducted under the assumption of whole-body exposure. The more realistic model in the scenario of a radiological event would be partial-body exposure, as most individuals would be partially shielded from the blast by structural materials. With the information on the cell-to-cell distributions of MNi or binned γ-H2AX fluorescence provided by the RABiT, this partial-body exposure can be modeled using the “contaminated Poisson” method (53). The performance of RABiT is currently being evaluated under various partial-body irradiation scenarios, using a mouse model.
CONCLUSION
We have developed software for automated scoring of the frequency of MNi in binucleated lymphocytes. This software is designed to function as an image analysis module for the MN assay in the Rapid Automated Biodosimetry Tool. The analysis is based on the dual staining of the samples and separate processing of cytoplasmic and nuclear images. We have demonstrated that the developed software is suitable for the estimation of the dose response using micronuclei frequency as a biomarker for radiation doses up to 6 Gy. Dose-response curves from seven individuals were generated for doses of 0–8 Gy and 10 Gy, a greater range than that used in most of the recently available systems. The information about the fractions of mononucleated cells and binucleated cells may be helpful for detection of high doses as an additional indicator of cell proliferation. In our current study, we established three dose responses that show dose dependence of this quantity.
The ability of our image analysis system to estimate individual dose at fast operating speed is consistent with the primary goal of the RABiT. Automated analysis of micronuclei frequencies in irradiated lymphocytes is estimated to be 40–100 times faster than manual scoring. Compared to commercial systems, our system demonstrated similar or superior performance and is considerably less expensive with the added benefit that it can work as a standalone image analysis tool.
Currently the image analysis system is being integrated into the RABiT and future work will focus on the development of extended analysis of the MN assay samples, such as detection of nucleoplasmic bridges and necrotic and apoptotic cells.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Charles Geard for his invaluable advice. This work was supported by grant no. U19 AI067773 to the Center for High-Throughput Minimally Invasive Radiation Biodosimetry from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
REFERENCES
- 1.Pellmar TC, Rockwell S. Priority list of research areas for radiological nuclear threat countermeasures. Radiat Res. 2005;163:115–123. doi: 10.1667/rr3283. [DOI] [PubMed] [Google Scholar]
- 2.Salerno A, Zhang J, Bhatla A, Lyulko OV, Dutta A, Garty G, et al. Design considerations for a minimally invasive high-throughput automation system for radiation biodosimetry. Automation Science and Engineering. CASE; Proceedings of the 3rd IEEE International Conference on Automation Science and Engineering; 2007 Sept 22–25; Scottsdale, AZ. 2007. pp. 846–852. [Google Scholar]
- 3.Garty G, Chen Y, Salerno A, Turner HC, Zhang J, Lyulko OV, et al. The RABiT: A rapid automated biodosimetry tool for radiological triage. Health Phys. 2010;98:209–217. doi: 10.1097/HP.0b013e3181ab3cb6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garty G, Chen Y, Turner HC, Zhang J, Lyulko OV, Bertucci A, et al. The RABiT: A Rapid Automated BIodosimetry Tool For Radiological Triage II. Technological Developments. Int J Rad Biol. 2011;87:776–790. doi: 10.3109/09553002.2011.573612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Zhang J, Wang H, Garty G, Xu Y, Lyulko OV, et al. Design and preliminary validation of a rapid automated biodosimetry tool for high-throughput radiological triage. MESA; Proceedings of the 5th ASME/IEEE International Conference on Mechatronic and Embedded Systems and Applications; 2009 Aug 30 – Sept 2; San Diego, CA. 2009. pp. 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garty G, Karam A, Brenner DJ. Infrastructure to support ultra high throughput biodosimetry screening after a radiological event. Int J Radiat Biol. 2011;87:754–765. doi: 10.3109/09553002.2011.583317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura A, Sedelnikova OA, Redon C, Pilch DR, Sinogeeva NI, Shroff R, et al. Techniques for gamma-H2AX detection. Meth Enzymol. 2006;409:266–250. doi: 10.1016/S0076-6879(05)09014-2. [DOI] [PubMed] [Google Scholar]
- 8.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 9.Fenech M. Cytokinesis-block micronucleus cytome assay. Nat Protoc. 2007;2:1084–1104. doi: 10.1038/nprot.2007.77. [DOI] [PubMed] [Google Scholar]
- 10.Turner HC, Brenner DJ, Chen Y, Bertucci A, Zhang J, Wang H, et al. Adapting the γ-H2AX assay for automated processing in human lymphocytes. 1. Technological aspects. Radiat Res. 2011;175:282–290. doi: 10.1667/RR2125.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gough AH, Johnston PA. Requirements, features, and performance of high content screening platforms. Methods Mol Biol. 2007;656:41–61. doi: 10.1385/1-59745-217-3:41. [DOI] [PubMed] [Google Scholar]
- 12.Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol. 2006;7:690–696. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 13.Fenech M, Morley AA. Measurement of micronuclei in lymphocytes. Mutat Res. 1985;147:29–36. doi: 10.1016/0165-1161(85)90015-9. [DOI] [PubMed] [Google Scholar]
- 14.Heddle JA. A rapid in vivo test for chromosomal damage. Mutat Res. 1973;18:187–190. doi: 10.1016/0027-5107(73)90035-3. [DOI] [PubMed] [Google Scholar]
- 15.Schmid W. Chemical mutagen testing on in vivo somatic mammalian cells. Agents Actions. 1973;3:77–85. doi: 10.1007/BF01986538. [DOI] [PubMed] [Google Scholar]
- 16.Kirsch-Volders M. Towards a validation of the micronucleus test. Mutat Res. 1997;392:1–4. doi: 10.1016/s0165-1218(97)00039-6. [DOI] [PubMed] [Google Scholar]
- 17.Cytogenetic analysis for radiation dose assessment: a manual. Vienna: IAEA; 2001. International Atomic Energy Agency. [Google Scholar]
- 18.Amundson SA, Bittner M, Meltzer P, Trent J, Fornace AJ., Jr Biological indicators for the identification of ionizing radiation exposure in humans. Expert Rev Mol Diagn. 2001;1:211–219. doi: 10.1586/14737159.1.2.211. [DOI] [PubMed] [Google Scholar]
- 19.Fenech M, Chang WP, Kirsch-Volders M, Holland N, Bonassi S, Zeiger E. HUMN project: detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat Res. 2003;534:65–75. doi: 10.1016/s1383-5718(02)00249-8. [DOI] [PubMed] [Google Scholar]
- 20.Iarmarcovai G, Bonassi S, Botta A, Baan RA, Orsière T. Genetic polymorphisms and micronucleus formation: A review of the literature. Mutat Res. 2008;658:215–233. doi: 10.1016/j.mrrev.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Fenech M, Holland N, Zeiger E, Chang WP, Burgaz S, Thomas P, et al. The HUMN and HUMNxL international collaboration projects on human micronucleus assays in lymphocytes and buccal cells—past, present and future. Mutagenesis. 2011;26:239–245. doi: 10.1093/mutage/geq051. [DOI] [PubMed] [Google Scholar]
- 22.Fenech M, Jarvis LR, Morley AA. Preliminary studies on scoring micronuclei by computerised image analysis. Mutat Res. 1988;203:33–38. doi: 10.1016/0165-1161(88)90005-2. [DOI] [PubMed] [Google Scholar]
- 23.Böcker W, Müller W-U, Streffer C. Image processing algorithms for the automated micronucleus assay in binucleated human lymphocytes. Cytometry. 1995;19:283–294. doi: 10.1002/cyto.990190402. [DOI] [PubMed] [Google Scholar]
- 24.Varga D, Johannes T, Jainta S, Schuster S, Schwarz-Boeger U, Kiechle M, et al. An automated scoring procedure for the micronucleus test by image analysis. Mutagenesis. 2004;19:391–397. doi: 10.1093/mutage/geh047. [DOI] [PubMed] [Google Scholar]
- 25.Böcker W, Streffer C, Müller W-U, Yu C. Automated scoring of micronuclei in binucleated human lymphocytes. Int J Radiat Biol. 1996;70:529–537. doi: 10.1080/095530096144734. [DOI] [PubMed] [Google Scholar]
- 26.Castelain P, Van Hummelen P, Deleener A, Kirsch-Volders M. Automated detection of cytochalasin-B blocked binucleated lymphocytes for scoring micronuclei. Mutagenesis. 1993;8:285–293. doi: 10.1093/mutage/8.4.285. [DOI] [PubMed] [Google Scholar]
- 27.Decordier I, Papine A, Plas G, Roesems S, Vande Loock K, Moreno-Palomo J, et al. Automated image analysis of cytokinesis-blocked micronuclei: an adapted protocol and a validated scoring procedure for biomonitoring. Mutagenesis. 2009;24:85–93. doi: 10.1093/mutage/gen057. [DOI] [PubMed] [Google Scholar]
- 28.Szirmai S, Berces J, Koteles GJ. Computerized Image Analysis for Determining Micronucleus Frequency. Environ Health Perspect. 1993;101(Suppl 3):S57–S60. doi: 10.1289/ehp.93101s357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tates AD, Van Welie MT, Ploem JS. The present state of the automated micronucleus test for lymphocytes. Int J Radiat Biol. 1990;58:813–825. doi: 10.1080/09553009014552191. [DOI] [PubMed] [Google Scholar]
- 30.Diaz D, Scott A, Carmichael P, Shi W, Costales C. Evaluation of an automated in vitro micronucleus assay in CHO-K1 cells. Mutat Res. 2007;630:1–13. doi: 10.1016/j.mrgentox.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Donnelly EH, Nemhauser JB, Smith JM, Kazzi ZN, Farfán EB, Chang AS, et al. Acute radiation syndrome: assessment and management. South Med J. 2010;103:541–544. doi: 10.1097/SMJ.0b013e3181ddd571. [DOI] [PubMed] [Google Scholar]
- 32.Diagnosis and treatment of radiation injuries. Vienna: IAEA; 1998. International Atomic Energy Agency. ( http://www-pub.iaea.org/MTCD/publications/PDF/P040_scr.pdf) [Google Scholar]
- 33.Hall EJ, Giaccia AJ. Radiobiology for the radiologist. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2006. [Google Scholar]
- 34.Armed Forces Radiobiology Research Institute. Medical management of radiological casualties. Bethesda: AFFRI; 2003. [Google Scholar]
- 35.Baranov A, Gale RP, Guskova A, Piatkin E, Selidovkin G, Muravyova L, et al. Bone marrow transplantation after the Chernobyl nuclear accident. N Engl J Med. 1989;321:205–212. doi: 10.1056/NEJM198907273210401. [DOI] [PubMed] [Google Scholar]
- 36.Müller W-U, Rode A. The micronucleus assay in human lymphocytes after high radiation doses (5–15 Gy) Mutat Res. 2002;502:47–51. doi: 10.1016/s0027-5107(02)00022-2. [DOI] [PubMed] [Google Scholar]
- 37.Romm H, Barnard S, Boulay-Greene H, De Amicis A, De Sanctis S, Franco M, et al. Laboratory intercomparison of the cytokinesis-block micronucleus assay. Radiat Res. 2013;180:120–128. doi: 10.1667/RR3234.1. [DOI] [PubMed] [Google Scholar]
- 38.Yao B, Jiang BR, Ai HS, Li YF, Liu GX, Man QH, et al. Biological dose estimation for two severely exposed patients in a radiation accident in Shandong Jining, China, in 2004. Int J Radiat Biol. 2010;86:800–808. doi: 10.3109/09553002.2010.481320. [DOI] [PubMed] [Google Scholar]
- 39.Yao B, Li Y, Liu G, Guo M, Bai J, Man Q, et al. Estimation of the biological dose received by five victims of a radiation accident using three different cytogenetic tools. Mutat Res. 2013;751:66–72. doi: 10.1016/j.mrgentox.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 40.Thomas P, Fenech M. Cytokinesis-block micronucleus cytome assay in lymphocytes. In: Didenko VV, editor. DNA damage detection in situ, ex vivo, and in vivo: Methods and protocols, Methods Mol Biol. Vol. 682. 2011. pp. 217–234. [DOI] [PubMed] [Google Scholar]
- 41.Slowinski J, Mazurek U, Bierzynska-Macyszyn G, Widel M, Latocha M, Glogowska-Ligus J, et al. Cell proliferative activity estimated by histone H2B mRNA level correlates with cytogenetic damage induced by radiation in human glioblastoma cell lines. J Neurooncol. 2005;71:237–243. doi: 10.1007/s11060-004-2030-6. [DOI] [PubMed] [Google Scholar]
- 42.Wu H, Hada M, Meador J, Hu X, Rusek A, Cucinotta FA. Induction of micronuclei in human fibroblasts across the Bragg curve of energetic heavy ions. Radiat Res. 2006;166:583–589. doi: 10.1667/RR0535.1. [DOI] [PubMed] [Google Scholar]
- 43.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9:62–66. [Google Scholar]
- 44.Ridler TW, Calvard S. Picture thresholding using an iterative selection method. IEEE Trans Syst Man Cybern. 1978;8:630–632. [Google Scholar]
- 45.McNamee JP, Flegal FN, Greene HB, Marro L, Wilkins R. Validation of the cytokinesis-block micronucleus (CBMN) assay for use as a triage biological dosimetry tool. Radiat Prot Dosim. 2009;135:232–242. doi: 10.1093/rpd/ncp119. [DOI] [PubMed] [Google Scholar]
- 46.Fenech M. Important variables that influence base-line micronucleus frequency in cytokinesis-blocked lymphocytes—a biomarker for DNA damage in human populations. Mutat Res. 1998;404:155–165. doi: 10.1016/s0027-5107(98)00109-2. [DOI] [PubMed] [Google Scholar]
- 47.Kato TA, Wilson PF, Nagasaw H, Peng Y, Weil MM, Little JB, et al. Variations in radiosensitivity among individuals: a potential impact on risk assessment? Health Phys. 2009;97:470–480. doi: 10.1097/HP.0b013e3181b08eee. [DOI] [PubMed] [Google Scholar]
- 48.Kacprzak J, Kuszewski T, Lankoff A, Müller W-U, Wojcik A, Lisowska H. Individual variations in the micronucleus assay for biological dosimetry after high dose exposure. Mutat Res. 2013;756:196–200. doi: 10.1016/j.mrgentox.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 49.Di Giorgio C, De Méo MP, Laget M, Guiraud H, Botta A, Duménil G. The micronucleus assay in human lymphocytes: screening for inter-individual variability and application to biomonitoring. Carcinogenesis. 1994;15:313–317. doi: 10.1093/carcin/15.2.313. [DOI] [PubMed] [Google Scholar]
- 50.Duffaud F, Orsière T, Villlani P, Pelissier AL, Volot F, Favre E, et al. Comparison between micronucleated lymphocyte rates observed in healthy subjects and cancer patients. Mutagenesis. 1997;12:227–231. doi: 10.1093/mutage/12.4.227. [DOI] [PubMed] [Google Scholar]
- 51.Swartz H, Flood AB, Gougelet RM, Rea ME, Nicolalde RJ, Williams BB. A critical assessment of biodosimetry methods for large-scale incidents. Health Phys. 2010;98:95–108. doi: 10.1097/HP.0b013e3181b8cffd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willems P, August L, Slabbert J, Romm H, Oestreicher U, Thierens H, et al. Automated micronucleus (MN) scoring for population triage in case of large scale radiation events. Int J Radiat Biol. 2010;86:2–11. doi: 10.3109/09553000903264481. [DOI] [PubMed] [Google Scholar]
- 53.Dolphin GW. Handling of Radiation Accidents. Vienna: IAEA; 1969. Biological dosimetry with particular reference to chromosome aberration analysis. A review of methods; pp. 215–224. [Google Scholar]