Abstract
This chapter provides an overview of DNA microarrays. Microarrays are a technology in which 1000’s of nucleic acids are bound to a surface and are used to measure the relative concentration of nucleic acid sequences in a mixture via hybridization and subsequent detection of the hybridization events. We first cover the history of microarrays and the antecedent technologies that led to their development. We then discuss the methods of manufacture of microarrays and the most common biological applications. The chapter ends with a brief discussion of the limitations of microarrays and discusses how microarrays are being rapidly replaced by DNA sequencing technologies.
Keywords: microarrays, genomics, gene expression, genotyping
Overview
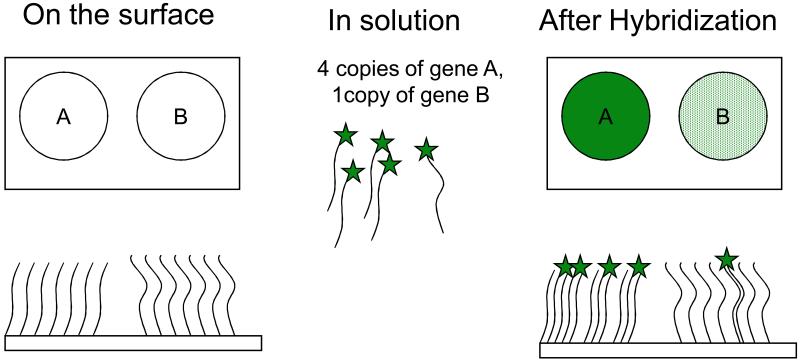
This chapter covers the use of nucleic acid arrays. Nucleic acid arrays or more simply DNA arrays are a group of technologies in which specific DNA sequences are either deposited or synthesized in a 2-D (or sometimes 3-D) array on a surface in such a way that the DNA is covalently or non-covalently attached to the surface. Figure 1 shows a simplified view of a DNA array. In typical use, a DNA array is used to probe a solution of a mixture of labeled nucleic acids and the binding (by hybridization) of these “targets” to the “probes” on the array is used to measure the relative concentrations of the nucleic acid species in solution. By generalizing to a very large number of spots of DNA, an array can be used to quantify an arbitrarily large number of different nucleic acid sequences in solution. There are other means of quantifying different nucleic acid sequences in a sample, including quantitative PCR (Units 15.7 and 15.8), “digital PCR” (Sykes et al., 1992; Vogelstein and Kinzler, 1999), and hybridization to optically tagged “probe sequences” (Geiss et al., 2008). These are not reviewed here. Also, it should be noted that the use of microarrays to measure the relative concentrations of nucleic acid sequences in solution is rapidly being supplanted by high throughput sequencing methods such as those discussed in Unit 7.8.
Figure 1.
Simplified view of a DNA array. The upper rectangles show two spots of DNA on a solid surface (sequences “A” and “B”) prior to and after hybridization. The lower rectangles show highly idealized side views of the same surfaces.
The early history of DNA arrays
One could argue that the original DNA array was created with the colony hybridization method of Grunstein and Hogness (Grunstein and Hogness, 1975). In this procedure, DNA of interest was randomly cloned into E. coli plasmids that were plated onto agar petri plates covered with nitrocellulose filters. Replica plating was used to produce additional agar plates. The colonies on the filters were lysed and their DNA’s were denatured and fixed to the filter to produce a random and unordered collection of DNA spots that represented the cloned fragments. Hybridization of a radiolabeled probe of an DNA or RNA of interest was used to rapidly screen 1000’s of colonies to identify clones containing DNA that was complimentary to the probe (Unit 6.3).
In 1979, this approach was adapted to create ordered arrays by Gergen et. al. (Gergen et al., 1979) who picked colonies into 144 well microplates. They created a mechanical 144 pin device and a jig that allowed them to replicate multiple microtiter plates on agar and produce arrays of 1728 different colonies in a 26 × 38 cm region. An additional transfer of colonies to squares of Whatman filter paper followed by a growth, lysis, denaturation and fixing of the DNA to the filter, allowed the production of DNA arrays on filters that could be re-used multiple times. During the next decade, filter based arrays and protocols similar to these were used in a variety of applications including: cloning genes of specific interest, identifying SNP’s (Miller and Barnes, 1986), cloning genes that are differentially expressed between two samples (Crampton et al., 1980) and physical mapping(Craig et al., 1990).
In the late 1980’s and early 1990’s Hans Lehrach’s group automated these processes by using robotic systems to rapidly array clones from microtiter plates onto filters(Craig et al., 1990; Lennon and Lehrach, 1991). The concomitant development of cDNA cloning in the late 1970’s and early 80’s (Auffray et al., 1980; Auffray and Rougeon, 1980a; Auffray and Rougeon, 1980b; Humphries et al., 1977) combined with international programs to fully sequence both the human genome (Barnhart, 1989; Watson and Jordan, 1989) and the human transcriptome(Aaronson et al., 1996; Dias Neto et al., 2000) led to efforts to create reference sets of cDNAs and cDNA filter arrays for human(Lennon et al., 1996) and other genomes(Bonaldo et al., 1996) By the late 1990’s and early 2000’s, sets of non-redundant cDNA’s became widely available and the complete genome sequences of some organisms allowed for sets of PRC products representing all the known open reading frames (ORFs) in small genomes (Lashkari et al., 1997; Richmond et al., 1999). These sets, combined with readily available robotics, allowed individual labs to make their own cDNA or ORF arrays that containing gene content that represented the vast majority of genes in a genome.
The birth of the modern DNA array
In the late 90’s and 2000’s, DNA array technology progressed rapidly as both new methods of production and fluorescent detection were adapted to the task. In addition, increases in our knowledge of the DNA sequences of multiple genomes provided the raw information necessary to assure that arrays could be made which fully represented the genes in a genome, all the sequence in a genome or a large fraction of the sequence variation in a genome. It should also be noted that during this time, there was a gradual transition from spotting relatively long DNA’s on arrays to producing arrays using 25-60bp oligos. The transition to oligo arrays was made possible by the increasing amounts of publicly available DNA sequence information. The use of oligos (as opposed to longer sequences) also provided an increase in specificity for the intended binding target as oligos could be designed to target regions of genes or the genome that were most dissimilar from other genes or regions. Three basic types of arrays came into play during this time frame, spotted arrays on glass, in-situ synthesized arrays and self assembled arrays (Figure 2).
Figure 2.
Three basic types of microarrays: (A) Spotted arrays on glass, (B) self assembled arrays and (C) in-situ synthesized arrays.
A. With spotted arrays, a “pen” (or multiple pens) are dipped into solutions containing the DNA of interest and physically deposited on a 1“x 3” glass microscope slide. Typically the glass slide surface is coated with something to help retain the DNA such as polylysine {DeRisi, 1997 #28191}, a silane {Call, 2001 #28277} or a chemically reactive surface {Rogers, 1999 #28278} (to which chemically reactive oligos or PCR products would be added).
B. Self assembled arrays can be created by applying a collection of beads containing a diverse set of oligos to a surface with pits the size of the beads. After the array is constructed a series of hybridizations determine which oligo is in what position on each unique array (Ferguson et al., 2000; Michael et al., 1998; Steemers et al., 2000; Walt, 2000) (Gunderson et al., 2004).
C1 and C2. In-situ synthesized arrays can be produced by inkjet oligo synthesis methods (C1) or by photolithographic methods such as used by Affymetrix (C2).
Spotted arrays
In 1996 Derisi et. al. published a method which allowed very high-density DNA arrays to be made on glass substrates(DeRisi et al., 1996). Poly-lysine coated glass microscope slides provided good binding of DNA and a robotic spotter was designed to spot multiple glass slide arrays from DNA stored in microtiter dishes. By using slotted pins (similar to fountain pens in design) a single dip of a pin in DNA solution could spot multiple slides. Spotting onto glass, allowed one to fluorescently label the sample. Fluorescent detection provided several advantages relative to the radioactive or chemilluminescent labels common to filter based arrays. First, fluorescent detection is quite sensitive and has a fairly large dynamic range. Second, fluorescent labeling is generally less expensive and less complicated than radioactive or chemilluminescent labeling. Third, fluorescent labeling allowed one to label two (or potentially more) samples in different colors and cohybridize the samples to the same array. As it was very difficult to reproducibly produce spotted arrays, comparisons of individually hybridized samples to ostensibly identical arrays would result in false differences due to array-to-array variation. However, a two-color approach in which the ratio of signals on the same array are measured is much more reproducible.
In-situ, Synthesized arrays
In 1991 Fodor et.al. published a method for light directed, spatially addressable chemical synthesis which combined photolabile protecting groups with photolithography to perform chemical synthesis on a solid substrate(Fodor et al., 1991). In this initial work, the authors demonstrated the production of arrays of 10-amino acid peptides and, separately, arrays of di-nucleotides. In 1994, Fodor et.al. at the recently formed company of Affymetrix demonstrated the ability to use this technology to generate DNA arrays consisting of 256 different octa-nucleotides (Pease et al., 1994). By 1995-1996, Affymetrix arrays were being used to detect mutations in the reverse transcriptase and protease genes of the highly polymorphic HIV-1 genome(Lipshutz et al., 1995) and to measure variation in the human mitochondrial genome(Chee et al., 1996). Eventually, Affymetrix used this technology to develop a wide catalogue of DNA arrays for use in expression analysis(Lockhart et al., 1996; Wodicka et al., 1997), genotyping (Chee et al., 1996; Hacia et al., 1996) and sequencing (G Wallraff, 1997)(see www.Affymetrix.com for the current catalog of arrays).
A major advantage of the Affymetrix technology is that because the DNA sequences are directly synthesized on the surface, only a small collection of reagents (the 4 modified nucleotides, plus a small handful of reagents necessary for the de-blocking and coupling steps) are needed to construct an arbitrarily complex array. This contrasts with the spotted array technologies in which one needed to construct or obtain all the sequences that one wished to deposit on the array in advance of array construction. However, the initial Affymetrix technology was limited in flexibility as each model of array required the construction of a unique set of photolithographic masks in order to direct the light to the array at each step of the synthesis process. In 2002, authors from Nimblegen Systems Inc., published a method in which the photo-deprotection step of Fodor et. al (Fodor et al., 1991; Lipshutz et al., 1999) is accomplished using micro-mirrors (similar to those in video computer projectors) to direct light at the pixels on the array(Nuwaysir et al., 2002). This allows for custom arrays to be manufactured in small volumes at much lower cost than by photolithographic methods using masks to direct light (which are cheaper for large volume production). One constraint with this method is that the total number of addressable pixels (e.g. unit oligos that can be synthesized) is limited to the number of addressable positions in the micro-mirror device (of order 1M).
In 1996, Blanchard et.al. proposed a method use inkjet printing technology and standard oligo synthesis chemistry to produce oligo arrays(Blanchard et al., 1996). In brief, inkjet printer heads were adapted to deliver to the four different nucleotide phosphoramidites to a glass slide that was pre-patterned to contain regions containing hydrophilic regions (with exposed hydroxyl groups) surrounded by hydrophobic regions. The hydroxylated regions provided a surface to which the phosphoramidites could couple, while the surrounding hydrophobic regions contained the droplet(s) emitted by the inkjets to defined regions. This technology was eventually commercialized by Rosetta Inpharmatics (Hughes et al., 2001) and licensed to Agilent Technologies who produces these arrays at present. The inkjet array approach shares the advantage of the Affymetrix/Nimblegen approach in that one only need to have available a small number of reagents to produce an array. In addition, similar to the Nimblegen approach, the production of a new type of array only requires that a different set of sequence information is delivered to the printer. Hence, the inkjet array technology has been particularly useful for the design of custom arrays that are produced in low volume.
Self assembled arrays
An alternative approach to the construction of arrays was created by the group of David Walt at Tufts University(Ferguson et al., 2000; Michael et al., 1998; Steemers et al., 2000; Walt, 2000) and ultimately licensed to Illumina. Their method involved synthesizing DNA on small polystryrene beads and depositing those beads on the end of a fiber optic array in which the ends of the fibers were etched to provide a well that is slightly larger than one bead. Different types of DNA would be synthesized on different beads and applying a mixture of beads to the fiber optic cable would result in a randomly assembled array. In early versions of these arrays, the beads were optically encoded with different fluorophore combinations in order to allow one to determine which oligo was in which position on the array (referred to as “decoding the array”)(Ferguson et al., 2000; Michael et al., 1998; Steemers et al., 2000; Walt, 2000). Optical decoding by fluorescent labeling limited the total number of unique beads that could be distinguished. Hence, the later and present day methods for decoding the beads involve hybridizing and detecting a number of short, fluorescently labeled oligos in a sequential series of steps(Gunderson et al., 2004). This not only allows for an extremely large number of different types of beads to be used on a single array but also functionally tests the array prior to its use in a biological assay. Later versions of the Illumina arrays used a pitted glass surface to contain the beads instead of a fiber option arrays.
The above is not intended to be a comprehensive history or survey of all DNA microarray technologies. However, it does cover the major advances in the field and the predominate methods of manufacture of arrays.
Applications of microarrays
Gene expression analysis
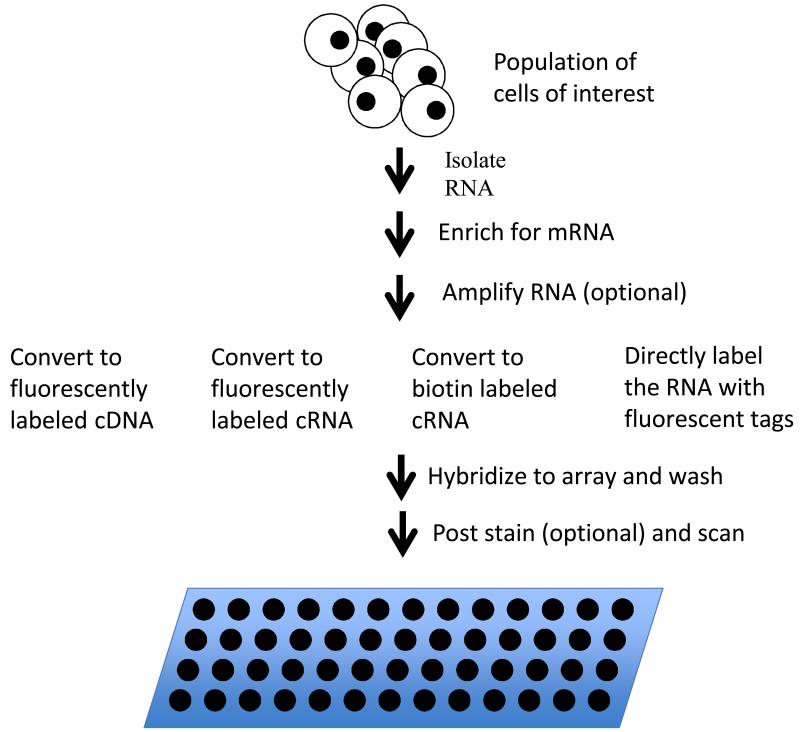
The predominate application of DNA microarrays has been to measure gene expression levels (Figure 3). In this application, RNA is extracted from the cells of interest and either, labeled directly, converted to a labeled cDNA or converted to a T7 RNA promoter tailed cDNA which is further converted to cRNA through the Eberwine amplification process (Van Gelder et al., 1990). A wide variety of methods have been developed for labeling of the cDNA or cRNA including: incorporation of fluorescently labeled nucleotides during the synthesis, incorporation of biotin labeled nucleotide which is subsequently stained fluorescently labeled streptavidin, incorporation of a modified reactive nucleotide to which a fluorescent tag is added later, and a variety of signal amplification methods (an early review of different labeling methods is provided in (Richter et al., 2002)). The two most frequently used methods are the incorporation of fluorescently labeled nucleotides in the cRNA or cDNA synthesis step or the incorporation of a biotin labeled nucleotide in the cRNA synthesis step (as is done by Affymetrix).
Figure 3.
Gene expression analysis via microarrays. RNA is isolated from the sample of interest and enriched for messenger RNA. In eucaryotes, poly-A tailed mRNA’s are typically enriched using affinity purification with oligo dT beads or columns. In procaryotes, unselected RNA is typically depleted for ribosomal sequences using bead or columns coated with sequences complementary to 16s. After message enriched RNA is in hand, it is optionally amplified and labeled by any one of a number of methods and the resulting labeled sample is hybridized to a microarray. The array is washed to remove unbound sample. If the sample was labeled with biotin, the array is post stained with fluorescently labeled streptavidin and washed again. The array is then scanned to measure fluorescence signal at each spot on the array.
The labeled cRNA or cDNA are then hybridized to the microarray, the array is washed and the signal is detected by measuring fluorescence at each spot. In the case of biotin labeled samples, the array is stained post-hybridization with fluorescently labeled streptavidin. Laser induced fluorescence is typically measured with a scanning confocal microscope. The intensity of the signal(s) on each spot is taken as a measure of the expression level of the corresponding gene. Gene expression analysis is described in more detail in Units 22.2-22.4.
Transcription factor binding analysis
Microarrays have also been used in combination with chromatin immunoprecipitation (Solomon et al., 1988) to determine the binding sites of transcription factors (Horak and Snyder, 2002; Iyer et al., 2001). In brief, transcription factors (TFs) are cross linked to DNA with formaldehyde and the DNA is fragmented. The TF(s) of interest (with the DNA to which they were boud still attached) are affinity purified using either an antibody to the TF or by tagging the transcription factor with peptide that’s amenable to affinity chromatography (for example a FLAG-, HIS-, myc or HA-tag). After purification, the DNA is released from the TF, amplified, labeled and hybridized to the array. This technique is commonly referred to as “ChIP-chip” for Chromatin Immuno-Precipitation on a “chip” or microarray.
As TF’s often bind quite a distance away from the genes that they regulate, the design of the array and size distribution of the fragment length are interrelated. E.g. the array must contain probes that will interrogate the region of DNA bound to the transcription factor. For bacteria or yeast, the intergenic regions are fairly small and the same arrays used for gene expression work can be applied to ChIP-chip. For mammalian genomes, the intergenic regions are large and the TF often bind many kbp away from the gene of interest. Hence, for mammalian genomes, oligo arrays with oligo’s spaced evenly across the entire genome are typically used for ChIP-chip experiments. Buck et. al. provide a good review(Buck and Lieb, 2004) of the considerations for the design and analysis of ChIP-chip experiments and the technique is discussed in detail in Units 21.9 and 21.13.
Genotyping
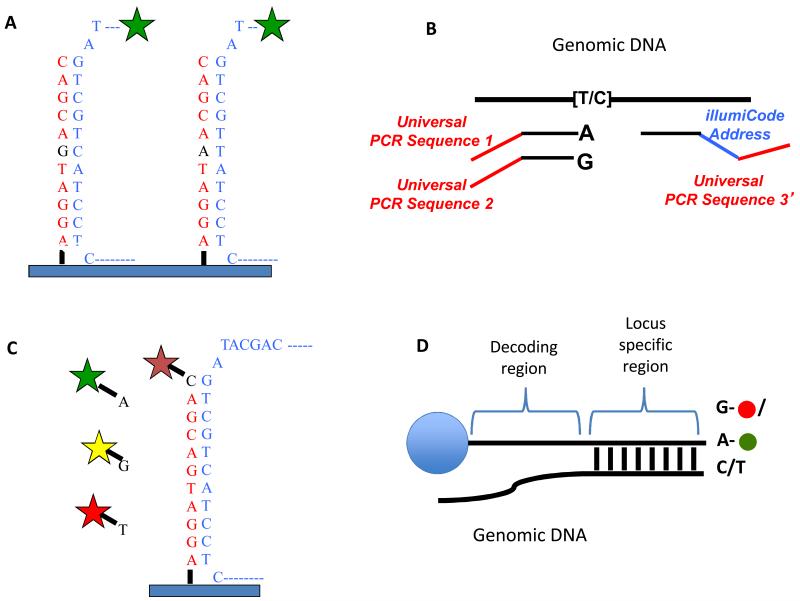
Microarrays have been widely used as single-nucleotide-polymorphism (SNP) genotyping platforms. Several alternative approaches have been used to detect SNP’s but the most commonly used are allele discrimination by hybridization as used by Affymetrix (Wang et al., 1998), allele specific extension and ligation to a “bar-code” oligo which is hybridized to a universal array (the Illumina “Golden Gate Assay”(Fan et al., 2003)) or approaches in which the arrayed DNA is extended across the SNP in a single nucleotide extension reaction (the Arrayed Primer Extension assay of Kurg et.al. (Kurg et al., 2000) or the Infinium Assay of Illumina (Gunderson et al., 2006)). Figure 4 explains the detection approaches in more detail. Allelic discrimination by hybridization suffers background due to non-specific hybridization in complex genomes. In order to reduce this background, Affymetrix developed a PCR based approach to reduce genomic complexity.
Figure 4.
SNP detections strategies for arrays. A) Allele discrimination by hybridization – Oligos that are complimentary to each allele are placed on the array and labeled genomic DNA is hybridized to the array. The variant position is placed in the center of the oligo (typically 25bp on Affymetrix arrays) as this position has the greatest affect on hybridization. Typically, multiple array positions are used for each allele to improve signal to noise. B) Illumina’s “Golden Gate Assay”- two allele specific oligos are each tailed with a different universal primer (1 and 2) and hybridized in solution to genomic DNA. A third oligo that is complementary to the same locus is tailed with a “barcode” sequence and a third universal primer (3). Polymerase is used to extend the allele specific primers across the genomic sequence and the extended products are ligated to the third oligo. PCR is performed using primers complimentary to universal sequences 1, 2 and 3. The PCR primers complimentary to the universal sequences 1 and 2 are labeled with a unique fluorophore. The barcode sequence on the third oligo allows the PCR product to be uniquely detected on an array containing oligos complimentary to the barcode sequence. The use of multiple barcodes (one for each locus of interest) allows the assay to be multiplexed to sample many loci. C) Arrayed primer extension (APEX) – In this assay, the array contains DNA oriented with the 5′ end attached to the array and the 3′ end stopping one nucleotide short of the SNP. Genomic DNA is fragmented and hybridized to the array and the oligo on the array is extended in single nucleotide dye terminator sequencing reaction. D) Illumina’s Infinium assay – This assay is similar to the APEX assay except that the oligo to be extended is on a bead and the single nucleotide that is added is labeled with a nucleotide specific hapten as opposed to a fluorophore. The haptens are then detected by staining with fluorescently labeled proteins that bind each hapten.
In brief, SNPs for their assay are selected to be between restriction sites that are <1kb apart. Genomic DNA is fragmented with a restriction enzyme, end repaired and adapters for PCR are ligated to the fragments. PCR is performed under conditions that selectively amplify products of <1kb in size. This method reduces genomic complexity by approximately 50-fold and results in a corresponding increase in signal to noise on the array(Matsuzaki et al., 2004).
Both the Affymetrix and the Illumina methods for SNP genotyping have been highly successful and are highly used. Today SNP arrays capable of detecting >1M different human SNPs are available from both vendors. Call rates (the fraction of SNPs on the array that can be reliable called) and reproducibility of SNP calls exceed 99.5%. In addition, the same arrays or variations thereof, can also be used to detect copy number variants(Bignell et al., 2004; Peiffer et al., 2006).
Data standards and data exchange
With the exception of DNA sequencing, microarrays were perhaps the earliest technology that allowed biologists to vast amounts of complex digital data. As the technology came into use, it rapidly became apparent that in order for others to be able to reproduce a given microarray experiment a detailed description of the array, the sample, the protocols and the data analysis methods needed to be available. Moreover, it also became apparent that access to the raw and processed data would allow others to perform analyses and meta analyses (on combinations of data) that the original data producers had not conceived. To address these issues of reproducible science and data exchange, members of the Microarray Gene Expression Data Society (now the Function Genomics Data Society – www.FGED.org) created the MIAME (Minimum Information About a Microarray Experiment) standards for the description of microarray experiments(Ball and Brazma, 2006; Brazma et al., 2001) and for the exchange of microarray data(Rayner et al., 2006; Spellman et al., 2002). These efforts influenced the creation public databases for microarray data (Barrett et al., 2007; Brazma et al., 2006; Brazma et al., 2003) and subsequent standards efforts in other areas (Deutsch et al., 2008; Field et al., 2008; Taylor et al., 2007).
Limitations of DNA microarrays
At their core, microarrays are simply devices to simultaneously measure the relative concentrations of many different DNA or RNA sequences. While they have been incredibly useful in a wide variety of applications, they have a number of limitations. First, arrays provide an indirect measure of relative concentration. That is the signal measured at a given position on a microarray is typically assumed to be proportional to the concentration of a presumed single species in solution that can hybridize to that location. However, due to the kinetics of hybridization, the signal level at a given location on the array is not linearly proportional to concentration of the species hybridizing to the array. At high concentrations the array will become saturated and at low concentrations, equilibrium favors no binding. Hence, the signal is linear only over a limited range of concentrations in solution. Second, especially for complex mammalian genomes, it is often difficult (if not impossible) to design arrays in which multiple related DNA/RNA sequences will not bind to the same probe on the array. A sequence on an array that was designed to detect “gene A”, may also detect “genes B, C and D” if those genes have significant sequence homology to gene A. This can particularly problematic for gene families and for genes with multiple splice variants. It should be noted that it is possible to design arrays specifically to detect splice variants either by making array probes to each exon in the genome(Gardina et al., 2006) or to exon junctions(Castle et al., 2003). However, it is difficult to design arrays that will uniquely detect every exon or gene in genomes with multiple related genes.
Finally, a DNA array can only detect sequences that the array was designed to detect. That is, if the solution being hybridized to the array contains RNA or DNA species for which there is no complimentary sequence on the array, those species will not be detected. For gene expression analysis, this typically means that genes that have not yet been annotated in a genome will not be represented on the array. In addition, non-coding RNA’s that are not yet recognized as expressed are typically not represented on an array. Moreover, for highly variable genomes such as those from bacteria, arrays are typically designed using information from the genome of a reference strain. Such arrays may be missing a large fraction of the genes present in a given isolate of the same species. For example, in the bacterial species Aggregatibacter actinomycetemcomitans, the gene content differs by as much as 20% between any two isolates (Kittichotirat et al., 2011). Hence an array designed using gene annotation from a “reference isolate” will not contain many of the genes found in other isolates.
The Future of DNA arrays
Given the limitations of arrays mentioned above, it would be far preferable to have an unbiased method to directly measure all the DNA or RNA species present in a particular sample. The advent of next generation sequencing technologies combined with the rapid decrease in the cost of sequencing (http://www.genome.gov/sequencingcosts/) has now made sequencing cost competitive with microarrays for all assays with the possible exception of genotyping. When the cost is similar, sequencing has many advantages relative to microarrays. Sequencing is a direct measurement of which nucleic acids are present in solution. One need only count the number of a given type of sequences present to determine it’s abundance. Counting sequences is linear with concentration and the signal to noise one can obtain by sequencing is only limited by the number of reads used for each sample.
Sequencing is a relatively unbiased approach to measuring which nucleic acids are present in solution. While sample preparation or different enzymes may bias sequencing counts, unlike DNA arrays, sequencing is not dependent on prior knowledge of which nucleic acids may be present. Sequencing is also able to independently detect closely related gene sequences, novel splice forms or RNA editing that may be missed due to cross hybridization on DNA microarrays. As a result of these advantages and the decreasing cost of sequencing, DNA arrays are being rapidly replaced by sequencing for nearly every assay that has been previously performed on microarrays (see for example (Wold and Myers, 2008)). As the cost of sequencing is currently dropping by a factor of two every five months, it’s likely that DNA arrays will be fully replaced by sequencing methods within the next 5-10 years.
Acknowledgements
Dr. Bumgarner is partially supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1 RR025014. The content is solely the responsibility of the author and does not necessarily represent the official views of the NIH
Literature Cited
- Aaronson JS, Eckman B, Blevins RA, Borkowski JA, Myerson J, Imran S, Elliston KO. Toward the development of a gene index to the human genome: an assessment of the nature of high-throughput EST sequence data. Genome research. 1996;6:829–845. doi: 10.1101/gr.6.9.829. [DOI] [PubMed] [Google Scholar]
- Auffray C, Nageotte R, Chambraud B, Rougeon F. Mouse immunoglobulin genes: a bacterial plasmid containing the entire coding sequence for a pre-gamma 2a heavy chain. Nucleic acids research. 1980;8:1231–1241. doi: 10.1093/nar/8.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C, Rougeon F. Nucleotide sequence of a cloned cDNA corresponding to secreted mu chain of mouse immunoglobulin. Gene. 1980a;12:77–86. doi: 10.1016/0378-1119(80)90017-7. [DOI] [PubMed] [Google Scholar]
- Auffray C, Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980b;107:303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Ball CA, Brazma A. MGED standards: work in progress. OMICS. 2006;10:138–144. doi: 10.1089/omi.2006.10.138. [DOI] [PubMed] [Google Scholar]
- Barnhart BJ, The Department of Energy (DOE) Human Genome Initiative Genomics. 1989;5:657–660. doi: 10.1016/0888-7543(89)90041-4. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic Acids Res. 2007;35:D760–765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Huang J, Greshock J, Watt S, Butler A, West S, Grigorova M, Jones KW, Wei W, Stratton MR, Futreal PA, Weber B, Shapero MH, Wooster R. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome research. 2004;14:287–295. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard AP, Kaiser RJ, Hood LE. High-density oligonucleotide arrays. Biosensors and Bioelectronics. 1996;11:687–690. [Google Scholar]
- Bonaldo MF, Lennon G, Soares MB. Normalization and subtraction: two approaches to facilitate gene discovery. Genome research. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature genetics. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Brazma A, Kapushesky M, Parkinson H, Sarkans U, Shojatalab M. Data storage and analysis in ArrayExpress. Methods Enzymol. 2006;411:370–386. doi: 10.1016/S0076-6879(06)11020-4. [DOI] [PubMed] [Google Scholar]
- Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M, Kemmeren P, Lara GG, Oezcimen A, Rocca-Serra P, Sansone SA. ArrayExpress--a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–360. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Castle J, Garrett-Engele P, Armour CD, Duenwald SJ, Loerch PM, Meyer MR, Schadt EE, Stoughton R, Parrish ML, Shoemaker DD, Johnson JM. Optimization of oligonucleotide arrays and RNA amplification protocols for analysis of transcript structure and alternative splicing. Genome Biology. 2003;4:R66. doi: 10.1186/gb-2003-4-10-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, WInkler J, Lockhart DJ, Morris MS, Fodor SPA. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- Craig AG, Nizetic D, Hoheisel JD, Zehetner G, Lehrach H. Ordering of cosmid clones covering the herpes simplex virus type I (HSV-I) genome: a test case for fingerprinting by hybridisation. Nucleic acids research. 1990;18:2653–2660. doi: 10.1093/nar/18.9.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton J, Humphries S, Woods D, Williamson R. The isolation of cloned cDNA sequences which are differentially expressed in human lymphocytes and fibroblasts. Nucleic acids research. 1980;8:6007–6017. doi: 10.1093/nar/8.24.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nature genetics. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- Deutsch EW, Ball CA, Berman JJ, Bova GS, Brazma A, Bumgarner RE, Campbell D, Causton HC, Christiansen JH, Daian F, Dauga D, Davidson DR, Gimenez G, Goo YA, Grimmond S, Henrich T, Herrmann BG, Johnson MH, Korb M, Mills JC, Oudes AJ, Parkinson HE, Pascal LE, Pollet N, Quackenbush J, Ramialison M, Ringwald M, Salgado D, Sansone SA, Sherlock G, Stoeckert CJ, Jr., Swedlow J, Taylor RC, Walashek L, Warford A, Wilkinson DG, Zhou Y, Zon LI, Liu AY, True LD. Minimum information specification for in situ hybridization and immunohistochemistry experiments (MISFISHIE) Nat Biotechnol. 2008;26:305–312. doi: 10.1038/nbt1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias Neto E, Correa RG, Verjovski-Almeida S, Briones MR, Nagai MA, da Silva W, Jr., Zago MA, Bordin S, Costa FF, Goldman GH, Carvalho AF, Matsukuma A, Baia GS, Simpson DH, Brunstein A, de Oliveira PS, Bucher P, Jongeneel CV, O’Hare MJ, Soares F, Brentani RR, Reis LF, de Souza SJ, Simpson AJ. Shotgun sequencing of the human transcriptome with ORF expressed sequence tags. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3491–3496. doi: 10.1073/pnas.97.7.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, Galver L, Hunt S, McBride C, Bibikova M, Rubano T, Chen J, Wickham E, Doucet D, Chang W, Campbell D, Zhang B, Kruglyak S, Bentley D, Haas J, Rigault P, Zhou L, Stuelpnagel J, Chee MS. Highly parallel SNP genotyping. Cold Spring Harbor symposia on quantitative biology. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- Ferguson JA, Steemers FJ, Walt DR. High-density fiber-optic DNA random microsphere array. Analytical chemistry. 2000;72:5618–5624. doi: 10.1021/ac0008284. [DOI] [PubMed] [Google Scholar]
- Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, Ashburner M, Axelrod N, Baldauf S, Ballard S, Boore J, Cochrane G, Cole J, Dawyndt P, De Vos P, DePamphilis C, Edwards R, Faruque N, Feldman R, Gilbert J, Gilna P, Glockner FO, Goldstein P, Guralnick R, Haft D, Hancock D, Hermjakob H, Hertz-Fowler C, Hugenholtz P, Joint I, Kagan L, Kane M, Kennedy J, Kowalchuk G, Kottmann R, Kolker E, Kravitz S, Kyrpides N, Leebens-Mack J, Lewis SE, Li K, Lister AL, Lord P, Maltsev N, Markowitz V, Martiny J, Methe B, Mizrachi I, Moxon R, Nelson K, Parkhill J, Proctor L, White O, Sansone SA, Spiers A, Stevens R, Swift P, Taylor C, Tateno Y, Tett A, Turner S, Ussery D, Vaughan B, Ward N, Whetzel T, San Gil I, Wilson G, Wipat A. The minimum information about a genome sequence (MIGS) specification. Nature biotechnology. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor SPA, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Wallraff G, brock P, Dipietro R, Nguyen T, Huynh T, Hinsberg W, McGall G. DNA sequencing on a chip. Chemtech. 1997:22–32. J.L. [Google Scholar]
- Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, Schweitzer A, Awad T, Sugnet C, Dee S, Davies C, Williams A, Turpaz Y. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- Gergen JP, Stern RH, Wensink PC. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic acids research. 1979;7:2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M, Hogness DS. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson KL, Kruglyak S, Graige MS, Garcia F, Kermani BG, Zhao C, Che D, Dickinson T, Wickham E, Bierle J, Doucet D, Milewski M, Yang R, Siegmund C, Haas J, Zhou L, Oliphant A, Fan JB, Barnard S, Chee MS. Decoding randomly ordered DNA arrays. Genome research. 2004;14:870–877. doi: 10.1101/gr.2255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson KL, Steemers FJ, Ren H, Ng P, Zhou L, Tsan C, Chang W, Bullis D, Musmacker J, King C, Lebruska LL, Barker D, Oliphant A, Kuhn KM, Shen R. Whole-genome genotyping. Methods in enzymology. 2006;410:359–376. doi: 10.1016/S0076-6879(06)10017-8. [DOI] [PubMed] [Google Scholar]
- Hacia JG, Brody LC, Chee MS, Fodor SP, Collins FS. Detection of heterozygous mutations in BRCA1 using high density oligonucleotide arrays and two-colour fluorescence analysis. Nature genetics. 1996;14:441–447. doi: 10.1038/ng1296-441. [DOI] [PubMed] [Google Scholar]
- Horak CE, Snyder M. ChIP-chip: a genomic approach for identifying transcription factor binding sites. Methods in enzymology. 2002;350:469–483. doi: 10.1016/s0076-6879(02)50979-4. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, Kobayashi S, Davis C, Dai H, He YD, Stephaniants SB, Cavet G, Walker WL, West A, Coffey E, Shoemaker DD, Stoughton R, Blanchard AP, Friend SH, Linsley PS. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nature biotechnology. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- Humphries P, Cochet M, Krust A, Gerlinger P, Kourilsky P, Chambon P. Molecular cloning of extensive sequences of the in vitro synthesized chicken ovalbumin structural gene. Nucleic acids research. 1977;4:2389–2406. doi: 10.1093/nar/4.7.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Kittichotirat W, Bumgarner RE, Asikainen S, Chen C. Identification of the pangenome and its components in 14 distinct Aggregatibacter actinomycetemcomitans strains by comparative genomic analysis. PLoS ONE. 2011;6:e22420. doi: 10.1371/journal.pone.0022420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurg A, Tonisson N, Georgiou I, Shumaker J, Tollett J, Metspalu A. Arrayed primer extension: solid-phase four-color DNA resequencing and mutation detection technology. Genet Test. 2000;4:1–7. doi: 10.1089/109065700316408. [DOI] [PubMed] [Google Scholar]
- Lashkari DA, DeRisi JL, McCusker JH, Namath AF, Gentile C, Hwang SY, Brown PO, Davis RW. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- Lennon GG, Lehrach H. Hybridization analyses of arrayed cDNA libraries. Trends in genetics: TIG. 1991;7:314–317. doi: 10.1016/0168-9525(91)90420-u. [DOI] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SPA, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nature Genetics. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Lipshutz RJ, Morris D, Chee M, Hubbell E, Kozal MJ, Shah N, Shen N, Yang R, Fodor SP. Using oligonucleotide probe arrays to access genetic diversity. Biotechniques. 1995;19:442–427. [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density iligonucleotide arrays. Nature Biotechnology. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Loi H, Dong S, Tsai YY, Fang J, Law J, Di X, Liu WM, Yang G, Liu G, Huang J, Kennedy GC, Ryder TB, Marcus GA, Walsh PS, Shriver MD, Puck JM, Jones KW, Mei R. Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome research. 2004;14:414–425. doi: 10.1101/gr.2014904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael KL, Taylor LC, Schultz SL, Walt DR. Randomly ordered addressable high-density optical sensor arrays. Analytical chemistry. 1998;70:1242–1248. doi: 10.1021/ac971343r. [DOI] [PubMed] [Google Scholar]
- Miller JK, Barnes WM. Colony probing as an alternative to standard sequencing as a means of direct analysis of chromosomal DNA to determine the spectrum of single-base changes in regions of known sequence. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:1026–1030. doi: 10.1073/pnas.83.4.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwaysir EF, Huang W, Albert TJ, Singh J, Nuwaysir K, Pitas A, Richmond T, Gorski T, Berg JP, Ballin J, McCormick M, Norton J, Pollock T, Sumwalt T, Butcher L, Porter D, Molla M, Hall C, Blattner F, Sussman MR, Wallace RL, Cerrina F, Green RD. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome research. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SPA. Light generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl. Acad. Sci., USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, Haden K, Li J, Shaw CA, Belmont J, Cheung SW, Shen RM, Barker DL, Gunderson KL. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome research. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner TF, Rocca-Serra P, Spellman PT, Causton HC, Farne A, Holloway E, Irizarry RA, Liu J, Maier DS, Miller M, Petersen K, Quackenbush J, Sherlock G, Stoeckert CJ, Jr., White J, Whetzel PL, Wymore F, Parkinson H, Sarkans U, Ball CA, Brazma A. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond CS, Glasner JD, Mau R, Jin H, Blattner FR. Genome-wide expression profiling in Escherichia coli K-12. Nucleic acids research. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A, Schwager C, Hentze S, Ansorge W, Hentze MW, Muckenthaler M. Comparison of fluorescent tag DNA labeling methods used for expression analysis by DNA microarrays. BioTechniques. 2002;33:620–628. 630. doi: 10.2144/02333rr05. [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Miller M, Stewart J, Troup C, Sarkans U, Chervitz S, Bernhart D, Sherlock G, Ball C, Lepage M, Swiatek M, Marks WL, Goncalves J, Markel S, Iordan D, Shojatalab M, Pizarro A, White J, Hubley R, Deutsch E, Senger M, Aronow BJ, Robinson A, Bassett D, Stoeckert CJ, Jr., Brazma A. Design and implementation of microarray gene expression markup language (MAGE-ML) Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0046. RESEARCH0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steemers FJ, Ferguson JA, Walt DR. Screening unlabeled DNA targets with randomly ordered fiber-optic gene arrays. Nature biotechnology. 2000;18:91–94. doi: 10.1038/72006. [DOI] [PubMed] [Google Scholar]
- Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- Taylor CF, Paton NW, Lilley KS, Binz PA, Julian RK, Jr., Jones AR, Zhu W, Apweiler R, Aebersold R, Deutsch EW, Dunn MJ, Heck AJ, Leitner A, Macht M, Mann M, Martens L, Neubert TA, Patterson SD, Ping P, Seymour SL, Souda P, Tsugita A, Vandekerckhove J, Vondriska TM, Whitelegge JP, Wilkins MR, Xenarios I, Yates JR, 3rd, Hermjakob H. The minimum information about a proteomics experiment (MIAPE) Nature biotechnology. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Digital PCR. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walt DR. Techview: molecular biology. Bead-based fiber-optic arrays. Science. 2000;287:451–452. doi: 10.1126/science.287.5452.451. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris MS, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson TJ, Lipshutz R, Chee M, Lander ES. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Watson JD, Jordan E. The Human Genome Program at the National Institutes of Health. Genomics. 1989;5:654–656. doi: 10.1016/0888-7543(89)90040-2. [DOI] [PubMed] [Google Scholar]
- Wodicka L, Dong H, Mittmann M, Ho M-H, Lockhart DJ. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nature Biotechnology. 1997;15:1359. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- Wold B, Myers RM. Sequence census methods for functional genomics. Nature methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]