Summary
NusG/Spt5 is a transcription elongation factor that assists in DNA-templated RNA synthesis by cellular RNA polymerases (RNAP). The modular domain composition of NusG/Spt5 and the way it binds to RNAP are conserved in all three domains of life. NusG/Spt5 closes RNAP around the DNA binding channel, thereby increasing transcription processivity. Recruitment of additional factors to elongating RNAP may be another conserved function of this ubiquitous protein. Eukaryotic Spt5 couples RNA processing and chromatin modification to transcription elongation, whereas bacterial NusG participates in a wide variety of processes, including RNAP pausing and Rho-dependent termination. Elongating RNAP forms a transcriptional bubble in which ~12 bp of the two DNA strands are locally separated. Within this transcription bubble the growing 3’-end of nascent RNA forms an 8-9 bp long hybrid with the template DNA strand. Because of their location in the transcriptional bubble, NusG and its paralog RfaH recognize specific sequences in the nontemplate DNA strand and regulate transcription elongation in response to these signals.
Five core subunits of RNA polymerases (RNAP) and the only ubiquitous transcription elongation factor, NusG/Spt5, are conserved in all three domains of life. Bacterial NusG and archaeal Spt5 proteins consist of an N-terminal domain (NGN) and a C-terminal Kyprides– Onzonis–Woese domain (KOW) [1,2]. These two domains are separated by a flexible linker. Eukaryotic Spt5 contains several copies of the KOW domain and additional N- and C-terminal sequences that are absent in prokaryotic homologues [3]. Archaeal and eukaryotic Spt5 forms a heterodimeric complex with a small zinc-binding protein, Spt4 (also known as RpoE” in archaea), through its NGN domain [4]. Bacteria lack a Spt4-like protein. NusG/Spt5 binds to RNAP through its NGN domain, whereas the β barrel KOW domain(s) recruit additional regulatory factors to RNAP (Fig. 1) [5]. Multiple copies of the KOW domain in eukaryotic Spt5 may allow the recruitment of a larger number of transcription factors.
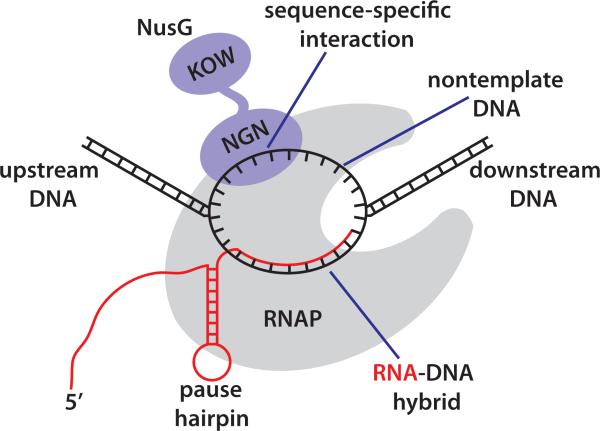
Fig. 1. Scheme of NusG interaction with elongating RNAP.
RNAP (gray) separates the template and nontemplate DNA strands in the transcription bubble. The template strand forms an RNA-DNA duplex with the nascent transcript (red) near the active site of RNAP. Binding of the NGN domain of NusG (purple) to RNAP increases transcription processivity. The NGN domain also recognizes sequence-specific signals in the displaced nontemplate DNA strand, leading to pause stabilization. The pause hairpin further increases the pause duration. The KOW domain recruits additional transcription factors to RNAP.
NusG and the sigma subunit, a specificity factor that participates in transcription initiation at promoters, compete for RNAP binding [6]. NusG is recruited to elongating E. coli RNAP in a stochastic fashion following sigma factor release and the ratio of NusG/RNAP is greater in the distal portions of some long transcription units [7]. NusG participates in transcription termination through interaction of its C-terminal KOW domain with the termination factor Rho [8,9]. Together with Rho, E. coli NusG inhibits expression of horizontally acquired AT-rich operons [10] and antisense transcripts [11•]. Some specific sequences in nascent transcripts of ribosomal RNA operons and bacteriophage lambda lytic operons direct formation of multiprotein complexes that contain NusG. Such protein-RNA complexes modify RNAP to a fast-transcribing termination-resistant state [12,13]. Paradoxically, bacteriophage HK022 orchestrates formation of a NusG-containing protein complex of similar composition that causes transcription termination at specific sequences [14]. Interestingly, another component of these termination and antitermination complexes is NusE, which is identical to ribosomal protein S10. NusG retains the ability to bind S10 as an integral part of the small ribosomal subunit. This interaction may recruit a ribosome to the nascent mRNA, thereby coupling transcription and translation, a hallmark of bacterial gene expression [8]. Although Rho binds NusG with much higher affinity, competition between S10 and Rho for NusG is apparently sufficient to protect translated transcripts from Rho-mediated termination. Transcribing RNAP is prone to random or programmed pausing that may be followed by backward sliding along DNA and RNA (backtracking) [15,16,17]. Translation positively affects transcription by reducing pausing and backtracking of RNAP [18]. The eukaryotic Spt4-Spt5 complex appears to couple RNA processing and chromatin modification to transcription elongation [19].
Rho is essential for E. coli survival [20], while participation in Rho-dependent silencing of the toxic kil gene from an integrated rac prophage is the only essential function of NusG in this organism. Although deletion of the kil gene from the chromosome makes NusG dispensable, the nusG knockout strain exhibits a growth defect [10]. Both NusG and Rho are dispensable in B. subtilis [21]. Evidently, coupling of transcription and translation and facilitation of Rho-dependent termination are not critical functions of NusG.
E. coli and other proteobacteria contain a NusG paralog called RfaH. In contrast to NusG, RfaH is an operon-specific antitermination factor. RfaH binds to RNAP transcribing horizontally transferred genes that contain the 12 nt-long sequence called ops (operon polarity suppressor) [22]. Although amino acid sequences of the C-terminal domains of RfaH and NusG are similar, the C-terminal domain of RfaH forms a different α helix structure that lacks flexibility and blocks the NGN domain from interaction with RNAP. However, interaction of the RfaH NGN domain with the ops sequence releases the C-terminal domain, which then completely refolds into a β barrel identical to that of the NusG KOW domain [23••]. The refolded KOW domain of RfaH can interact with ribosomal protein S10 but not with Rho. Thus, RfaH-mediated coupling of transcription and translation and reduced Rho-dependent termination leads to higher expression of otherwise poorly expressed ops operons [23••]. No other specialized NusG/Spt5 paralog has been identified in other organisms in which the single protein functions as a general transcription elongation factor and may fulfill some sequence-specific responses.
It is apparent that NusG/Spt5 can function as a positive or negative transcription elongation factor. In Drosophila, the Spt4-Spt5 complex, also known as DSIF, participates in widespread promoter-proximal pausing of RNA polymerase II as a component of a multiprotein complex that includes the negative elongation factor NELF. Although such interacting partners may obscure the direct effect of NusG/Spt5, the large Drosophila Spt5 protein was shown to directly contact the nascent transcript [24]. The smaller bacterial NusG and archaeal Spt4-Spt5 proteins may be capable of contacting nascent transcripts that have reached 25-30 nucleotides in length such that they have emerged well beyond the RNA exit channel [25].
NusG accelerates transcription by suppressing RNAP pausing, and this activity is viewed as a general mechanism of increasing processivity of the enzyme [26]. Archaeal Spt4-Spt5 also stimulates transcription processivity by binding to the clamp domain of RNAP [27]. The archaeal Spt4-Spt5 complex with RNAP revealed that binding of Spt4-Spt5 on the RNAP clamp domain would completely encircle the DNA binding channel of RNAP, thereby providing an explanation for how NusG/Spt5 is able to enhance transcription processivity [28]. Similar conclusions were reached in another structural study with archaeal Spt4-Spt5 bound to the isolated clamp domain [25]. In stark contrast to its general antipausing activity, NusG dramatically stimulates pausing at two pause sites in the untranslated leader of the B. subtilis trp operon [29••]. These two regulatory pause sites participate in transcription attenuation and translational control mechanisms, respectively [30,31]. Both pause sites have similar upstream sequences that dictate the precise position of pausing [32]. Transcription elongation complexes reconstituted in vitro with nucleic acid scaffolds revealed that NusG makes sequence-specific contacts with a T-rich sequence in the nontemplate DNA strand within the paused transcription bubble (A.V.Y. and P.B., unpublished results) (Fig. 1). This finding is consistent with a cryoelectron microscopy structure of archaeal Spt4-Spt5 bound to RNAP, which placed the Spt5 NGN domain in close proximity to the nontemplate DNA in the transcription bubble [28]. This pattern of sequence recognition is reminiscent of RfaH-stimulated pausing at ops sites in E. coli, which results in suppression of downstream pause signals [33]. However, it is not known whether B. subtilis RNAP becomes qualitatively modified once it escapes from the long-lived trp leader pause sites.
NusG is thought to suppress pausing by stabilizing the closed conformation of the RNAP clamp domain [25,28,34•]. E. coli NusG interacts with the clamp helices of the β' subunit of RNAP. Since NusG from B. subtilis and E. coli compete for RNAP binding, it appears that both proteins interact with RNAP similarly [29••]. Therefore, stabilizing the closed conformation of the clamp domain of RNAP does not explain the pause-enhancing activity of B. subtilis NusG. We envision a mechanism in which B. subtilis NusG binds simultaneously to RNAP and the nontemplate DNA strand of the paused transcription bubble (Fig. 1). As these two components must move with respect to one another for elongation to resume, simultaneous interaction of NusG with both components would inhibit elongation, leading to a long-lived pause. This model is reminiscent of a translocation barrier model as a step towards entrance into a pause state [35•]. Interestingly, archaeal Spt5 and the Spt4-Spt5 complex also compete with NusG for B. subtilis RNAP binding (A.V.Y. and P. B., unpublished results), further highlighting the evolutionary conservation of NusG/Spt5 binding to RNAP. Perhaps NusG/Spt5 proteins from all organisms stimulate pausing at certain sites by interacting with specific sequences found within the nontemplate DNA strand of the transcription bubble (Fig. 1).
The NGN domains of E. coli NusG and RfaH are sufficient to fulfill the antipausing effect of the full-length proteins [5,36]. Similarly, the NGN domain of B. subtilis NusG is capable of stimulating pausing (Fig. 1). Our preliminary studies indicate that the amino acid sequences of two short regions within the NGN domain are primarily responsible for specific recognition of the trp pause signals by B. subtilis NusG (A.V.Y. and P.B., unpublished results). The finding that these amino acid sequences are not conserved in E. coli NusG may explain why the E. coli protein is not capable of stimulating pausing at the B. subtilis trp pause sites.
The structure of RNAP is under strong evolutionary pressure to maintain different conformations that participate in transcription initiation and elongation, as well as for proper transitions between these conformations. Transcription elongation factors like NusG recognize and bind only elongating RNAP. Evidently NusG, as a part of the elongation complex, is readily distinguishable from free NusG making it well suited to conduct signals and execute regulation specific for transcription elongation. Not surprisingly, many additional elongation and termination factors are recruited to RNAP through NusG/Spt5, which may obscure or override the direct effect of NusG/Spt5 on transcription.
Recently published results provided indirect evidence of NusG participation in genome stability. RNAP backtracking-mediated DNA double-strand breaks can be prevented by translation of nascent RNA, Rho-dependent termination of untranslated transcripts, and increased processivity of transcription [37]. NusG stimulates all three of these activities. Another conserved role of NusG/Spt5 in maintaining genome stability may be protection of the nontemplate DNA strand in the transcription bubble (Fig. 1). Crystallographic studies of transcription elongation complexes did not resolve the position of the nontemplate DNA strand in the transcription bubble, indicating that this DNA segment is highly flexible. However, biophysical and computational studies resulted in a structural model containing all nucleic acid regions within the elongation complex [38]. NusG binds near the upstream end of the transcription bubble ([29••] and A.V.Y. and P.B., unpublished results) and the upstream half of the nontemplate DNA is normally exposed and sensitive to digestion by a single strand-specific nuclease [39]. Perhaps NusG/Spt5 protects this DNA from undesirable activation of a variety of DNA repair systems that are stimulated by single-stranded DNA. Therefore, NusG/Spt5 may partially release the transcription-associated stress on genome stability. In this case, one would expect that NusG becomes especially important under genotoxic conditions.
Highlights.
NusG/Spt5 is the only universally conserved transcription elongation factor.
NusG/Spt5 recruits other elongation and termination factors to RNAP.
NusG increases the processivity of transcription by suppressing RNAP pausing.
Bacterial NusG recognizes specific sequences in the nontemplate DNA.
Bacterial NusG stabilizes certain paused transcription elongation complexes.
Acknowledgements
The authors would like to thank Katsuhiko Murakami for critical reading of the manuscript. This work was supported by grant GM098399 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.Kyrpides NC, Woese CR, Ouzounis CA. KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem Sci. 1996;21:425–426. doi: 10.1016/s0968-0004(96)30036-4. [DOI] [PubMed] [Google Scholar]
- 2.Steiner T, Kaiser JT, Marinkovic S, Huber R, Wahl MC. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 2002;21:4641–4653. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo M, Xu F, Yamada J, Egelhofer T, Gao Y, Hartzog GA, Teng M, Niu L. Core structure of the yeast spt4-spt5 complex: a conserved module for regulation of transcription elongation. Structure. 2008;16:1649–1658. doi: 10.1016/j.str.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou H, Liu Q, Gao Y, Teng M, Niu L. Crystal structure of NusG N-terminal (NGN) domain from Methanocaldococcus jannaschii and its interaction with rpoE”. Proteins. 2009;76:787–793. doi: 10.1002/prot.22465. [DOI] [PubMed] [Google Scholar]
- 5.Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R. Two structurally independent domains of E coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol. 2009;391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sevostyanova A, Artsimovitch I. Functional analysis of Thermus thermophilus transcription factor NusG. Nucleic Acids Res. 2010;38:7432–7445. doi: 10.1093/nar/gkq623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, Rosch P. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 9.Chalissery J, Muteeb G, Kalarickal NC, Mohan S, Jisha V, Sen R. Interaction surface of the transcription terminator Rho required to form a complex with the C-terminal domain of the antiterminator NusG. J Mol Biol. 2011;405:49–64. doi: 10.1016/j.jmb.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 10.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26:2621–2633. doi: 10.1101/gad.196741.112. [This work revealed that a major function of Rho and NusG in E. coli was suppression of ubiquitous antisense transcription genome-wide.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zellars M, Squires CL. Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol Microbiol. 1999;32:1296–1304. doi: 10.1046/j.1365-2958.1999.01442.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y, Filter JJ, Court DL, Gottesman ME, Friedman DI. Requirement for NusG for transcription antitermination in vivo by the lambda N protein. J Bacteriol. 2002;184:3416–3418.. doi: 10.1128/JB.184.12.3416-3418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burova E, Hung SC, Chen J, Court DL, Zhou JG, Mogilnitskiy G, Gottesman ME. Escherichia coli nusG mutations that block transcription termination by coliphage HK022 Nun protein. Mol Microbiol. 1999;31:1783–1793. doi: 10.1046/j.1365-2958.1999.01315.x. [DOI] [PubMed] [Google Scholar]
- 15.Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 17.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartzog GA, Fu J. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta. 2013;1829:105–115. doi: 10.1016/j.bbagrm.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das A, Court D, Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci USA. 1976;73:1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingham CJ, Dennis J, Furneaux PA. Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol Microbiol. 1999;31:651–663. doi: 10.1046/j.1365-2958.1999.01205.x. [DOI] [PubMed] [Google Scholar]
- 22.Belogurov GA, Mooney RA, Svetlov V, Landick R, Artsimovitch I. Functional specialization of transcription elongation factors. EMBO J. 2009;28:112–122. doi: 10.1038/emboj.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Burmann BM, Knauer SH, Sevostyanova A, Schweimer K, Mooney RA, Landick R, Artsimovitch I, Rösch P. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. doi: 10.1016/j.cell.2012.05.042. [The released C-terminal domain of RfaH completely refolded from an a helix to a P barrel structure that was virtually identical to the KOW domain of NusG. The refolded KOW domain formed a complex with ribosomal protein S10.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Missra A, Gilmour DS. Interactions between DSIF (DR B sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci USA. 2010;107:11301–11306. doi: 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herbert KM, Zhou J, Mooney RA, Porta AL, Landick R, Block SM. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 2010;399:17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirtreiter A, Damsma GE, Cheung AC, Klose D, Grohmann D, Vojnic E, Martin AC, Cramer P, Werner F. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010;38:4040–4051. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci USA. 2011;108:546–550. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Yakhnin AV, Yakhnin H, Babitzke P. Function of the Bacillus subtilis transcription elongation factor NusG in hairpin-dependent RNA polymerase pausing in the trp leader. Proc Natl Acad Sci USA. 2008;105:16131–16136. doi: 10.1073/pnas.0808842105. [B. subtilis NusG was shown to greatly stimulate RNAP pausing and protect the nontemplate DNA strand within the paused transcription bubble. NusG-stimulated pausing was also shown to reduce expression of the trp operon.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yakhnin AV, Babitzke P. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism in vitro. Proc Natl Acad Sci USA. 2002;99:11067–11072. doi: 10.1073/pnas.162373299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yakhnin AV, Yakhnin H, Babitzke P. RNA polymerase pausing regulates translation initiation by providing additional time for TRAP-RNA interaction. Mol Cell. 2006;24:547–557. doi: 10.1016/j.molcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 32.Yakhnin AV, Babitzke P. Mechanism of NusG-stimulated pausing, hairpin-dependent pause site selection and intrinsic termination at overlapping pause and termination sites in the Bacillus subtilis trp leader. Mol Microbiol. 2010;76:690–705. doi: 10.1111/j.1365-2958.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 34•.Weixlbaumer A, Leon K, Landick R, Darst SA. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–441. doi: 10.1016/j.cell.2012.12.020. [Crystal structures of Thermus RNAP elongation complexes paused at the E. coli his pause site revealed an open-clamp RNAP conformation with a sterically blocked active site.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Imashimizu M, Kireeva ML, Lubkowska L, Gotte D, Parks AR, Strathern JN, Kashlev M. Intrinsic translocation barrier as an initial step in pausing by RNA polymerase II. J Mol Biol. 2013;425:697–712. doi: 10.1016/j.jmb.2012.12.002. [A three-step mechanism for pausing was proposed. A prolonged dwell time of RNA polymerase II in the pre-translocated state increases the likelihood of misalignment of the RNA 3′ end in the active center, resulting in RNA polymerase backtracking.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrecka J, Treutlein B, Arcusa MA, Muschielok A, Lewis R, Cheung AC, Cramer P, Michaelis J. Nano positioning system reveals the course of upstream and nontemplate DNA within the RNA polymerase II elongation complex. Nucleic Acids Res. 2009;37:5803–5809. doi: 10.1093/nar/gkp601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Landick R. Nuclease cleavage of the upstream half of the nontemplate strand DNA in an Escherichia coli transcription elongation complex causes upstream translocation and transcriptional arrest. J Biol Chem. 1997;272:5989–5994. doi: 10.1074/jbc.272.9.5989. [DOI] [PubMed] [Google Scholar]