Abstract
The mir-34 family was originally cloned and characterized in 2007 as a p53 target gene. Almost immediately it became clear that its major role is as a master regulator of tumor suppression. Indeed, when overexpressed, it directly and indirectly represses several oncogenes, resulting in an increase of cancer cell death (including cancer stem cells), and in an inhibition of metastasis. Moreover, its expression is deregulated in several human cancers. In 2013, a miR-34 mimic has become the first microRNA to reach phase 1 clinical trials. Here we review the miR-34 family and their role in tumor biology, and discuss the potential therapeutic applications of miR-34a mimic.
Keywords: microRNA, miR-34 family, cancer, therapy
INTRODUCTION
The battle against cancer has just recruited a new potential weapon. Indeed, the first microRNA (miRNA) has now reached phase I studies (http://clinicaltrials.gov/ct2/show/NCT01829971). In April 2013, a study was started to evaluate the safety of MRX34 in patients with unresectable primary liver cancer and advanced or metastatic cancer with liver involvement. The “drug” is given intravenously as a single agent, twice per week for three weeks and then one week off.
miRNAs form one family of small non-coding regulatory RNAs [1]. Several studies have implicated miRNAs in a number of biological processes including cell proliferation, differentiation and the control of developmental timing. They are also involved in pathological conditions such as cancer [2-7] and neurodegeneration [8-11]. The canonical biogenesis of miRNAs involves two fundamental events. The first takes place in the nucleus, where the primary transcript (pri-miRNA), is processed into a precursor (pre-miRNA) by a nuclear RNase III enzyme (DROSHA). The second event occurs in the cytoplasm. The pre-miRNA is exported by exportin V from the nucleus and is cleaved by Dicer into a short-lived dsRNA of about 20-25 nucleotides. This double strand becomes unwound and one strand (forming the mature miR) becomes incorporated into an Argonaut (Ago)-protein containing complex called the RNA induced silencing complex (RISC). Generally, the mature miRNA within the RISC recognises complementary sites in the 3'-UTR of target genes, resulting in translational inhibition or destabilisation of the target mRNAs and downregulation of expression of the encoded protein [12]. Recently, however, some observations have demonstrated that miRNAs can also regulate their targets by binding to the 5'-UTR [13, 14]. In contrast to this classical inhibitory pathway, miRNAs can also stimulate the expression of target genes. This indicates that miRNAs can regulate gene expression not only through base pairing with mRNA targets but also through a decoy activity that interferes with the function of regulatory proteins [15].
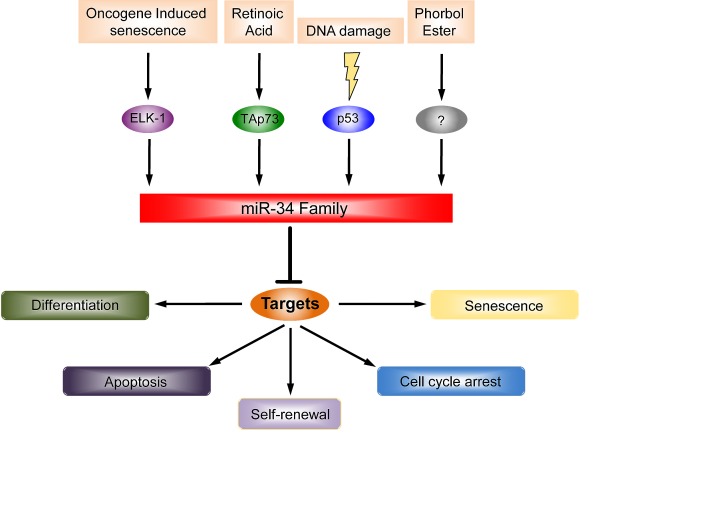
The miR-34 family, which consists of miR-34a, b and c, has attracted a lot of attention since it plays a key role as a tumor suppressor in several cancers [16-18]. Indeed, it is a direct target of the tumor suppressor gene p53 [19-23] and when up-regulated, it induces apoptosis [24], cell cycle arrest [25-28] and senescence. It also negatively influences the viability of cancer stem cells and inhibits metastasis formation [29-32]. Moreover, systemic delivery of miR-34 in a mouse model of hepatocellular carcinoma resulted in a reduced tumor burden and prolonged survival [33]. As a result of these and other studies, a miR-34 analogue has become the first microRNA to enter the clinic after a surprisingly swift 6 years passage from the bench to bedside. In addition, since a considerable number of oncogenes are direct targets of miR-34, and cancer is now considered a multipathway disease [34-36], this therapeutic approach would allow the use of only one bullet to hit more than one pathway deregulated by the loss of miR-34.
The miR-34 Family: origin, regulation and function
The miR-34 gene was first identified in C. elegans where it encodes a single miR that is evolutionarily conserved in several invertebrates [37, 38]. In mammals, the miR-34 family consists of three homologous transcripts miR-34a, miR-34b and miR-34c. In man, the miR-34a gene maps to chromosome 1p36.22 and is located within the second exon of its non-coding host gene. It is significant that 1p36 region deletions are frequently observed in a variety of human cancers including neuroblastoma, glioma, breast cancer, non-small cell lung cancer, small cell lung cancer, colorectal cancer and melanoma [39]. However, the genes coding for both miR-34b and miR-34c map to chromosome 11q23.1 and are located within intron 1 and exon 2 respectively, of the same primary transcript. Deletion of this region has been detected in breast, lung, cervical and prostate cancers [40]. Moreover, the 11q23 region is frequently rearranged (translocated, inserted and inverted) in hematological malignancies.[41] In the mouse, miR-34a is located on chromosome 4, while miR-34b/c are located on chromosome 9.
Analysis of miR-34a tissue distribution in the mouse shows that it is ubiquitously expressed but with the highest levels of expression in the brain, while miR-34b/c are mainly expressed in the lung [23], although, in general, the basal expression of miR-34a is higher than that of miR-34b/c. In man, also, miR-34a is ubiquitously expressed with high levels in the ovary, prostate and testes. Intermediate levels are found in brain, lung, thymus and kidney, while liver and heart show low levels of miR-34a. In contrast, miR-34b/c are mainly expressed in the ovary, testes, trachea and lung (http://mirnamap.mbc.nctu.edu.tw).
Although, as mentioned above, the miR-34 family is regulated by p53, it would be more correct to say the p53 family [42]. Retinoic acid induces the expression of miR-34a [43] and we have shown that, at least in the context of terminal differentiation of neuroblastoma cells, is driven by the p53 family member TAp73 [44]. TAp73 is a direct transcriptional activator of miR-34a, since it binds to p53 consensus elements in the miR-34a promoter, but TAp73 does not activate miR-34b and c. This role of the TAp73/miR-34a axis in neuronal differentiation is consistent with the predominantly neuronal phenotype of TAp73 null mice. However, unlike p53, TAp73 activation of miR-34a does not lead to apopotosis – and more work is clearly needed to understand how two members of the p53 family can activate the same miR but with very different biological effects.
Ectopic expression of the members of the miR-34 family can recapitulate some biological functions of p53 such as apoptosis [20, 45] and cell cycle arrest [46, 47], at least in some cell types, although other studies have failed to demonstrate an apoptotic effect of overexpressed miR-34 [44, 48]. Thus, the direct effect on apoptosis by miR-34a, and possibly the absolute requirement for miR-34a for p53-mediated apoptosis, is cell context-dependent. Figure 1 summarizes regulators and functions of the miR-34 family.
Figure 1. The miR-34 family regulators and their functions.
In the last few years, the miR-34a family has emerged as a pleiotropic microRNA. It was originally identified as a p53 target after DNA damage. The outcome of this upregulation is the induction of apoptosis, cell cycle arrest and senescence. Lately, p53 independent regulation has been observed. For instance, TAp73 is able to drive the expression of miR-34a and in turn, it regulates neuronal differentiation. miR-34a is also regulated by phorbol ester during megakaryocytic differentiation but the detailed molecular mechanisms have not been defined (?).
Oncogene-induced senescence is another stimulus, which increases expression of miR-34a. Specifically, after constitutive activation of B-RAF in TIG3 fibroblasts, the upregulation of miR-34a is mediated by the transcription factor ELK1 [49]. Thus, like other miRs, the transcriptional regulation of miR-34a expression is promiscuous.
The miR-34 family acts on apoptosis and cell cycle through the repression of many proteins involved in the regulation of these two biological processes. In particular, the miR-34 family binds to the 3'-UTRs of genes such as CDK4 and CDK6 [50, 51] (cell cycle) [19], Bcl-2 [24, 52] (apoptosis), SNAIL [29, 32] (epithelial mesenchymal transition) [53] and CD44 (migration and metastasis) [54], and the miR-34 family thus represses their expression. A detailed list of miR-34 family targets is provided in Table 1.
Table 1. miR-34 Targets list in Cancer.
| Direct Target | miR-34 member | Biological effect | Cancer cell lines/Cancer | Reference |
|---|---|---|---|---|
| CDK4 | miR-34a, b, c | Cell cycle arrest | IMR90, A549, HCT116 | 19 |
| CDK6 | miR-34a, b | Cell cycle arrest | SW480, PC3, Colorectal adenocarcinoma, Prostate cancer, NSCL | 23,47, 51 |
| CCNE2 | miR-34a, b, c | Cell cycle arrest | SW480, IMR90, A549, HCT116, Lung carcinoma, Colorectal Cancer | 19, 23, 63 |
| CCND1 | miR-34a | Cell cycle arrest | PC3 A549 | 47, 51 |
| c-MYC | miR-34b, c | Cell cycle arrest | Burkitt Lymphoma, Raji, Ramos, LCL, SIHN-011B | 17, 25 |
| N-MYC | miR-34a | Cell cycle arrest | NLF, IMR32, LAN-5, Neuroblastoma | 24, 28 |
| E2F5 | miR-34a | Cell cycle arrest | SW480 | 23 |
| CREB | miR-34b | Inhibition of proliferation | K652, HL60, ML2, NB4, NOMO1 | 92 |
| E2F3 | miR-34a, c | Inhibition of proliferation, senescence | PC3, IMR5, SK-N-BE, NLF, HCT116, RKO | 17, 24, 43, 48 |
| DLL1 | miR-34a | Inhibition of proliferation | Medulloblastoma | 100 |
| Notch-2 | miR-34a | Inhibition of growth | Glioblastoma | 16 |
| YY1 | miR-34a | Inhibition of growth | Glioblastoma cells | 76 |
| PDGFRA | miR-34a | Inhibition of growth | Glioblastoma cells | 27 |
| BCL2 | miR-34a | Apoptosis | SW480, PC3, NLF, Kato III | 23, 24, 52 |
| SIRT1 | miR-34a | Apoptosis/p53 activity | PC3, chronic lymphocytic leukaemia | 69, 70 |
| Survivin | miR-34a | Apoptosis | Hep-2/ Laryngeal squamous cell carcinoma | 18 |
| AXIN2 | miR-34a | Inhibition EMT | HCT116 | 30 |
| MET | miR-34a, b, c | Inhibition of invasion and migration | IMR90, A549, HCT116, Lung carcinoma, Colorectal Cancer | 19 |
| AXL | miR-34a | Inhibition migration and invasion | NSCLC cell lines, BRC cell lines, CRC cell lines | 31 |
| Fra-1 | miR-34a/c | Inhibition of invasion and migration | Human primary breast tumors, breast cancer cell lines | 94, 95 |
| SNAIL | miR-34a, b, c | Regulation of cancer EMT | H1299, HCT116, SW480, HCT-15 | 29 |
| CD44 | miR-34a | Inhibition of CSC and metastasis | Prostate | 54 |
| ARHGAP1 | miR-34a | Inhibition of invasion and migration | Human lung cancer cells human lung adenocarcinoma | 32 |
| NANOG | miR-34a | Cancer cell stemness | 56 | |
| SOX2 | miR-34a | Cancer cell stemness | 56 | |
| Notch-1 | miR-34a | Cancer cell stemness | Colon cancer | 72 |
| LDHA | miR-34a | Glucose metabolism | SW480 | 86 |
| IMPDH | miR-34a | Purine metabolism | H1299, HCT116 | 96 |
| HK1 | miR-34a | Glucose metabolism | H1299, HCT116 | 97 |
| HK2 | miR-34a | Glucose metabolism | H1299, HCT116 | 97 |
| GP1 | miR-34a | Glucose metabolism | H1299, HCT116 | 97 |
| PDK1 | miR-34a | Glucose metabolism | H1299, HCT116 | 97 |
| SIRT6 | miR-34a | Cell differentiation | Squamous cell carcinoma (SCCs) | 98 |
| WNT1 | miR-34a, b, c | Inhibition WNT signalling | MCF-7, A549, SW480, SW620, LoVo, SNU-81 | 99 |
| WNT3 | miR-34a, b, c | Inhibition WNT signalling | MCF-7, A549, SW480, SW620, LoVo, SNU-81 | 99 |
| LRP6 | miR-34a, b, c | Inhibition WNT signalling | MCF-7, A549, SW480, SW620, LoVo, SNU-81 | 99 |
| β-Catenin | miR-34a, b, c | Inhibition WNT signalling | MCF-7, A549, SW480, SW620, LoVo, SNU-81 | 99 |
| LEF1 | miR-34a | Inhibition WNT signalling | SW480 | 86 |
| MTA2 | miR-34a | p53 activity | SW480 | 86 |
CDK, Cycli-dependent kinase; CCNE, Cyclin E2; CCND1, Cylin D1; CREB, cAMP response element-binding protein; DLL1, Detlta-like protein 1; YY1, transcription factor Ying Yang 1;
GP1, glycoprotein 1; PDGFRA,platelet-derived growth factor receptor, alpha polypeptide; BCL2,B-cell lymphoma 2; SIRT1, Sirtuin; AXIN2, Axin-like protein 2; AXL, Tyrosine-Protein Kinase Receptor; HK, hexokinase; Fra-1, Fos-related antigen 1, SNAIL1, Snail Family Zinc Finger 1; ARHGAP1, Rho GTPase Activating Protein 1; NANOG, Homeobox Transcription Factor Nanog; SOX2, SRY(sex determining region Y)-box 2; LDHA, lactate dehydrogenase A; IMPDH, IMP (inosine 5'-monophosphate) dehydrogenase; PDK1, pyruvate dehydrogenase kinase, isozyme 1; WNT, wingless-type MMTV integration site family, member 1; LRP, low density lipoprotein receptor-related protein 6; LEF1, Lymphoid enhancer binding factor 1.
EMT, Epithelial–mesenchymal transition; CSC, Cancer stem cells; NSCLC Non-small-cell lung carcinoma; BRC, Breast Cancer; CRC, Colorectal Cancer
The absolute requirement for p53 to drive miR-34 family expression, and for miR-34 family members to mediate the p53 phenotype has, however, recently been questioned. Thus, while, miR-34 expression is reduced in some tissues in p53 null mice, in others it remains unaffected, confirming the promiscuity and cell context dependency referred to above. In particular, miR-34a expression remains high in the brains of p53−/− animals. Moreover, miR-34 knockout mice are born with the normal Mendelian ratio, are fertile, and are not, as might be expected, a phenocopy of the p53 knockout. In particular, miR-34 null mice do not show increased spontaneous or irradiation-induced tumorigenesis, and show only small and subtle differences from wild-type mice in other p53-dependent functions such as replicative senescence and the DNA damage response [55, 56].
miR34 expression in human cancer
Despite the lack of spontaneous tumours in miR-34 knockout mice, there is evidence, at least in some cancers, for miR-34 dysregulation. Thus, as mentioned above, the miR-34a locus on 1p36 is frequently lost in cancer [57]. Moreover, miR-34a has been found to be downregulated in neuroblastoma [43] and glioblastoma [58, 59], and its expression is frequently reduced in pancreatic cancer cell lines [20, 60]. CpG promoter methylation with miR-34a silencing has also been reported in several cancers including prostate, pancreatic, colorectal, ovarian cancer and melanoma [61, 62]. Mir-34b/c is also downregulated in colorectal cancer (CRC). This down-regulation is associated with hypermethylation of the neighboring CpG island; and DAC (5-aza-2'-deoxycytidine) treatment rapidly restores miR-34b/c expression. Methylation of the miR-34b/c CpG island was frequently observed in CRC cell lines and in primary CRC tumors (101 of 111, 90%), but not in normal colonic mucosa [63, 64].
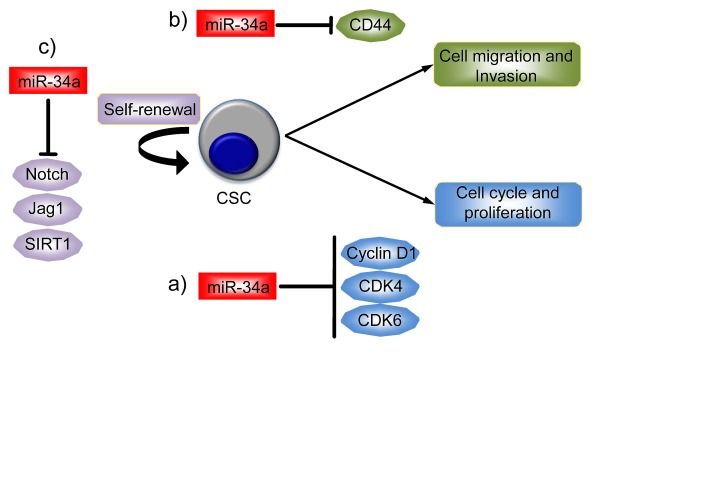
There is also some evidence for the involvement of the miR-34 family, again particularly miR-34a, in cancer stem cells (CSCs). CSCs are self-renewing cells within a tumor that have the capacity to regenerate the phenotypic diversity of the original tumor [65-68]. Nalls et al reported the first experimental evidence implicating miR-34a in CSCs [60]. First, they found that the expression of miR-34a was reduced in pancreatic CSCs and in pancreatic tumor cells independently of their p53 status when compared to normal pancreatic ductal epithelial cells. Importantly, the expression of miR-34a was restored by treatment with chromatin modifier agents such as the histone deacetylase inhibitor, Verinostat, in a p53 independent manner. The treatment also inhibited cell growth and induced apoptosis. At the molecular level, the well-known targets of miR-34a (such as SIRT-1 [69-71], Cyclin D1, Bcl-2, VEGF and CDK6) were downregulated and these effects were rescued by miR-34a inhibition (Figure 2a).
Figure 2. miR-34a as a regulator of cancer stem cell biology.
Cancer stem cells (CSC) have the capacity to self-renew and differentiate as well as the ability to regenerate tumors. miR-34a has been found to be dysregulated in CSC, particularly in pancreatic, prostatic cancer and in glioblastoma. a) In pancreatic CSC, miR-34a is able to regulate the proliferation of CSC, targeting Cyclin D1, CDK4 and CDK6. b) In prostatic CSC, miR-34a inhibits cell migration and invasion through the inhibition of CD44 expression. c) Finally, in glioblastoma, miR-34a regulates CSC self-renewal through the inhibition of Notch signaling and SIRT1.
Prostate CSCs with tumor initiating and metastatic potential are enriched in the CD44+ subpopulation. In this subset of cells, including CD44+ cells from individual patients tumours, expression of miR-34a, but not miR-34b/c, is also reduced and this does correlate with p53 status [54]. Ectopic expression of miR-34a either in prostate cancer cells or in the CD44+ fraction leads to inhibition of clonogenic expansion, tumor regeneration, and metastasis in vivo. In contrast, all these neoplastic phenotypes were promoted when expression of miR-34a was inhibited. Moreover, intravenous delivery of miR-34a inhibited lung metastasis and extended the survival of mice bearing human prostate cancer xenografts. At the molecular level CD44 has been identified and validated as a direct and functional target of miR-34a. Indeed, the inhibition of CD44 expression itself phenocopied miR-34a overexpression by inhibiting prostate tumor development and metastasis (Figure 3b).
Figure 3. Survival correlation of miR-34 family in several human cancer datasets.
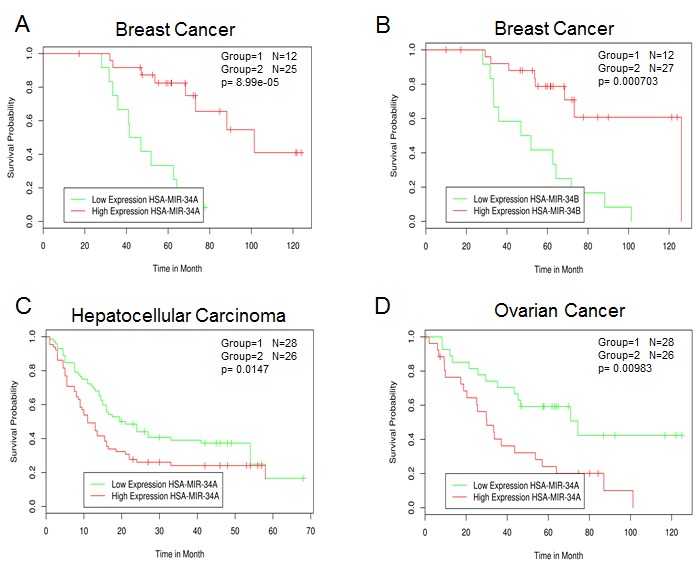
A and B) GEO dataset Title: Global microRNA expression profiling of high-risk ER+ breast cancers from patients receiving adjuvant Tamoxifen mono-therapy: a DBCG study. C) GEO dataset Title: MicroRNA expression profile in human hepatocellular carcinoma. D) GEO dataset Title: MicroRNA profiling of advanced serous ovarian carcinoma.
miR-34a expression is also reduced in human glioblastoma tissue when compared with normal brain [16], although this downregulation of miR-34a was only seen in tumours with mutant p53 and not in glioblastomas with wild-type p53. Transfection of precursor miR-34a in glioblastoma cells, as well as in glioblastoma CSCs, induced cell cycle arrest, apoptosis and also inhibited xenograft growth. The effects of miR-34a on glioma cells are partially mediated by the inhibition of c-Met and Notch expression, and levels of miR-34a are inversely correlated with the levels of c-Met in human gliomas. A role of miR-34a has been also shown in colon cancer stem cells (CCSCs) [72]. In this cellular context, miR-34a controls the decision of CCSCs to perform either symmetric or asymmetric division. Mechanistically, high levels of miR-34a reduce Notch1 signaling and promote asymmetric division. In contrast, low miR-34a levels upregulate Notch1 signaling and promote symmetric division. Promoting cell differentiation is another mechanism by which miR-34a exerts its tumor suppressor function in CRC (Figure 3c).
miR-34 family survival analysis in cancer
Overall, miR-34a and its family are tumor suppressors. Therefore, we would predict that the reduction of miR-34 expression is associated with poor prognosis and survival. Using MIRUMIR [73, 74] (http://www.bioprofiling.de/GEO/MIRUMIR/mirumir.html), an online tool that provides an analysis of miRs as potential biomarkers to predict survival of cancer patients, the following picture has emerged. Several datasets (breast, prostate and lung cancer, ovarian, hepatocellular and nasopharyngeal carcinoma) are currently available although statistical significance was only reached in three of them. Low expression of miR-34a/b was associated with poor outcome in breast cancer (Figure 3A and 3B) confirming its role as tumor suppressor. Moreover, the expression of several miR-34a/b validated targets including BCL-2 [75], CCNE2 [52], CCND1 [50], E2F3 [24, 43], MET [19], CD44 [54] and YY1 [76, 77] correlates with survival across different breast cancer datasets. In contrast, low levels of miR-34a are positive prognostic factors in human hepatocellular and advanced serous ovarian carcinoma (Figure 3C and 3D). Although this is at first sight surprising, this correlation analysis of the human hepatocellular carcinoma dataset is in agreement with a previous report, and may be further evidence for the cell context dependency of the biological effects of miR-34. Indeed, Pineau et al observed that miR-34a expression was increased in hepatocellular carcinoma and was linked to disease progression from normal liver through cirrhosis to full-blown hepatocellular carcinoma [78]. In human ovarian cancer (83 samples) miR-34 family expression was found to be reduced when compared to six (apparently mouse) ovarian surface epithelium cell samples. However, there were no significant differences when the expression of miR-34a was compared between stage III and stage IV distant metastatic disease [79]. Clearly, future studies are required in order to have a more coherent picture of the miR-34 family regulation in cancer and whether this family can be used as a prognostic biomarker [80, 81].
Perspectives and Conclusions
The last 7 years of studies have clearly shown that the miR-34 family is a master regulator of tumor biology. The family is frequently deregulated in cancer as discussed above, and preclinical in vivo studies have highlighted its therapeutic potential [82-84]. Overall, miR-34a (or a mimic) would seem a perfect candidate to enter clinical trials. However, it should be remembered that miR-34 and p53 have independent functions. Thus, miR34 null mice do not develop spontaneous tumors like p53 knockout mice [55]. In addition, the correlation between miR-34 family expression and patient survival would not always support its tumor suppressor role. In conclusion, more preclinical research on miR-34 is needed in order to better characterize its regulation and its downstream molecular pathways.
On the other hand, the phase 1 clinical trail that has recently started represents an important step forward not only for miR-34 itself, but forms a valuable proof of principle study for the rationale of using miRNAs as anticancer drugs. Indeed, although the endpoint of this clinical trial at this stage is to investigate the safety, pharmacokinetics and pharmacodynamics of the miR-34 mimetic in patients with unresectable primary liver cancer, it might shed light on two main challenges for miRNA-based therapies: i) delivery system and ii) potential off-target effects.
Tissues-specific delivery and cellular uptake of sufficient amounts of synthetic oligonucleotides to achieve sustained target inhibition is one of the major issues. Indeed, in miRNA-based therapy, two relevant obstacles need to be overcome, including the biological instability of the oligonucleotides in tissues and the poor cellular uptake [85]. MRX34 is a double stranded RNA, which is delivered by liposome. Since liposomes accumulate in the liver, liver cancer would theoretically be the main target organ affected by MRX34. Moreover, tumor uptake should be enhanced by the particular chemical composition of the liposomes. These liposomes are anionic at normal body pH, but in the tumor microenvironment, which has a lower pH, they become cationic form. This characteristic should therefore provide tumor specificity and prevent uptake by normal tissues.
One of the main advantages of miRNA-based therapy is the fact that microRNAs have the ability to simultaneously regulate several cellular pathways [86-89]. This makes them suitable “drugs” for the treatment of a multipathway disease such as cancer [90, 91]. In contrast, this multi-target property of the microRNA could potentially result in off-target side effects. Systemic overexpression of miR-34, which is broadly expressed and regulates physiological processes, could target genes in healthy tissues and cause side effects such as cardiovascular disease, although this may be minimised by the use of the particular liposome formulation [92].
In conclusion, while we should celebrate the entry of miR therapy into the oncologists drug cupboard, we should use the opportunity to learn as much about the disadvantages and qualifications of this new approach in order to optimize its therapeutic application in the future.
Acknowledgments
We apologize for those whose contributions could not be cited due to space constraints. This work has been supported by the Medical Research Council, UK
Footnotes
The authors declare no conflict of interest
REFERENCES
- 1.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nature reviews Molecular cell biology. 2008;9(3):219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 2.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annual review of medicine. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 3.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 4.Kinoshita T, Hanazawa T, Nohata N, Kikkawa N, Enokida H, Yoshino H, Yamasaki T, Hidaka H, Nakagawa M, Okamoto Y, Seki N. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma. Oncotarget. 2012;3(11):1386–1400. doi: 10.18632/oncotarget.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lezina L, Purmessur N, Antonov AV, Ivanova T, Karpova E, Krishan K, Ivan M, Aksenova V, Tentler D, Garabadgiu AV, Melino G, Barlev NA. miR-16 and miR-26a target checkpoint kinases Wee1 and Chk1 in response to p53 activation by genotoxic stress. Cell death & disease. 2013;4:e953. doi: 10.1038/cddis.2013.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH, Muller PA, Dotsch V, Kehrloesser S, Sayan BS, Giaccone G, Lowe SW, Takahashi N, Vandenabeele P, Knight RA, Levine AJ, et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci U S A. 2012;109(38):15312–15317. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ell B, Kang Y. MicroRNAs as regulators of tumor-associated stromal cells. Oncotarget. 2013;4(12):2166–2167. doi: 10.18632/oncotarget.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartl M, Grunwald Kadow IC. New roles for “old” microRNAs in nervous system function and disease. Frontiers in molecular neuroscience. 2013;6:51. doi: 10.3389/fnmol.2013.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parisi C, Arisi I, D'Ambrosi N, Storti AE, Brandi R, D'Onofrio M, Volonte C. Dysregulated microRNAs in amyotrophic lateral sclerosis microglia modulate genes linked to neuroinflammation. Cell death & disease. 2013;4:e959. doi: 10.1038/cddis.2013.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sano T, Reynolds JP, Jimenez-Mateos EM, Matsushima S, Taki W, Henshall DC. MicroRNA-34a upregulation during seizure-induced neuronal death. Cell death & disease. 2012;3:e287. doi: 10.1038/cddis.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volvert ML, Rogister F, Moonen G, Malgrange B, Nguyen L. MicroRNAs tune cerebral cortical neurogenesis. Cell Death Differ. 2012;19(10):1573–1581. doi: 10.1038/cdd.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340(6128):82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 13.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 14.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, Becker H, Chandler JC, Andino R, Cortes J, Hokland P, Huettner CS, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140(5):652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, Lopes B, Schiff D, Purow B, Abounader R. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69(19):7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105(36):13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Z, Zhan G, Ye D, Ren Y, Cheng L, Wu Z, Guo J. MicroRNA-34a affects the occurrence of laryngeal squamous cell carcinoma by targeting the antiapoptotic gene survivin. Medical oncology. 2012;29(4):2473–2480. doi: 10.1007/s12032-011-0156-x. [DOI] [PubMed] [Google Scholar]
- 19.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26(5):731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 22.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6(13):1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 23.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Current biology : CB. 2007;17(15):1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 24.Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, Maris JM. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6(5):735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leucci E, Cocco M, Onnis A, de Falco G, van Cleef P, Bellan C, van Rijk A, Nyagol J, Byakika B, Lazzi S, Tosi P, van Krieken H, Leoncini L. MYC translocation-negative classical Burkitt lymphoma cases: an alternative pathogenetic mechanism involving miRNA deregulation. J Pathol. 2008;216(4):440–450. doi: 10.1002/path.2410. [DOI] [PubMed] [Google Scholar]
- 26.Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG, Hamilton TL, Meijer HA, Dobbyn HC, Stoneley M, Spriggs KA, Willis AE, Bushell M. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci U S A. 2008;105(26):8866–8871. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silber J, Jacobsen A, Ozawa T, Harinath G, Pedraza A, Sander C, Holland EC, Huse JT. miR-34a repression in proneural malignant gliomas upregulates expression of its target PDGFRA and promotes tumorigenesis. PLoS One. 2012;7(3):e33844. doi: 10.1371/journal.pone.0033844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT, Tsang P, Zhang Q, Thiele CJ, Slack A, Shohet J, Khan J. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27(39):5204–5213. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, Rowe RG, Lee S, Maher CA, Weiss SJ, Yook JI. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. The Journal of cell biology. 2011;195(3):417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim NH, Cha YH, Kang SE, Lee Y, Lee I, Cha SY, Ryu JK, Na JM, Park C, Yoon HG, Park GJ, Yook JI, Kim HS. p53 regulates nuclear GSK-3 levels through miR-34-mediated Axin2 suppression in colorectal cancer cells. Cell Cycle. 2013;12(10):1578–1587. doi: 10.4161/cc.24739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mudduluru G, Ceppi P, Kumarswamy R, Scagliotti GV, Papotti M, Allgayer H. Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene. 2011;30(25):2888–2899. doi: 10.1038/onc.2011.13. [DOI] [PubMed] [Google Scholar]
- 32.Ahn YH, Gibbons DL, Chakravarti D, Creighton CJ, Rizvi ZH, Adams HP, Pertsemlidis A, Gregory PA, Wright JA, Goodall GJ, Flores ER, Kurie JM. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. The Journal of clinical investigation. 2012;122(9):3170–3183. doi: 10.1172/JCI63608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Frontiers in genetics. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 35.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q, Wang X, He C, Pan H. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell death & disease. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sotgia F, Martinez-Outschoorn UE, Lisanti MP. Cancer metabolism: new validated targets for drug discovery. Oncotarget. 2013;4(8):1309–1316. doi: 10.18632/oncotarget.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Chen D, He Y, Melendez A, Feng Z, Hong Q, Bai X, Li Q, Cai G, Wang J, Chen X. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age (Dordr) 2013;35(1):11–22. doi: 10.1007/s11357-011-9324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henrich KO, Schwab M, Westermann F. 1p36 tumor suppression--a matter of dosage? Cancer Res. 2012;72(23):6079–6088. doi: 10.1158/0008-5472.CAN-12-2230. [DOI] [PubMed] [Google Scholar]
- 40.Laake K, Odegard A, Andersen TI, Bukholm IK, Karesen R, Nesland JM, Ottestad L, Shiloh Y, Borresen-Dale AL. Loss of heterozygosity at 11q23.1 in breast carcinomas: indication for involvement of a gene distal and close to ATM. Genes Chromosomes Cancer. 1997;18(3):175–180. [PubMed] [Google Scholar]
- 41.Bernard OA, Berger R. Molecular basis of 11q23 rearrangements in hematopoietic malignant proliferations. Genes Chromosomes Cancer. 1995;13(2):75–85. doi: 10.1002/gcc.2870130202. [DOI] [PubMed] [Google Scholar]
- 42.Seeliger MA, Moll UM. p73 - constitutively open for business. Cell Death Differ. 2013;20(8):972–973. doi: 10.1038/cdd.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26(34):5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 44.Agostini M, Tucci P, Killick R, Candi E, Sayan BS, Rivetti di Val Cervo P, Nicotera P, McKeon F, Knight RA, Mak TW, Melino G. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci U S A. 2011;108(52):21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kracikova M, Akiri G, George A, Sachidanandam R, Aaronson SA. A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death Differ. 2013;20(4):576–588. doi: 10.1038/cdd.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghawanmeh T, Thunberg U, Castro J, Murray F, Laytragoon-Lewin N. miR-34a expression, cell cycle arrest and cell death of malignant mesothelioma cells upon treatment with radiation, docetaxel or combination treatment. Oncology. 2011;81(5-6):330–335. doi: 10.1159/000334237. [DOI] [PubMed] [Google Scholar]
- 47.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582(10):1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 48.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104(39):15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund AH. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17(2):236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 50.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582(10):1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 51.Fujita Y, Kojima K, Hamada N, Ohhashi R, Akao Y, Nozawa Y, Deguchi T, Ito M. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun. 2008;377(1):114–119. doi: 10.1016/j.bbrc.2008.09.086. [DOI] [PubMed] [Google Scholar]
- 52.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brabletz T. MiR-34 and SNAIL: another double-negative feedback loop controlling cellular plasticity/EMT governed by p53. Cell Cycle. 2012;11(2):215–216. doi: 10.4161/cc.11.2.18900. [DOI] [PubMed] [Google Scholar]
- 54.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17(2):211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D'Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8(7):e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C, Ozturk A, Hicks GG, Hannon GJ, He L. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13(11):1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68(8):2551–2556. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao H, Zhao H, Xiang W. Expression level of human miR-34a correlates with glioma grade and prognosis. Journal of neuro-oncology. 2013;113(2):221–228. doi: 10.1007/s11060-013-1119-1. [DOI] [PubMed] [Google Scholar]
- 59.Li WB, Ma MW, Dong LJ, Wang F, Chen LX, Li XR. MicroRNA-34a targets notch1 and inhibits cell proliferation in glioblastoma multiforme. Cancer biology & therapy. 2011;12(6):477–483. doi: 10.4161/cbt.12.6.16300. [DOI] [PubMed] [Google Scholar]
- 60.Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS One. 2011;6(8):e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogt M, Munding J, Gruner M, Liffers ST, Verdoodt B, Hauk J, Steinstraesser L, Tannapfel A, Hermeking H. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458(3):313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 62.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7(16):2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 63.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68(11):4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 64.Siemens H, Neumann J, Jackstadt R, Mansmann U, Horst D, Kirchner T, Hermeking H. Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and beta-catenin predicts distant metastasis of colon cancer. Clin Cancer Res. 2013;19(3):710–720. doi: 10.1158/1078-0432.CCR-12-1703. [DOI] [PubMed] [Google Scholar]
- 65.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nature reviews Cancer. 2012;12(2):133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 66.Fareh M, Turchi L, Virolle V, Debruyne D, Almairac F, de-la-Forest Divonne S, Paquis P, Preynat-Seauve O, Krause KH, Chneiweiss H, Virolle T. The miR 302-367 cluster drastically affects self-renewal and infiltration properties of glioma-initiating cells through CXCR4 repression and consequent disruption of the SHH-GLI-NANOG network. Cell Death Differ. 2012;19(2):232–244. doi: 10.1038/cdd.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu H, Zhang W, Jia Y, Yu Q, Grau GE, Peng L, Ran Y, Yang Z, Deng H, Lou J. Single-cell clones of liver cancer stem cells have the potential of differentiating into different types of tumor cells. Cell death & disease. 2013;4:e857. doi: 10.1038/cddis.2013.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bier A, Giladi N, Kronfeld N, Lee HK, Cazacu S, Finniss S, Xiang C, Poisson L, de Carvalho AC, Slavin S, Jacoby E, Yalon M, Toren A, Mikkelsen T, Brodie C. MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget. 2013;4(5):665–676. doi: 10.18632/oncotarget.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Audrito V, Vaisitti T, Rossi D, Gottardi D, D'Arena G, Laurenti L, Gaidano G, Malavasi F, Deaglio S. Nicotinamide blocks proliferation and induces apoptosis of chronic lymphocytic leukemia cells through activation of the p53/miR-34a/SIRT1 tumor suppressor network. Cancer Res. 2011;71(13):4473–4483. doi: 10.1158/0008-5472.CAN-10-4452. [DOI] [PubMed] [Google Scholar]
- 70.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105(36):13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8(5):712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- 72.Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ, Goerger JP, Sun J, Witherspoon M, Rakhilin N, Li J, Yang H, Milsom J, Lee S, Zipfel W, Jin MM, et al. A microRNA miR-34a-regulated bimodal switch targets notch in colon cancer stem cells. Cell stem cell. 2013;12(5):602–615. doi: 10.1016/j.stem.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Antonov AV, Knight RA, Melino G, Barlev NA, Tsvetkov PO. MIRUMIR: an online tool to test microRNAs as biomarkers to predict survival in cancer using multiple clinical data sets. Cell Death Differ. 2013;20(2):367. doi: 10.1038/cdd.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Antonov AV. BioProfiling.de: analytical web portal for high-throughput cell biology. Nucleic acids research. 2011;39(Web Server issue):W323–327. doi: 10.1093/nar/gkr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, Maris JM. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6(5):735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yin D, Ogawa S, Kawamata N, Leiter A, Ham M, Li D, Doan NB, Said JW, Black KL, Phillip Koeffler H. miR-34a functions as a tumor suppressor modulating EGFR in glioblastoma multiforme. Oncogene. 2013;32(9):1155–1163. doi: 10.1038/onc.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen QR, Yu LR, Tsang P, Wei JS, Song YK, Cheuk A, Chung JY, Hewitt SM, Veenstra TD, Khan J. Systematic proteome analysis identifies transcription factor YY1 as a direct target of miR-34a. J Proteome Res. 2011;10(2):479–487. doi: 10.1021/pr1006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107(1):264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH, Hermeking H, Nikitin AY. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16(4):1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kobayashi N, Uemura H, Nagahama K, Okudela K, Furuya M, Ino Y, Ito Y, Hirano H, Inayama Y, Aoki I, Nagashima Y, Kubota Y, Ishiguro H. Identification of miR-30d as a novel prognostic maker of prostate cancer. Oncotarget. 2012;3(11):1455–1471. doi: 10.18632/oncotarget.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Visone R, Veronese A, Balatti V, Croce CM. MiR-181b: new perspective to evaluate disease progression in chronic lymphocytic leukemia. Oncotarget. 2012;3(2):195–202. doi: 10.18632/oncotarget.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, Bader AG. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70(14):5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, Maitra A. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Molecular cancer therapeutics. 2011;10(8):1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Molecular therapy : the journal of the American Society of Gene Therapy. 2010;18(9):1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao X, Pan F, Holt CM, Lewis AL, Lu JR. Controlled delivery of antisense oligonucleotides: a brief review of current strategies. Expert opinion on drug delivery. 2009;6(7):673–686. doi: 10.1517/17425240902992894. [DOI] [PubMed] [Google Scholar]
- 86.Kaller M, Liffers ST, Oeljeklaus S, Kuhlmann K, Roh S, Hoffmann R, Warscheid B, Hermeking H. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol Cell Proteomics. 2011;10(8):M111 010462. doi: 10.1074/mcp.M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karaayvaz M, Zhai H, Ju J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell death & disease. 2013;4:e659. doi: 10.1038/cddis.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu X, Zhen Y, Yang H, Wang H, Zhou Y, Wang E, Marincola FM, Mai C, Chen Y, Wei H, Song Y, Lyu X, Ye Y, Cai L, Wu Q, Zhao M, et al. Loss of connective tissue growth factor as an unfavorable prognosis factor activates miR-18b by PI3K/AKT/C-Jun and C-Myc and promotes cell growth in nasopharyngeal carcinoma. Cell death & disease. 2013;4:e634. doi: 10.1038/cddis.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin J, Teo S, Lam DH, Jeyaseelan K, Wang S. MicroRNA-10b pleiotropically regulates invasion, angiogenicity and apoptosis of tumor cells resembling mesenchymal subtype of glioblastoma multiforme. Cell death & disease. 2012;3:e398. doi: 10.1038/cddis.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell death & disease. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sarvaiya PJ, Guo D, Ulasov I, Gabikian P, Lesniak MS. Chemokines in tumor progression and metastasis. Oncotarget. 2013;4(12):2171–2185. doi: 10.18632/oncotarget.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495(7439):107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 93.Pigazzi M, Manara E, Baron E, Basso G. miR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer Res. 2009;69(6):2471–8. doi: 10.1158/0008-5472.CAN-08-3404. [DOI] [PubMed] [Google Scholar]
- 94.Yang S, Li Y, Gao J, Zhang T, Li S, Luo, Chen H, Ding F, Wang X, Liu Z. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. 2013;32(36):4294–303. doi: 10.1038/onc.2012.432. [DOI] [PubMed] [Google Scholar]
- 95.Wu J, Wu G, Lv L, Ren YF, Zhang XJ, Xue YF, Li G, Lu X, Sun Z, Tang KF. MicroRNA-34a inhibits migration and invasion of colon cancer cells via targeting to Fra-1. Carcinogenesis. 2012;33(3):519–28. doi: 10.1093/carcin/bgr304. [DOI] [PubMed] [Google Scholar]
- 96.Kim HR, Roe JS, Lee JE, Hwang IY, Cho EJ, Youn HD. A p53-inducible microRNA-34a downregulates Ras signaling by targeting IMPDH. Biochem Biophys Res Commun. 2012;418(4):682–8. doi: 10.1016/j.bbrc.2012.01.077. [DOI] [PubMed] [Google Scholar]
- 97.Kim HR, Roe JS, Cho EJ, Youn HD. p53 regulates glucose metabolism by miR-34a. Biochem Biophys Res Commun. 2013;437(2):225–31. doi: 10.1016/j.bbrc.2013.06.043. [DOI] [PubMed] [Google Scholar]
- 98.Lefort K, Brooks Y, Ostano P, Cario-Andre M, Calpini V, Guinea-Viniegra J, Albinger-Hegyi A, Hoetzenecker W, Kolfschoten I, Wagner EF, Werner S, Dotto G P. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J. 2013;32(16):2248–63. doi: 10.1038/emboj.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim NH, Kim HS, Kim NG, Lee I, Choi HS, Li XY, Kang SE, Cha SY, Ryu JK, Na JM, Park C, Kim K, Lee S, Gumbiner BM, Yook JI, Weiss SJ. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci Signal. 2011;4(197):ra71. doi: 10.1126/scisignal.2001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.De Antonellis P, Medaglia C, Cusanelli E, Andolfo I, Liguori L, de Vita G, Carotenuto M, Bello A, Formiggini F, Galeone A, de Rosa G, Virgilio A, Scognamiglio I, Sciro M, Basso G, et al. MiR-34a targeting of Notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS One. 2011;6(9):e24584. doi: 10.1371/journal.pone.0024584. [DOI] [PMC free article] [PubMed] [Google Scholar]