Abstract
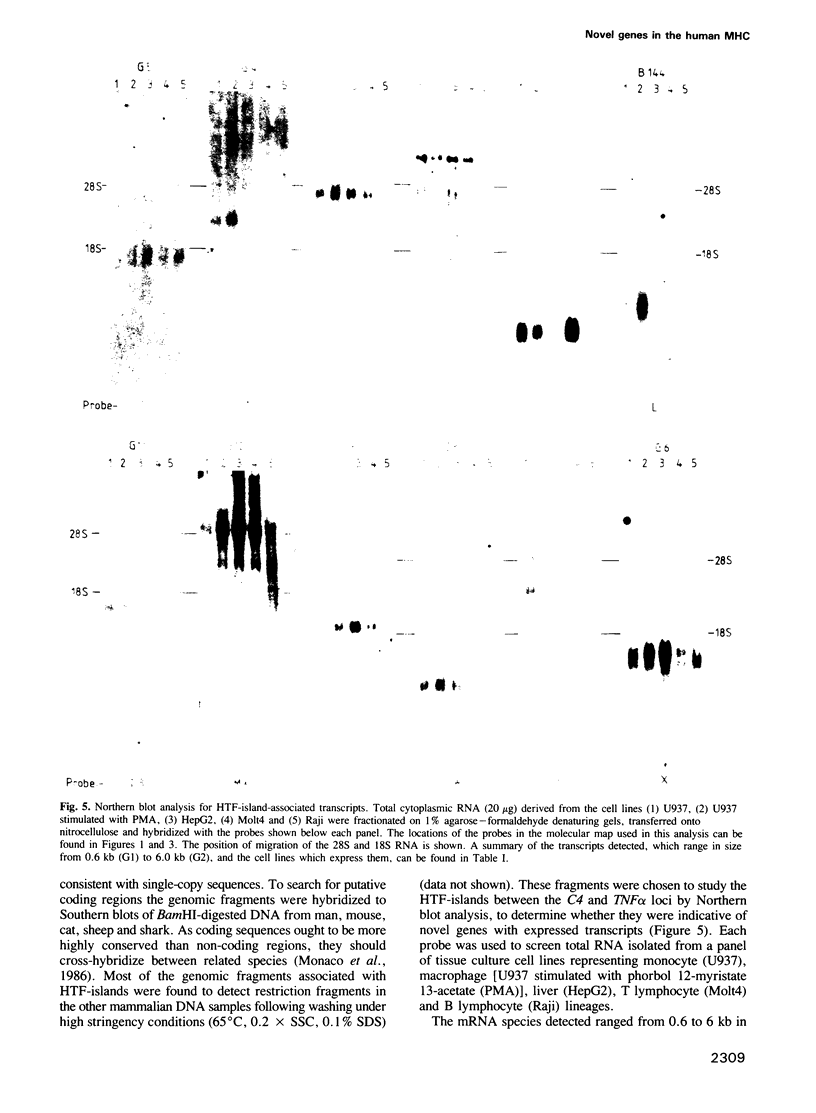
Chromosome walking in the major histocompatibility complex (MHC) class III region has resulted in the isolation of 541 kb of genomic DNA in two sets of overlapping cosmid clones. These two sets encompass the 340 kb separating the C2 and tumour necrosis factor (TNF) alpha and beta genes, except for a 22 kb gap 108 kb centromeric to the TNF alpha gene. The genomic DNA inserts have been characterized for the presence of clusters of restriction sites with CpG dinucleotides in their recognition sequence. In conjunction with pulsed field gel electrophoresis the exact sites which cleave in chromosomal DNA have been established and this has suggested the presence of a number of HTF-islands. Genomic probes flanking the HTF-islands have been hybridized to Northern blots of RNA from a number of cell lines. Transcripts ranging in size from 0.6 to 6 kb corresponding to the products of 12 novel, single copy genes have been identified. In addition the human equivalent of the murine B144 gene was mapped approximately 10 kb centromeric of the TNF alpha gene. The location of so many new genes in this region raises the question as to whether they play any role in the observed HLA associations with an individual's susceptibility to develop autoimmune disease.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Batchelor J. R., McMichael A. J. Progress in understanding HLA and disease associations. Br Med Bull. 1987 Jan;43(1):156–183. doi: 10.1093/oxfordjournals.bmb.a072169. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985 May 24;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. R., Bird A. P. Long-range restriction site mapping of mammalian genomic DNA. 1986 Jul 31-Aug 6Nature. 322(6078):477–481. doi: 10.1038/322477a0. [DOI] [PubMed] [Google Scholar]
- Carroll M. C., Campbell R. D., Bentley D. R., Porter R. R. A molecular map of the human major histocompatibility complex class III region linking complement genes C4, C2 and factor B. Nature. 1984 Jan 19;307(5948):237–241. doi: 10.1038/307237a0. [DOI] [PubMed] [Google Scholar]
- Carroll M. C., Campbell R. D., Porter R. R. Mapping of steroid 21-hydroxylase genes adjacent to complement component C4 genes in HLA, the major histocompatibility complex in man. Proc Natl Acad Sci U S A. 1985 Jan;82(2):521–525. doi: 10.1073/pnas.82.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M. C., Katzman P., Alicot E. M., Koller B. H., Geraghty D. E., Orr H. T., Strominger J. L., Spies T. Linkage map of the human major histocompatibility complex including the tumor necrosis factor genes. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8535–8539. doi: 10.1073/pnas.84.23.8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Comings D. E. Mechanisms of chromosome banding and implications for chromosome structure. Annu Rev Genet. 1978;12:25–46. doi: 10.1146/annurev.ge.12.120178.000325. [DOI] [PubMed] [Google Scholar]
- Davis M. M., Bjorkman P. J. T-cell antigen receptor genes and T-cell recognition. Nature. 1988 Aug 4;334(6181):395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Drmanac R., Petrović N., Glisin V., Crkvenjakov R. A calculation of fragment lengths obtainable from human DNA with 78 restriction enzymes: an aid for cloning and mapping. Nucleic Acids Res. 1986 Jun 11;14(11):4691–4692. doi: 10.1093/nar/14.11.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I., Sargent C. A., Dawkins R. L., Campbell R. D. Direct observation of the gene organization of the complement C4 and 21-hydroxylase loci by pulsed field gel electrophoresis. J Exp Med. 1989 May 1;169(5):1803–1818. doi: 10.1084/jem.169.5.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I., Sargent C. A., Trowsdale J., Campbell R. D. Molecular mapping of the human major histocompatibility complex by pulsed-field gel electrophoresis. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7237–7241. doi: 10.1073/pnas.84.20.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Fischel-Ghodsian N., Nicholls R. D., Higgs D. R. Long range genome structure around the human alpha-globin complex analysed by PFGE. Nucleic Acids Res. 1987 Aug 11;15(15):6197–6207. doi: 10.1093/nar/15.15.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischel-Ghodsian N., Nicholls R. D., Higgs D. R. Unusual features of CpG-rich (HTF) islands in the human alpha globin complex: association with non-functional pseudogenes and presence within the 3' portion of the zeta gene. Nucleic Acids Res. 1987 Nov 25;15(22):9215–9225. doi: 10.1093/nar/15.22.9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987 Jul 20;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Goldman M. A., Holmquist G. P., Gray M. C., Caston L. A., Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984 May 18;224(4650):686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Grosveld F. G., Lund T., Murray E. J., Mellor A. L., Dahl H. H., Flavell R. A. The construction of cosmid libraries which can be used to transform eukaryotic cells. Nucleic Acids Res. 1982 Nov 11;10(21):6715–6732. doi: 10.1093/nar/10.21.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist G., Gray M., Porter T., Jordan J. Characterization of Giemsa dark- and light-band DNA. Cell. 1982 Nov;31(1):121–129. doi: 10.1016/0092-8674(82)90411-1. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott V., Rees D. J., Cheng Z., Brownlee G. G. Randomly picked cosmid clones overlap the pyrB and oriC gap in the physical map of the E. coli chromosome. Nucleic Acids Res. 1988 Mar 25;16(6):2601–2612. doi: 10.1093/nar/16.6.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenberg J. R., Rykowski M. C. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell. 1988 May 6;53(3):391–400. doi: 10.1016/0092-8674(88)90159-6. [DOI] [PubMed] [Google Scholar]
- Lavia P., Macleod D., Bird A. Coincident start sites for divergent transcripts at a randomly selected CpG-rich island of mouse. EMBO J. 1987 Sep;6(9):2773–2779. doi: 10.1002/j.1460-2075.1987.tb02572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay S., Bird A. P. Use of restriction enzymes to detect potential gene sequences in mammalian DNA. 1987 May 28-Jun 3Nature. 327(6120):336–338. doi: 10.1038/327336a0. [DOI] [PubMed] [Google Scholar]
- Lévi-Strauss M., Carroll M. C., Steinmetz M., Meo T. A previously undetected MHC gene with an unusual periodic structure. Science. 1988 Apr 8;240(4849):201–204. doi: 10.1126/science.3353717. [DOI] [PubMed] [Google Scholar]
- Monaco A. P., Neve R. L., Colletti-Feener C., Bertelson C. J., Kurnit D. M., Kunkel L. M. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986 Oct 16;323(6089):646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- Morley B. J., Campbell R. D. Internal homologies of the Ba fragment from human complement component Factor B, a class III MHC antigen. EMBO J. 1984 Jan;3(1):153–157. doi: 10.1002/j.1460-2075.1984.tb01776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedospasov S. A., Shakhov A. N., Turetskaia R. L., Mett V. A., Georgiev G. P. Molekuliarnoe klonirovanie genov cheloveka, kodiruiushchikh faktory nekroza opukholei: tandemnoe raspolozhenie al'fa- i beta-genov v korotkom segmente (6 tys. par nukleotidov) genoma cheloveka. Dokl Akad Nauk SSSR. 1985;285(6):1487–1490. [PubMed] [Google Scholar]
- Nedospasov S. A., Shakhov A. N., Turetskaya R. L., Mett V. A., Azizov M. M., Georgiev G. P., Korobko V. G., Dobrynin V. N., Filippov S. A., Bystrov N. S. Tandem arrangement of genes coding for tumor necrosis factor (TNF-alpha) and lymphotoxin (TNF-beta) in the human genome. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):611–624. doi: 10.1101/sqb.1986.051.01.073. [DOI] [PubMed] [Google Scholar]
- Olaisen B., Sakaguchi A. Y., Naylor S. L. Report of the committee on the genetic constitution of chromosomes 5 and 6. Cytogenet Cell Genet. 1987;46(1-4):147–169. doi: 10.1159/000132475. [DOI] [PubMed] [Google Scholar]
- Pontarotti P., Chimini G., Nguyen C., Boretto J., Jordan B. R. CpG islands and HTF islands in the HLA class I region: investigation of the methylation status of class I genes leads to precise physical mapping of the HLA-B and -C genes. Nucleic Acids Res. 1988 Jul 25;16(14B):6767–6778. doi: 10.1093/nar/16.14.6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent C. A., Dunham I., Trowsdale J., Campbell R. D. Human major histocompatibility complex contains genes for the major heat shock protein HSP70. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1968–1972. doi: 10.1073/pnas.86.6.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealey P. G., Whittaker P. A., Southern E. M. Removal of repeated sequences from hybridisation probes. Nucleic Acids Res. 1985 Mar 25;13(6):1905–1922. doi: 10.1093/nar/13.6.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. I., Golembieski W., Gilbert J. D., Kizyma L., Miller O. J. Overabundance of rare-cutting restriction endonuclease sites in the human genome. Nucleic Acids Res. 1987 Feb 11;15(3):1173–1184. doi: 10.1093/nar/15.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M., Anand R., Brown W. R., Fletcher D. S. A model for the separation of large DNA molecules by crossed field gel electrophoresis. Nucleic Acids Res. 1987 Aug 11;15(15):5925–5943. doi: 10.1093/nar/15.15.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies T., Blanck G., Bresnahan M., Sands J., Strominger J. L. A new cluster of genes within the human major histocompatibility complex. Science. 1989 Jan 13;243(4888):214–217. doi: 10.1126/science.2911734. [DOI] [PubMed] [Google Scholar]
- Spies T., Morton C. C., Nedospasov S. A., Fiers W., Pious D., Strominger J. L. Genes for the tumor necrosis factors alpha and beta are linked to the human major histocompatibility complex. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8699–8702. doi: 10.1073/pnas.83.22.8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan T. Molecular genetics and polymorphism of class I HLA antigens. Br Med Bull. 1987 Jan;43(1):1–14. doi: 10.1093/oxfordjournals.bmb.a072166. [DOI] [PubMed] [Google Scholar]
- Todd J. A., Acha-Orbea H., Bell J. I., Chao N., Fronek Z., Jacob C. O., McDermott M., Sinha A. A., Timmerman L., Steinman L. A molecular basis for MHC class II--associated autoimmunity. Science. 1988 May 20;240(4855):1003–1009. doi: 10.1126/science.3368786. [DOI] [PubMed] [Google Scholar]
- Todd J. A., Bell J. I., McDevitt H. O. A molecular basis for genetic susceptibility to insulin-dependent diabetes mellitus. Trends Genet. 1988 May;4(5):129–134. doi: 10.1016/0168-9525(88)90135-7. [DOI] [PubMed] [Google Scholar]
- Todd J. A., Bell J. I., McDevitt H. O. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987 Oct 15;329(6140):599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- Trowsdale J. Genetics and polymorphism: class II antigens. Br Med Bull. 1987 Jan;43(1):15–36. doi: 10.1093/oxfordjournals.bmb.a072168. [DOI] [PubMed] [Google Scholar]
- Tsuge I., Shen F. W., Steinmetz M., Boyse E. A. A gene in the H-2S:H-2D interval of the major histocompatibility complex which is transcribed in B cells and macrophages. Immunogenetics. 1987;26(6):378–380. doi: 10.1007/BF00343709. [DOI] [PubMed] [Google Scholar]
- Tykocinski M. L., Max E. E. CG dinucleotide clusters in MHC genes and in 5' demethylated genes. Nucleic Acids Res. 1984 May 25;12(10):4385–4396. doi: 10.1093/nar/12.10.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P. C., Grossberger D., Onufer B. J., Chaplin D. D., New M. I., Dupont B., Strominger J. L. Two genes encoding steroid 21-hydroxylase are located near the genes encoding the fourth component of complement in man. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1089–1093. doi: 10.1073/pnas.82.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Hunt C., Morimoto R. Structure and expression of the human gene encoding major heat shock protein HSP70. Mol Cell Biol. 1985 Feb;5(2):330–341. doi: 10.1128/mcb.5.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. Y., Campbell R. D. Definitive RFLPs to distinguish between the human complement C4A/C4B isotypes and the major Rodgers/Chido determinants: application to the study of C4 null alleles. Immunogenetics. 1987;25(6):383–390. doi: 10.1007/BF00396104. [DOI] [PubMed] [Google Scholar]