Abstract
Purpose of Review
Despite progress towards understanding the molecular pathogenesis of Rheumatoid Arthritis (RA), its etiology remains elusive. Genes are important but rather insufficient to explain the majority of RA cases. This review describes novel data supporting the microbiome and its interactions with the human host as potential en(‘in’)vironmental factors in RA pathogenesis.
Recent Findings
Animal models of inflammatory arthritis have shown that the presence of bacteria in mucosal surfaces is sufficient to alter local and systemic host immune responses and elicit joint inflammation. Human RA studies have focused on three mucosal sites: the gut, the gingival, and the respiratory tree. The oral microbiome, and specifically Porphyromonas gingivalis (P. gingivalis), has long been implicated. Novel sequencing technologies have allowed investigations into the role of the gut microbiome in the development of autoimmune arthritis. Most recently, the pulmonary parenchyma has also been described as yet another possible mucosal site of initiation of autoimmunity in RA.
Summary
Emerging data implicates the microbiome in RA pathogenesis. Mucosal sites exposed to a high load of bacterial antigens - such as the periodontium, lung, and gut - may represent the initial site of autoimmune generation. If validated, these findings could lead to the discovery of potential biomarkers and therapeutic approaches in the pre-clinical and clinical phases of RA.
Keywords: Rheumatoid, Arthritis, Microbiome, Peridontal, Porphyromonas
Introduction
The notion that the human genome and its encoded machinery are sufficient for our evolutionary success has recently been questioned. In fact, humans are vastly overshadowed by their bacterial companions. Literally trillions of bacterial organisms form unique ecological niches in almost all body surfaces. From the moment we are born, humans are populated with a complex, constantly changing community of bacteria that both helps shape the mucosal immune system and provides essential nutrients [1, 2]. Despite their overwhelming antigenic load, most of these microorganisms have long been ignored as potential determinants in the pathogenesis of human disease. Novel technological tools such as culture-independent 16S ribosomal RNA pyrosequencing, advances in computational biology, and initiatives such as the NIH Human Microbiome Project (HMP) have all reenergized this field of study [3]. Microbiome is the term utilized to describe the sum of ecological bacterial communities (and their genes) that populate human skin, oral cavity, airways, gastrointestinal tract (GI), and genitourinary tract (GU). These bacteria and their genomes are classically identified as commensal, symbiotic, or pathogenic; however, the utility of these labels has grown less clear and the interactions between host and bacteria are becoming increasingly complex.
To date, a nearly complete catalogue of the diverse microbial communities in the human body has been revealed [4, 5**]. The largest bacterial burden lies in the intestines, where nearly three pounds of bacteria and over three million bacterial genes outnumber the human host genome 100 times over [6]. The two predominant microbial phyla of the human gut are the Firmicutes, which includes the important Clostridia class, and the Bacteroidetes. Also present but less pervasive are the phyla Proteobacteria, Actinobacteria, Fusobacteria, and Verrucomicrobia [7]. Interestingly, although debated, studies have identified three core human microbial enterotypes [8] that cluster subjects according to the relative abundance of the microbial genera Bacteroides, Prevotella, and Ruminococcus [9**].
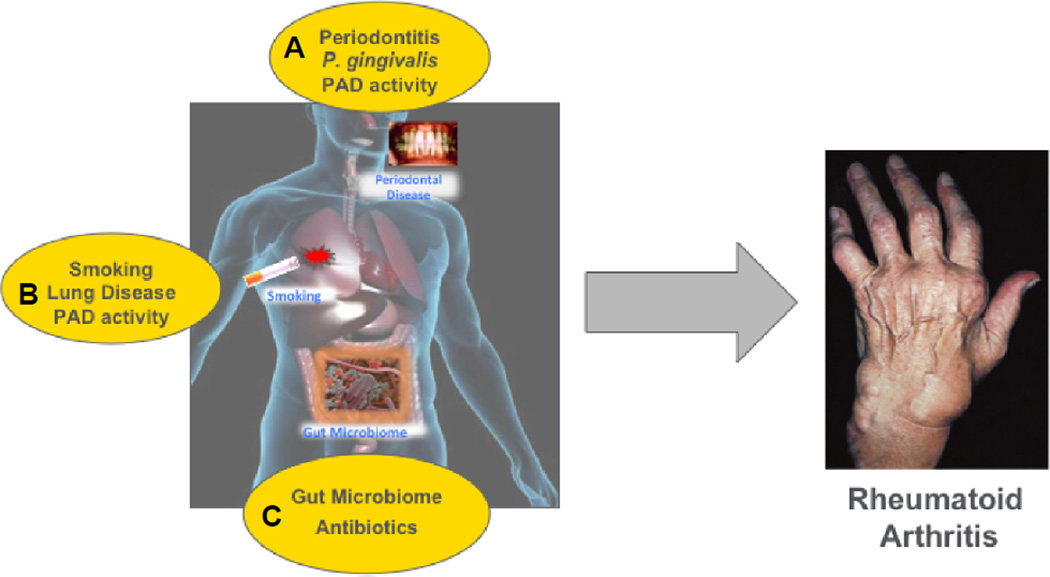
Despite the advent of genome-wide association studies (GWAS) and intense research into both MHC and non-MHC genetic polymorphisms, the etiologies of most rheumatic diseases remain elusive. Multiple genetic studies have shaped our understanding of RA susceptibility (i.e. shared epitope hypothesis) [10] and continue to elucidate pathways and function [11]; however, causal evidence is lacking, as illustrated by a relatively low concordance rate in monozygotic twins. Thus, much focus has shifted to the role of environmental factors and gene-environmental interaction in the pathogenesis of rheumatic diseases. This is particularly true for RA because evidence for autoimmunity in the form of circulating anti-cyclic citrullinated peptide (CCP) antibodies and/or rheumatoid factor (RF) can be found up to a decade before disease onset [12]. Interestingly, both antibody titers and epitope specificity rise just a few months before synovitis is apparent, and circulating pro-inflammatory cytokines are also elevated during this pre-clinical phase [13]. Moreover, synovial biopsies and joint MRIs of asymptomatic individuals with circulating anti-CCP fail to demonstrate signs of tissue damage [14*]. Taken together, these data are highly relevant because they suggests that autoimmunity in RA has an extra-articular origin (i.e. outside-in process) and that a “second hit” is necessary for arthritis development. Among possible non-genetic triggers, the microbiome has been identified as a possible inciting factor. This review will focus on the recent evidence implicating the gut, oral, and lung microbiomes as en(‘in’)vironmental factors in autoimmune arthritis (Figure).
Figure.
Potential sites of microbiome involvement and related risk factors in the pathogenesis of RA. A. Periodontal disease is linked to increased RA prevalence and disease severity. The oral microbiome, specifically P. gingivalis through PAD activity, has been implicated. B. The lung mucosa has increased PAD activity and airway inflammation is more prevalent in both seropositive, at-risk subjects (i.e. anti-CCP positive) and RA patients. C. An altered gut microbiome has been identified in multiple animal models and in human RA.
Microbiome and Murine models of inflammatory arthritis
The notion that gut microorganisms can modulate extra-intestinal autoimmunity is not novel. As the gut represents the largest reservoir from which the immune system actively samples antigens, it seems plausible that the microbial make-up of a susceptible individual could influence the initiation, progression, and/or intensity of local and systemic autoimmune disease. Seminal work in animal models conducted more than three decades ago have established a relationship between the development of inflammatory arthritis and the presence or absence of certain bacterial genera (Table). Rat models of adjuvant-induced and streptococcal cell wall-induced arthritis show protection against the development of arthritis in the presence of mucosal microbes. In both studies, germ-free reared rats showed increased vulnerability to arthritis [15, 16]. In contrast, a germ-free environment (sterile cages devoid of microorganisms) is protective against the development of arthritis in the spontaneous spondyloarthropathy model of HLA-B27 transgenic rats [17]. This may be explained by recent work showing that misfolded HLA-B27 in LPS-stimulated macrophages resulted in robust increases in proinflammatory cytokines IL-23 and IL-17, along with intestinal inflammation [18]. Additionally, HLA-B27 transgenic rats lack certain populations of dendritic cells important in maintaining tolerance to self-antigen in their mesenteric lymph nodes [19]. Similarly, both the IL-1 receptor antagonist knockout (IL-1RA −/−) and the K/BxN mouse models of arthritis remain healthy in germ-free environments. Gavaging these mice with Lactobacillus and segmented filamentous bacteria (SFB) respectively, is sufficient for development of autoimmunity and inflammatory arthritis via induction of a robust TH17 response [20, 21]. The ZAP-70 single-point mutation mouse model, SKG, also develops joint inflammation when reared in conventional cages. In these mice however, lung-residing fungal organisms appear to be responsible, as SKG mice harbor a larger respiratory fungal load and inflammatory arthritis can be induced by injection of beta-glucans, a component of fungal cell-wall [22]. Most recently, inflammatory arthritis was induced in rats by introducing oral antigens in the setting of mucosal barrier dysfunction [23]. Taken together, these studies reveal a role for the microbiome in various susceptible animal models and validate a mechanistic relationship between microbes, mucosal immunity, and joint inflammation.
TABLE.
Murine models of inflammatory arthritis, associated gut microbiota and potential immunological mechanisms involved. Four of the most recent animal models of inflammatory arthritis. The presence of bacteria seems to mainly act via inducing local and systemic Th17 cell response. Decreased production of Treg cells and increased gut permeability are also implicated.
| Animal Model | Identified taxa | Proposed Immune Mechanism |
|---|---|---|
| HLA-B27 transgenic rat | - Bacteroides | HLA-B27 misfolding; increased IL-23 and IL-17 |
| IL-1 receptor antagonist knockout mice (with TLR2/TLR4 double knock out) | - Lactobacillus | TLR2−/− leads to decreased Tregs and increased in IFN-γ production (more arthritis incidence/severity) TLR4−/− leads to decreased Th17 cells and IL-17 production (less arthritis incidence/severity) |
| K/BxN T-cell receptor transgenic mice | - Segmented filamentous bacteria (SFB) | Induction of Th17 cells in the intestinal lamina propria and periphery |
| Arthritis-susceptible *0401 transgenic mice | - Clostridium | Increased gut mucosal permeability with increased induction of Th17 gene transcripts |
A recent experiment investigated the microbiome of arthritis-susceptible *0401 transgenic mice and arthritis-resistant *0402 transgenic mice [24**]. The *0401 mice microbiome was dominated by Clostridium bacteria and bacterial diversity was not influenced by age or sex. In contrast, the resistant *0402 mice were enriched by the Porphyromonadaceae and Bifidobacteria families, and they displayed strong influences of age and sex. Additionally, the susceptible mice demonstrated increased mucosal permeability and an altered TH17 gene transcriptomic profile. These findings suggest the possibility that host genetics may indeed represent a predetermined factor for harboring a unique microbiome profile.
Gut dysbiosis and RA
Linking intestinal dysbiosis (i.e. alteration of the gut microbiome homeostasis) to RA pathogenesis is far from a novel concept. In fact, the toxemic factor hypothesis was originally formulated at the turn of the twentieth century. It proposed that the pathological abundance of gram negative bacteria in the intestinal canal led to an increase in toxic metabolites that entered the circulation and ultimately promoted joint inflammation. In fact, therapeutic regimens that theoretically target the intestinal flora and the gut-joint axis have been in use since the 1940s (Figure). Several of them even became disease-modifying antirheumatic drugs (DMARDs), such as sulfasalazine and minocycline. Since then, however, there have been relatively few studies directly evaluating the gut microbiota in human RA.
In one study, low-throughput 16S hybridization technique (only 8 probes included) was used to evaluate the fecal microbiota of patients with newly diagnosed RA [25]. RA patients showed overall fewer bacterial counts when compared to patients with fibromyalgia. A second study investigated the composition of fecal lactobacillus communities in early RA patients. Using specific lactobacillus primers, this study revealed a higher diversity of the genera in early disease [26]. Specifically, there were increased levels of L. salivarius, L. iners, and L. ruminis in the RA group compared to controls, and the presence of L. mucosae was unique to RA patients.
We have recently observed that the majority of new-onset, DMARD-naïve RA patients carry high levels of Prevotella copri, a unique species that defines the microbial genome of these patients (manuscript in press, eLife Journal 2013). Gut Prevotella, a gram-negative anaerobe, appears to be relevant in many inflammatory and autoimmune conditions. In addition to being considered one of the three recently described enterotypes, Prevotella has also been found in patients with inflammatory bowel disease and was sufficient to induce colitis in an inflammasome knock out mouse model (i.e. NLRP6 pathway) [27]. Most recently, Koeth et al. demonstrated that high abundance of gut Prevotella in healthy subjects correlated with increased plasma levels of trimethylamine N-oxide (TMAO), a substance predictive of cardiovascular events in humans. Metabolism by intestinal microbiota of dietary L-carnitine, a compound abundant in red meat, also produced TMAO and accelerated atherosclerosis in mice, but this did not occur if the intestinal microbiota was concurrently suppressed [28*]. These findings are relevant because they may partially help explain the higher incidence of cardiovascular disease in some RA patients.
Despite the recent animal and human evidence implicating the intestinal microbiome in RA development, it remains uncertain whether these states of gut dysbiosis represent a causal-etiologic factor or rather a secondary effect of local and systemic inflammation. Further work is required to better understand the enzimatic and metabolic effects underlying the changes in gut bacterial communities in RA patients.
Evidence for the RA-periodontal disease association
Periodontal disease (PD) is a common polybacterial infection of the gingival tissue [29]. RA and PD are known to share pathogenic mechanisms and immunologic pathways of bone and tissue destruction. In both diseases, adaptive and innate immune cells promote secretion of pro-inflammatory cytokines (i.e. IL-1, IL-6, and TNF) and other degrading substances such as matrix metalloproteinases and reactive oxygen species [30–31]. Moreover, although proposed more than a century ago, an epidemiologic association between RA and PD has recently been validated. A number of studies have now demonstrated an increased incidence and severity of PD in RA [32, 33*, 34]. A recent population-based effort reinforced an association, albeit rather weak, between a prior history of PD and new onset RA, suggesting a causal role for PD in RA pathogenesis [35]. These findings are not universal, however, and a recent study utilizing patient-reported data failed to demonstrate the association [36]. Additional longitudinal studies are therefore needed to establish a role for PD in RA pathogenesis. Intriguingly, however, treating PD appears to improve RA severity and concomitantly decrease pro-inflammatory biomarkers of disease activity such as CRP, ESR, TNF, and IL-1beta [37–38]. RA patients with active PD are also less responsive to anti-TNF-α inhibition, indicating a potential role for the co-treatment of PD in TNF refractory patients [39].
Much research has focused on the specific bacteria associated with PD. The healthy oral microbiome is second in diversity only to the gut and is dominated by the bacterial phyla Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Spirochaetes, and Fusobacteria [40]. PD has traditionally been linked to the “red-complex” bacteria, which include P. gingivalis, Treponema denticola, and Tannerella forsythia [41]. New technologies, particularly the aforementioned 16S pyrosequencing, have confirmed these results. Meanwhile, other taxa have also been associated with both PD development and other aggravating environmental risk factors such as cigarette smoking [42 – 43*].
Despite the established knowledge that PD is actually a polymicrobial disorder, RA research has focused mostly on P. gingivalis. This bacterium is presumably the only known prokaryote with a gene encoding for peptidylarginine deiminase (PAD), an enzyme that converts arginine residues into citrulline [44 – 45]. The citrullination of mucosal protein-peptides such as vimentin, keratin, and α-enolase reportedly generates neo-epitopes that can then foster loss of immune tolerance and eventually the production of anti-citriullinated protein antibodies (ACPAs). ACPAs are the most specific biomarkers for RA to date, are found in 70–80% of patients, and are known to confer worse prognosis [46 – 47]. The prevalent hypothesis posits that Pg (and the humoral response to it) represents an antigenic source for the creation of ACPAs and subsequent progression to RA [30]. This was further validated by studies showing that P. gingivalis-derived PAD was capable of citrullinating the human fibrinogen and α-enolase isoforms [48], and that human α-enolase could also induce autoimmunity and inflammatory arthritis in DR4-transgenic mice [49]. Additionally, treatment of PD can decrease the levels of circulating ACPAs and anti- P. gingivalis antibodies [50].
Exposure to P. gingivalis has been indirectly assessed serologically in both clinical RA and in subjects considered to be at-risk for RA development (i.e., first-degree relatives, Native Americans or those with ACPA positivity without synovitis). Anti- P. gingivalis antibodies correlated with the presence of ACPAs in both groups when compared to controls [51, 52*, 53]. Utilizing pyrosequencing to assess the oral microbiome of patients with new onset, DMARD-naive RA, we showed a direct presence of P. gingivalis in slightly more than half the cohort. The relative abundance of P. gingivalis, although not statistically significant, was two-fold higher in patients with new-onset RA compared to non-RA controls. Most microbial differences, however, could be attributed to the presence of concomitant advanced PD [54**]. The presence of other taxa, such as Anaerglobus geminatus and Prevotella/Leptotrichia were intriguingly associated with ACPA seropositivity and clinical RA. Overall, the cumulative data argues for a potential involvement of PD and P. gingivalis in the development of autoimmunity and clinical RA. Additional population and basic investigations are required to elucidate this complex picture involving altered bacterial diversity in periodontal tissues and the consequent microbe-host interactions in the form of local and systemic inflammatory responses.
Airway inflammation and RA – a role for the microbiome?
The human airways constitute a vast mucosal surface constantly challenged by a high microbial-derived antigenic diversity. Similarly to the oral cavity and the intestinal tract, the respiratory mucosa is home to its own characteristic microbiome [55–57]. Interestingly, cigarette smoking (arguably the best studied environmental risk factor for RA) is not definitively responsible for producing an altered lung microbiome [55–56, 58]. Although studies found no association between smoking and modified bacterial diversity, Segal et al. [58] did note a correlation between increased airway inflammation and transposition of traditionally supraglottic bacteria into the lungs, particularly with Prevotella and Porphyromonas.
Although there are no reports directly evaluating the lung microbiome in patients with RA, several studies have suggested a possible role in pathogenesis. Chief among those is the evidence that the lung is a site for local citrullination. Cigarette smoking has been shown to generate increased PAD expression and protein citrullination in bronchoalveolar lavage (BAL) cells of smokers without arthritis [59], and a subset of patients with lung disease in the absence of existing RA test positive for plasma anti-CCP antibodies. Moreover, PAD polymorphisms, such as PADI4, have also been shown to predispose male smokers to RA [60]. This association appears to be cigarette smoking specific since non-smoking particulate matter (i.e. a measure of air pollution) was not associated with increased risk of progression to RA in asymptomatic seropositive patients [61].
Recent studies examined early RA patients and at-risk individuals (i.e. autoantibody positive) for signs of airway abnormalities using high resolution CT [62**]. Signs of inflammation, such as bronchial wall thickening, bronchiectasis, and centrilobular opacities, were common in at-risk subjects and were similar to airway abnormalities seen in patients with early RA. In a separate report, anti-CCP antibodies and RF were found in induced sputa of several patients from the same cohort [63**]. Interestingly, enrichment of the lung microbiome with supraglottic taxa (including oral Porphyromonas and Prevotella) was associated with increased airway inflammation. Taken together, these data suggest that the lung may be an early site of autoimmune-related injury and/or potentially a site of generation of RA-related autoimmunity, perhaps due to the presence of periodontal tissue-derived pro-inflammatory microbiota. These data suggests that the lungs participate in the citrullination of proteins and consequent generation of autoimmunity. The potential role of PD-associated bacteria in inciting those inflammatory responses, perhaps due to microaspiration, has only recently been described and requires further investigation.
Conclusions
A growing body of evidence linking the microbiome with RA pathogenesis is now becoming available. Animal models set a solid foundation for linking bacterial antigenicity to the generation of inflammatory arthritis. However, the application of these studies to human populations is complex, and further investigations into bacterial-host mucosal interactions are needed. The microbiome is extensive and multiple human body sites are home to a wide diversity of bacterial populations. The PD-associated bacteria are the most extensively studied. Strong evidence confirms the ability of Pg to citrullinate host proteins, but the relevance of this one bacterium is less obvious when assessed in isolation. A polymicrobial effect in periodontal inflammation and RA pathogenesis seems more likely. The gut carries the largest bacterial burden and is the most directly linked to RA when evaluating animal studies. To date, only a handful of human studies have investigated the gut microbiome in RA patients. Results are encouraging but mechanistic insights to address causality are still lacking. The notion that the respiratory mucosa could play a role in RA pathogenesis is relatively new. Advanced imaging techniques indicate that airway inflammation and autoimmunity predate joint disease, as they are often present in the pre-clinical phases of RA. Taken together, in vitro, animal, and human data suggest that host-bacterial interactions may play a role in autoimmunity initiation.
One possibility is that bacteria sharing similar pro-inflammatory properties may serve as triggering factors in various mucosal sites, particularly in genetically predisposed individuals. “Pan-microbiome” studies will need to be conducted as they may hold an important key to a deeper understanding of RA pathogenesis. The validation of recent findings while studying the microbiome in the various phases of RA (and in different at-risk populations) will be of the utmost importance. Synergistic in vivo and in vitro work will also be required to advance our knowledge in this field, which is promising for its discovery potential in biomarker identification and new therapeutic targets.
Key Points.
Animal models of inflammatory arthritis demonstrate the importance of the microbiome in the development of local and systemic autoimmunity.
There is validated evidence that the autoimmune process in RA begins many years before the emergence of clinical synovitis. The source of autoantibodies such as RF and anti-CCP appear to be extra-articular in nature since seropositive individuals without RA have normal synovial tissue.
Mucosal surfaces constantly exposed to bacterial antigens are being studied as possible sites of autoimmunity initiation.
Peridontal disease is strongly linked to RA, and an altered oral microbiome has been identified in patients with early RA. P. gingivalis and its enzymatic machinery for peptide citrullination appears to play a role in pathogenesis.
The community bacterial composition in the gut is altered in mouse models of inflammatory arthritis and in human patients with RA.
Airway inflammation and autoimmunity (i.e. anti-CCP and RF antibodies) appear to be present even at the pre-clinical phase of disease. Studies of the RA lung microbiome are currently underway.
Footnotes
Disclosures: None
No conflicts of interest
References
- 1.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-Bacterial Mutualism in the Human Intestine. Science. 2005 Mar 25;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Hooper LV, Midtvedt T, Gordon JI. How Host-Microbial Interactions Shape the Nutrient Environment of the Mammalian Intestine. Annual Review of Nutrition. 2002;22(1):283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 3.Zoetendal EG, Akkermans ADL, Vos WMD. Temperature Gradient Gel Electrophoresis Analysis of 16S rRNA from Human Fecal Samples Reveals Stable and Host-Specific Communities of Active Bacteria. Appl Environ Microbiol. 1998 Oct 1;64(10):3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010 Mar 4;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012 Jun 14;486(7402):207–214. doi: 10.1038/nature11234. The Human Microbiome Project (HMP) has investigated a large number of humans in order to identify the “core” human flora and their enzymatic functions.
- 6.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol. 2011 Oct;7(10):569–578. doi: 10.1038/nrrheum.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the Human Intestinal Microbial Flora. Science. 2005 Jun 10;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011 May 12;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, et al. A Guide to Enterotypes across the Human Body: Meta-Analysis of Microbial Community Structures in Human Microbiome Datasets. PLoS Comput Biol. 2013 Jan 10;9(1):e1002863. doi: 10.1371/journal.pcbi.1002863. A deep computational analysis comparing multiple large human cohorts in order to evaluate for the presence of core microbial enterotypes across various body habitats.
- 10.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis & Rheumatism. 1987;30(11):1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 11.Eyre S, Bowes J, Diogo D, Lee A, Barton A, Martin P, et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012 Dec;44(12):1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielen MMJ, van Schaardenburg D, Reesink HW, van de Stadt RJ, van der Horst-Bruinsma IE, de Koning MHMT, et al. Specific autoantibodies precede the symptoms of rheumatoid arthritis: A study of serial measurements in blood donors. Arthritis & Rheumatism. 2004;50(2):380–386. doi: 10.1002/art.20018. [DOI] [PubMed] [Google Scholar]
- 13.Sokolove J, Bromberg R, Deane KD, Lahey LJ, Derber LA, Chandra PE, et al. Autoantibody Epitope Spreading in the Pre-Clinical Phase Predicts Progression to Rheumatoid Arthritis. PLoS ONE. 2012 May 25;7(5):e35296. doi: 10.1371/journal.pone.0035296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van de Sande MGH, de Hair MJH, van der Leij C, Klarenbeek PL, Bos WH, Smith MD, et al. Different stages of rheumatoid arthritis: features of the synovium in the preclinical phase. Ann Rheum Dis. 2011 May;70(5):772–777. doi: 10.1136/ard.2010.139527. [DOI] [PubMed] [Google Scholar]
- 15.Kohashi O, Kuwata J, Umehara K, Uemura F, Takahashi T, Ozawa A. Susceptibility to adjuvant-induced arthritis among germfree, specific-pathogen-free, and conventional rats. Infect Immun. 1979 Dec 1;26(3):791–794. doi: 10.1128/iai.26.3.791-794.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van den Broek MF, van Bruggen MC, Koopman JP, Hazenberg MP, van den Berg WB. Gut flora induces and maintains resistance against streptococcal cell wall-induced arthritis in F344 rats. Clin Exp Immunol. 1992 May;88(2):313–317. doi: 10.1111/j.1365-2249.1992.tb03079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernández-Sueiro JL, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994 Dec 1;180(6):2359–2364. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLay ML, Turner MJ, Klenk EI, Smith JA, Sowders DP, Colbert RA. HLA-B27 Misfolding and the Unfolded Protein Response Augment IL-23 Production and are Associated with Th17 Activation in Transgenic Rats. Arthritis Rheum. 2009 Sep;60(9):2633–2643. doi: 10.1002/art.24763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Utriainen L, Firmin D, Wright P, Cerovic V, Breban M, McInnes I, et al. Expression of HLA-B27 Causes Loss of Migratory Dendritic Cells in a Rat Model of Spondylarthritis. Arthritis Rheum. 2012 Oct;64(10):3199–3209. doi: 10.1002/art.34561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdollahi-Roodsaz S, Joosten LAB, Koenders MI, Devesa I, Roelofs MF, Radstake TRDJ, et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest. 2008 Jan 2;118(1):205–216. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H-J, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010 Jun 25;32(6):815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshitomi H, Sakaguchi N, Kobayashi K, Brown GD, Tagami T, Sakihama T, et al. A role for fungal β-glucans and their receptor Dectin-1 in the induction of autoimmune arthritis in genetically susceptible mice. J Exp Med. 2005 Mar 21;201(6):949–960. doi: 10.1084/jem.20041758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu D, Liu X, Su H, Chen X, Zhang H, Hu D, et al. Oral antigens induce rheumatoid arthritis-like inflammation in a rat model. Inflamm Res. 2013 Mar 1;62(3):291–297. doi: 10.1007/s00011-012-0577-9. [DOI] [PubMed] [Google Scholar]
- 24. Gomez A, Luckey D, Yeoman CJ, Marietta EV, Berg Miller ME, Murray JA, et al. Loss of Sex and Age Driven Differences in the Gut Microbiome Characterize Arthritis-Susceptible *0401 Mice but Not Arthritis-Resistant *0402 Mice. [cited 2013 Apr 13];PLoS One [Internet] 2012 Apr 24;7(4) doi: 10.1371/journal.pone.0036095. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3338357/. Murine evidence that host genes can predispose to specific bacterial communities.
- 25.Vaahtovuo J, Munukka E, Korkeamäki M, Luukkainen R, Toivanen P. Fecal Microbiota in Early Rheumatoid Arthritis. J Rheumatol. 2008 Aug 1;35(8):1500–1505. [PubMed] [Google Scholar]
- 26.Liu X, Zou Q, Zeng B, Fang Y, Wei H. Analysis of Fecal Lactobacillus Community Structure in Patients with Early Rheumatoid Arthritis. Curr Microbiol. 2013 Aug 1;67(2):170–176. doi: 10.1007/s00284-013-0338-1. [DOI] [PubMed] [Google Scholar]
- 27.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome is a regulator of colonic microbial ecology and risk for colitis. Cell. 2011 May 27;145(5):745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013 May;19(5):576–585. doi: 10.1038/nm.3145. The intestinal microbiome is implicated in predisposing carnivores to the development of atherosclerosis. The increased heart disease observed in RA may share a similar mechanism.
- 29.Kawar N, Gajendrareddy PK, Hart TC, Nouneh R, Maniar N, Alrayyes S. Periodontal Disease for the Primary Care Physician. Disease-a-Month. 2011 Apr;57(4):174–183. doi: 10.1016/j.disamonth.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Liao F, Li Z, Wang Y, Shi B, Gong Z, Cheng X. Porphyromonas gingivalis may play an important role in the pathogenesis of periodontitis-associated rheumatoid arthritis. Medical Hypotheses. 2009 Jun;72(6):732–735. doi: 10.1016/j.mehy.2008.12.040. [DOI] [PubMed] [Google Scholar]
- 31.Berthelot J-M, Le Goff B. Rheumatoid arthritis and periodontal disease. Joint Bone Spine. 2010 Dec;77(6):537–541. doi: 10.1016/j.jbspin.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 32.De Pablo P, Dietrich T, McAlindon TE. Association of periodontal disease and tooth loss with rheumatoid arthritis in the US population. Journal of Rheumatology. 2008;35(1):70–76. [PubMed] [Google Scholar]
- 33. Potikuri D, Dannana KC, Kanchinadam S, Agrawal S, Kancharla A, Rajasekhar L, et al. Periodontal disease is significantly higher in non-smoking treatment-naive rheumatoid arthritis patients: results from a case-control study. Ann Rheum Dis. 2012 Sep 1;71(9):1541–1544. doi: 10.1136/annrheumdis-2011-200380. This study demonstrates that the association between RA and PD remains positive even when the common risk factor, cigarette smoking, is excluded.
- 34.Pischon N, Pischon T, Kröger J, Gülmez E, Kleber B-M, Bernimoulin J-P, et al. Association Among Rheumatoid Arthritis, Oral Hygiene, and Periodontitis. Journal of Periodontology. 2008 Jun;79(6):979–986. doi: 10.1902/jop.2008.070501. [DOI] [PubMed] [Google Scholar]
- 35.Chen H-H, Huang N, Chen Y-M, Chen T-J, Chou P, Lee Y-L, et al. Association between a history of periodontitis and the risk of rheumatoid arthritis: a nationwide, population-based, case-control study. [cited 2013 May 30];Annals of the Rheumatic Diseases [Internet] 2012 Aug 31; doi: 10.1136/annrheumdis-2012-201593. Available from: http://ard.bmj.com/cgi/doi/10.1136/annrheumdis-2012-201593. [DOI] [PubMed] [Google Scholar]
- 36.Arkema EV, Karlson EW, Costenbader KH. A Prospective Study of Periodontal Disease and Risk of Rheumatoid Arthritis. J Rheumatol. 2010 Sep 1;37(9):1800–1804. doi: 10.3899/jrheum.091398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erciyas K, Sezer U, Üstün K, Pehlivan Y, Kısacık B, Şenyurt S, et al. Effects of periodontal therapy on disease activity and systemic inflammation in rheumatoid arthritis patients. Oral Diseases. 2013 May;19(4):394–400. doi: 10.1111/odi.12017. [DOI] [PubMed] [Google Scholar]
- 38.Bıyıkoğlu B, Buduneli N, Aksu K, Nalbantsoy A, Lappin DF, Evrenosoğlu E, et al. Periodontal therapy in chronic periodontitis lowers gingival crevicular fluid interleukin-1beta and DAS28 in rheumatoid arthritis patients. Rheumatol Int. :1–10. doi: 10.1007/s00296-013-2781-5. [DOI] [PubMed] [Google Scholar]
- 39.Savioli C, Ribeiro ACM, Fabri GMC, Calich AL, Carvalho J, Silva CA, et al. Persistent Periodontal Disease Hampers Anti–Tumor Necrosis Factor Treatment Response in Rheumatoid Arthritis. Journal of Clinical Rheumatology. 2012 May;1 doi: 10.1097/RHU.0b013e31825828be. [DOI] [PubMed] [Google Scholar]
- 40.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu W-H, et al. The Human Oral Microbiome. J Bacteriol. 2010 Oct 1;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wade WG. The oral microbiome in health and disease. Pharmacological Research. 2013 Mar;69(1):137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New Bacterial Species Associated with Chronic Periodontitis. J DENT RES. 2003 May 1;82(5):338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 43. Bizzarro S, Loos BG, Laine ML, Crielaard W, Zaura E. Subgingival microbiome in smokers and non-smokers in periodontitis: an exploratory study using traditional targeted techniques and a next-generation sequencing. Journal of Clinical Periodontology. 2013;40(5):483–492. doi: 10.1111/jcpe.12087. This study shows that cigarette smoking leads to an altered oral microbiome. Lower bacterial diversity was associated with increased PD severity.
- 44.McGraw WT, Potempa J, Farley D, Travis J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infection and Immunity. 1999;67(7):3248–3256. doi: 10.1128/iai.67.7.3248-3256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenstein ED, Greenwald RA, Kushner LJ, Weissmann G. Hypothesis: the humoral immune response to oral bacteria provides a stimulus for the development of rheumatoid arthritis. Inflammation. 2004 Dec;28(6):311–318. doi: 10.1007/s10753-004-6641-z. [DOI] [PubMed] [Google Scholar]
- 46.McInnes IB, Schett G. The Pathogenesis of Rheumatoid Arthritis. New England Journal of Medicine. 2011;365(23):2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 47.Barra L, Scinocca M, Saunders S, Bhayana R, Rohekar S, Racapé M, et al. Anti–Citrullinated Protein Antibodies in Unaffected First-Degree Relatives of Rheumatoid Arthritis Patients. Arthritis & Rheumatism. 2013;65(6):1439–1447. doi: 10.1002/art.37911. [DOI] [PubMed] [Google Scholar]
- 48.Wegner N, Wait R, Sroka A, Eick S, Nguyen K-A, Lundberg K, et al. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α-enolase: Implications for autoimmunity in rheumatoid arthritis. Arthritis & Rheumatism. 2010;62(9):2662–2672. doi: 10.1002/art.27552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kinloch AJ, Alzabin S, Brintnell W, Wilson E, Barra L, Wegner N, et al. Immunization with Porphyromonas gingivalis enolase induces autoimmunity to mammalian α-enolase and arthritis in DR4-IE-transgenic mice. Arthritis Rheum. 2011 Dec;63(12):3818–3823. doi: 10.1002/art.30639. [DOI] [PubMed] [Google Scholar]
- 50.Okada M, Kobayashi T, Ito S, Yokoyama T, Abe A, Murasawa A, et al. Periodontal Treatment Decreases Levels of Antibodies to Porphyromonas Gingivalis and Citrulline in Patients With Rheumatoid Arthritis and Periodontitis. Journal of Periodontology. 2013 May 23;:1–12. doi: 10.1902/jop.2013.130079. [DOI] [PubMed] [Google Scholar]
- 51.Mikuls TR, Payne JB, Reinhardt RA, Thiele GM, Maziarz E, Cannella AC, et al. Antibody responses to Porphyromonas gingivalis (P. gingivalis) in subjects with rheumatoid arthritis and periodontitis. International Immunopharmacology. 2009 Jan;9(1):38–42. doi: 10.1016/j.intimp.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mikuls TR, Thiele GM, Deane KD, Payne JB, O’Dell JR, Yu F, et al. Porphyromonas gingivalis and disease-related autoantibodies in individuals at increased risk of rheumatoid arthritis. Arthritis & Rheumatism. 2012;64(11):3522–3530. doi: 10.1002/art.34595. This study found a significant association between the presence of ACPA and anti-Pg antibodies in patients at risk for RA development (shared epitope, first degree relatives with RA).
- 53.Hitchon CA, Chandad F, Ferucci ED, Willemze A, Ioan-Facsinay A, Woude D van der, et al. Antibodies to Porphyromonas gingivalis Are Associated with Anticitrullinated Protein Antibodies in Patients with Rheumatoid Arthritis and Their Relatives. J Rheumatol. 2010 Jun 1;37(6):1105–1112. doi: 10.3899/jrheum.091323. [DOI] [PubMed] [Google Scholar]
- 54. **Scher JU, Ubeda C, Equinda M, Khanin R, Buischi Y, Viale A, et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis Arthritis & Rheumatism 201264103083–3094. New-onset and chronic RA patients demonstrate increased PD severity. Using high-throughput DNA sequencing, RA patients were found to have increased relative abundance of several PD-associated taxa.
- 55.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the Respiratory Microbiome in Healthy Nonsmokers and Smokers. American Journal of Respiratory and Critical Care Medicine. 2013 May 15;187(10):1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, et al. Analysis of the Lung Microbiome in the “Healthy” Smoker and in COPD. PLoS ONE. 2011 Feb 22;6(2):e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered Microbial Communities in Asthmatic Airways. PLoS ONE. 2010 Jan 5;5(1):e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Chen H, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013 Jul 1;1(1):19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Makrygiannakis D, Hermansson M, Ulfgren A-K, Nicholas AP, Zendman AJW, Eklund A, et al. Smoking increases peptidylarginine deiminase 2 enzyme expression in human lungs and increases citrullination in BAL cells. Ann Rheum Dis. 2008 Oct 1;67(10):1488–1492. doi: 10.1136/ard.2007.075192. [DOI] [PubMed] [Google Scholar]
- 60.Kochi Y, Thabet MM, Suzuki A, Okada Y, Daha NA, Toes REM, et al. PADI4 polymorphism predisposes male smokers to rheumatoid arthritis. Ann Rheum Dis. 2011 Mar 1;70(3):512–515. doi: 10.1136/ard.2010.130526. [DOI] [PubMed] [Google Scholar]
- 61.Gan RW, Deane KD, Zerbe GO, Demoruelle MK, Weisman MH, Buckner JH, et al. Relationship between air pollution and positivity of RA-related autoantibodies in individuals without established RA: a report on SERA. [cited 2013 May 30];Ann Rheum Dis [Internet] 2013 Apr 9; doi: 10.1136/annrheumdis-2012-202949. Available from: http://ard.bmj.com/content/early/2013/04/08/annrheumdis-2012-202949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Demoruelle MK, Weisman MH, Simonian PL, Lynch DA, Sachs PB, Pedraza IF, et al. Brief Report: Airways abnormalities and rheumatoid arthritis–related autoantibodies in subjects without arthritis: Early injury or initiating site of autoimmunity? Arthritis & Rheumatism. 2012;64(6):1756–1761. doi: 10.1002/art.34344. Utilizing high resolution CT, this study demonstrates the presence of airway inflammation prior to joint synovitis in antibody positive patients.
- 63. Willis VC, Demoruelle MK, Derber LA, Chartier-Logan CJ, Parish MC, Pedraza IF, et al. Sputa autoantibodies in patients with established rheumatoid arthritis and subjects at-risk for future clinically apparent disease. Arthritis & Rheumatism. 2013 doi: 10.1002/art.38066. n/a–n/a. This study shows that patients at-risk for RA development have measurable auto-antibodies in sputa.