SUMMARY
Activating mutations in the EGF receptor (EGFR) are associated with clinical responsiveness to EGFR tyrosine kinase inhibitors (TKIs), such as erlotinib and gefitinib. However, resistance eventually arises, often due to a second EGFR mutation, most commonly T790M. Through a genome-wide siRNA screen in a human lung cancer cell line and analyses of murine mutant EGFR-driven lung adenocarcinomas, we found that erlotinib resistance was associated with reduced expression of neurofibromin, the RAS GTPase activating protein encoded by the NF1 gene. Erlotinib failed to fully inhibit RAS-ERK signaling when neurofibromin levels were reduced. Treatment of neurofibromin-deficient lung cancers with a MEK inhibitor restored sensitivity to erlotinib. Low levels of NF1 expression were associated with primary and acquired resistance of lung adenocarcinomas to EGFR TKIs in patients. These findings identify a subgroup of patients with EGFR mutant lung adenocarcinoma who might benefit from combination therapy with EGFR and MEK inhibitors.
Keywords: NF1, EGFR, KRAS, MAPK pathway, drug resistance, erlotinib, gefitinib, lung adenocarcinoma
INTRODUCTION
Lung cancer is the most frequently diagnosed cancer and leading cause of cancer-related mortality worldwide, accounting for nearly 1.4 million deaths per year (1, 2). Lung adenocarcinoma is the most common histological subtype of lung cancer, with 10–40% displaying activating mutations in the epidermal growth factor receptor gene (EGFR), occurring most frequently in never-smokers and in East Asian populations (3, 4). The presence of activating EGFR mutations strongly correlates with clinical responsiveness to the EGFR tyrosine kinase inhibitors (TKIs) erlotinib and gefitinib. However, while these drugs are initially very effective, resistance eventually arises in almost all patients, resulting in a modest overall survival benefit (3).
Several groups have investigated the mechanisms that underlie resistance to EGFR TKIs. One of the first resistance mechanisms identified in tumors in treated patients was a secondary mutation (T790M) in the EGFR gene (3, 5, 6), which enhances the affinity of EGFR for ATP and reduces binding of the inhibitor (5, 7–9) and accounts for about 50–60% of cases (5, 6, 10–12).
Additional mechanisms of resistance identified in experimental settings include activation of IGF1R, amplification of MET, HER2 or MAPK1, upregulation of the AXL receptor or its ligand, or activating mutations in PIK3CA (13–18). The majority of these genetic alterations have been confirmed in human EGFR mutant TKI-resistant lung tumor samples, varying in frequency from 5–20%. Histological transformation involved in resistance has also been observed in clinical samples, most prominently the conversion of EGFR inhibitor sensitive lung adenocarcinomas to drug-resistant small cell lung cancer (SCLC), described in about 5% of cases of acquired resistance to EGFR TKI (10, 12, 19). Less frequently, epithelial-to-mesenchymal transition, potentially related to loss of MED12 or upregulation of AXL, has been reported to result in a broad treatment resistance, including to EGFR TKIs (10, 18, 20, 21). Importantly, the mechanism of acquired resistance is still unknown for about one third of TKI-resistant lung adenocarcinomas (10, 20). In addition, it is evident that multiple mechanisms may contribute to resistance within one tumor (10, 15, 18, 22). Understanding the heterogeneity of molecular mechanisms involved in the evolution of resistance is therefore necessary to optimize the treatment of individual patients with mutant EGFR-driven tumors.
With the aim of improving therapy for EGFR mutant TKI-resistant lung cancer, we have set out to identify previously unknown mechanisms of resistance to TKIs in this disease. We first performed an in vitro systematic genome-wide analysis to screen for genes whose silencing by short interference RNA (siRNA) confers resistance to the EGFR inhibitor erlotinib in a human lung cancer cell line which is sensitive to this drug due to the presence of an activating mutation in EGFR. We also took an in vivo approach using a mutant EGFR-driven mouse lung cancer model, analyzing gene expression in tumors associated with the acquisition of resistance to erlotinib. As described previously, these mice develop lung adenocarcinomas that are initially responsive to EGFR TKIs, but develop resistance following repeated cycles of treatment (23). Analysis of the erlotinib-resistant mouse tumors revealed the T790M mutation in about 20% of cases, and occasional amplification of Met (23), suggesting that the erlotinib-resistant mouse tumors recapitulate the molecular changes identified in human lung tumors that acquire resistance to EGFR TKIs.
One gene emerging from these two approaches is the negative regulator of RAS proteins, NF1. We show that reduced expression of neurofibromin, the RAS GTPase activating protein (GAP) product of this gene, is associated with decreased sensitivity of human lung cancer cells to EGFR inhibitory drugs, due presumably to enhanced RAS signaling. Treatment of EGFR mutant lung cancer cells expressing low levels of neurofibromin with inhibitors of MEK, a RAS effector pathway component, restores their sensitivity to EGFR inhibitors. Moreover, the majority of erlotinib-resistant EGFR mutant mouse lung tumors that do not express the T790M mutation respond to co-treatment with a MEK inhibitor. Finally, we observed reduced NF1 expression in two independent datasets of paired pre- and post-treatment lung adenocarcinomas that acquired resistance to EGFR TKI treatment. We also found that low levels of NF1 expression in pre-treatment clinical specimens correlate with poor overall survival in EGFR-mutant lung cancer patients treated with EGFR TKI.
Collectively, our data identified low neurofibromin expression as a novel mechanism by which tumor cells are intrinsically less sensitive or acquire resistance to EGFR TKIs and provide a rationale for using drugs targeting MEK in combination with EGFR inhibitors as a therapeutic approach for the treatment of T790M-negative EGFR TKI-resistant lung cancer.
RESULTS
Genome-wide siRNA screen identifies determinants of erlotinib resistance
To identify novel determinants of resistance to the EGFR TKI erlotinib, we performed a genome-wide RNA interference screen examining cell viability in the absence or presence of the drug. We transfected the EGFR mutant human lung adenocarcinoma-derived PC9 cell line that is exquisitely sensitive to EGFR TKIs with a library of siRNA pools targeting ~21,000 unique human transcripts. Forty-eight hours after transfection, culture medium was replaced and cells were incubated for an additional 72hrs, in the presence or absence of erlotinib (Figure 1A). The experiment was performed in triplicate for both conditions. We used an erlotinib concentration slightly above the IC50 determined for PC9 cells, favoring identification of siRNAs that confer resistance to the drug, but still allowing detection of siRNAs that enhance killing in the same screen.
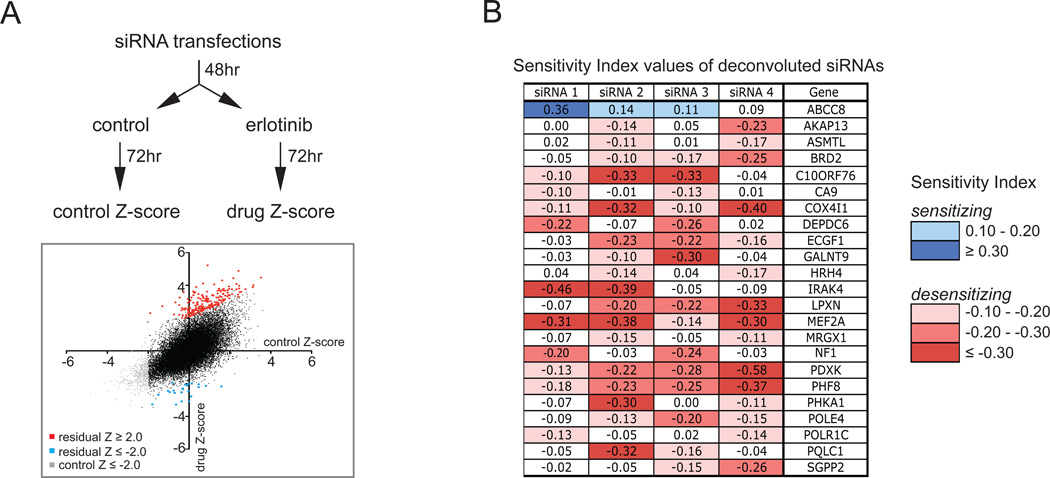
Figure 1. Genome-wide siRNA screen identifies determinants of resistance to erlotinib treatment.
A. Schematic diagram of the genome-wide siRNA screens performed in PC9 cells. Two sets of 384-well plates were reverse transfected with the genome-wide siRNA library and after 48hr treated with 30 nM erlotinib or left untreated (control) for 72 hours. Cells were fixed, stained with DAPI, and cell number was quantified on a microplate cytometer to obtain a Z-score for each siRNA in both conditions. Median Z-scores from the triplicate experiments are presented on the graph. siRNAs desensitizing cells to the drug are indicated in red (residual Z-score ≥ 2.0) and sensitizing siRNAs are indicated in blue (residual Z-score ≤ −2.0), whereas siRNAs that killed cells in the absence of the drug (control Z-score ≤ −2.0) are indicated in grey, and siRNAs that have no effect on modulating the response of cells to erlotinib (-2.0 < residual Z-score < 2.0, with control Z-score > 2.0) are indicated in black.
B. Tabulated sensitivity index-values of the deconvoluted siRNAs targeting 23 genes, of which two out of the four deconvoluted siRNAs had a substantial sensitizing (≥ 0.10) or desensitizing (≤ −0.10) effect on cell viability when cultured in the presence of erlotinib.
The effect of the individual siRNA pools on cell survival was analyzed in drug treated versus untreated conditions (Table S1) and we selected siRNAs that showed a substantial differential effect (Residual Z-score ≥ 2.0 or ≤ −2.0) without killing the untreated cells (Control Z-score ≥ −2.0; the Z-score corresponds to how many standard deviations away from the mean of the population an individual siRNA lies, see Experimental Procedures). Using these criteria, we identified 242 siRNA pools, of which 212 enhanced and 30 decreased cell survival in the presence of erlotinib (Table S2). To test the reproducibility of our findings, we repeated the genome-wide siRNA screen in an independent experiment. Ranking the siRNA pools based on their Residual Z-scores from this repeat screen revealed that 106 of the previously identified 242 siRNA pools were in the top 5% of the repeat screen list. Almost all of these 106 siRNA pools enhanced survival in the presence of erlotinib (Figure S1 and Table S2). The validated 106 siRNA pools were then taken forward to a deconvolution screen, where the four individual siRNA oligos targeting each gene were analyzed separately.
We established the influence of each individual siRNA-induced silencing on erlotinib sensitivity by determining the Sensitivity Index (SI) that takes into account the individual effects of erlotinib and siRNA-induced knockdown on cell viability (24). Using a cut-off value of ≥ 0.10 for sensitizing siRNAs and ≤ −0.10 for desensitizing siRNAs, for 23 siRNA pools a reproducible effect of at least two out of four of their individual deconvoluted siRNAs could be found (Figure 1B).
Nf1 down-regulation in a mouse model of erlotinib-resistant EGFR mutant lung cancer
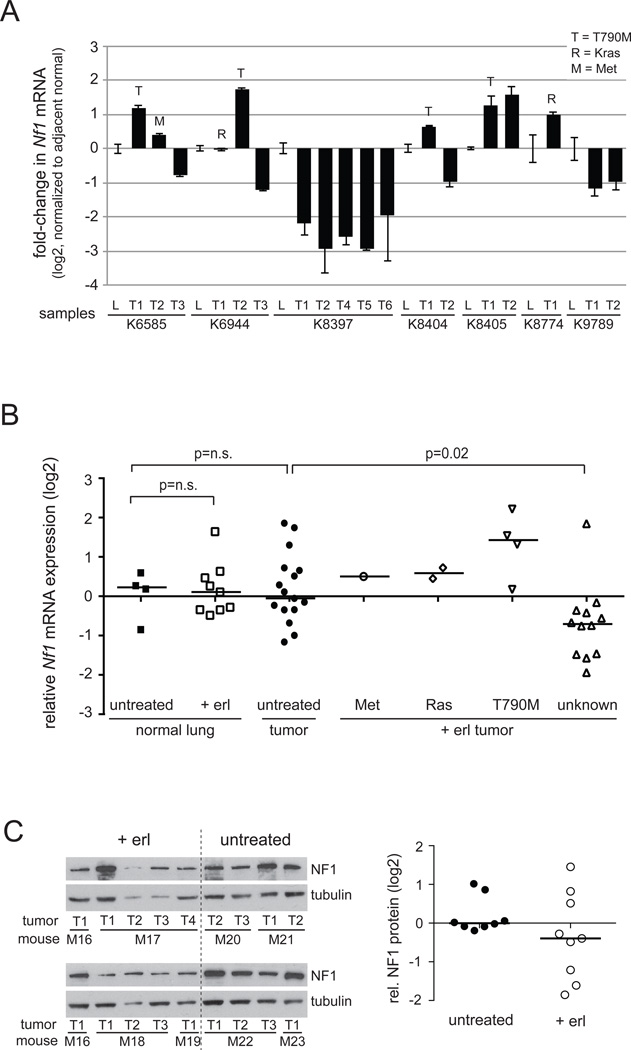
One of these genes, NF1, stood out since its gene product neurofibromin has a recognized negative regulatory role in signaling downstream of EGFR due to its function as a RAS GTPase activating protein (25, 26), suggesting a possible mechanistic rationale for its association with acquisition of resistance to EGFR inhibitory drugs. To assess the possible relevance of these in vitro results on resistance to EGFR inhibitory drugs in vivo we made use of an inducible mouse model of EGFR-driven lung cancer (23, 27). In this model, expression of mutant EGFR leads to the development of lung adenocarcinomas that are sensitive to erlotinib treatment. Long-term intermittent treatment of these mice with erlotinib, however, leads to the outgrowth of resistant tumors. To assess whether the genes identified in our siRNA screen showed altered expression between untreated (erlotinib-sensitive) tumors and erlotinib-resistant tumors, we compared the expression levels of Nf1 in erlotinib-resistant tumors and corresponding adjacent normal lung using quantitative RT-PCR analysis. These experiments revealed a decrease in Nf1 mRNA levels compared to normal lung in ten of eighteen erlotinib-resistant tumors, of which seven showed a more than 2-fold decrease (Figure 2A). Comparable results were obtained with two additional quantitative RT-PCR assays using different primer sets (Figure S2). Interestingly, tumors bearing the EGFR-T790M gatekeeper mutation, Kras mutations or Met amplification did not show decreased Nf1 expression, suggesting that loss of neurofibromin could be selected for by EGFR TKIs in the absence of other mechanisms of resistance. Gene expression profiling of erlotinib sensitive and resistant tumors from these mice failed to show significant differential expression of any of the other genes emerging from the siRNA screen of PC9 cells listed in Figure 1B.
Figure 2. Reduced Nf1 expression in erlotinib-resistant EGFR-driven lung tumors.
A. Quantitative RT-PCR for Nf1 in erlotinib-resistant mutant EGFR-induced murine lung tumors. cDNA extracted from tumor nodules and adjacent lung from each individual mouse was amplified using Taqman assays to Nf1 and the lung epithelial marker Surfactant Protein C to normalize for epithelial content in each samples. The fold-change in Nf1 expression vs. adjacent lung for each tumor is shown in log2. Tumors harbouring the EGFR T790M mutation, MET amplification and a Kras mutation are indicated. On the X-axis, L=lung and T=Tumor and the individual mouse numbers are indicated.
B. Nf1 expression levels of untreated (erlotinib sensitive) and erlotinib-treated (+ erl; erlotinib resistant) tumors and adjacent normal lung from EGFR mutant mice are plotted in log2 scale. Erlotinib-resistant tumors are subdivided according to known secondary lesions present in the tumors as indicated. Mann-Whitney U-test was performed to determine p-values.
C. NF1 protein levels in untreated and erlotinib-treated tumors were analyzed by Western blotting. NF1 expression was first normalized towards the loading control, tubulin, and then towards sample M1T1 that was present on both blots. NF1 expression relative to the median expression in the untreated tumors is plotted in the right graph in log2 scale. Mouse (M) and tumor (T) samples are indicated.
Since Nf1 is a known tumor suppressor, neurofibromin expression might be decreased during tumor progression independent of erlotinib treatment. To directly address this, one would ideally compare erlotinib-sensitive versus erlotinib-resistant tumors derived from the same animal. Regrettably, such material is not available. Instead, we analyzed the relative amounts of Nf1 mRNA in EGFR-induced lung tumors and adjacent normal lung from untreated and erlotinib-treated animals. This analysis showed no significant differences in Nf1 expression between normal lung samples and untreated tumors, whereas erlotinib-resistant tumors with an unknown resistance mechanism did express significantly lower levels of Nf1 compared to untreated (erlotinib-sensitive) tumors (Figure 2B). In an independent set of tumors, we evaluated NF1 protein expression in erlotinib-treated tumors versus untreated tumors, and confirmed lower NF1 protein expression in a subset of erlotinib-resistant tumors relative to those that had not been exposed to the drug (Figure 2C).
Overall, these observations with mouse lung tumors confirm our data with human lung cancer cell lines, showing that low Nf1 expression is associated with erlotinib resistance in EGFR-driven lung tumors that lack known resistance mechanisms such as the EGFRT790M mutation or MET amplification.
Reduced NF1 expression confers resistance of lung cancer cell lines to erlotinib
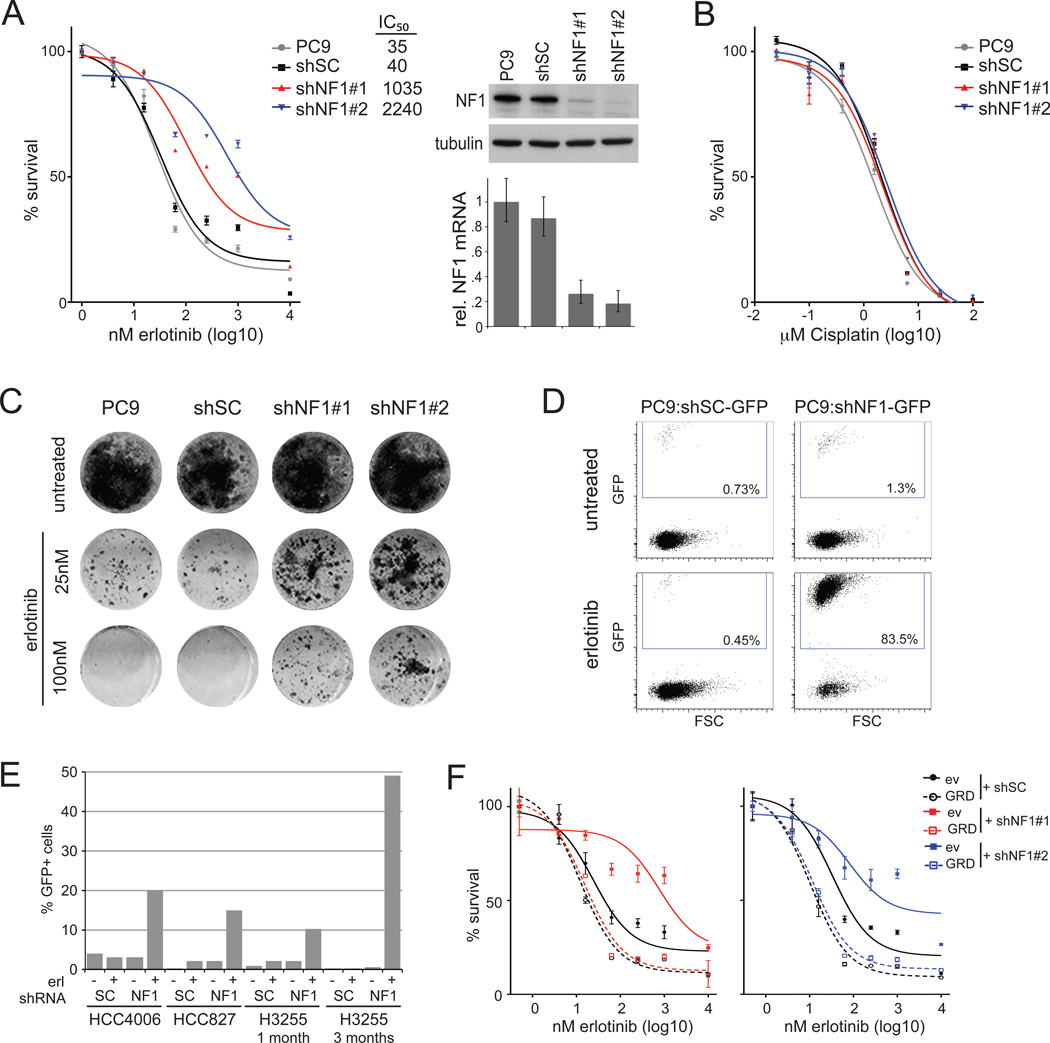
To validate the possible role of neurofibromin in erlotinib resistance, we introduced into PC9 cells two individual shRNAs (#1 and #2) targeting different non-overlapping regions of the NF1 coding sequence, that are distinct to the previously used siRNA sequences. A non-silencing scrambled shRNA (shSC) was used as a control throughout the study. Both NF1 targeting shRNA constructs decreased sensitivity to erlotinib, with a 26-fold increase in the drug concentration required for an absolute survival inhibition of 50% (IC50) for shNF1#1 and 56-fold for shNF1#2 (Figure 3A) as determined by Cell Titer Blue measurement. The shRNA constructs efficiently suppressed neurofibromin mRNA and protein expression, with shNF1#2 having stronger effects. Whereas neurofibromin silencing conferred resistance of PC9 cells to erlotinib or another EGF receptor kinase inhibitor, gefitinib, the response to chemotherapeutic agents such as cisplatin or docetaxel was not affected (Figure 3B and Figure S3A).
Figure 3. NF1 silencing confers resistance of PC9 lung adenocarcinoma cells to erlotinib.
A. Silencing of NF1 increases cell survival in the presence of erlotinib. PC9 cells are uninfected (PC9) or stably infected with shRNA containing a non-silencing control sequence (shSC) or an NF1-targeting sequence (shNF1#1 or #2). Cells were treated with the indicated concentrations of erlotinib for 72hr before cell viability was measured by Cell Titer Blue and normalized to untreated controls. Error bars denote standard error of the mean (SEM). Drug concentrations inducing 50% inhibition in survival (IC50-values; nM) are indicated for each cell line. Knockdown ability of each shRNA is demonstrated both at mRNA level and protein level using quantitative RT-PCR and immunoblotting, respectively. Error bars denote standard deviation (SD).
B. Silencing of NF1 does not affect the sensitivity to cisplatin. Cells were treated with the indicated concentrations of cisplatin for 72hr before cell viability was measured and normalized to untreated controls. Error bars denote SEM.
C. NF1 downregulation enhances cell survival in longer-term colony formation assays. Cells were fixed and stained after 14 days treatment with indicated concentrations of erlotinib. Uninfected PC9 cells or control shRNA-infected cells (shSC) were used as control.
D. Selective outgrowth of NF1-silenced cells in the presence of erlotinib. GFP negative PC9 cells were spiked with approximately 1% GFP positive shRNA-infected cells, non-silencing control (shSC) or targeting NF1 (shNF1#2), and grown for 4 weeks in the presence or absence of 30nM erlotinib. Cells were collected and analyzed for GFP expression by FACS (x-axis). The percentage of GFP positive cells is indicated.
E. Selective outgrowth of NF1-silenced cells in the presence of erlotinib in NSCLC cell lines HCC4006, HCC827 and H3255. GFP negative cells were spiked with approximately 1% GFP positive shRNA-infected cells, non-silencing control (shSC) or targeting NF1 (shNF1#2), and grown for 1 or 3 months in the presence or absence of 30nM erlotinib. Cells were collected and analyzed for GFP expression by FACS (plots presented in Figure S3D). The percentage of GFP positive cells is indicated.
F. Expression of NF1-GRD restores sensitivity to erlotinib. Cells were transfected with the NF1-GRD (dotted lines) or empty vector (ev) control (solid lines) prior to infection with control (shSC) or NF1-targeting shRNAs #1 or #2 (left and right panels respectively). Cells were treated with the indicated concentrations of erlotinib for 72hr before cell viability was measured and normalized to untreated controls. Error bars denote SEM. Expression of the NF1-GRD and silencing of endogenous NF1 are shown by quantitative RT-PCR and immunoblot in Figure S3E.
To test longer-term effects of NF1 silencing, we performed colony formation assays in which cells were cultured in the presence of erlotinib for 10 days. NF1 silencing substantially enhanced survival in these assays (Figure 3C). Prolonged treatment for four weeks in a competition assay, in which unlabeled parental PC9 cells were mixed in a ratio of 100:1 with GFP-labeled cells that expressed an shRNA targeting NF1 or a non-silencing control shRNA, showed substantial outgrowth of the shNF1 cells (Figure 3D). The percentage of GFP-positive cells remained approximately 1% in the absence of erlotinib, suggesting that NF1 silencing has little or no effect on the basal proliferation of PC9 cells. Selective outgrowth of NF1 knockdown cells was also seen in long-term assays with other lung adenocarcinoma cell lines harboring activating EGFR mutations that are sensitive to erlotinib treatment (Figure 3E, Figures S3B, S3C and S3D). Thus, our data provide evidence to suggest that NF1 silencing reduces the sensitivity of lung adenocarcinoma cells to erlotinib-induced cell death and/or growth arrest.
To exclude off-target effects of the shRNAs, cells were stably transfected with the GAP-related domain (GRD) of neurofibromin or a control empty vector prior to the shRNA infections. Whereas NF1 silencing desensitized the control cells to erlotinib treatment, it did not affect erlotinib sensitivity of the neurofibromin GRD expressing cells, in which the GRD was not targeted by the shNF1 constructs (Figure S3E). In fact, expression of neurofibromin GRD slightly increased erlotinib sensitivity (Figure 3F). These data confirm a role for neurofibromin in erlotinib response in lung adenocarcinoma cells. Moreover, the observation that the GAP-related domain of neurofibromin could restore sensitivity suggests that neurofibromin influences this sensitivity through its function as a negative regulator of RAS proteins, rather than through RAS-independent pathways.
NF1 silencing activates the MAPK pathway in presence of erlotinib
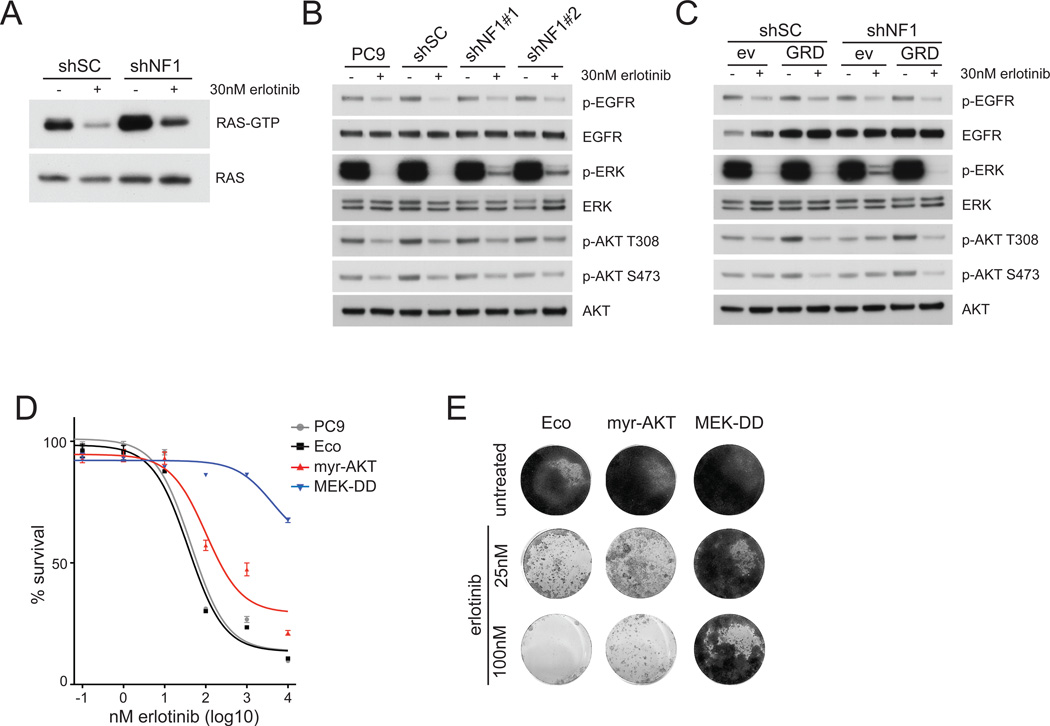
To investigate whether NF1 silencing promotes RAS activation in PC9 cells, we analyzed the amounts of active, GTP-bound RAS in the absence and presence of erlotinib. Although erlotinib reduced the amount of active RAS both in control and NF1 knockdown cells, cells retain substantially higher levels of active RAS in the presence of erlotinib when NF1 expression is reduced (Figure 4A).
Figure 4. NF1 downregulation activates MAPK pathway signaling.
A. NF1 silencing leads to increased levels of RAS activity in untreated and erlotinib treated conditions. Relative levels of RAS activation were assessed in cells infected with a control shRNA (shSC) or an shRNA targeting NF1 (shNF1#2). RAS immunoblots from RAS pull-down assays are shown (RAS-GTP), along with a RAS immunoblot from total cell lysates as loading control. HRAS, KRAS and NRAS isoforms are detected in this assay.
B. NF1 silencing prevents complete erlotinib-induced ERK dephosphorylation. Parental PC9 cells, and cells expressing control shRNA (shSC) or NF1 targeting shRNAs (NF1#1 or NF1#2) were left untreated or treated with 30nM erlotinib for 1 hr. Cell extracts were immunoblotted to detect the indicated proteins.
C. NF1 silenced cells maintain ERK phosphorylation in the presence of erlotinib, which is abolished by re-expression of the NF1-GRD. Cells were transfected with the NF1-GRD (GRD) or empty vector control (ev) prior to infection with control (shSC) or NF1-targeting shRNAs (shNF1#2). Cells were treated with 30nM erlotinib for 1hr, and cell extracts were immunoblotted to detect the indicated proteins.
D. Active MEK and active AKT de-sensitizes cells to erlotinib in a short-term survival assay. PC9 cells expressing the ecotropic receptor (Eco) were stably transfected with myristylated AKT (myr-AKT) or active MEK (MEK-DD) and treated with indicated concentrations of erlotinib for 72hrs before cell viability was measured using Cell Titer Blue. Cell survival is normalized to untreated controls. Error bars denote SEM.
E. Constitutively active MEK and to a lesser extend AKT enhance cell survival in longer-term colony formation assays. Cells were fixed and stained after 14 days treatment with indicated concentrations of erlotinib.
To determine if this increased RAS activity affects downstream signaling pathways, we examined the phosphorylation status of several downstream signaling proteins (Figure 4B). Erlotinib reduced the phosphorylation of EGFR and AKT similarly in all cells. Importantly, while erlotinib completely abolished ERK phosphorylation in the parental and control shRNA-infected cells, remaining phosphorylated ERK could still be detected in shNF1 cells (Figure 4B and Figure S4A), also at 1µM (Figure S4B), which is around the steady state plasma concentration found in patients treated with erlotinib (28, 29).
Examination of cells expressing the GAP-related domain of NF1 revealed that ERK phosphorylation was also completely abolished in these cells in the presence of erlotinib (Figure 4C). We confirmed an important role for active MAPK pathway by studying PC9 cells expressing constitutively active forms of MEK (MEK-DD) or AKT (myristylated-AKT; Figure S4C). MEK-DD caused a strong decrease in erlotinib sensitivity, while the effect of myr-AKT in cell survival assays is more moderate (Figure 4D, 4E). The ability of MEK-DD expressing cells to resist the inhibitory effects of erlotinib can be reversed by treatment with the MEK inhibitor AZD-6244 (Figure S4D). Thus, neurofibromin downregulation increases RAS activity and attenuates the effect of erlotinib on the downstream MAPK pathway, thereby decreasing the sensitivity to EGFR inhibitory drugs.
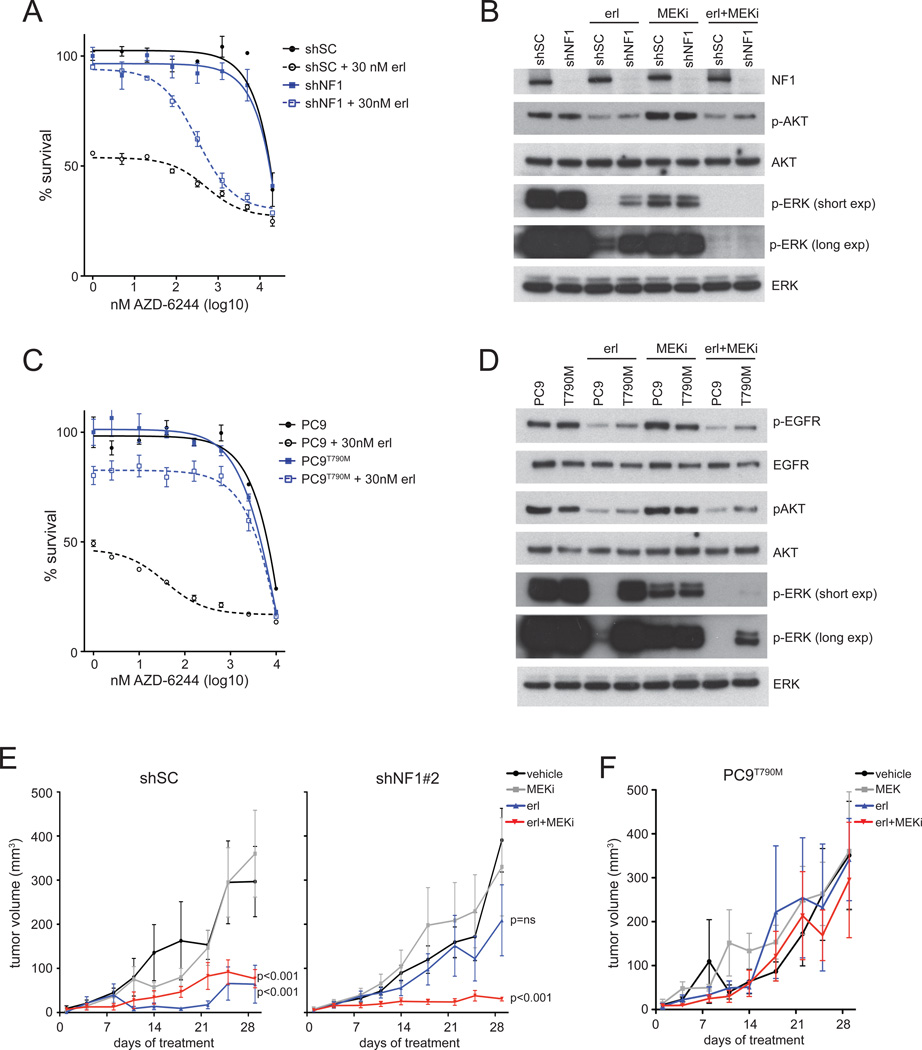
Cells expressing reduced neurofibromin respond to erlotinib in combination with a MEK inhibitor
Given the likely importance of the residual ERK phosphorylation for cell survival in the presence of erlotinib, we reasoned that EGFR mutant cells expressing low neurofibromin levels could be sensitive to treatment with a MEK inhibitor combined with a TKI. Indeed, analyzing a dose response curve in the presence or absence of erlotinib revealed that the NF1 knockdown cells are resistant to the MEK inhibitor AZD-6244 or erlotinib alone, but do respond to the combination of both inhibitors (Figure 5A). As expected, the control shSC cells did respond to single-agent erlotinib as evidenced by the decreased survival at the beginning of the experiment. Similar results were obtained with the MEK inhibitors CI-1040 and PD0325901 (Figure S5A). Whereas EGFR or MEK inhibition alone is insufficient, these drugs abolished ERK phosphorylation completely in the shNF1 cells when used in combination (Figure 5B). Similar assays were performed with a clone of PC9 cells that had acquired erlotinib resistance in vitro following prolonged drug exposure that resulted in the emergence of cells with the EGFR gatekeeper mutation T790M (PC9T790M) (Figure S5B). These cells could not be re-sensitized to erlotinib by the addition of a MEK inhibitor (Figure 5C), and maintain low levels of phosphorylated ERK in the presence of both drugs, in contrast to parental PC9 cells (Figure 5D).
Figure 5. Low neurofibromin expressing cells respond to erlotinib when combined with MEK inhibition.
A. NF1 silenced cells are insensitive to EGFR or MEK inhibitor alone but sensitive to combined drug treatment. Cells expressing control shRNA (shSC) or shRNA targeting NF1 (shNF1#2) were treated with indicated concentrations of AZD-6244 alone (solid line) or in combination with 30 nM erlotinib (dotted line) for 72hrs before cell viability was measured using Cell Titer Blue. Cell survival is shown normalized to untreated controls. Error bars denote SEM.
B. Levels of pERK and pAKT are analyzed by western blotting after treating the shSC- or shNF1#2-infected cells with 30nM erlotinib or 1µM MEK inhibitor AZD-6244 (MEKi) alone or in combination for 1hr. A short and long exposure is shown of pERK. Total ERK and AKT are shown as loading control.
C. T790M-positive erlotinib resistant PC9 (PC9T790M) cells remain insensitive to erlotinib when treatment is combined with the MEK inhibitor. PC9T790M and parental PC9 cells are treated with indicated concentrations of MEK inhibitor AZD-6244 alone (straight line) or in combination with 30nM erlotinib (dotted line) for 72hrs before cell viability was measured and survival normalized to untreated controls. Error bars denote SEM.
D. Levels of pERK and pAKT are analyzed by western blotting after PC9 and PC9T790M cells were treated with 30nM erlotinib or 1µM AZD-6244 (MEKi) alone or in combination for 1hr. A short and long exposure is shown of pERK. Total ERK and AKT are shown as loading control.
E. NF1 silenced xenograft tumors are sensitive to erlotinib in combination with MEK inhibition, but insensitive to either of both drugs alone. Mice were subcutaneously injected with shSC- or shNF1#2-infected PC9 cells. Once tumors were detectable, mice were randomized to 4 different treatment regimens: vehicle, erlotinib, AZD-6244 (MEKi) or combined erlotinib and AZD-6244 (erl + MEKi) as described in Experimental Procedures. Data in the figure represent the mean ± SEM tumor volumes for each treatment group over time. P-values are indicated for the mice receiving erlotinib alone or combined with AZD-6244 and analyzed against the vehicle control group.
F. PC9T790M xenograft tumors are insensitive to erlotinib in combination with MEK inhibition. Mice were injected with PC9T790M cells. Once tumors were detectable, mice were treated by 4 different treatment regimens as in Figure 5E.
While the above studies revealed that MEK inhibition in combination with erlotinib re-sensitizes erlotinib-resistant shNF1-infected cells in vitro, we continued to examine the sensitizing effect of MEK inhibition to erlotinib in vivo using tumor xenografts. We established xenografts of PC9 cells stably infected with a control (shSC) or NF1 targeting (shNF1#2) shRNA. Once tumors were detectable, mice were treated with either erlotinib or AZD-6244 or a combination of both drugs. Combined erlotinib and AZD-6244 treatment for 30 days indeed effectively reduced the tumor growth of both the NF1 silenced and the control tumors. As expected, we did not see a response to erlotinib in the NF1 knockdown tumors, while the control tumors did clearly respond (Figure 5E). By contrast, xenograft tumors of PC9T790M cells failed to respond to the combination treatment (Figure 5F). These data suggest that modest doses of a MEK inhibitor may resensitize tumors with reduced neurofibromin expression to erlotinib in the absence of the T790M mutation.
Erlotinib-resistant mouse lung adenocarcinomas respond to combined EGFR and MEK inhibition
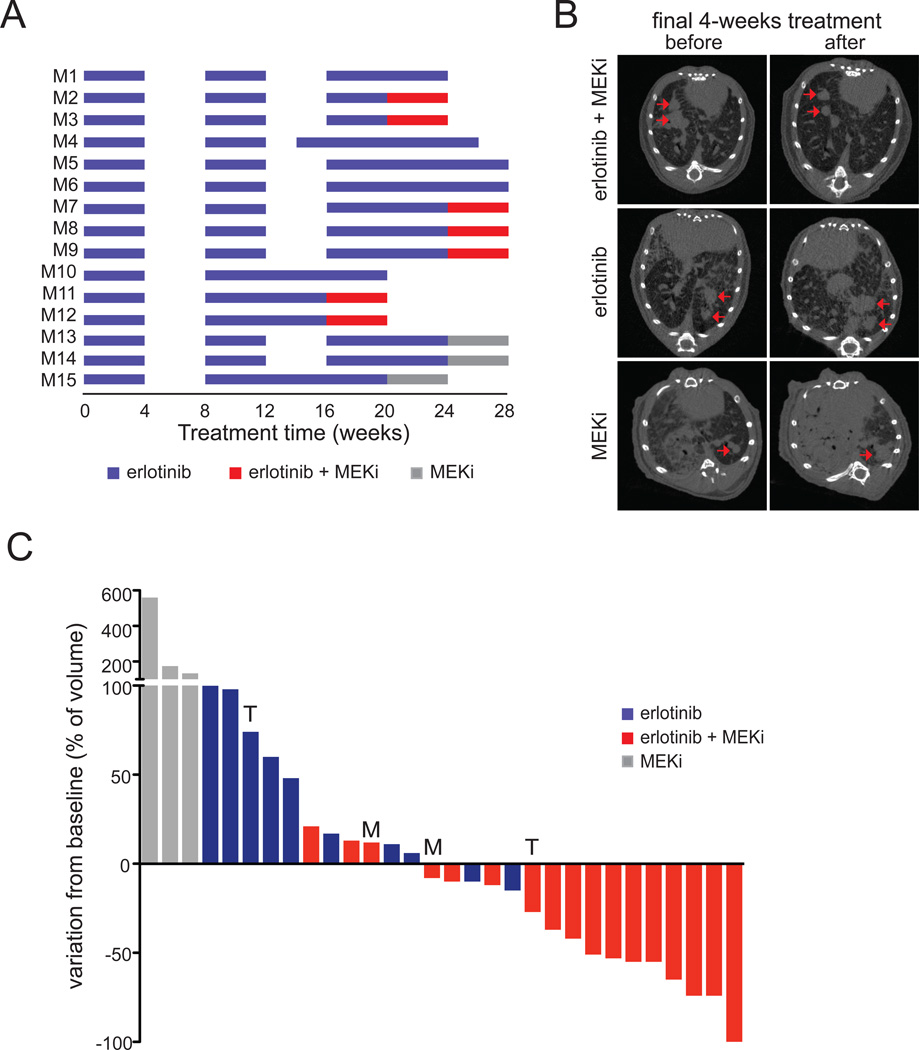
While the above findings suggest a possible treatment opportunity for lung adenocarcinoma cells that express reduced levels of neurofibromin, the effect of combination EGFR and MEK inhibition in established EGFR-driven lung adenocarcinomas that developed resistance to erlotinib remained unknown. We therefore used our tetracycline-inducible mouse model of EGFR-dependent lung cancer, driven by the EGFRL858R point mutation, to generate erlotinib-resistant tumors (23). While maintaining the mice on a diet containing doxycycline to ensure continued expression of the transgene, the mice were treated with erlotinib for three treatment rounds of four weeks long each, followed by four weeks without drug treatment. Tumors were monitored using micro-CT at the start and end of each treatment period. In a few mice, the tumor response was relatively minor during the second round of erlotinib, and these mice continued on erlotinib (Figure 6A, M10, 11, 12, 15).
Figure 6. Erlotinib-resistant murine EGFRL858R lung adenocarcinomas respond to combined EGFR and MEK inhibition.
A. Line chart depicting the schedule used to treat individual mice with erlotinib alone, MEK inhibitor alone or both inhibitors combined. Mice were treated 5 days per week for 4 weeks (blue horizontal bars indicate treatment with 25 mg/kg/day of erlotinib) after which treatment was interrupted for 4 weeks (no bars). Erlotinib administration was continued or started sooner in animals that became cachectic during this period (see, for example, mice M4 and M10). The treatment cycle was repeated up to three times, and followed by erlotinib administration alone (blue bars), the MEK inhibitor GSK-1120212 alone (grey bars), or combined (red bars indicate treatment with 25 mg/kg/day erlotinib combined with 2.5 mg/kg/day GSK-1120212).
B. Representative transaxial micro-CT images of lungs of mice before and after the final 4-weeks treatment round. The left image is from a mouse treated with combined erlotinib and MEK inhibitor, demonstrating a reduction in tumor volume (mouse M11), the middle image is from a mouse treated with erlotinib alone (mouse M6) and the right from a mouse treated with MEK inhibitor alone (mouse M13); both showing increased tumor volume. Red arrows point measurable tumor nodules.
C. Waterfall plot showing tumor response of the final 4 weeks of combined treatment with EGFR and MEK inhibitors (red bars), the EGFR inhibitor alone (blue bars) or MEK inhibitor alone (grey). Each bar represents one individual tumor, with data expressed relative to the tumor volume measured prior to the 4-weeks treatment round, showing progressive disease (PD), stable disease (SD), partial response (PR) or complete response (CR). Tumor mRNA was sequenced for the presence of EGFR-T790M and analysed for Met expression; T790M-positive tumors are marked with an T, and tumors showing a >4-fold Met expression are marked with an M. T790M and high Met expression were not detected in the other tumors, or not determined (n.d.) in cases where no frozen tumor material was available, as indicated in Table S3.
As described previously, a diminished response during the third round of erlotinib treatment was commonly observed (23). When possible, we maintained the mice on erlotinib for another four-week round of treatment after the third round, to confirm that the tumors were indeed growing and resistant to long-term erlotinib treatment. Subsequent to the emergence of erlotinib-resistant tumors, mice were treated with erlotinib and the MEK inhibitor GSK-1120212, also known as trametinib, for another four weeks, or as control with either inhibitor alone (Figure 6A). We scanned the animals before and after these final four weeks of treatment and quantified the tumor volumes (Figure 6B and Figure S6). As seen in Figures 6B and C, the combined treatment of EGFR and MEK inhibitors had a striking effect on most erlotinib resistant mouse lung adenocarcinomas. On the contrary, erlotinib or MEK inhibitor alone failed to induce significant tumor regression, confirming the resistant nature of these tumors (Figure 6C). Importantly, combined erlotinib and MEK inhibitor treatment was well tolerated and mice did not show any sign of weight loss (data not shown).
To assess whether secondary mutations in EGFR were associated with a reduced response to the combined erlotinib and MEK inhibitor treatment, we generated cDNA from RNA that was extracted from individual tumor nodules, and sequenced part of the human EGFR transgene cDNA spanning the transgene (L858R) and T790M region, as described previously (23). We detected the T790M amino acid substitution in two tumors and relatively high Met expression in two other tumours; three of these tumours were treated with combined erlotinib and MEK inhibitor, and these tumours showed a minimal response (Figure 6C; Table S3). As expected, all tumors did express the L858R driver mutation (data not shown). In addition, we evaluated Nf1 mRNA expression in these tumors and found that tumors responding well to erlotinib combined with MEK inhibitor more often express relatively lower levels of Nf1, but some tumors with higher levels do also respond (Table S3), indicating that these tumors might have found alternative mechanisms to activate MEK (16). We furthermore confirmed that all tumors expressed wild-type Kras, Hras and Nras (codons 12, 13 and 61; data not shown).
Collectively, these results demonstrate that erlotinib combined with a licensed MEK inhibitor substantially affects T790M-negative erlotinib-resistant lung adenocarcinomas in mouse models.
Reduced NF1 expression in human lung adenocarcinoma samples with resistance to EGFR inhibitors
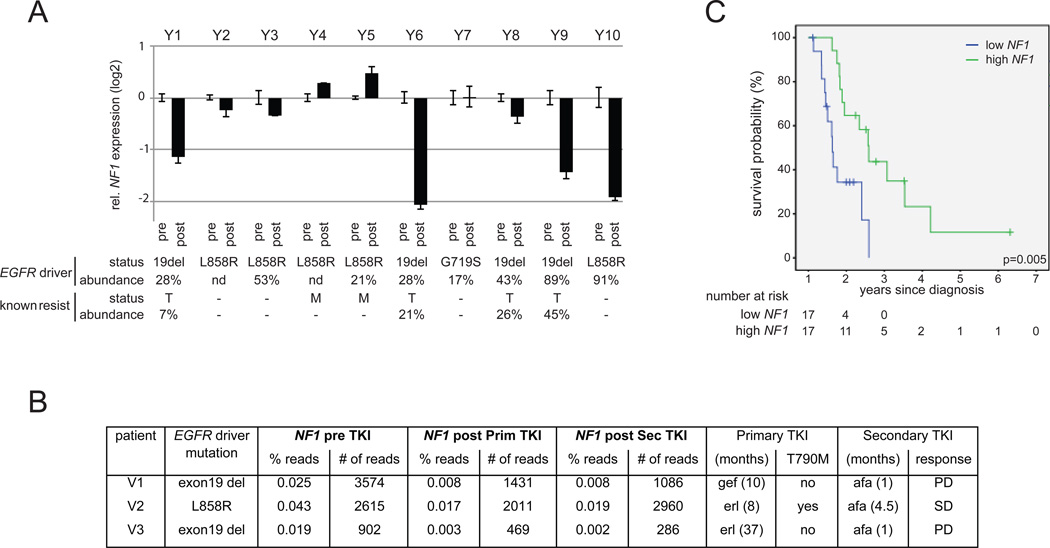
To assess the clinical relevance of this resistance mechanism, we evaluated NF1 expression in 13 human EGFR mutant lung adenocarcinoma samples that acquired resistance to EGFR TKI treatment compared to matched pre-treatment samples (Figures 7A and 7B). We were able to extract sufficient RNA from ten sample pairs to perform NF1 mRNA expression analyses by quantitative RT-PCR (Table S4 provides tumor purity and treatment response for each sample presented in Figure 7A). Four of the erlotinib-resistant samples showed a >2-fold decrease in NF1 compared to their matched pre-treatment sample, with relatively stable expression in the remaining six pairs (Figure 7A). Importantly, one of the erlotinib-resistant tumors with reduced NF1 did not harbor an EGFR T790M mutation nor amplified MET (sample pair Y10). In addition, using RNA sequencing data from a separate set of three human lung adenocarcinomas treated with EGFR TKIs (erlotinib or gefitinib), we identified reduced NF1 expression in all three post-treatment tumours, with the strongest reduction in the two EGFR-TKI resistant samples that did not express T790M (V1 and V3) and failed to respond to a second-line treatment with afatinib (Figure 7B). Experiments with the PC9 cell line confirmed that NF1 silencing strongly reduced sensitivity to afatinib as well (Figure S7A). Together, these two clinical data sets indicate that low NF1 might be driving erlotinib resistance in these tumors.
Figure 7. Reduced NF1 expression in erlotinib-resistant human NSCLC samples.
A. RNA extracted from matched pre- and post-treatment FFPE samples of human lung adenocarcinomas was amplified using quantitative RT-PCR assays to NF1 and three housekeeping genes (ESD, POLR2A and ActB). NF1 expression levels were normalized to the average of the housekeeping genes for each sample, and the fold-change in NF1 expression in the resistant sample compared to the paired pre-treatment sample is shown in the graph. For each tumor, the activating EGFR mutation is indicated, and the presence of T790M mutation (T) or MET amplification (M) in the post-treatment sample. The percentages of EGFR alleles harboring the activating mutation or the T790M mutation in the post-treatment sample as detected by pyrosequencing is indicated
B. NF1 expression is determined by RNA-seq in paired pre- and post-EGFR TKI samples of three NSCLC patients. The percentage of the total amount of protein-coding reads assigned to NF1 (% reads) as well as the absolute number of NF1 sequencing reads (# of reads) is listed for all three patients in the pre-treatment samples (pre TKI) and in samples obtained upon resistance to the first line of treatment with erlotinib or gefitinib (primary TKI) and to the second line of treatment with afatinib (secondary TKI). Duration of both treatments is listed, and response to afatinib is indicated as progressive disease (PD) or stable disease (SD).
C. Effects of NF1 expression on overall survival in patients treated with EGFR TKI as first (n=5) or second (n=29) line of treatment, starting from time of diagnosis when the samples were obtained. Patients were divided into two groups, low and high NF1, using the median NF1 expression as cut-off value. The log rank p-value is indicated.
Some post-treatment samples showed reduced NF1 mRNA in the post-treatment tumor co-occurring with an EGFR T790M mutation (sample pairs Y1, Y6 and Y9). Similar observations have been published for other resistance mechanisms (10, 18, 22), which could indicate that multiple mechanisms may contribute to resistance to EGFR TKIs. In order to address if heterogeneity in resistance mechanisms could explain our observations, we assessed the abundance of the T790M mutation by pyrosequencing and compared this to the abundance of the original TKI-sensitivity-conferring EGFR driver mutation. In the post-treatment tumor of Y1, 28% of the EGFR present in the post-treatment sample harbored the exon19 deletion while only 7% harbored the T790M mutation. Since a minority of cells in the resistant tumor cells expressed T790M, one can speculate that heterogeneity in resistance might occur in this sample with T790M expression in part of the tumor cells and low NF1 in other cells. A similar heterogeneity is seen in the post-treatment sample of Y9. However, one sample (pair Y6) showed almost similar abundance of the driver mutation and T790M in the post-treatment sample, 28% and 21% respectively. We therefore hypothesized that low NF1 and expression of T790M might have additive effects on drug resistance. Indeed, long-term treatment of erlotinib-resistant PC9T790M cells infected with an shRNA targeting NF1 (shNF1) or a non-silencing control shRNA (shSC) resulted in selective outgrowth of the shNF1 cells when cultured in the presence of erlotinib or afatinib (Figure S7B). These data suggest that NF1 silencing and T790M have additive effects on the resistance of lung adenocarcinoma cells to EGFR inhibitor-induced death. Combined, the RNA sequencing and the qRT-PCR analyses on human samples confirm a clinical relevance of NF1 downregulation in acquired resistance to EGFR TKIs.
To evaluate whether NF1 might be involved in primary resistance as well, we examined NF1 expression in a cohort of 34 NSCLC samples taken at diagnosis from patients who were then treated with erlotinib as first (n=5) or second (n=29) line of therapy; the EGFR mutation status of these tumors is unknown. Using the median as cut-off, we found that low NF1 expression was strongly associated with decreased overall survival with a median survival time of 7.6 months (95% confidence interval: 6.8–8.4) compared to 19.1 months (95% confidence interval: 14.0–24.2) for patients with high NF1 expression in their tumor (Figure 7C). A multivariate Cox regression analysis with NF1 expression, gender and morphology as input variables confirmed NF1 expression as an independent prognostic factor with a relative risk of 4.1 (CI 1.6–10.7, p=0.004).
Overall, these clinical data suggest that the level of NF1 expression can determine the responsiveness to EGFR TKI, and provide rationale for testing a MEK inhibitor in combination with an EGFR TKI in EGFR-mutant lung cancer patients.
DISCUSSION
We have used a functional genomic in vitro screening approach together with a genetically modified mouse lung cancer model system to investigate mechanisms of acquired resistance to erlotinib in EGFR mutant lung adenocarcinoma, identifying NF1 as a gene whose loss of function is capable of causing EGFR TKI drug resistance in both settings. Our analyses of two independent sets of paired EGFR mutant lung adenocarcinoma samples from patients treated with EGFR inhibitors confirms that downregulation of NF1 expression at the time of TKI resistance is a common occurrence in the clinic. Furthermore, NF1 levels may influence the initial response to TKIs, as low neurofibromin in pre-treatment specimens is strongly associated with reduced overall survival for patients with EGFR-mutant lung adenocarcinomas treated with EGFR TKI.
Several studies have shown that neurofibromin expression and function can be altered at a number of levels in lung and other cancers. Sequence analysis of the NF1 gene in 188 lung adenocarcinomas revealed mutations in nearly 10% of tumors, most of which lacked coincident KRAS mutations (30). A detailed genomic analysis of human glioblastoma by the Cancer Genome Atlas Research Network showed heterozygous deletion of the NF1 gene resulting in reduced neurofibromin expression, but also low NF1 mRNA without evidence of genomic alterations (31). Our RNA sequencing data did not reveal mutations in NF1 in the resistant samples of the three patients analyzed (data not shown). We have also sequenced the DNA coding region of NF1 in 10 additional paired samples from patients with mutant EGFR-expressing lung tumors before and after the acquisition of resistance to EGFR inhibitor that do not express EGFR-T790M, and found no evidence for somatic mutations in these samples either (data not shown). In addition, four of these matched biopsies had sufficient DNA material to analyze the methylation state of the NF1 promoter region, but we found no evidence for differences (data not shown). In our study, 6 out of a total of 13 paired samples show significant downregulation of NF1 mRNA upon TKI resistance. Future studies should evaluate neurofibromin expression at protein level, as additional post-transcriptional regulation could occur as described in glioma cells (32, 33). We have screened multiple antibodies for immunohistochemistry, but none showed the specificity required to detect neurofibromin in human tissue (data not shown).
Some of the human lung cancer samples analyzed in our study showed reduced NF1 expression co-occurring with the T790M mutation, suggesting that multiple mechanisms may contribute to resistance to EGFR TKIs. Such heterogeneity in mechanisms of resistance is consistent with other studies that have described T790M co-occurring with other resistance mechanisms (10, 18, 22). The abundance of transcripts containing the T790M mutation in comparison with those containing only the EGFR driver mutation in these resistant tumors suggests that heterogeneity in resistance mechanisms can either be in different cells of a tumor (Figure 6, samples Y1 and Y9) or in an additive manner within the same cells (Figure 6, sample Y6 and Figure S7).
We provide several lines of evidence that reduced neurofibromin levels in lung adenocarcinoma cells affect drug response through activating RAS and the downstream ERK-MAPK pathway. First, we show increased levels of GTP-bound RAS in cells where NF1 expression has been reduced by RNA interference, when treated with erlotinib. Second, phosphorylated ERK could be detected in NF1 silenced cells in the presence of erlotinib. Although reduced, such low levels of continued flux through ERK appeared sufficient to maintain cell survival in the presence of erlotinib, as inhibition of the upstream MEK kinase with a small-molecule inhibitor abolished ERK phosphorylation completely and resulted in cell death. Furthermore, xenograft tumors of shNF1 cells failed to grow when the animals were treated with erlotinib in combination with a MEK inhibitor, whereas either drug on its own had no effect. In line with our findings, the MAPK pathway is directly activated by point mutation of KRAS in about 20% of human lung tumors (34), which is associated with primary resistance to drugs targeting EGFR in colon and lung cancer (35, 36). In addition, a recently published study identified MAPK1 amplification in 5% of erlotinib-resistant samples (16), also indicating a prominent role for active MAPK signaling as a mediator of EGFR TKI resistance. Research on acquired resistance to EGFR inhibitory drugs in lung cancer has mainly focused on the PI3K-AKT pathway, activated by MET amplification or PTEN loss of function (14, 15, 37). In line with these studies, introduction of an active AKT construct into PC9 cells indeed reduced erlotinib sensitivity (Figure 4D). An active MEK construct showed, however, a more pronounced effect on drug resistance, providing evidence that the MAPK pathway can play a crucial role in resistance to EGFR TKIs in lung adenocarcinomas.
Importantly, our in vivo studies show that more than half (59%) of the EGFRL858R-driven mouse tumors that acquired resistance to erlotinib did respond to combination treatment with EGFR and MEK inhibitory drugs. At the moment, treatment options for tumors with acquired resistance to EGFR inhibitors are limited (3, 10, 20). Our experiments provide a molecular basis for the combination of EGFR and MEK inhibitors for the treatment of patients with mutant EGFR-driven tumors that have acquired resistance to TKI but lack the T790M gatekeeper mutation. A recent clinical trial showed improved progression-free survival in BRAF mutant melanoma patients when a BRAF inhibitor is combined with the MEK inhibitor trametinib (38), which we used in our studies in mouse models. Interestingly, resistance to BRAF kinase inhibitors is often associated with reactivation of the MAPK pathway (39–41), and two recent studies found NF1 mutations to be associated with reduced response to BRAF inhibition in BRAF mutant melanoma (42, 43). Given the effect of EGFR inhibitors on EGFR mutant lung cancer, the combination of MEK and EGFR inhibition in the setting of EGFR mutant disease may be very effective. A combination of MEK and EGFR inhibitors is currently being evaluated in clinical trials for lung cancer (NCI-10-C-0218 and NCT01192165). In these trials, increased toxicity has been reported in patients that received both inhibitors (44, 45). Nevertheless, it is expected that novel EGFR inhibitors that specifically inhibit the mutant forms of EGFR while sparing the wild type protein (46) will reduce such toxicity.
Collectively, the present study provides a molecular basis for the combination of EGFR TKI with clinically available MEK inhibitors for the treatment of patients with mutant EGFR driven lung adenocarcinomas that fail to respond to EGFR TKIs either due to primary or acquired resistance.
METHODS
Cell culture
H3255, HCC827 and HCC4006 cells were purchased from the ATCC Cell Biology collection, and PC9 cells were kindly provided by Jeff Settleman. Cell lines were authenticated by the Cancer Research UK Central Cell Services facility using short tandem repeat profiling. Cells were cultured in RPMI, supplemented with 10% Fetal Bovine Serum (FBS), 100 µg/ml Streptomycin and 100 U/ml Penicillin, at 37°C and 10% CO2. PC9T790M cells were generated by continued culturing of PC9 cells with 1µM erlotinib. Surviving clones were picked and cultured in the presence of 1µM erlotinib for 3 months, and thereafter without erlotinib.
Genome-wide siRNA screen
The genome-wide siRNA library (21,121 siRNAs) was purchased from Dharmacon as the siGENOME SMARTpool collection. siRNA pools targeting PLK1 or UBB1 were used as positive controls for the transfection, and siGENOME RISC-Free Control siRNA, siGENOME Non-Targeting siRNA Pool #2 (scrambled) and siRNA pools targeting Luciferase GL3 were used as negative controls.
PC9 cells were seeded in 384 well plates (500 cells/well) and reverse transfected with 37.5nM siRNA using DharmaFECT transfection reagent #2. After 48hr, the culture medium was replaced by RPMI with 30nM erlotinib (drug screen) or without erlotinib (control screen). As a control for drug treatment, we left two columns of the control siRNAs untreated in the erlotinib-treated plates. Cells were incubated for 72hr before being fixed with 80% ethanol, stained with 1ug/ml DAPI (Roche) for 1.5 hr, and stored in PBS at 4°C. The number of cells in each well was quantified using an Acumen Explorer microplate cytometer (TTP LabTech).
We performed triplicate experiments for both conditions: control and drug screens. We performed an independent genome-wide screen using the same conditions in order to validate the reproducibility of the hits.
The follow-up deconvolution siRNA screen was performed using Dharmacon’s collection siGENOME set of 4 individual siRNAs targeting a single gene. The deconvolution siRNA screen was performed using the same conditions as in the genome-wide screen.
Details of data analysis of the genome-wide and deconvolution siRNA screens are in Supplemental Experimental Procedures.
Quantitative RT-PCR paired human NSCLC samples
Samples for quantitative RT-PCR analysis were obtained from patients with EGFR mutant lung adenocarcinoma who developed acquired resistance to erlotinib under Human Investigations Protocol # 111000928 (Yale Cancer Center). Pre- and post-treatment specimens were reviewed by a pathologist to ensure adequate tumor content. Tumor areas were circled and manual microdissection was performed to enrich for tumor content in downstream applications. Pyrosequencing was used to determine the abundance of the EGFR L858R and exon 19 DEL mutations as well as the T790M mutation using the EGFR Pyro Kit (Qiagen)
Total RNA was isolated from FFPE tissue (5×5µm slides) using the FFPE RNeasy Kit (Qiagen). Quantitative RT-PCR assays were performed in triplicate with 4.5ng RNA input per reaction, using Taqman RNA-to-Ct 1 step kit, and Taqman primers to amplify NF1 and the housekeeping genes ActB, ESD and POLR2A, described to be relatively stably expressed in human NSCLC (47). NF1 mRNA levels were analyzed using the comparative CT method and normalized to the average of the three housekeeping genes for individual samples, before the fold-change relative to the pre-treatment sample was determined.
RNA sequencing analysis of human NSCLC samples
RNA sequencing was performed as previously described (21), using RNA extracted from FFPE material, with at least 50% tumor content (areas were circled and manual microdissection was performed to enrich for tumor content), from paired pre- and post-treatment biopsies of three NSCLC patients. These patients were treated with either erlotinib or gefitinib followed by afatinib at the Department of Pulmonary Diseases, VU University Medical Center, Amsterdam, The Netherlands. Tumor biopsies were obtained as part of routine medical care after informed consent had been obtained according to local ethical committee regulations. Tumor EGFR mutational status was determined as previously described (48).
Overall survival analysis of NSCLC patients treated with EGFR TKI
The samples analyzed were retrieved from the diagnostic biobank of Pathology Service from the Hospital Universitario Marques de Valdecilla. For this study, primary tumors were obtained from 34 patients with stage IV non-small cell lung cancer (NSCLC) at time of diagnosis. None of the patients received chemo- or radiotherapy prior to sampling, and patients received EGFR TKI as primary (n=5) or secondary (n=29) treatment upon diagnosis. Presence of an EGFR mutation was not an inclusion criterion, and EGFR mutation status is therefore not available. The study was approved by the local ethics committee.
Total RNA was extracted using the FFPE RNeasy Kit (Qiagen). Quantitative RT-PCR assays were performed in triplicate using Taqman RNA-to-Ct 1 step kit and Taqman primers to amplify NF1 and the housekeeping gene ESD. NF1 mRNA levels were normalized to ESD. Patients were divided into two groups, low and high NF1, using the median NF1 expression as cut-off value. Kaplan Meier analysis was carried out using SPSS Statistics for probability of survival (overall survival) in both groups starting from time of diagnosis.
Supplementary Material
SIGNIFICANCE.
The emergence of resistance to EGFR TKIs is a major clinical challenge in the treatment of lung adenocarcinomas driven by mutations in EGFR. This study suggests that, in a subset of patients, resistance is caused by reduced neurofibromin expression and that in these cases there may be clinical benefit to combining EGFR TKIs with MEK inhibitors.
ACKNOWLEDGEMENTS
We thank Rachael Instrell and Michael Howell from the LRI High Throughput Screening laboratory for their support performing the screen. We thank Stuart Horswell from the Bioinformatics and Biostatistics department at the London Research Institute for analyzing the xenograft tumor measurements, and Francois Lassailly from the In Vivo Imaging department for assistance with the micro-CT imaging. We thank Francesco Mauri from the Department of Histopathology at Hammersmith Hospital, London for evaluating NF1 antibodies for immunohistochemistry. We thank Aldona K. Fonfara from the Division of Molecular Carcinogenesis at the NKI for help with RNA extractions, and the members of the NKI Genomics Core Facility for the RNA-seq processing and analysis. We thank Don Nguyen from the Department of Pathology and Yale Cancer Center for analyzing mouse gene expression data. We are grateful for the support given by the Equipment Park, Biological Resources, and the FACS Lab at the London Research Institute. E.B. was financially supported by a KWF (Dutch Cancer Society) fellowship and has received funding from the European Commission’s Seventh Framework Programme (FP7/2007–2013) under the grant agreement Lungtarget (project 259770). This work was supported by grants R00 CA131488 (KP), R01 CA120247 (KP), and P01 CA129243 (ML) from the National Cancer Institute, NIH and a Pilot grant from the Section of Medical Oncology, Yale University School of Medicine (SG and KP), and by Cancer Research UK.
Footnotes
Conflict of interest: the authors have no conflicts of interest to declare.
Further experimental protocols are described in the Supplemental Methods online.
REFERENCES
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blencke S, Ullrich A, Daub H. Mutation of threonine 766 in the epidermal growth factor receptor reveals a hotspot for resistance formation against selective tyrosine kinase inhibitors. J Biol Chem. 2003;278:15435–15440. doi: 10.1074/jbc.M211158200. [DOI] [PubMed] [Google Scholar]
- 9.Kwak EL, Sordella R, Bell DW, Godin-Heymann N, Okimoto RA, Brannigan BW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002003. 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lind JS, Dingemans AM, Groen HJ, Thunnissen FB, Bekers O, Heideman DA, et al. A multicenter phase II study of erlotinib and sorafenib in chemotherapy-naive patients with advanced non-small cell lung cancer. Clin Cancer Res. 2010;16:3078–3087. doi: 10.1158/1078-0432.CCR-09-3033. [DOI] [PubMed] [Google Scholar]
- 12.Hidalgo M, Siu LL, Nemunaitis J, Rizzo J, Hammond LA, Takimoto C, et al. Phase I and pharmacologic study of OSI-774, an epidermal growth factor receptor tyrosine kinase inhibitor, in patients with advanced solid malignancies. J Clin Oncol. 2001;19:3267–3279. doi: 10.1200/JCO.2001.19.13.3267. [DOI] [PubMed] [Google Scholar]
- 13.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 16.Ercan D, Xu C, Yanagita M, Monast CS, Pratilas CA, Montero J, et al. Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer Discov. 2012;2:934–947. doi: 10.1158/2159-8290.CD-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takezawa K, Pirazzoli V, Arcila ME, Nebhan CA, Song X, de Stanchina E, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2:922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zakowski MF, Ladanyi M, Kris MG. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med. 2006;355:213–215. doi: 10.1056/NEJMc053610. [DOI] [PubMed] [Google Scholar]
- 20.Oxnard GR, Arcila ME, Chmielecki J, Ladanyi M, Miller VA, Pao W. New strategies in overcoming acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer. Clin Cancer Res. 2011;17:5530–5537. doi: 10.1158/1078-0432.CCR-10-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang S, Holzel M, Knijnenburg T, Schlicker A, Roepman P, McDermott U, et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Politi K, Fan PD, Shen R, Zakowski M, Varmus H. Erlotinib resistance in mouse models of epidermal growth factor receptor-induced lung adenocarcinoma. Dis Model Mech. 2010;3:111–119. doi: 10.1242/dmm.003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swanton C, Marani M, Pardo O, Warne PH, Kelly G, Sahai E, et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007;11:498–512. doi: 10.1016/j.ccr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Le LQ, Parada LF. Tumor microenvironment and neurofibromatosis type I: connecting the GAPs. Oncogene. 2007;26:4609–4616. doi: 10.1038/sj.onc.1210261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becerra C, Infante JR, Garbo LE, Gordon MS, Smith DA, Braiteh FS, et al. A five-arm, open-label, phase I/lb study to assess safety and tolerability of the oral MEK1/MEK2 inhibitor trametinib (GSK1120212) in combination with chemotherapy or erlotinib in patients with advanced solid tumors. J Clin Oncol. 2012;30 : (suppl; abstr 3023). [Google Scholar]
- 29.Carter CA, Rajan A, Szabo E, Khozin S, Thomas A, Brzezniak CE, et al. Two parallel randomized phase II studies of selumetinib (S) and erlotinib (E) in advanced non-small cell lung cancer selected by KRAS mutations. J Clin Oncol. 2013;31 : suppl; abstr 8026. [Google Scholar]
- 30.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.TCGA_Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cichowski K, Santiago S, Jardim M, Johnson BW, Jacks T. Dynamic regulation of the Ras pathway via proteolysis of the NF1 tumor suppressor. Genes Dev. 2003;17:449–454. doi: 10.1101/gad.1054703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16:44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mao C, Qiu LX, Liao RY, Du FB, Ding H, Yang WC, et al. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: a meta-analysis of 22 studies. Lung Cancer. 2010;69:272–278. doi: 10.1016/j.lungcan.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 36.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sos ML, Koker M, Weir BA, Heynck S, Rabinovsky R, Zander T, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009;69:3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A. 2009;106:20411–20416. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–362. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becerra CIJR, Garbo LE, Gordon MS, Smith DA, Braiteh FS, Gandara DR. A five-arm, open-label, phase I/Ib study to assess safety and tolerability of the oral MEK1/MEK2 inhibitor trametinib (GSK1120212) in combination with chemotherapy or erlotinib in patients with advanced solid tumors. J Clin Oncol. 2012;30 suppe; abstract 3023. [Google Scholar]
- 45.Carter CARA, Szabo E, Kozin S, Thomas A, Brzezniak CE, Guha U. Two parallel randomized phase II studies of selumetinib (S) and erlotinib (E) in advanced non-small cell lung cancer selected by KRAS mutations. J Clin Oncol. 2013;31 suppl; abstr 8026. [Google Scholar]
- 46.Walter AO, Sjin RT, Haringsma HJ, Ohashi K, Sun J, Lee K, et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov. 2013;3:1404–1415. doi: 10.1158/2159-8290.CD-13-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saviozzi S, Cordero F, Lo Iacono M, Novello S, Scagliotti GV, Calogero RA. Selection of suitable reference genes for accurate normalization of gene expression profile studies in non-small cell lung cancer. BMC Cancer. 2006;6:200. doi: 10.1186/1471-2407-6-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heideman DA, Thunnissen FB, Doeleman M, Kramer D, Verheul HM, Smit EF, et al. A panel of high resolution melting (HRM) technology-based assays with direct sequencing possibility for effective mutation screening of EGFR and K-ras genes. Cell Oncol. 2009;31:329–333. doi: 10.3233/CLO-2009-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.