Abstract
Backgroud
The XPG (xeroderma pigmentosum type G) Asp1104His and XPF (xeroderma pigmentosum type F) Arg415Gln polymorphisms had been implicated in cancer susceptibility. The previous published data on the association between XPG Asp1104His and XPF Arg415Gln polymorphisms and cancer risk remained controversial.
Methodology/Principal Findings
To derive a more precise estimation of the association between the XPG Asp1104His and XPF Arg415Gln polymorphisms and overall cancer risk, we performed a meta-analysis to investigate the association between cancer susceptibility and XPG Asp1104His (32,162 cases and 39,858 controls from 66 studies) and XPF Arg415Gln polymorphisms (17,864 cases and 20,578 controls from 32 studies) in different inheritance models. We used odds ratios with 95% confidence intervals to assess the strength of the association. Overall, significantly elevated cancer risk was found when all studies were pooled into the meta-analysis of XPG Asp1104His (dominant model: OR = 1.05, 95% CI = 1.00–1.10; Asp/His vs. Asp/Asp: OR = 1.06, 95% CI = 1.01–1.11). In the further stratified and sensitivity analyses, significantly decreased lung cancer risk was found for XPF Arg415Gln (dominant model: OR = 0.82, 95% CI = 0.71–0.96; Arg/Gln versus Arg/Arg: OR = 0.83, 95% CI = 0.71–0.97; additive model: OR = 0.83, 95% CI = 0.72–0.95) and significantly increased other cancer risk was found among hospital-based studies for XPG Asp1104His (dominant model: OR = 1.23, 95% CI = 1.02–1.49).
Conclusions/Significance
In summary, this meta-analysis suggests that XPF Arg415Gln polymorphism may be associated with decreased lung cancer risk and XPG Asp1104His may be a low-penetrant risk factor in some cancers development. And larger scale primary studies are required to further evaluate the interaction of XPG Asp1104His and XPF Arg415Gln polymorphisms and cancer risk in specific populations.
Introduction
DNA repair systems play critical roles in protecting cells against mutations and are essential for maintaining the genome integrity. Certain common genetic polymorphisms within the genes involved in DNA damage responses may contribute to the development of cancer and be associated with an increased risk of the disease. Because reduced DNA repair capacity may cause genetic instability and carcinogenesis, genes involved in DNA repair have been proposed as candidate cancer susceptibility genes [1]. Nucleotide excision repair (NER) is a crucial DNA repair mechanism, which counteracts the consequences of mutagenic exposure of cells [2].
The NER pathway consists of >30 proteins involved in DNA damage recognition, incision, DNA ligation and resynthesis. Seven XP(xeroderma pigmentosum) complementation groups have been identified, from XPA to XPG, representing the malfunctioning proteins in the NER mechanism [3]. The XPG (xeroderma pigmentosum type G), one important component of the NER pathway, encodes a structure-specific endonuclease catalyzing 3′ incision and involves the subsequent 5′ incision by ERCC1-XPF heterodimer [4], [5]. It has been observed that there is a relationship between the SNP in exon 15 (G3507C, Asp1104His) and cancer susceptibility. ERCC4/XPF (Arg-to-Gln substitution in codon 415 of exon 8, rs1800067) forms a tight complex with ERCC1 to incise 5′ to the damage site recognized and repaired by NER [6]. The XPF gene encodes a protein which, together with ERCC1, creates the 5′ endonuclease [7].
To date, a number of molecular epidemiological studies have been done to evaluate the association between XPG Asp1104His and XPF Arg415Gln polymorphisms and different types of cancer risk in diverse populations [8]–[83]. However, the results were inconsistent or even contradictory, partially because of the possible small effect of the polymorphism on cancer risk and the relatively small sample size in each of published study. In addition, two recent meta-analyses have studied the association between XPG Asp1104His and XPF Arg415Gln and risk of cancer. However, many published studies were not included in the two recent meta-analyses [84], [85]. Therefore, we performed a comprehensive meta-analysis by including the most recent and relevant articles to identify statistical evidence of the association between XPG Asp1104His and XPF Arg415Gln polymorphisms and risk of all cancers that have been investigated. Meta-analysis is an outstanding tool for summarizing the different studies. It can not only overcome the problem of small size and inadequate statistical power of genetic studies of complex traits, but also can provide more reliable results than a single case–control study.
Materials and Methods
Identification and eligibility of relevant studies
A comprehensive literature search was performed using the PubMed and Medline database for relevant articles published (the last search update was Sep 5, 2013) with the following key words “XPG”, “ERCC5”, “XPF”, “ERCC4”, “polymorphism”, “Variant” or “Mutation”, and “Cancer” or “Carcinoma.” In addition, studies were identified by a manual search of the reference lists of reviews and retrieved studies. We included all the case–control studies and cohort studies that investigated the association between XPG Asp1104His and XPF Arg415Gln polymorphisms and cancer risk with genotype data. All eligible studies were retrieved, and their bibliographies were checked for other relevant publications. When the same sample was used in several publications, only the most complete study was considered for further analysis.
Inclusion criteria
The included studies needed to have met the following criteria:: (1) only the case–control studies or cohort studies were considered, (2) evaluated the XPG Asp1104His and XPF Arg415Gln polymorphisms and the risk of cancer, and (3) the genotype distribution of the polymorphisms in cases and controls were described in details and the results were expressed as odds ratio (OR) and corresponding 95% confidence interval (95% CI). Major reasons for exclusion of studies were as follows: (1) not for cancer research, (2) only case population, and (3) duplicate of previous publication.
Data extraction
Information was carefully extracted from all eligible studies independently by two investigators according to the inclusion criteria listed above. The following data were collected from each study: first author's name, year of publication, country of origin, ethnicity, source of controls, sample size, and numbers of cases and controls in the XPG Asp1104His and XPF Arg415Gln genotypes whenever possible. Ethnicity was categorized as “Caucasian,” “African,” (including African Americans) and “Asian.” Two studies were carried out with Hispanic ethnic groups. When one study did not state which ethnic groups was included or if it was impossible to separate participants according to phenotype, the sample was termed as “mixed population.” Meanwhile, studies investigating more than one kind of cancer were counted as individual data set only in subgroup analyses by cancer type. We did not define any minimum number of patients to include in this meta-analysis. In case of articles reported different ethnic groups and different countries or locations, we considered them different study samples for each category cited above.
Statistical analysis
Crude odds ratios (ORs) together with their corresponding 95% CIs were used to assess the strength of association between the XPG Asp1104His and XPF Arg415Gln polymorphisms and the risk of cancer. The pooled ORs were performed for co-dominant model (XPG Asp1104His: His/His versus Asp/Asp and Asp/His versus Asp/Asp, XPF Arg415Gln: Gln/Gln versus Arg/Arg and Arg/Gln versus Arg/Arg); dominant model (XPG Asp1104His: Asp/His+His/His versus Asp/Asp, XPF Arg415Gln: Arg/Gln+Gln/Gln versus Arg/Arg); recessive model (XPG Asp1104His: His/His versus Asp/His+Asp/Asp, XPF Arg415Gln: Gln/Gln versus Arg/Gln+Arg/Arg); and additive model (XPG Asp1104His: His versus Asp, XPF Arg415Gln: Gln versus Arg), respectively. Between-study heterogeneity was assessed by calculating Q-statistic (Heterogeneity was considered statistically significant if P<0.10) [86] and quantified using the I2 value, a value that describes the percentage of variation across studies that are due to heterogeneity rather than chance, where I2 = 0% indicates no observed heterogeneity, with 25% regarded as low, 50% as moderate, and 75% as high [87]. If results were not heterogeneous, the pooled ORs were calculated by the fixed-effect model (we used the Q-statistic, which represents the magnitude of heterogeneity between-studies) [88]. Otherwise, a random-effect model was used (when the heterogeneity between-studies were significant) [89]. In addition to the comparison among all subjects, we also performed stratification analyses by cancer type (if one cancer type contained less than three individual studies, it was combined into the “other cancers” group), Moreover, the extent to which the combined risk estimate might be affected by individual studies was assessed by consecutively omitting every study from the meta-analysis (leave-one-out sensitivity analysis). This approach would also capture the effect of the oldest or first positive study (first study effect). In addition, we also ranked studies according to sample size, and then repeated this meta-analysis. Sample size was classified according to a minimum of 200 participants and those with fewer than 200 participants. The cite criteria were previously described [90]. Last, sensitivity analysis was also performed, excluding studies whose allele frequencies in controls exhibited significant deviation from the Hardy–Weinberg equilibrium (HWE), given that the deviation may denote bias. HWE was calculated by using the goodness-of-fit test, and deviation was considered when P<0.05. Begg's funnel plots [91] and Egger's linear regression test [92] were used to assess publication bias. If publication bias existed, the Duval and Tweedie nonparametric “trim and fill” method was used to adjust for it [93]. A meta-regression analysis was carried out to identify the major sources of between-studies variation in the results, using the log of the ORs from each study as dependent variables, and cancer type, ethnicity, sample size, HWE, and source of controls as the possible sources of heterogeneity. All of the calculations were performed using STATA version 10.0 (STATA Corporation, College Station, TX).
Results
Eligible studies and meta-analysis databases
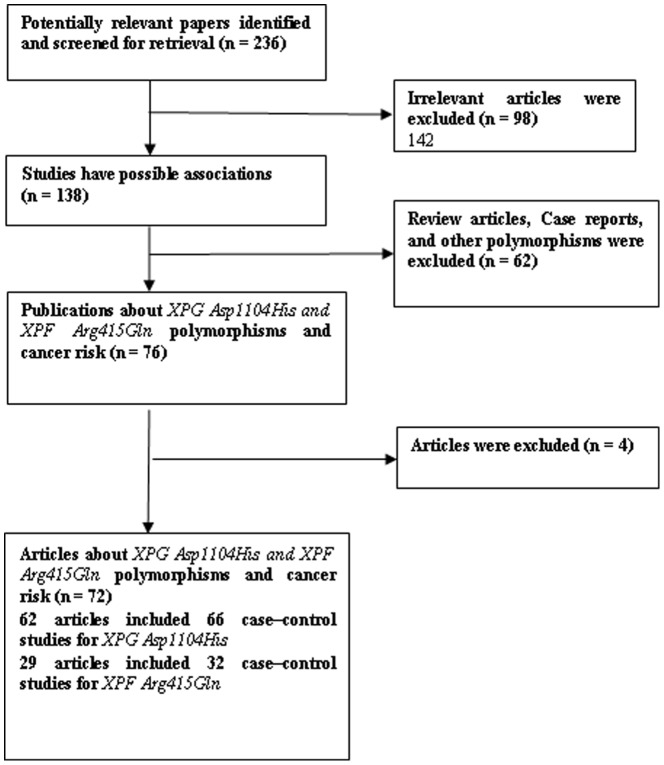
Fig. 1 graphically illustrates the trial flow chart. A total of 236 articles regarding XPG Asp1104His and XPF Arg415Gln polymorphisms with respect to cancer were identified. After screening the titles and abstracts, 160 articles were excluded because they were review articles, case reports, other polymorphisms of CYP1A1, or irrelevant to the current study. In addition, of these published articles, 4 publications [76]–[79] were excluded because of their populations overlapped with another 3 included studies [40], [44], [68]. Five publications [17], [20], [40], [41], [57] including different case–control groups should be considered as two separate studies each. As summarized in Table 1 , 72 publications with 98 case–control studies were selected among the meta-analysis, including 32,162 cases and 39,858 controls for XPG Asp1104His (66 studies from 62 publications) and 17,864 cases and 20,578 controls for XPF Arg415Gln (32 studies from 29 publications). Among these studies, for XPG Asp1104His, there were 7 bladder cancer studies, 11 breast cancer studies, 7 colorectal cancer studies, 5 head and neck cancer studies, 7 lung cancer studies, 4 non-Hodgkin lymphoma studies, 3 glioma studies, 8 melanoma studies, and 14 studies with the “other cancers”. There were 10 breast cancer studies, 3 lung cancer studies, 4 head and neck cancer studies, 4 colorectal cancer, 3 glioma studies, and 8 studies with the “other cancers” for XPF Arg415Gln. All of the cases were pathologically confirmed.
Figure 1. Study flow chart explaining the selection of the 72 eligible articles included in the meta-analysis.
Table 1. Main characteristics of all studies included in the meta-analysis.
| First author/year | Country | Ethnicity | Cancer type | SC | XPG Asp1104His (Case/control) | XPF Arg415Gln (Case/control) | HWE | ||||
| Smith [8] 2003 | USA | Caucasian | Breast | HB | NA | NA | NA | 217/236 | 29/32 | 7/0 | Yes |
| Kumar [9] 2003 | Filand | Caucasian | Breast | HB | 108/182 | 96/107 | 16/19 | NA | NA | NA | Yes |
| Jeon [10] 2003 | Korea | Asian | Lung | HB | 58/90 | 164/132 | 88/89 | NA | NA | NA | No |
| Sanyal [11] 2004 | Swede | Caucasian | Bladder | NA | 182/173 | 109/91 | 8/20 | NA | NA | NA | Yes |
| Blankenburg [12] 2005 | German | Caucasian | Melanoma | HB | 184/232 | 100/124 | 9/18 | NA | NA | NA | Yes |
| Weiss [13] 2005 | USA | Mixed | Endometrial | PB | 215/250 | 134/148 | 22/22 | 316/369 | 54/49 | 1/2 | Yes |
| Shen [14] 2005 | China | Asian | Lung | PB | 26/25 | 52/46 | 38/38 | NA | NA | NA | Yes |
| Bigler [15] 2005 | USA | Mixed | Colorectal | PB | 440/353 | 243/226 | 36/37 | NA | NA | NA | Yes |
| Sakiyama [16] 2005 | Japan | Asian | Lung | HB | 300/228 | 500/333 | 202/124 | NA | NA | NA | Yes |
| Cui [17] 2006 | USA | Mixed | Lung | PB | 244/468 | 212/356 | 41/78 | NA | NA | NA | Yes |
| Cui [17] 2006 | USA | Mixed | Multiple | PB | 214/474 | 194/357 | 35/80 | NA | NA | NA | Yes |
| Zienolddiny [18] 2006 | Norway | Caucasian | Lung | HB | NA | NA | NA | 195/178 | 26/21 | 3/1 | Yes |
| Millikan [19] 2006 | USA | Caucasian | Melanoma | PB | 731/1513 | 389/780 | 73/115 | 1026/2073 | 173/360 | 9/12 | Yes |
| Mechanic [20] 2006 | USA | Caucasian | Breast | PB | 771/661 | 409/412 | 69/60 | 1049/980 | 185/150 | 12/3 | Yes |
| Mechanic [20] 2006 | USA | African | Breast | PB | 231/231 | 387/320 | 139/123 | 738/642 | 18/31 | 1/0 | Yes |
| Huang [21] 2006 | USA | Mixed | Colorectal | PB | 407/403 | 243/265 | 29/29 | 624/623 | 78/86 | 1/7 | Yes |
| García-Closas [22] 2006 | Spain | Caucasian | Bladder | HB | 629/607 | 434/445 | 78/84 | 885/824 | 203/182 | 14/19 | Yes |
| Moreno [23] 2006 | Spain | Caucasian | Colorectal | HB | NA | NA | NA | 282/257 | 71/61 | 7/5 | Yes |
| Shen [24] 2006 | USA | Mixed | NHL | PB | 260/352 | 170/169 | 34/29 | NA | NA | NA | Yes |
| Shen [25] 2006 | USA | Mixed | Breast | FB | 83/82 | 63/62 | 8/7 | NA | NA | NA | Yes |
| Wen [26] 2006 | China | Asian | HNC | PB | 55/129 | 81/296 | 39/100 | NA | NA | NA | No |
| Li [27] 2006 | USA | Caucasian | Melanoma | HB | 373/370 | 206/206 | 23/27 | NA | NA | NA | Yes |
| Wu [28] 2006 | USA | Caucasian | Bladder | HB | 364/371 | 225/211 | 26/18 | NA | NA | NA | Yes |
| Sugimura [29] 2006 | Japan | Asian | HNC | HB | 20/52 | 59/112 | 43/77 | NA | NA | NA | Yes |
| Thirumaran [30] 2006 | Multiple | Caucasian | Skin | HB | 325/330 | 172/173 | 32/30 | NA | NA | NA | Yes |
| Hill [31] 2006 | Multiple | Mixed | NHL | PB | 599/521 | 425/331 | 77/71 | NA | NA | NA | Yes |
| Crew [32] 2007 | USA | Mixed | Breast | PB | 562/571 | 371/409 | 66/71 | 859/888 | 156/167 | 3/10 | Yes |
| Jorgensen [33] 2007 | USA | Caucasian | Breast | PB | 159/165 | 93/95 | 12/15 | 221/231 | 37/43 | 1/1 | Yes |
| Romanowicz [34] 2007 | Poland | Caucasian | Breast | NA | NA | NA | NA | 31/21 | 40/48 | 29/37 | Yes |
| Povey [35] 2007 | UK | Caucasian | Melanoma | PB | 314/252 | 169/162 | 24/27 | NA | NA | NA | Yes |
| Wang [36] 2007 | USA | Mixed | Skin | HB | 146/200 | 89/119 | 11/10 | NA | NA | NA | Yes |
| Shen [37] 2007 | Australia | Caucasian | NHL | PB | 340/294 | 170/163 | 30/27 | NA | NA | NA | Yes |
| McWilliams [38] 2008 | USA | Mixed | Pancreatic | HB | NA | NA | NA | 411/481 | 59/111 | 0/4 | Yes |
| Hooker [39] 2008 | USA | African | Prostate | HB | 74/100 | 119/141 | 61/60 | NA | NA | NA | Yes |
| Smith [40] 2008 | USA | Caucasian | Breast | HB | 195/256 | 113/124 | 12/28 | 278/358 | 39/47 | 7/1 | Yes |
| Smith [40] 2008 | USA | African | Breast | HB | 13/18 | 32/37 | 7/20 | 51/73 | 2/2 | 0/0 | Yes |
| Chang [41] 2008 | USA | Hispanic | Lung | HB | 60/138 | 44/127 | 9/34 | 97/267 | 16/31 | 0/1 | Yes |
| Chang [41] 2008 | USA | African | Lung | HB | 68/93 | 119/138 | 68/49 | NA | NA | NA | Yes |
| Rajaraman [42] 2008 | USA | Caucasian | Breast | PB | 482/674 | 288/352 | 49/53 | 714/922 | 124/147 | Yes | |
| Frei˘din [43] 2008 | Russia | Caucasian | Multiple | HB | 38/92 | 12/36 | 2/12 | NA | NA | NA | No |
| Hung [44] 2008 | Multiple | Mixed | Lung | NA | 1852/2485 | 1155/1510 | 209/286 | 2201/2208 | 306/390 | 13/21 | No for Asp1104His |
| He [45] 2008 | China | Asian | Cervical | HB | 35/53 | 94/80 | 71/67 | NA | NA | NA | No |
| Pardini [46] 2008 | Czech | Caucasian | Colorectal | HB | 334/356 | 177/153 | 21/23 | NA | NA | NA | Yes |
| Joshi [47] 2009 | USA | Caucasian | Colorectal | FB | 183/213 | 125/148 | 265/313 | 40/47 | NA | ||
| El-Zein [48] 2009 | USA | Mixed | NHL | HB | 104/127 | 78/80 | 16/12 | NA | NA | NA | Yes |
| Wen [49] 2009 | China | Asian | Bladder | HB | 15/45 | 57/233 | NA | NA | NA | NA | |
| Narter [50] 2009 | Turkey | Caucasian | Bladder | NA | 25/18 | 28/19 | 3/3 | NA | NA | NA | Yes |
| Abbasi [51] 2009 | German | Caucasian | HNC | PB | 137/380 | 103/230 | 8/37 | 203/554 | 44/90 | 1/3 | Yes |
| Hussain [52] 2009 | China | Asian | Gastric | PB | 38/90 | 104/180 | 39/91 | NA | NA | NA | Yes |
| McKean-Cowdin [53] 2009 | USA | Caucasian | Glioma | PB | 499/989 | 348/657 | 157/311 | NA | NA | NA | No |
| Pan [54] 2009 | USA | Caucasian | esophageal | HB | 222/287 | 145/155 | 15/15 | NA | NA | NA | Yes |
| Han [55] 2009 | USA | Caucasian | Breast | PB | 142/285 | 80/167 | 17/20 | 200/401 | 38/69 | 0/2 | Yes |
| Liu [56] 2009 | USA | Caucasian | Glioma | PB | 353/351 | 20/13 | NA | NA | NA | NA | |
| Agalliu [57] 2010 | USA | Caucasian | Prostate | PB | NA | NA | NA | 1025/1012 | 183/202 | 13/5 | Yes |
| Agalliu [57] 2010 | USA | African | Prostate | PB | NA | NA | NA | 136/78 | 8/3 | 0/0 | Yes |
| Rajaraman [58] 2010 | USA | Caucasian | Glioma | HB | 206/286 | 123/156 | 13/26 | 280/405 | 56/62 | 1/4 | Yes |
| Ming-Shiean [59] 2010 | China | Asian | Breast | HB | 134/159 | 191/243 | 76/129 | NA | NA | NA | Yes |
| Li [60] 2010 | China | Asian | Liver | HB | 174/151 | 233/265 | 93/91 | NA | NA | NA | Yes |
| Canbay [61] 2010 | Turkey | Caucasian | Gastric | NA | 25/148 | 12/83 | 3/16 | NA | NA | NA | Yes |
| Figl [62] 2010 | Multiple | Caucasian | Melanoma | HB | 703/725 | 409/465 | 74/84 | NA | NA | NA | Yes |
| Rouissi [63] 2011 | Tunis | African | Bladder | HB | 95/87 | 70/86 | 28/20 | NA | NA | NA | Yes |
| Liu [64] 2011 | China | Asian | Colorectal | HB | 233/329 | 603/537 | 192/219 | NA | NA | NA | Yes |
| Canbay [65] 2011 | Turkey | Caucasian | Colorectal | NA | 43/148 | 34/83 | 2/16 | NA | NA | NA | Yes |
| Gonçalves [66] 2011 | Braze | Caucasian | Melanoma | HB | 105/109 | 77/74 | 10/25 | NA | NA | NA | Yes |
| Ibarrola-Villava [67] 2011 | Spain | Caucasian | Melanoma | HB | 412/242 | 222/140 | 50/24 | 560/316 | 117/87 | 7/3 | Yes |
| Doherty [68] 2011 | USA | Mixed | Endometrial | PB | 418/408 | 254/248 | 42/47 | 593/620 | 107/89 | 3/5 | Yes |
| Biason [69] 2011 | Italy | Caucasian | Osteosarcoma | HB | 75/141 | 39/94 | 16/15 | NA | NA | NA | Yes |
| Krupa [70] 2011 | Poland | Caucasian | HNC | HB | NA | NA | NA | 221/224 | 26/29 | 6/0 | Yes |
| Yu [71] 2011 | USA | Caucasian | HNC | HB | NA | NA | NA | 837/829 | 195/209 | 8/8 | Yes |
| Ma [72] 2012 | USA | Caucasian | HNC | HB | 648/654 | 359/350 | 52/62 | NA | NA | NA | Yes |
| Gil [73] 2012 | Poland | Caucasian | Colorectal | HB | 86/64 | 35/31 | 11/5 | 119/83 | 14/15 | 0/0 | Yes |
| Berhane [74] 2012 | India | Asian | Prostate | HB | 58/128 | 72/146 | 20/26 | NA | NA | NA | Yes |
| Paszkowska-Szczur [75] 2013 | Poland | Caucasian | Melanoma | PB | 412/869 | 200/404 | 28/85 | NA | NA | NA | Yes |
| Wen [80] 2013 | China | Asian | Bladder | HB | 40/172 | 46/62 | 26/44 | NA | NA | NA | No |
| Wang [81] 2013 | China | Asian | Glioma | HB | NA | NA | NA | 265/609 | 59/36 | 6/7 | No |
| Santos [82] 2013 | Portugal | Caucasian | HNC | HB | 51/106 | 50/85 | 4/21 | 77/168 | 23/38 | 2/4 | No |
| Cheng [83] 2013 | China | Asian | Glioma | HB | NA | NA | NA | 149/182 | 41/43 | 17/11 | Yes |
HNC head and neck cancer, PB population-based study, HB hospital-based study.
XPG Asp1104His
The evaluations of the association of XPG Asp1104His polymorphism with cancer risk are shown in Table 2 . Overall, significantly increased risk of cancer was observed in dominant model (OR = 1.05, 95% confidence interval [CI] = 1.00–1.10, P value of heterogeneity test [Ph] = 0.001, I 2 = 40.4) and in Asp/His versus Asp/Asp (OR = 1.06, 95% CI = 1.01–1.11, P h<0.001, I 2 = 43.3) when all the eligible studies were pooled into the meta-analysis. Then we performed subgroup analysis by cancer type. No significant association was found in any cancer type, such as breast cancer (dominant model: OR = 1.01, 95% CI = 0.94–1.09, P h = 0.128, I 2 = 33.8, recessive model: OR = 0.95, 95% CI = 0.83–1.09, P h = 0.173, I 2 = 28.6; additive model: OR = 1.00, 95% CI = 0.93–1.09, P h = 0.098, I 2 = 37.8; His/His versus Asp/Asp: OR = 0.99, 95% CI = 0.86–1.14, P h = 0.185, I 2 = 27.2; Asp/His versus Asp/Asp: OR = 1.02, 95% CI = 0.94–1.10, P h = 0.136, I 2 = 32.8), lung cancer (dominant model: OR = 1.13, 95% CI = 0.98–1.31, P h = 0.045, I 2 = 53.4, recessive model: OR = 1.04, 95% CI = 0.93–1.17, P h = 0.212, I 2 = 28.4; additive model: OR = 1.08, 95% CI = 0.98–1.19, P h = 0.073, I 2 = 48.0; His/His versus Asp/Asp: OR = 1.15, 95% CI = 0.94–1.42, P h = 0.071, I 2 = 48.3; Asp/His versus Asp/Asp: OR = 1.13, 95% CI = 0.98–1.31, P h = 0.077, I 2 = 47.3), and so on.
Table 2. Stratified analysis of XPG Asp1104His and XPF Arg415Gln polymorphisms on cancer risk. 1 .
| Genetic model | N | Recessive model | Dominant model | Homozygote | Heterozygote | Additive model | |||||
| OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | ||
| XPG Asp1104His | |||||||||||
| Overall | 66 (32162/39858) | 1.00 (0.94–1.07)* | 0.073/21.2 | 1.05 (1.00–1.10)* | 0.001/40.4 | 1.04 (0.96–1.12)* | 0.012/30.9 | 1.06 (1.01–1.11)* | <0.001/43.3 | 1.03 (0.99–1.06)* | 0.008/32.8 |
| Cancer type | |||||||||||
| Bladder cancer | 7 (2488/2809) | 1.06 (0.72–1.56)* | 0.041/56.8 | 1.10 (0.85–1.44)* | 0.001/74.9 | 1.11 (0.69–1.80)* | 0.006/69.7 | 2 | <0.001/77.5 | 2 | <0.001/77.7 |
| Breast cancer | 11 (5474/6157) | 0.95 (0.83–1.09) | 0.173/28.6 | 1.01 (0.94–1.09) | 0.128/33.8 | 0.99 (0.86–1.14) | 0.185/27.2 | 1.02 (0.94–1.10) | 0.136/32.8 | 1.00 (0.93–1.09)* | 0.098/37.8 |
| Colorectal cancer | 7 (3471/3638) | 0.91 (0.77–1.08) | 0.696/0.0 | 1.07 (0.88–1.29)* | 0.004/69.1 | 1.08 (0.89–1.30) | 0.411/0.7 | 1.11 (0.86–1.42)* | <0.001/78.0 | 1.03 (0.95–1.12) | 0.169/35.7 |
| Glioma | 3 (1719/2789) | 0.98 (0.81–1.19) | 0.262/25.3 | 1.03 (0.90–1.18) | 0.984/0.0 | 0.97 (0.78–1.19) | 0.322/0.0 | 1.06 (0.92–1.23) | 0.810/0.0 | 1.01 (0.91–1.12) | 0.774/0.0 |
| HNC | 5 (1709/2691) | 0.92 (0.74–1.15) | 0.114/46.4 | 1.01 (0.89–1.16) | 0.244/26.6 | 0.86 (0.67–1.10) | 0.257/24.6 | 1.05 (0.83–1.31)* | 0.087/50.8 | 0.99 (0.90–1.10) | 0.735/0.0 |
| NHL | 4 (2303/2176) | 1.06 (0.84–1.35) | 0.389/0.6 | 1.12 (0.99–1.26) | 0.117/49.2 | 1.11 (0.88–1.42) | 0.279/22.0 | 1.12 (0.99–1.27) | 0.194/36.3 | 1.11 (0.95–1.29)* | 0.087/54.4 |
| Lung cancer | 7 (5509/6867) | 1.04 (0.93–1.17) | 0.212/28.4 | 1.13 (0.98–1.31)* | 0.045/53.4 | 1.15 (0.94–1.42)* | 0.071/48.3 | 1.13 (0.98–1.31)* | 0.077/47.3 | 1.08 (0.98–1.19)* | 0.073/48.0 |
| Melanoma | 8 (5297/7072) | 0.87 (0.69–1.12)* | 0.050/50.3 | 0.97 (0.90–1.04) | 0.762/0.0 | 0.87 (0.68–1.11)* | 0.059/48.4 | 0.98 (0.90–1.06) | 0.854/0.0 | 0.97 (0.91–1.03) | 0.336/12.1 |
| Other cancer | 14 (4192/5659) | 1.07 (0.93–1.22) | 0.578/0.0 | 1.06 (0.97–1.15) | 0.406/4.1 | 1.12 (0.96–1.30) | 0.533/0.0 | 1.05 (0.96–1.15) | 0.290/14.9 | 1.05 (0.98–1.12) | 0.675/0.0 |
| XPF Arg415Gln | |||||||||||
| Overall | 32 (17864/20578) | 1.11 (0.81–1.52)* | 0.068/30.5 | 1.04 (0.93–1.15)* | <0.001/62.6 | 1.10 (0.79–1.54)* | 0.035/35.7 | 1.02 (0.91–1.14)* | <0.001/62.5 | 1.05 (0.94–1.16)* | <0.001/66.7 |
| Cancer type | |||||||||||
| Breast cancer | 10 (5086/5542) | 1.22 (0.82–1.83)* | 0.017/58.9 | 1.03 (0.92–1.15) | 0.167/30.2 | 1.18 (0.76–1.83)* | 0.007/63.8 | 0.99 (0.87–1.12) | 0.277/18.6 | 1.01 (0.83–1.22)* | 0.034/52.0 |
| Lung cancer | 3 (2857/3118) | 0.75 (0.40–1.41) | 0.491/0.0 | 0.82 (0.71–0.96) | 0.104/55.7 | 0.73 (0.39–1.37) | 0.466/0.0 | 0.83 (0.71–0.97) | 0.132/50.7 | 0.83 (0.72–0.95)* | 0.091/58.4 |
| HNC | 4 (1643/2156) | 1.47 (0.72–2.98) | 0.364/5.8 | 1.04 (0.88–1.23) | 0.359/6.9 | 1.48 (0.73–3.00) | 0.370/4.5 | 1.02 (0.86–1.21) | 0.323/13.9 | 1.05 (0.90–1.23) | 0.302/17.7 |
| Colorectal cancer | 4 (1501/1497) | 0.51 (0.06–4.35)* | 0.069/69.7 | 0.93 (0.76–1.14) | 0.605/0.0 | 0.51 (0.06–4.45)* | 0.067/70.3 | 0.93 (0.74–1.18) | 0.526/0.0 | 0.90 (0.72–1.11) | 0.315/13.4 |
| Glioma | 3 (874/1359) | 1.51 (0.83–2.74) | 0.368/0.0 | 2 | <0.001/87.0 | 1.61 (0.88–2.93) | 0.357/3.0 | 2 | <0.001/88.0 | 2 | 0.001/86.0 |
| Other cancer | 8 (5903/6906) | 1.03 (0.69–1.53) | 0.239/24.9 | 0.95 (0.82–1.10)* | 0.048/50.6 | 1.02 (0.68–1.52) | 0.254/23.0 | 0.95 (0.82–1.11)* | 0.040/52.3 | 0.96 (0.84–1.09)* | 0.067/47.0 |
All summary ORs were calculated using fixed-effects models. In the case of significant heterogeneity (indicated by *), ORs were calculated using random-effects models.
The results were excluded due to high heterogeneity. The bold values indicate that the results are statistically significant.
We further examined the association of the XPG Asp1104His polymorphism and cancer risk according to cancer type and ethnicity ( Table 3 ). For samples of Caucasians, significant association was only be found in head and neck cancer (His/His vs. Asp/His+Asp/Asp: OR = 0.71, 95% CI = 0.51–0.97, P h = 0.271, I 2 = 23.5%) but not bladder cancer (dominant model: OR = 0.99, 95% CI = 0.88–1.12, P h = 0.673, I 2 = 0.0, recessive model: OR = 0.84, 95% CI = 0.50–1.41, P h = 0.078, I 2 = 56.0; additive model: OR = 0.98, 95% CI = 0.89–1.08, P h = 0.433, I 2 = 0.0; His/His versus Asp/Asp: OR = 0.85, 95% CI = 0.51–1.42, P h = 0.090, I 2 = 53.8; Asp/His versus Asp/Asp: OR = 1.01, 95% CI = 0.89–1.15, P h = 0.688, I 2 = 0.0), breast cancer (dominant model: OR = 1.07, 95% CI = 0.92–1.24, P h = 0.065, I 2 = 51.8, recessive model: OR = 1.07, 95% CI = 0.86–1.32, P h = 0.221, I 2 = 28.6; additive model: OR = 1.03, 95% CI = 0.95–1.12, P h = 0.113, I 2 = 43.8; His/His versus Asp/Asp: OR = 1.08, 95% CI = 0.87–1.34, P h = 0.215, I 2 = 29.3; Asp/His versus Asp/Asp: OR = 1.07, 95% CI = 0.91–1.26, P h = 0.048, I 2 = 55.2), and so on. For samples of Asians, significant association was found in lung cancer (dominant model: OR = 1.27, 95% CI = 1.06–1.51, P h = 0.133, I 2 = 50.5%; His/His versus Asp/Asp: OR = 1.28, 95% CI = 1.02–1.60, P h = 0.516, I 2 = 0.0%; additive model: OR = 1.13, 95% CI = 1.02–1.26, P h = 0.130, I 2 = 50.9%).
Table 3. Summary ORs (95% CI) categorized by ethnicity for the XPG Asp1104His and XPF Arg415Gln polymorphisms under different genetic models and cancer type. 1 .
| Ethnicity | Cancer type | N | Recessive model | Dominant model | Homozygote | Heterozygote | Additive model | |||||
| OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | |||
| XPG Asp1104His | ||||||||||||
| Caucasian | Bladder cancer | 4 (2111/2060) | 0.84 (0.50–1.41)* | 0.078/56.0 | 0.99 (0.88–1.12) | 0.673/0.0 | 0.85 (0.51–1.42)* | 0.090/53.8 | 1.01 (0.89–1.15) | 0.688/0.0 | 0.98 (0.89–1.08) | 0.433/0.0 |
| Breast cancer | 6 (3111/3675) | 1.07 (0.86–1.32) | 0.221/28.6 | 1.07 (0.92–1.24)* | 0.065/51.8 | 1.08 (0.87–1.34) | 0.215/29.3 | 1.07 (0.91–1.26)* | 0.048/55.2 | 1.03 (0.95–1.12) | 0.113/43.8 | |
| Colorectal cancer | 4 (1051/1240) | 0.92 (0.57–1.48) | 0.262/25.2 | 1.11 (0.93–1.31) | 0.688/0.0 | 0.97 (0.59–1.58) | 0.372/0.0 | 1.20 (0.96–1.49) | 0.397/0.0 | 1.10 (0.93–1.31) | 0.940/0.0 | |
| Glioma | 3 (1719/2789) | 0.98 (0.81–1.19) | 0.262/25.3 | 1.03 (0.90–1.18) | 0.984/0.0 | 0.97 (0.78–1.19) | 0.322/0.0 | 1.06 (0.92–1.23) | 0.810/0.0 | 1.01 (0.91–1.12) | 0.774/0.0 | |
| HNC | 3 (1412/1925) | 0.71 (0.51–0.97) | 0.271/23.5 | 1.04 (0.90–1.20) | 0.739/0.0 | 0.73 (0.53–1.02) | 0.378/0.0 | 1.10 (0.95–1.28) | 0.543/0.0 | 0.98 (0.87–1.10) | 0.819/0.0 | |
| Melanoma | 8 (5297/7072) | 0.87 (0.69–1.12)* | 0.050/50.3 | 0.97 (0.90–1.04) | 0.762/0.0 | 0.87 (0.68–1.11)* | 0.059/48.4 | 0.98 (0.90–1.06) | 0.854/0.0 | 0.97 (0.91–1.03) | 0.336/12.1 | |
| Other cancer | 5 (1133/1627) | 1.21 (0.86–1.70) | 0.345/10.7 | 1.04 (0.89–1.22) | 0.599/0.0 | 1.20 (0.85–1.69) | 0.422/0.0 | 1.02 (0.86–1.20) | 0.522/0.0 | 1.06 (0.93–1.21) | 0.501/0.0 | |
| Asian | Lung cancer | 3 (1428/1105) | 1.07 (0.88–1.29) | 0.673/0.0 | 1.27 (1.06–1.51) | 0.133/50.5 | 1.28 (1.02–1.60) | 0.516/0.0 | 1.35 (0.93–1.96)* | 0.073/61.9 | 1.13 (1.01–1.26) | 0.559/0.0 |
| Other cancer | 4 (1031/1368) | 1.04 (0.85–1.28) | 0.350/8.6 | 1.14 (0.82–1.60)* | 0.029/66.9 | 1.12 (0.88–1.43) | 0.176/39.3 | 1.15 (0.79–1.67)* | 0.017/70.7 | 1.03 (0.92–1.16) | 0.187/37.5 | |
| XPF Arg415Gln | ||||||||||||
| Caucasian | Breast cancer | 7 (3258/3729) | 2.17 (0.68–6.88)* | 0.022/61.9 | 1.10 (0.96–1.25) | 0.396/3.9 | 2.07 (0.56–7.62)* | 0.008/68.2 | 1.05 (0.89–1.23) | 0.522/0.0 | 1.10 (0.89–1.35)* | 0.094/46.8 |
| HNC | 4 (1643/2156) | 1.47 (0.72–2.98) | 0.364/5.8 | 1.04 (0.88–1.23) | 0.359/6.9 | 1.48 (0.73–3.00) | 0.370/4.5 | 1.02 (0.86–1.21) | 0.323/13.9 | 1.05 (0.90–1.23) | 0.302/17.7 | |
| Colorectal cancer | 3 (798/781) | 1.26 (0.40–4.01) | – | 0.99 (0.76–1.30) | 0.519/0.0 | 1.28 (0.40–4.07) | – | 0.97 (0.69–1.36) | 0.271/17.6 | 1.00 (0.74–1.36) | 0.253/23.5 | |
| Other cancer | 4 (4215/5095) | 1.20 (0.77–1.87) | 0.168/40.6 | 0.95 (0.85–1.06) | 0.549/0.0 | 1.19 (0.77–1.86) | 0.184/38.0 | 0.94 (0.84–1.05) | 0.406/0.0 | 0.96 (0.87–1.07) | 0.666/0.0 | |
All summary ORs were calculated using fixed-effects models. In the case of significant heterogeneity (indicated by *), ORs were calculated using random-effects models. The bold values indicate that the results are statistically significant.
We also examined the association of the XPG Asp1104His polymorphism and cancer risk according to cancer type and source of controls ( Table 4 ). For the population-based studies, no significant association was found between XPG Asp1104His polymorphism and cancer risk according to cancer type and source of controls. For the hospital-based studies, significant association was observed among breast cancer (recessive model: OR = 0.71, 95% CI = 0.55–0.92, P h = 0.262, I 2 = 24.9%; His/His versus Asp/Asp: OR = 0.74, 95% CI = 0.55–0.98, P h = 0.213, I 2 = 33.3%), colorectal cancer (dominant model: OR = 1.33, 95% CI = 1.15–1.55, P h = 0.188, I 2 = 0.0%; additive model: OR = 1.13, 95% CI = 1.02–1.25, P h = 0.971, I 2 = 0.0%), and other cancer (His/His versus Asp/Asp: OR = 1.22, 95% CI = 1.01–1.47, P h = 0.322, I 2 = 13.5%) but not lung cancer (dominant model: OR = 1.22, 95% CI = 0.91–1.63, P h = 0.030, I 2 = 66.4, recessive model: OR = 1.15, 95% CI = 0.96–1.37, P h = 0.105, I 2 = 51.1; additive model: OR = 1.13, 95% CI = 0.95–1.35, P h = 0.057, I 2 = 60.1; His/His versus Asp/Asp: OR = 1.32, 95% CI = 0.95–1.85, P h = 0.095, I 2 = 53.5; Asp/His versus Asp/Asp: OR = 1.21, 95% CI = 0.89–1.63, P h = 0.035, I 2 = 65.2) and head and neck cancer (dominant model: OR = 1.04, 95% CI = 0.89–1.22, P h = 0.548, I 2 = 0.0, recessive model: OR = 0.88, 95% CI = 0.66–1.16, P h = 0.135, I 2 = 50.1; additive model: OR = 1.00, 95% CI = 0.88–1.13, P h = 0.441, I 2 = 0.0; His/His versus Asp/Asp: OR = 0.90, 95% CI = 0.66–1.22, P h = 0.115, I 2 = 53.2; Asp/His versus Asp/Asp: OR = 1.08, 95% CI = 0.91–1.27, P h = 0.591, I 2 = 0.0), and so on.
Table 4. Summary ORs (95% CI) and value of value of the heterogeneity of XPG Asp1104His and XPF Arg415Gln polymorphisms for studies according to source of controls and cancer type 1 .
| Source of control | Cancer type | N | Recessive model | Dominant model | Homozygote | Heterozygote | Additive model | |||||
| OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | |||
| XPG Asp1104His | ||||||||||||
| PB | Breast cancer | 6 (4327/4684) | 1.06 (0.91–1.24) | 0.642/0.0 | 1.00 (0.92–1.09) | 0.130/41.4 | 1.09 (0.92–1.29) | 0.579/0.0 | 0.99 (0.91–1.08) | 0.130/41.3 | 1.01 (0.95–1.08) | 0.130/41.3 |
| Melanoma | 3 (2340/4207) | 0.91 (0.58–1.42)* | 0.036/70.0 | 1.00 (0.90–1.11) | 0.212/35.5 | 0.90 (0.56–1.43) | 0.372/0.0 | 1.00 (0.89–1.12) | 0.372/0.0 | 0.97 (0.83–1.13)* | 0.073/61.7 | |
| NHL | 3 (2105/1957) | 1.03 (0.80–1.31) | 0.345/6.1 | 1.11 (0.89–1.38)* | 0.062/64.0 | 1.07 (0.83–1.38) | 0.238/30.4 | 1.11 (0.90–1.37) | 0.100/56.7 | 1.08 (0.90–1.30)* | 0.053/66.0 | |
| Other cancer | 4 (1709/2395) | 0.89 (0.71–1.12) | 0.847/0.0 | 1.08 (0.95–1.23) | 0.646/0.0 | 0.97 (0.76–1.24) | 0.900/0.0 | 1.11 (0.96–1.26) | 0.522/0.0 | 1.02 (0.93–1.13) | 0.840/0.0 | |
| HB | Bladder cancer | 5 (2133/2485) | 1.16 (0.92–1.46) | 0.219/32.3 | 2 | <0.001/83.2 | 1.39 (0.86–2.23)* | 0.022/68.8 | 2 | <0.001/86.4 | 2 | <0.001/85.5 |
| Breast cancer | 4 (993/1322) | 0.71 (0.55–0.92) | 0.262/24.9 | 1.06 (0.89–1.26)* | 0.100/51.9 | 0.74 (0.55–0.98) | 0.213/33.3 | 1.16 (0.96–1.39) | 0.247/27.4 | 0.97 (0.77–1.22)* | 0.039/64.2 | |
| Colorectal cancer | 3 (1692/1717) | 0.93 (0.76–1.13) | 0.525/0.0 | 1.33 (1.15–1.55) | 0.188/0.0 | 1.21 (0.96–1.53) | 0.668/0.0 | 1.29 (0.97–1.72)* | 0.072/62.1 | 1.13 (1.02–1.25) | 0.971/0.0 | |
| HNC | 3 (1286/1519) | 0.88 (0.66–1.16) | 0.135/50.1 | 1.04 (0.89–1.22) | 0.548/0.0 | 0.90 (0.66–1.22) | 0.115 | 1.08 (0.91–1.27) | 0.591/0.0 | 1.00 (0.88–1.13) | 0.441/0.0 | |
| Lung cancer | 4 (1680/1575) | 1.15 (0.96–1.37) | 0.105/51.1 | 1.22 (0.91–1.63)* | 0.030/66.4 | 1.32 (0.95–1.85)* | 0.092/53.5 | 1.21 (0.89–1.63)* | 0.035/65.2 | 1.13 (0.95–1.35)* | 0.057/60.1 | |
| Melanoma | 5 (2957/2865) | 0.88 (0.70–1.09) | 0.145/41.5 | 0.94 (0.85–1.04) | 0.981/0.0 | 0.86 (0.69–1.08) | 0.213/31.3 | 0.95 (0.85–1.06) | 0.915/0.0 | 0.94 (0.86–1.02) | 0.766/0.0 | |
| Other cancer | 9 (2443/3017) | 1.18 (0.99–1.41) | 0.576/0.0 | 1.05 (0.94–1.18) | 0.171/31.0 | 1.22 (1.01–1.47) | 0.322/13.5 | 1.02 (0.90–1.15) | 0.155/32.9 | 1.07 (0.98–1.16) | 0.361/8.9 | |
| XPF Arg415Gln | ||||||||||||
| PB | Breast cancer | 6 (4356/4687) | 1.05 (0.29–3.77)* | 0.098/49.0 | 1.02 (0.90–1.16) | 0.158/37.3 | 1.05 (0.29–3.81)* | 0.093/49.7 | 1.00 (0.87–1.15) | 0.133/43.2 | 0.96 (0.77–1.20)* | 0.069/54.0 |
| Other cancer | 5 (3647/4879) | 1.48 (0.84–2.60) | 0.354/7.9 | 1.03 (0.91–1.17) | 0.477/0.0 | 1.48 (0.84–2.60) | 0.386/1.2 | 1.02 (0.90–1.15) | 0.286/20.2 | 1.05 (0.93–1.17) | 0.731/0.0 | |
| HB | Breast cancer | 4 (730/855) | 3.66 (0.38–34.9)* | 0.009/78.7 | 1.04 (0.78–1.39) | 0.178/38.9 | 3.39 (0.26–43.9)* | 0.003/82.8 | 0.92 (0.68–1.25) | 0.463/0.0 | 1.13 (0.73–1.73)* | 0.054/60.7 |
| Other cancer | 3 (2256/2027) | 0.70 (0.39–1.25) | 0.341/6.9 | 0.79 (0.59–1.07)* | 0.035/70.1 | 0.69 (0.38–1.24) | 0.347/5.6 | 0.81 (0.59–1.10)* | 0.033/70.8 | 0.80 (0.61–1.05)* | 0.045/67.7 | |
All summary ORs were calculated using fixed-effects models. In the case of significant heterogeneity (indicated by *), ORs were calculated using random-effects models.
The results were excluded due to high heterogeneity. The bold values indicate that the results are statistically significant. PB Population-based studies, HB Hospital-based studies, the bold values indicate that the results are statistically significant.
There was significant heterogeneity among these studies for dominant model comparison (P h = 0.001), recessive model comparison (P h = 0.073), additive model comparison (P h = 0.008), homozygote model comparison (P h = 0.012), and heterozygote model comparison (P h<0.001). Then, we assessed the source of heterogeneity by ethnicity, cancer type, source of controls, HWE, and sample size. The results indicated that sample size (recessive model: P = 0.038) but not cancer type (dominant model: P = 0.782; recessive model: P = 0.208; His/His versus Asp/Asp: P = 0.336; Asp/His versus Asp/Asp: P = 0.825; additive model: P = 0.556), ethnicity (dominant model: P = 0.298; recessive model: P = 0.119; His/His versus Asp/Asp: P = 0.066; Asp/His versus Asp/Asp: P = 0.449; additive model: P = 0.241), source of controls (dominant model: P = 0.433; recessive model: P = 0.821; His/His versus Asp/Asp: P = 0.634; Asp/His versus Asp/Asp: P = 0.358; additive model: P = 0.429), and HWE (dominant model: P = 0.126; recessive model: P = 0.660; His/His versus Asp/Asp: P = 0.272; Asp/His versus Asp/Asp: P = 0.123; additive model: P = 0.217) contributed to substantial heterogeneity among the meta-analysis. Examining genotype frequencies in the controls, significant deviation from HWE was detected in the eight studies [10], [26], [43], [44], [45], [53], [80], [81]. When these studies were excluded, the results were changed among overall cancer (dominant model: OR = 1.03, 95% CI = 0.99–1.08), Asians of lung cancer (dominant model: OR = 1.15, 95% CI = 0.95–1.41; His/His versus Asp/Asp: OR = 1.20, 95% CI = 0.92–1.55; additive model: OR = 1.10, 95% CI = 0.96–1.25), and hospital-based studies of other cancer (recessive model: OR = 1.23, 95% CI = 1.02–1.49; His/His versus Asp/Asp: OR = 1.20, 95% CI = 0.97–1.48), as shown in Table 5 . In addition, when the meta-analysis was performed excluding studies with small sample sizes, the results did not change among overall cancer studies and any subgroup analysis, as shown in Table 6 . Last, a single study involved in the meta–analysis was deleted each time to reflect the influence of individual data set to the pooled ORs, the results were changed among Caucasians of head and neck cancer (recessive model: OR = 0.75, 95% CI = 0.53–1.06), hospital-based studies of breast cancer (recessive model: OR = 1.22, 95% CI = 0.98–1.52; Gln/Gln versus Arg/Arg: OR = 0.79, 95% CI = 0.51–1.24), hospital-based studies of colorectal cancer (dominant model: OR = 1.15, 95% CI = 0.92–1.45; additive model: OR = 1.12, 95% CI = 0.92–1.35).
Table 5. Summary ORs (95% CI) and value of the heterogeneity of XPG Asp1104His and XPF Arg415Gln polymorphisms under different genetic models according to studies with HWE on cancer risk. 1 .
| Genetic model | No. comparisons (SZ case/control) | Recessive model | Dominant model | Homozygote | Heterozygote | Additive model | |||||
| OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | ||
| XPG Asp1104His | |||||||||||
| Overall | 58 (26988/31954) | 0.99 (0.92–1.07)* | 0.068/22.9 | 1.03 (0.99–1.08)* | 0.092/20.6 | 1.02 (0.94–1.11)* | 0.066/23.4 | 1.04 (1.00–1.09)* | 0.055/24.5 | 1.02 (0.99–1.05) | 0.139/17.3 |
| Cancer type | |||||||||||
| Bladder cancer | 6 (2376/2531) | 0.95 (0.62–1.47)* | 0.065/54.9 | 0.97 (0.87–1.09) | 0.724/0.0 | 0.94 (0.73–1.20) | 0.112/46.6 | 0.98 (0.87–1.11) | 0.517/0.0 | 0.98 (0.89–1.08) | 0.599/0.0 |
| Glioma | 2 (715/832) | 0.99 (0.61–1.60) | 0.102/62.6 | 1.04 (0.78–1.38) | – | 0.69 (0.35–1.38) | – | 1.09 (0.81–1.47) | – | 0.97 (0.77–1.24) | – |
| HNC | 3 (1429/1954) | 0.88 (0.67–1.16) | 0.240/29.9 | 1.06 (0.92–1.23) | 0.454/0.0 | 0.90 (0.67–1.22) | 0.194/39.0 | 1.10 (0.95–1.28) | 0.462/0.0 | 1.02 (0.91–1.14) | 0.537/0.0 |
| Lung cancer | 5 (1983/2275) | 1.12 (0.95–1.34) | 0.139/42.4 | 1.12 (0.98–1.28) | 0.348/10.2 | 1.19 (0.98–1.44) | 0.117/45.8 | 1.11 (0.96–1.27) | 0.694/0.0 | 1.08 (0.94–1.24)* | 0.098/48.9 |
| Other cancer | 12 (3940/5319) | 1.08 (0.93–1.24) | 0.532/0.0 | 1.05 (0.96–1.14) | 0.665/0.0 | 1.10 (0.94–1.29) | 0.667/0.0 | 1.04 (0.95–1.14) | 0.459/0.0 | 1.05 (0.98–1.12) | 0.835/0.0 |
| Ethnicity and cancer type | |||||||||||
| Lung cancer/Asian | 2 (1118/794) | 1.10 (0.88–1.38) | 0.463/0.0 | 1.15 (0.95–1.41) | 0.710/0.0 | 1.20 (0.92–1.55) | 0.517/0.0 | 1.14 (0.92–1.40) | 0.894/0.0 | 1.10 (0.96–1.25) | 0.484/0.0 |
| Other cancer/Caucasian | 4 (1081/1487) | 1.30 (0.92–1.85) | 0.473/0.0 | 1.07 (0.90–1.26) | 0.679/0.0 | 1.29 (0.91–1.85) | 0.618/0.0 | 1.03 (0.87–1.23) | 0.418/0.0 | 1.09 (0.95–1.25) | 0.811/0.0 |
| Other cancer/Asian | 3 (831/1168) | 1.03 (0.81–1.30) | 0.199/38.1 | 0.96 (0.70–1.17) | 0.109/54.8 | 1.02 (0.78–1.34) | 0.240/30.0 | 1.01 (0.71–1.44)* | 0.071/62.1 | 0.99 (0.87–1.13) | 0.269/23.8 |
| Source of controls and cancer type | |||||||||||
| Bladder cancer/HB | 4 (2021/2207) | 1.08 (0.84–1.40) | 0.254/27.1 | 0.97 (0.85–1.10) | 0.425/0.0 | 1.04 (0.80–1.36) | 0.299/17.2 | 0.96 (0.84–1.10) | 0.296/17.9 | 1.00 (0.90–1.10) | 0.352/4.1 |
| Lung cancer/HB | 3 (1370/1264) | 1.20 (0.80–1.79) | 0.077/61.0 | 1.13 (0.96–1.34) | 0.112/54.3 | 1.23 (0.76–2.00)* | 0.050/66.5 | 1.09 (0.91–1.30) | 0.347/5.5 | 1.09 (0.85–1.40)* | 0.029/71.8 |
| Other cancer/HB | 7 (2191/2677) | 1.23 (1.02–1.49) | 0.595/0.0 | 1.03 (0.92–1.16) | 0.375/7.0 | 1.20 (0.97–1.48) | 0.394/4.3 | 0.99 (0.87–1.12) | 0.324/13.9 | 1.07 (0.97–1.17) | 0.515/0.0 |
| XPF Arg415Gln | |||||||||||
| Overall | 30 (17432/19716) | 1.09 (0.78–1.54)* | 0.047/34.6 | 0.99 (0.91–1.07)* | 0.026/36.4 | 1.07 (0.74–1.53)* | 0.027/38.6 | 0.97 (0.89–1.05)* | 0.059/31.4 | 1.00 (0.91–1.08) | 0.003/47.8 |
| Cancer type | |||||||||||
| Glioma | 2 (544/707) | 1.44 (0.71–2.93) | 0.161/49.2 | 1.28 (0.96–1.70) | 0.868/0.0 | 1.49 (0.73–3.03) | 0.163/48.5 | 1.25 (0.92–1.69) | 0.716/0.0 | 1.28 (0.99–1.65) | 0.525/0.0 |
| HNC | 3 (1541/1946) | 1.58 (0.72–3.46) | 0.204/37.1 | 1.02 (0.85–1.21) | 0.277/22.1 | 1.57 (0.72–3.45) | 0.206/36.6 | 0.99 (0.83–1.19) | 0.264/25.0 | 1.04 (0.88–1.22) | 0.201/37.7 |
All summary ORs were calculated using fixed-effects models. In the case of significant heterogeneity (indicated by *), ORs were calculated using random-effects models. The bold values indicate that the results are statistically significant.
Table 6. Summary ORs (95% CI) and value of the heterogeneity of XPG Asp1104His and XPF Arg415Gln polymorphisms under different genetic models according to studies with a minimum of 200 participants on cancer risk. 1 .
| Genetic model | No. comparisons (SZ case/control) | Recessive model | Dominant model | Homozygote | Heterozygote | Additive model | |||||
| OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | OR (95%CI) | Ph/I2 (%) | ||
| XPG Asp1104His | |||||||||||
| Overall | 63 (32002/39603) | 1.01 (0.94–1.07)* | 0.085/20.6 | 1.05 (1.01–1.10)* | <0.001/42.5 | 1.04 (0.97–1.13)* | 0.012/31.6 | 1.06 (1.01–1.11)* | <0.001/45.8 | 1.03 (0.99–1.06)* | 0.007/33.5 |
| Cancer type | |||||||||||
| Breast cancer | 10 (5422/6082) | 0.97 (0.85–1.11) | 0.265/19.3 | 1.03 (0.93–1.14)* | 0.089/40.3 | 1.00 (0.87–1.15) | 0.205/25.9 | 1.04 (0.93–1.16)* | 0.098/39.0 | 1.01 (0.93–1.09)* | 0.096/39.3 |
| Bladder cancer | 6 (2432/2769) | 1.08 (0.71–1.63) | 0.023/64.7 | 2 | <0.001/79.0 | 1.14 (0.68–1.91)* | 0.003/75.4 | 2 | <0.001/82.0 | 2 | <0.001/82.1 |
| Other cancer | 13 (4140/5519) | 1.08 (0.94–1.24) | 0.618/0.0 | 1.07 (0.98–1.16) | 0.425/2.1 | 1.13 (0.97–1.32) | 0.596/0.0 | 1.06 (0.96–1.15) | 0.252/18.9 | 1.06 (0.99–1.13) | 0.783/0.0 |
| XPF Arg415Gln | |||||||||||
| Overall | 31 (17811/20503) | 1.11 (0.81–1.52)* | 0.068/30.5 | 1.04 (0.93–1.15)* | <0.001/63.7 | 1.10 (0.79–1.54)* | 0.035/35.7 | 1.02 (0.91–1.14)* | <0.001/63.7 | 1.05 (0.94–1.16)* | <0.001/67.8 |
| Cancer type | |||||||||||
| Breast cancer | 9 (5033/5467) | 1.54 (0.59–3.99)* | 0.017/58.9 | 1.02 (0.91–1.15) | 0.119/37.5 | 1.49 (0.52–4.25) | 0.007/63.8 | 0.98 (0.87–1.12) | 0.207/27.8 | 1.00 (0.83–1.22)* | 0.021/57.7 |
All summary ORs were calculated using fixed-effects models. In the case of significant heterogeneity (indicated by *), ORs were calculated using random-effects models.
The results were excluded due to high heterogeneity. The bold values indicate that the results are statistically significant.
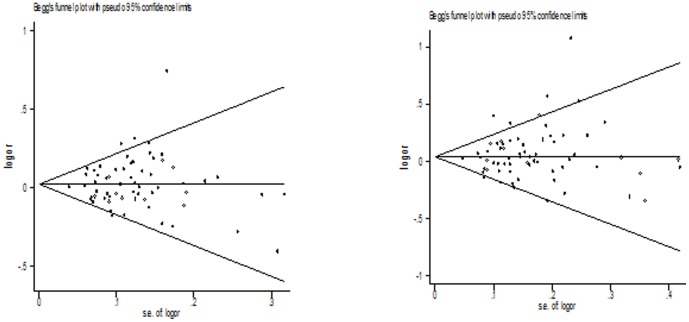
Both Begg's funnel plot and Egger's test were performed to assess the publication bias of literatures. The Egger's test results (dominant model: P = 0.245; recessive model: P = 0.482; additive model: P = 0.581; Homozygote model: P = 0.443; Heterozygote model: P = 0.148) and Begg's funnel plot ( Fig. 2 ) suggested no evidence of publication bias in the meta-analysis.
Figure 2. Begg's funnel plot for publication bias test between XPG Asp1104His polymorphism and cancer risk (additive model and dominant model).
XPF Arg415Gln
The evaluations of the association of XPF Arg415Gln polymorphism with cancer risk are shown in Table 2 . No significant association was observed between XPF Arg415Gln polymorphism and cancer risk when all the eligible studies were pooled into the meta-analysis (dominant model: OR = 1.04, 95% CI = 0.93–1.15, P h<0.001, I 2 = 62.6; recessive model: OR = 1.11, 95% CI = 0.81–1.52, P h = 0.068, I 2 = 30.5; additive model: OR = 1.05, 95% CI = 0.94–1.16, P h<0.001, I 2 = 66.7; Gln/Gln versus Arg/Arg: OR = 1.10, 95% CI = 0.79–1.54, P h = 0.035, I 2 = 35.7; Arg/Gln versus Arg/Arg: OR = 1.02, 95% CI = 0.91–1.14, P h<0.001, I 2 = 62.5). Then we performed subgroup analysis by cancer type. Significant association was found among lung cancer (dominant model: OR = 0.82, 95% CI = 0.71–0.96, P h = 0.104, I 2 = 55.7%; Arg/Gln versus Arg/Arg: OR = 0.83, 95% CI = 0.71–0.97, P h = 0.132, I 2 = 50.7%; additive model: OR = 0.83, 95% CI = 0.72–0.95, P h = 0.091, I 2 = 58.4%) but not breast cancer (dominant model: OR = 1.03, 95% CI = 0.92–1.15, P h = 0.167, I 2 = 30.2; recessive model: OR = 1.22, 95% CI = 0.82–1.83, P h = 0.017, I 2 = 58.9; additive model: OR = 1.01, 95% CI = 0.83–1.22, P h = 0.034, I 2 = 52.0; Gln/Gln versus Arg/Arg: OR = 1.18, 95% CI = 0.76–1.83, P h = 0.007, I 2 = 63.8; Arg/Gln versus Arg/Arg: OR = 0.99, 95% CI = 0.87–1.12, P h = 0.277, I 2 = 18.6), head and neck cancer (dominant model: OR = 1.04, 95% CI = 0.88–1.23, P h = 0.359, I 2 = 6.9; recessive model: OR = 1.47, 95% CI = 0.72–2.98, P h = 0.364, I 2 = 5.8; additive model: OR = 1.05, 95% CI = 0.90–1.23, P h = 0.302, I 2 = 17.7; Gln/Gln versus Arg/Arg: OR = 1.48, 95% CI = 0.73–3.00, P h = 0.370, I 2 = 4.5; Arg/Gln versus Arg/Arg: OR = 1.02, 95% CI = 0.86–1.21, P h = 0.323, I 2 = 13.9), and so on.
We further examined the association of the XPF Arg415Gln polymorphism and cancer risk according to cancer type and ethnicity ( Table 3 ). For the samples of Caucasians, no significant association was found among breast cancer (dominant model: OR = 1.10, 95% CI = 0.96–1.25, P h = 0.396, I 2 = 3.9; recessive model: OR = 2.17, 95% CI = 0.68–6.88, P h = 0.022, I 2 = 61.9; additive model: OR = 1.10, 95% CI = 0.89–1.35, P h = 0.094, I 2 = 46.8; Gln/Gln versus Arg/Arg: OR = 2.07, 95% CI = 0.56–7.62, P h = 0.008, I 2 = 68.2; Arg/Gln versus Arg/Arg: OR = 1.05, 95% CI = 0.89–1.23, P h = 0.522, I 2 = 0.0), head and neck cancer (dominant model: OR = 1.04, 95% CI = 0.88–1.23, P h = 0.359, I 2 = 6.9; recessive model: OR = 1.47, 95% CI = 0.72–2.98, P h = 0.364, I 2 = 5.8; additive model: OR = 1.05, 95% CI = 0.90–1.23, P h = 0.302, I 2 = 17.7; Gln/Gln versus Arg/Arg: OR = 1.48, 95% CI = 0.73–3.00, P h = 0.370, I 2 = 4.5; Arg/Gln versus Arg/Arg: OR = 1.02, 95% CI = 0.86–1.21, P h = 0.323, I 2 = 13.9), and so on.
We also examined the association of the XPF Arg415Gln polymorphism and cancer risk according to cancer type and source of controls ( Table 4 ). For the population-based studies, no significant association was found among breast cancer (dominant model: OR = 1.02, 95% CI = 0.90–1.16, P h = 0.158, I 2 = 37.3; recessive model: OR = 1.05, 95% CI = 0.29–3.77, P h = 0.098, I 2 = 49.0; additive model: OR = 0.96, 95% CI = 0.77–1.20, P h = 0.069, I 2 = 54.0; Gln/Gln versus Arg/Arg: OR = 1.05, 95% CI = 0.29–3.81, P h = 0.093, I 2 = 49.7; Arg/Gln versus Arg/Arg: OR = 1.00, 95% CI = 0.87–1.15, P h = 0.133, I 2 = 43.2) and other cancer (dominant model: OR = 1.03, 95% CI = 0.91–1.17, P h = 0.477, I 2 = 0.0; recessive model: OR = 1.48, 95% CI = 0.84–2.60, P h = 0.354, I 2 = 7.9; additive model: OR = 1.05, 95% CI = 0.93–1.17, P h = 0.731, I 2 = 0.0; Gln/Gln versus Arg/Arg: OR = 1.48, 95% CI = 0.84–2.60, P h = 0.386, I 2 = 1.2; Arg/Gln versus Arg/Arg: OR = 1.02, 95% CI = 0.90–1.15, P h = 0.286, I 2 = 20.2). For the hospital-based studies, no significant association was also observed among breast cancer (dominant model: OR = 1.04, 95% CI = 0.78–1.39, P h = 0.178, I 2 = 38.9; recessive model: OR = 3.66, 95% CI = 0.38–34.9, P h = 0.009, I 2 = 78.7; additive model: OR = 1.13, 95% CI = 0.73–1.73, P h = 0.054, I 2 = 60.7; Gln/Gln versus Arg/Arg: OR = 3.39, 95% CI = 0.26–43.9, P h = 0.003, I 2 = 82.8; Arg/Gln versus Arg/Arg: OR = 0.92, 95% CI = 0.68–1.25, P h = 0.463, I 2 = 0.0) and other cancer (dominant model: OR = 0.79, 95% CI = 0.59–1.07, P h = 0.035, I 2 = 70.1; recessive model: OR = 0.70, 95% CI = 0.39–1.25, P h = 0.341, I 2 = 6.9; additive model: OR = 0.80, 95% CI = 0.61–1.05, P h = 0.045, I 2 = 67.7; Gln/Gln versus Arg/Arg: OR = 0.69, 95% CI = 0.38–1.24, P h = 0.347, I 2 = 5.6; Arg/Gln versus Arg/Arg: OR = 0.81, 95% CI = 0.59–1.10, P h = 0.033, I 2 = 70.8).
There was significant heterogeneity among these studies for dominant model comparison (P h<0.001), recessive model comparison (P h = 0.068), additive model comparison (P h<0.001), homozygote model comparison (P h = 0.035), and heterozygote model comparison (P h<0.001). Then, we assessed the source of heterogeneity by ethnicity, cancer type, source of controls, HWE, and sample size. Meta-regression analysis indicated that HWE (Arg/Gln versus Arg/Arg: P<0.001; additive model: P = 0.001; dominant model: P<0.001) and ethnicity (Gln/Gln versus Arg/Arg: P = 0.001; recessive model: P = 0.001) but not cancer type (dominant model: P = 0.446; recessive model: P = 0.344; Gln/Gln versus Arg/Arg: P = 0.314; Arg/Gln versus Arg/Arg: P = 0.694; additive model: P = 0.456), source of controls (dominant model: P = 0.710; recessive model: P = 0.218; Gln/Gln versus Arg/Arg: P = 0.221; Arg/Gln versus Arg/Arg: P = 0.558; additive model: P = 0.962), and sample size (dominant model: P = 0.125; recessive model: P = 0.255; Gln/Gln versus Arg/Arg: P = 0.076; Arg/Gln versus Arg/Arg: P = 0.252; additive model: P = 0.153) contributed to substantial heterogeneity among the meta-analysis. Examining genotype frequencies in the controls, significant deviation from HWE was detected in the two studies [81], [82]. When these two studies were excluded, the results were not changed among overall cancer and any subgroup analysis, as shown in Table 5 . In addition, when the meta-analysis was performed excluding studies with small sample sizes, the results did not also change among overall cancer and any subgroup analysis, as shown in Table 6 . Last, a single study involved in the meta–analysis was deleted each time to reflect the influence of individual data set to the pooled ORs, the results did not also change among this meta-analysis, indicating that our results did not influenced statistically robust.
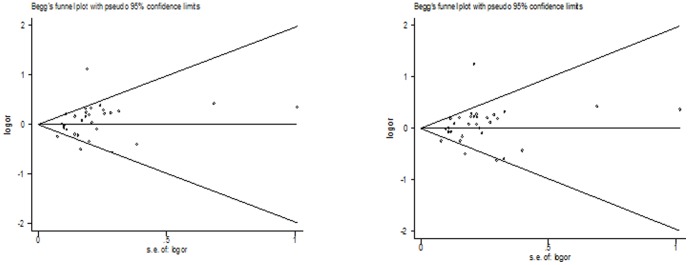
Both Begg's funnel plot and Egger's test were performed to assess the publication bias of literatures. The Egger's test results (P = 0.171; recessive model: P = 0.437; additive model: P = 0.114; Homozygote model: P = 0.425; Heterozygote model: P = 0.229) and Begg's funnel plot ( Fig. 3 ) suggested no evidence of publication bias in the meta-analysis.
Figure 3. Begg's funnel plot for publication bias test between XPF Arg415Gln polymorphism and cancer risk (additive model and dominant model).
Discussion
NER is a crucial DNA repair mechanism, which counteracts the consequences of mutagenic exposure of cell. XPF and XPG are both central players in the NER pathway, and involved in incision 5′ and 3′-ends, respectively, of the DNA lesion. A number of epidemiological studies have evaluated the association between XPG Asp1104His and XPF Arg415Gln polymorphisms and cancer risk, but the results remain inconclusive.
For instance, McWilliams et al. [38] reported a significantly decreased pancreatic cancer risk with XPF Arg415Gln polymorphism (P = 0.003). But Liu et al. [64] reported a significantly increased colorectal cancer risk associated with the variant allele of XPG Asp1104His. Goncalves et al. [66] found that significantly decreased melanoma cancer risk with the XPG 1104 His/His genotype (OR = 0.32; 95% CI = 0.13–0.75). However, Berhane et al. [74] found that statistically significant increased risk of prostate cancer was observed on individuals that posses His/His genotype of XPG (OR = 2.53, 95% CI = 0.99–6.56, P = 0.031). Ming-Shiean et al. [59] reported a significantly increased breast cancer risk with the variant allele of XPG Asp1104His (OR = 1.42; 95% CI = 1.08–1.97). He et al. [45] found that Women carrying homozygous Asp1104Asp genotypes had a significantly decreased risk of cervical or cervical squamous cell carcinoma compared to His1104Asp or His1104His genotypes. Smith et al. [8] reported a statistically significant difference in the XPF Arg415Gln genotype distributions between breast cancer cases and controls (P = 0.02). Furthermore, Kumar et al. [9] reported a marginally significant increase in breast cancer risk associated with the variant allele of XPG Asp1104His. What's more, more studies did not find obvious association among them. In order to resolve this conflict, a meta-analysis of 98 eligible studies including 32,162 cases and 39,858 controls for XPG Asp1104His and 17,864 cases and 20,578 controls for XPF Arg415Gln was performed to derive a more precise estimation of the association.
Overall, significantly elevated cancer risk was found when all studies were pooled into the meta-analysis of XPG Asp1104His (dominant model: OR = 1.05, 95% CI = 1.00–1.10; Asp/His versus Asp/His: OR = 1.06, 95% CI = 1.01–1.11). Based on biochemical properties described for XPG Asp1104His and XPF Arg415Gln polymorphisms, we would expect that the His or Gln alleles would be associated for all types of cancer. However, our results showed that such association was observed just among lung cancer (dominant model: OR = 0.82, 95% CI = 0.71–0.96; Asp/His versus Asp/Asp: OR = 0.83, 95% CI = 0.71–0.97; additive model: OR = 0.83, 95% CI = 0.72–0.95) for XPF Arg415Gln and hospital-based studies of other cancer (dominant model: OR = 1.23, 95% CI = 1.02–1.49) for XPG Asp1104His, suggesting that other factors may be modulating the XPG Asp1104His and XPF Arg415Gln polymorphisms functionality. However, the exact mechanism for association between different tumor sites and XPG Asp1104His and XPF Arg415Gln polymorphisms was not clear, carcinogenetic mechanism may differ by different tumor sites and the XPG Asp1104His and XPF Arg415Gln genetic variants may exert varying effects in different cancers. Hung et al. [44] reported a marginally significantly decreased lung cancer risk with the variant allele of XPF Arg415Gln (dominant model: OR = 0.78, 95% CI = 0.67–0.91). Our results seem to confirm and establish the trend in the meta-analysis of XPF Arg415Gln polymorphism and lung cancer risk that the data by Hung et al. [40] had indicated. However, at any case, the association between XPF Arg415Gln and lung cancer risk remain an open field, as the number of studies (n = 3 for Arg415Gln) is considerably smaller than that needed for the achievement of robust conclusions [94]. In the subgroup analysis by source of control and cancer type, significantly increased other cancer association was found among the hospital-based studies for the XPG Asp1104His polymorphism, but not the population-based studies. However, the hospital-based studies may have certain biases for such controls and may only represent a sample of an ill-defined reference population, and may not be representative of the general population or it may be that numerous subjects in the population-based controls were susceptible individuals. The results only indicate that participation of XPG Asp1104His may be a genetic susceptibility for other cancer. Therefore, the use of proper and representative population-based controls control subjects is important to reduce biases and in such genetic studies.
We noticed with great interest that 2 previous meta-analysis had been reported on the cancer risk with XPG Asp1104His and XPF Arg415Gln polymorphisms [84], [85]. Zhu et al. [84] had 49 case–control studies, in which a total of 23,490 cases and 27,168 controls were included. Their meta-analysis suggested that it was unlikely that the XPG Asp1104His polymorphism may contribute to individual susceptibility to cancer risk. Shi et al. [85] had 23 case-control studies, in which a total of 14,632 cancer cases and 15,545 controls. Their meta-analysis suggested that it was unlikely that the XPF Arg415Gln polymorphism may contribute to individual susceptibility to cancer risk. However, several published studies were not included in that meta-analysis [84], [85]. By analyzing a larger number of studies than the previous meta-analysis [84], [85], our meta-analysis included 32,162 cases and 39,858 controls (from 66 studies) for XPG Asp1104His and 17,864 cases and 20,578 controls (from 32 studies) for XPF Arg415Gln to perform the two gene polymorphisms and cancer risk. Our meta-analysis suggests that XPF Arg415Gln polymorphism may be associated with decreased lung cancer risk and XPG Asp1104His may be a low-penetrant risk factor in some cancer development. Our results seem to confirm and establish the trend in the meta-analysis of the XPG Asp1104His and XPF Arg415Gln polymorphisms according to the previous meta-analysis [84], [85].
In the present meta-analysis, between-studies heterogeneity was observed between XPG Asp1104His and XPF Arg415Gln polymorphisms and cancer of risk. Meta-regression analysis indicated that HWE contributed to substantial heterogeneity among the meta-analysis for XPF Arg415Gln polymorphism and sample size contributed to substantial heterogeneity among the meta-analysis for XPG Asp1104His. Deviation of HWE may reflect methodological problems such as genotyping errors, population stratification or selection bias. When these studies were excluded, the results were changed among overall cancer and some subgroup analyses for XPG Asp1104His, indicating that our meta-analysis was not statistically robust. Hence, significant association may be not existed in some cancer types when the results were changed. When the meta-analysis was performed excluding studies with small sample sizes, the results did not change among overall cancer studies and any subgroup analysis, indicating that small sample sizes did not influenced statistically robust.
Our meta-analysis has several strengths. First, a systematic review of the association of XPG Asp1104His and XPF Arg415Gln polymorphisms with cancer risk is statistically more powerful than any single study. Second, the quality of eligible studies included in current meta-analysis was satisfactory and met our inclusion criterion. Third, we did not detect any publication bias indicating that the whole pooled results should be unbiased. However, although we have put considerable efforts and resources into testing possible association between XPG Asp1104His and XPF Arg415Gln polymorphisms and cancer risk, there are still some limitations inherited from the published studies. First, our results were based on single-factor estimations without adjustment for other risk factors including alcohol usage, environmental factors and other lifestyles. At lower levels of alcohol consumption, the difference in cancer risk between the various gene carriers was less striking. And higher levels of alcohol consumption result in production of more acetaldehyde which then can exert its carcinogenic effect [95]. Second, in the subgroup analysis may have had insufficient statistical power to check an association. Third, the controls were not uniformly defined. Some studies used a healthy population as the reference group, whereas others selected hospital patients without organic cancer as the reference group. Therefore, non-differential misclassification bias is possible because these studies may have included the control groups who have different risks of developing cancer of various organs.
In conclusion, this meta-analysis suggests that XPF Arg415Gln polymorphism may be associated with decreased lung cancer risk and XPG Asp1104His may be a low-penetrant risk factor in some cancer development. However, it is necessary to conduct large sample studies using standardized unbiased genotyping methods, homogeneous cancer patients and well-matched controls. Moreover, further studies estimating the effect of gene–gene and gene–environment interactions may eventually lead to our better, comprehensive understanding of the association between the XPF Arg415Gln and XPG Asp1104His polymorphisms and cancer risk.
Supporting Information
PRISMA Checklist.
(DOC)
Funding Statement
The authors have no funding or support to report.
References
- 1. Wood RD, Mitchell M, Sgouros J, Lindahl T (2001) Human DNA repair genes. Science 291: 1284–1289. [DOI] [PubMed] [Google Scholar]
- 2. Friedberg EC (2001) How nucleotide excision repair protects against cancer. Nature Rev Cancer 1: 22–33. [DOI] [PubMed] [Google Scholar]
- 3. Cleaver JE (2000) Common pathways for ultraviolet skin carcinogenesis in the repair and replication defective groups of xeroderma pigmentosum. J Dermatol Sci 23: 1–11. [DOI] [PubMed] [Google Scholar]
- 4. O'Donovan A, Davies AA, Moggs JG, West SC, Wood RD (1994) XPG endonuclease makes the 30 incision in human DNA nucleotide excision repair. Nature 371: 432–435. [DOI] [PubMed] [Google Scholar]
- 5. Wakasugi M, Reardon JT, Sancar A (1997) The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J Biol Chem 272: 16030–16034. [DOI] [PubMed] [Google Scholar]
- 6. Araujo SJ, Nigg EA, Wood RD (2001) Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol Cell Biol 21: 2281–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gillet LC, Scharer OD (2006) Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev 106: 253–276. [DOI] [PubMed] [Google Scholar]
- 8. Smith TR, Levine EA, Perrier ND, Miller MS, Freimanis RI, et al. (2003) DNA-repair genetic polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev 12: 1200–1204. [PubMed] [Google Scholar]
- 9. Kumar R, Höglund L, Zhao C, Försti A, Snellman E, et al. (2003) Single nucleotide polymorphisms in the XPG gene: determination of role in DNA repair and breast cancer risk. Int J Cancer 103: 671–675. [DOI] [PubMed] [Google Scholar]
- 10. Jeon HS, Kim KM, Park SH, Lee SY, Choi JE, et al. (2003) Relationship between XPG codon 1104 polymorphism and risk of primary lung cancer. Carcinogenesis 24: 1677–1681. [DOI] [PubMed] [Google Scholar]
- 11. Sanyal S, Festa F, Sakano S, Zhang Z, Steineck G, et al. (2004) Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis 25: 729–734. [DOI] [PubMed] [Google Scholar]
- 12. Blankenburg S, König IR, Moessner R, Laspe P, Thoms KM, et al. (2005) No association between three xeroderma pigmentosum group C and one group G gene polymorphisms and risk of cutaneous melanoma. Eur J Hum Genet 13: 253–255. [DOI] [PubMed] [Google Scholar]
- 13. Weiss JM, Weiss NS, Ulrich CM, Doherty JA, Voigt LF, et al. (2005) Interindividual variation in nucleotide excision repair genes and risk of endometrial cancer. Cancer Epidemiol Biomarkers Prev 14: 2524–2530. [DOI] [PubMed] [Google Scholar]
- 14. Shen M, Berndt SI, Rothman N, Demarini DM, Mumford JL, et al. (2005) Polymorphisms in the DNA nucleotide excision repair genes and lung cancer risk in Xuan Wei, China. Int J Cancer 116: 768–773. [DOI] [PubMed] [Google Scholar]
- 15. Bigler J, Ulrich CM, Kawashima T, Whitton J, Potter JD (2005) DNA repair polymorphisms and risk of colorectal adenomatous or hyperplastic polyps. Cancer Epidemiol Biomarkers Prev 14: 2501–2508. [DOI] [PubMed] [Google Scholar]
- 16. Sakiyama T, Kohno T, Mimaki S, Ohta T, Yanagitani N, et al. (2005) Association of amino acid substitution polymorphisms in DNA repair genes TP53, POLI, REV1 and LIG4 with lung cancer risk. Int J Cancer 114: 730–737. [DOI] [PubMed] [Google Scholar]
- 17. Cui Y, Morgenstern H, Greenland S, Tashkin DP, Mao J, et al. (2006) Polymorphism of Xeroderma Pigmentosum group G and the risk of lung cancer and squamous cell carcinomas of the oropharynx, larynx and esophagus. Int J Cancer 118: 714–720. [DOI] [PubMed] [Google Scholar]
- 18. Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, et al. (2006) Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis 27: 560–567. [DOI] [PubMed] [Google Scholar]
- 19. Millikan RC, Hummer A, Begg C, Player J, de Cotret AR, et al. (2006) Polymorphisms in nucleotide excision repair genes and risk of multiple primary melanoma: the Genes Environment and Melanoma Study. Carcinogenesis 27: 610–618. [DOI] [PubMed] [Google Scholar]
- 20. Mechanic LE, Millikan RC, Player J, de Cotret AR, Winkel S, et al. (2006) Polymorphisms in nucleotide excision repair genes, smoking and breast cancer in African Americans and whites: a population-based case–control study. Carcinogenesis 27: 1377–1385. [DOI] [PubMed] [Google Scholar]
- 21. Huang WY, Berndt SI, Kang D, Chatterjee N, Chanock SJ, et al. (2006) Nucleotide excision repair gene polymorphisms and risk of advanced colorectal adenoma: XPC polymorphisms modify smoking-related risk. Cancer Epidemiol Biomarkers Prev 15: 306–311. [DOI] [PubMed] [Google Scholar]
- 22. García-Closas M, Malats N, Real FX, Welch R, Kogevinas M, et al. (2006) Genetic variation in the nucleotide excision repair pathway and bladder cancer risk. Cancer Epidemiol Biomarkers Prev 15: 536–542. [DOI] [PubMed] [Google Scholar]
- 23. Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, et al. (2006) Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res 12: 2101–2108. [DOI] [PubMed] [Google Scholar]
- 24. Shen M, Zheng T, Lan Q, Zhang Y, Zahm SH, et al. (2006) Polymorphisms in DNA repair genes and risk of non-Hodgkin lymphoma among women in Connecticut. Hum Genet 119: 659–668. [DOI] [PubMed] [Google Scholar]
- 25. Shen J, Desai M, Agrawal M, Kennedy DO, Senie RT, et al. (2006) Polymorphisms in nucleotide excision repair genes and DNA repair capacity phenotype in sisters discordant for breast cancer. Cancer Epidemiol Biomarkers Prev 15: 1614–1619. [DOI] [PubMed] [Google Scholar]
- 26. Wen SX, Tang PZ, Zhang XM, Zhao D, Guo YL, et al. (2006) Association between genetic polymorphism in xeroderma pigmentosum G gene and risks of laryngeal and hypopharyngeal carcinomas. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 28: 703–706. [PubMed] [Google Scholar]
- 27. Li C, Hu Z, Liu Z, Wang LE, Strom SS, et al. (2006) Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case–control analysis. Cancer Epidemiol Biomarkers Prev 15: 2526–2532. [DOI] [PubMed] [Google Scholar]
- 28. Wu X, Gu J, Grossman HB, Amos CI, Etzel C, et al. (2006) Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet 78: 464–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sugimura T, Kumimoto H, Tohnai I, Fukui T, Matsuo K, et al. (2006) Gene-environment interaction involved in oral carcinogenesis: molecular epidemiological study for metabolic and DNA repair gene polymorphisms. J Oral Pathol Med 35: 11–18. [DOI] [PubMed] [Google Scholar]
- 30. Thirumaran RK, Bermejo JL, Rudnai P, Gurzau E, Koppova K, et al. (2006) Single nucleotide polymorphisms in DNA repair genes and basal cell carcinoma of skin. Carcinogenesis 27: 1676–1681. [DOI] [PubMed] [Google Scholar]
- 31. Hill DA, Wang SS, Cerhan JR, Davis S, Cozen W, et al. (2006) Risk of non-Hodgkin lymphoma (NHL) in relation to germline variation in DNA repair and related genes. Blood 108: 3161–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crew KD, Gammon MD, Terry MB, Zhang FF, Zablotska LB, et al. (2007) Polymorphisms in nucleotide excision repair genes, polycyclic aromatic hydrocarbon-DNA adducts, and breast cancer risk. Cancer Epidemiol Biomarkers Prev 16: 2033–2041. [DOI] [PubMed] [Google Scholar]
- 33. Jorgensen TJ, Visvanathan K, Ruczinski I, Thuita L, Hoffman S, et al. (2007) Breast cancer risk is not associated with polymorphic forms of xeroderma pigmentosum genes in a cohort of women from Washington County, Maryland. Breast Cancer Res Treat 101: 65–71. [DOI] [PubMed] [Google Scholar]
- 34. Romanowicz-Makowska H, Smolarz B, Kulig A (2007) Polymorphisms in XRCC1 and ERCC4/XPF DNA repair genes and associations with breast cancer risk in women. Pol Merkur Lekarski 22: 200–203. [PubMed] [Google Scholar]
- 35. Povey JE, Darakhshan F, Robertson K, Bisset Y, Mekky M, et al. (2007) DNA repair gene polymorphisms and genetic predisposition to cutaneous melanoma. Carcinogenesis 28: 1087–1093. [DOI] [PubMed] [Google Scholar]
- 36. Wang LE, Li C, Strom SS, Goldberg LH, Brewster A, et al. (2007) Repair capacity for UV light induced DNA damage associated with risk of nonmelanoma skin cancer and tumor progression. Clin Cancer Res 13: 6532–6539. [DOI] [PubMed] [Google Scholar]
- 37. Shen M, Purdue MP, Kricker A, Lan Q, Grulich AE, et al. (2007) Polymorphisms in DNA repair genes and risk of non-Hodgkin's lymphoma in New South Wales, Australia. Haematologica 92: 1180–1185. [DOI] [PubMed] [Google Scholar]
- 38. McWilliams RR, Bamlet WR, Cunningham JM, Goode EL, de Andrade M, et al. (2008) Polymorphisms in DNA repair genes, smoking, and pancreatic adenocarcinoma risk. Cancer Res 68: 4928–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hooker S, Bonilla C, Akereyeni F, Ahaghotu C, Kittles RA (2008) NAT2 and NER genetic variants and sporadic prostate cancer susceptibility in African Americans. Prostate Cancer Prostatic Dis 11: 349–356. [DOI] [PubMed] [Google Scholar]
- 40. Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, et al. (2008) Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis 29: 2132–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, et al. (2008) Nucleotide excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African Americans. Int J Cancer 123: 2095–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rajaraman P, Bhatti P, Doody MM, Simon SL, Weinstock RM, et al. (2008) Nucleotide excision repair polymorphisms may modify ionizing radiation-related breast cancer risk in US radiologic technologists. Int J Cancer 123: 2713–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frei˘din MB, Ivanina PV, Takhauov RM, Goncharova IA, Dvornichenko MV, et al. (2008) The evaluation of association between polymorphisms of DNA excision repair enzyme genes and risk of malignant tumors development in Siberian Group of Chemical Enterprises workers. Radiats Biol Radioecol 48: 439–444. [PubMed] [Google Scholar]
- 44. Hung RJ, Christiani DC, Risch A, Popanda O, Haugen A, et al. (2008) International Lung Cancer Consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol Biomarkers Prev 17: 3081–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. He X, Ye F, Zhang J, Cheng Q, Shen J, et al. (2008) Susceptibility of XRCC3, XPD, and XPG genetic variants to cervical carcinoma. Pathobiology 75: 356–363. [DOI] [PubMed] [Google Scholar]
- 46. Pardini B, Naccarati A, Novotny J, Smerhovsky Z, Vodickova L, et al. (2008) DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res 638: 146–153. [DOI] [PubMed] [Google Scholar]
- 47. Joshi AD, Corral R, Siegmund KD, Haile RW, Le Marchand L, et al. (2009) Red meat and poultry intake, polymorphisms in the nucleotide excision repair and mismatch repair pathways and colorectal cancer risk. Carcinogenesis 30: 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. El-Zein R, Monroy CM, Etzel CJ, Cortes AC, Xing Y, et al. (2009) Genetic polymorphisms in DNA repair genes as modulators of Hodgkin disease risk. Cancer 115: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wen H, Ding Q, Fang ZJ, Xia GW, Fang J (2009) Population study of genetic polymorphisms and superficial bladder cancer risk in Han-Chinese smokers in Shanghai. Int Urol Nephrol 41: 855–864. [DOI] [PubMed] [Google Scholar]
- 50. Narter KF, Ergen A, Agaçhan B, Görmüs U, Timirci O, et al. (2009) Bladder cancer and polymorphisms of DNA repair genes (XRCC1, XRCC3, XPD, XPG, APE1, hOGG1). Anticancer Res 29: 1389–1393. [PubMed] [Google Scholar]
- 51. Abbasi R, Ramroth H, Becher H, Dietz A, Schmezer P, et al. (2009) Laryngeal cancer risk associated with smoking and alcohol consumption is modified by genetic polymorphisms in ERCC5, ERCC6 and RAD23B but not by polymorphisms in five other nucleotide excision repair genes. Int J Cancer 125: 1431–1439. [DOI] [PubMed] [Google Scholar]
- 52. Hussain SK, Mu LN, Cai L, Chang SC, Park SL, et al. (2009) Genetic variation in immune regulation and DNA repair pathways and stomach cancer in China. Cancer Epidemiol Biomarkers Prev 18: 2304–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McKean-Cowdin R, Barnholtz-Sloan J, Inskip PD, Ruder AM, Butler M, et al. (2009) Associations between Polymorphisms in DNA Repair Genes and Glioblastoma. Cancer Epidemiology Biomarkers & Prevention 18: 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pan J, Lin J, Izzo JG, Liu Y, Xing J, et al. (2009) Genetic susceptibility to esophageal cancer: the role of the nucleotide excision repair pathway. Carcinogenesis 30: 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Han J, Haiman C, Niu T, Guo Q, Cox DG, et al. (2009) Genetic variation in DNA repair pathway genes and premenopausal breast cancer risk. Breast Cancer Res Treat 115: 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu Y, Scheurer ME, El-Zein R, Cao Y, Do KA, et al. (2009) Association and Interactions between DNA Repair Gene Polymorphisms and Adult Glioma. Cancer Epidemiology Biomarkers & Prevention 18: 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Agalliu I, Kwon EM, Salinas CA, Koopmeiners JS, Ostrander EA, et al. (2010) Genetic variation in DNA repair genes and prostate cancer risk: results from a population-based study. Cancer Causes Control 21: 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rajaraman P, Hutchinson A, Wichner S, Black PM, Fine HA, et al. (2010) DNA repair gene polymorphisms and risk of adult meningioma, glioma, and acoustic neuroma. Neuro Oncol 12: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ming-Shiean H, Yu JC, Wang HW, Chen ST, Hsiung CN, et al. (2010) Synergistic effects of polymorphisms in DNA repair genes and endogenous estrogen exposure on female breast cancer risk. Ann Surg Oncol 17: 760–771. [DOI] [PubMed] [Google Scholar]
- 60. Li LM, Zeng XY, Ji L, Fan XJ, Li YQ, et al. (2010) Association of XPC and XPG polymorphisms with the risk of hepatocellular carcinoma. Zhonghua Gan Zang Bing Za Zhi 18: 271–275. [DOI] [PubMed] [Google Scholar]
- 61. Canbay E, Agachan B, Gulluoglu M, Isbir T, Balik E, et al. (2010) Possible associations of APE1 polymorphism with susceptibility and HOGG1 polymorphism with prognosis in gastric cancer. Anticancer Res 30: 1359–1364. [PubMed] [Google Scholar]
- 62. Figl A, Scherer D, Nagore E, Bermejo JL, Botella-Estrada R, et al. (2010) Single-nucleotide polymorphisms in DNA-repair genes and cutaneous melanoma. Mutation Research - Genetic Toxicology and Environmental Mutagenesis 702: 8–16. [DOI] [PubMed] [Google Scholar]
- 63. Rouissi K, Bahria IB, Bougatef K, Marrakchi R, Stambouli N, et al. (2011) The effect of tobacco, XPC, ERCC2 and ERCC5 genetic variants in bladder cancer development. BMC Cancer 11: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu D, Wu HZ, Zhang YN, Kang H, Sun MJ, et al. (2011) DNA repair genes XPC, XPG polymorphisms: Relation to the risk of colorectal carcinoma and therapeutic outcome with oxaliplatin-based adjuvant chemotherapy. Mol Carcinog doi:10.1002/mc.21862 [DOI] [PubMed] [Google Scholar]
- 65. Canbay E, Cakmakoglu B, Zeybek U, Sozen S, Cacina C, et al. (2011) Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population. Curr Med Res Opin 27: 1295–1302. [DOI] [PubMed] [Google Scholar]
- 66. Gonçalves FT, Francisco G, de Souza SP, Luiz OC, Festa-Neto C, et al. (2011) European ancestry and polymorphisms in DNA repair genes modify the risk of melanoma: a case–control study in a high UV index region in Brazil. J Dermatol Sci 64: 59–66. [DOI] [PubMed] [Google Scholar]
- 67. Ibarrola-Villava M, Peña-Chilet M, Fernandez LP, Aviles JA, Mayor M, et al. (2011) Genetic polymorphisms in DNA repair and oxidative stress pathways associated with malignant melanoma susceptibility. European Journal of Cancer 47: 2618–2625. [DOI] [PubMed] [Google Scholar]
- 68. Doherty JA, Weiss NS, Fish S, Fan W, Loomis MM, et al. (2011) Polymorphisms in nucleotide excision repair genes and endometrial cancer risk. Cancer Epidemiol Biomarkers Prev 20: 1873–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Biason P, Hattinger CM, Innocenti F, Talamini R, Alberghini M, et al. (2012) Nucleotide excision repair gene variants and association with survival in osteosarcoma patients treated with neoadjuvant chemotherapy. Pharmacogenomics J 12: 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Krupa R, Kasznicki J, Gajęcka M, Rydzanicz M, Kiwerska K, et al. (2011) Polymorphisms of the DNA repair genes XRCC1 and ERCC4 are not associated with smoking- and drinking-dependent larynx cancer in a Polish population. Exp Oncol 33: 55–56. [PubMed] [Google Scholar]
- 71. Yu H, Liu Z, Huang YJ, Yin M, Wang LE, et al. (2012) Association between single nucleotide polymorphisms in ERCC4 and risk of squamous cell carcinoma of the head and neck submitted. PLoS One 7: e41853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ma H, Yu H, Liu Z, Wang LE, Sturgis EM, et al. (2012) Polymorphisms of XPG/ERCC5 and risk of squamous cell carcinoma of the head and neck. Pharmacogenet Genomics 22: 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gil J, Ramsey D, Stembalska A, Karpinski P, Pesz KA, et al. (2012) The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual's susceptibility to sporadic colorectal cancer. Mol Biol Rep 39: 527–534. [DOI] [PubMed] [Google Scholar]
- 74. Berhane N, Sobti RC, Mahdi SA (2012) DNA repair genes polymorphism (XPG and XRCC1) and association of prostate cancer in a north Indian population. Mol Biol Rep 39: 2471–2479. [DOI] [PubMed] [Google Scholar]
- 75. Paszkowska-Szczur K, Scott RJ, Serrano-Fernandez P, Mirecka A, Gapska P, et al. (2013) Xeroderma pigmentosum genes and melanoma risk. Int J Cancer 133: 1094–1100. [DOI] [PubMed] [Google Scholar]
- 76. Weiss JM, Weiss NS, Ulrich CM, Doherty JA, Chen C (2006) Nucleotide excision repair genotype and the incidence of endometrial cancer: effect of other risk factors on the association. Gynecol Oncol 103: 891–896. [DOI] [PubMed] [Google Scholar]
- 77. Weiss JM, Weiss NS, Ulrich CM, Doherty JA, Voigt LF, et al. (2005) Interindividual variation in nucleotide excision repair genes and risk of endometrial cancer. Cancer Epidemiol Biomarkers Prev 14: 2524–2530. [DOI] [PubMed] [Google Scholar]
- 78. Smith TR, Liu-Mares W, Van Emburgh BO, Levine EA, Allen GO, et al. (2011) Genetic polymorphisms of multiple DNA repair pathways impact age at diagnosis and TP53 mutations in breast cancer. Carcinogenesis 32: 1354–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, et al. (2006) Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis 27: 560–567. [DOI] [PubMed] [Google Scholar]
- 80. Wen H, Feng CC, Fang ZJ, Xia GW, Jiang HW, et al. (2013) Study on Bladder Cancer Susceptibility and Genetic Polymorphisms of XPC, XPG, and CYP in Smokers and Non-Smokers. Actas Urol Esp 37: 259–265. [DOI] [PubMed] [Google Scholar]
- 81. Wang XF, Liu S, Shao ZK (2013) Effects of polymorphisms in nucleotide excision repair genes on glioma risk in a Chinese population. Gene doi:pii: S0378-1119(13)00896-2. 10.1016/j.gene.2013.07.025 [DOI] [PubMed] [Google Scholar]
- 82. Santos LS, Gomes BC, Gouveia R, Silva SN, Azevedo AP, et al. (2013) The role of CCNH Val270Ala (rs2230641) and other nucleotide excision repair polymorphisms in individual susceptibility to well-differentiated thyroid cancer. Oncol Rep 30: 2458–2466. [DOI] [PubMed] [Google Scholar]
- 83. Cheng HB, Xie C, Zhang RY, Hu SS, Wang Z, et al. (2013) Xeroderma pigmentosum complementation group of polymorphisms influence risk of glioma. Asian Pac J Cancer Prev 14: 4083–4087. [DOI] [PubMed] [Google Scholar]
- 84. Zhu ML, Wang M, Cao ZG, He J, Shi TY, et al. (2012) Association between the ERCC5 Asp1104His polymorphism and cancer risk: a meta-analysis. PLoS One 7: e36293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shi TY, He J, Qiu LX, Zhu ML, Wang MY, et al. (2012) Association between XPF polymorphisms and cancer risk: a meta-analysis. PLoS One 7: e38606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Davey SG, Egger M (1997) Meta-analyses of randomized controlled trials. Lancet 350: 1182. [DOI] [PubMed] [Google Scholar]
- 87. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analysis. Br Med J 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 89. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 90. Klug SJ, Ressing M, Koenig J, Abba MC, Agorastos T, et al. (2009) TP53 codon 72 polymorphism and cervical cancer: a pooled analysis of individual data from 49 studies. Lancet Oncol 10: 772–784. [DOI] [PubMed] [Google Scholar]
- 91. Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50: 1088–1101. [PubMed] [Google Scholar]
- 92. Egger M, Smith DG, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. Br Med J 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dual S, Tweedie R (2000) A nonparametric “trim and fill” method of accounting for publication bias in meta-analysis. J Am Stat Assoc 95: 89–98. [Google Scholar]
- 94.Higgins JPT, Green S (2008) Cochrane handbook for systematic reviews of interventions version 5.0.1. The Cochrane Collaboration, Oxford. [Google Scholar]
- 95. Visapää JP, Götte K, Benesova M, Li J, Homann N, et al. (2004) Increased cancer risk in heavy drinkers with the alcohol dehydrogenase 1C*1 allele, possibly due to salivary acetaldehyde. Gut 53: 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA Checklist.
(DOC)