Abstract
Purpose
To develop a novel imaging technique to reduce the number of excitations and required scan time for hyperpolarized 13C imaging.
Methods
A least-squares based optimization and reconstruction is developed to simultaneously solve for both spatial and spectral encoding. By jointly solving both domains, spectral imaging can potentially be performed with a spatially oversampled single echo spiral acquisition. Digital simulations, phantom experiments, and initial in vivo hyperpolarized [1-13C]pyruvate experiments were performed to assess the performance of the algorithm as compared to a multi-echo approach.
Results
Simulations and phantom data indicate that accurate single echo imaging is possible when coupled with oversampling factors greater than six (corresponding to a worst case of pyruvate to metabolite ratio < 9%), even in situations of substantial T2* decay and B0 heterogeneity. With lower oversampling rates, two echoes are required for similar accuracy. These results were confirmed with in vivo data experiments, showing accurate single echo spectral imaging with an oversampling factor of 7 and two echo imaging with an oversampling factor of 4.
Conclusion
The proposed k-t approach increases data acquisition efficiency by reducing the number of echoes required to generate spectroscopic images, thereby allowing accelerated acquisition speed, preserved polarization, and/or improved temporal or spatial resolution. Magn Reson Med
Keywords: metabolism, CSI, pyruvate, dynamic nuclear polarization
Magnetic resonance imaging (MRI) of 13C holds the potential to probe pathology at a molecular level. Unfortunately, signal from endogenous 13C is indistinguishable from noise due to low receptivity, sub millimolar in vivo concentrations, and scan time limitations. Recent advances in hyperpolarization (1,2) now allow for 100,000 fold increases in polarization, enabling the use of 13C-labeled molecules as tracers. With modern dynamic nuclear polarization, almost any endogenous, organic molecule can be polarized and subsequently imaged. For example, hyperpolarized (HP) [1-13C]pyruvate shows great potential as a molecular biomarker of cellular metabolism. In such experiments, a small sample of [1-13C]pyruvate is hyper-polarized and intravenously injected. In vivo it is transported into the cell and undergoes rapid conversion into its primary metabolic byproducts, [1-13C]lactate, [1-13C]alanine, and 13C bicarbonate (3). Preferential conversion of pyruvate to lactate is an indicator of increased glycolysis, a common feature of cancerous lesions associated with the Warburg Effect (4). Quantification of pyruvate metabolism promises to dramatically improve evaluation of prostate cancer and glioblastomas, where fluorodeoxyglucose positron emission tomography (5) shows limited prognostic value. Other promising HP compounds include 13C bicarbonate to measure extracellular pH (6) and [1,4-13C2]fumarate to measure cellular necrosis (7).
Imaging of HP 13C compounds is more challenging than other contrast agents. Immediately after the sample is removed from the polarizer, HP molecules undergo chemical and spin exchange with the endogenous environment. Polarization also decays at a rate governed by the nuclear spin–lattice relaxation time, T1. In vivo, chemical species with unique resonant frequencies are often produced, especially with injection of metabolically active molecules. As with pyruvate to lactate conversion, there is often great value in measuring the relative signal from each species, which can be achieved with spectral imaging techniques (8).
Spectral imaging with HP species presents substantially different challenges than endogenous, thermally polarized nuclei. While the signal-to-noise ratio (SNR) is quite high for a single radio frequency (RF) excitation, each excitation consumes a portion of the finite hyperpolarization (9). Due to perfusion and washout, 13C molecules are imaged in a nonequilibrium state and thus dynamic imaging is essential for potential quantification of apparent conversion rates (10). Conventional spectroscopic imaging techniques, such as chemical shift imaging (CSI) (3,11), provide excellent spectral resolution, but the long imaging time ( ~10s) and the need to phase encode both spatial dimensions limits the ability to measure dynamics. Rapid spectroscopic imaging techniques, such as echo planar spectroscopic imaging (EPSI) (12,13) and spiral CSI (14,15), greatly reduce the scan time and RF deposition but are often limited by a narrow spectral bandwidth, a consequence of gradient slew rate limitations and the reduced 13C gyromagnetic ratio (~4× lower than 1H). However, the sparse [1-13C]pyruvate spectrum is well suited to these techniques, as off-resonance peaks can be aliased without spectral overlap and corrected in the reconstruction (16).
A priori knowledge of the expected chemical species presents opportunities for model-based spectral reconstructions such as least squares estimation algorithms (e.g., iterative decomposition of water and fat with echo asymmetry and least squares estimation (IDEAL)) (17–19) and compressed sensing approaches (20). Unlike conventional Fourier-based spectroscopic imaging, optimally chosen echo times will prevent spectral aliasing without the need to fully sample the spectral dimension (21). Combined with efficient sampling strategies, such as centric phase encoding, variable flip-angle schemes (22), and spiral sampling (14,15,19), these techniques can further reduce RF decay while still providing high SNR data. Nevertheless, these sequences still require spatial or spectral interleaves to sufficiently sample kx-ky-kf space. This limits the acquisition efficiency and necessitates multiple RF excitations.
In this work, we propose a novel technique, hereafter referred to as k-t spiral, to generate spatially and spectrally resolved images with a single RF excitation. This technique oversamples spatial k-space to provide temporal sampling suitable for reconstruction of images with sparse spectra. We employ a reconstruction that jointly solves for both spatial frequency and spectral frequency dimensions (i.e., k-t sampling) utilizing a model-based approach. Using the phase evolution over the full k-t trajectory allows reconstruction from a single spiral shot. Digital simulations and phantom experiments were performed to verify theory and evaluate performance. Initial experiments are performed to show in-vivo feasibility. It is anticipated that this method can minimize RF excitations, increase acquisition speed, and mitigate polarization decay for HP 13C MRI applications.
Theory
k-t Reconstruction
In traditional approaches to spoiled gradient-echo CSI, several images are collected with the same k-space trajectory but at different echo times (TE). Ignoring the field map term, the signal at each TE can then be expressed in k-space as:
| [1] |
where S̃(,TE) is the spatial Fourier transform of the signal, is the spatial Fourier transform of the ith chemical species signal distribution with chemical shift δfi, and is the relative time between k-space acquisition and the echo time. For a forward spiral acquisition, is zero at the start of acquisition and increases throughout the readout. To obtain a fully sampled data-set, at least N echo times are required to generate N metabolite images. In order to maximize SNR efficiency for 13C applications, a long duration readout is typically used to take advantage of the extended (20–60 ms) T2* of HP 13C species (19). Using Eq. 1, data are reconstructed by first performing matrix inversion (e.g., IDEAL) or an inverse discrete Fourier transform (e.g., spiral CSI) at each k-space location to separate chemically shifted species (17). For example, with a Fourier transform:
| [2] |
where M is the number of collected echo times; , the signal distribution of the ith chemical species, can then be reconstructed by standard k-space reconstruction schemes. These reconstructions separate spectral and k-space reconstruction and are sensitive to errors from field map variations.
In a more complete form, the signal from N expected chemical species from an HP MRI experiment can be modeled:
| [3] |
where S(t) is the acquired signal, ()is the B0 field map, (t) is the k-space position, and δfi and ρi are the chemical shift and spatial signal distribution of the ith species, respectively. In principle, solving for ρi directly from Eq. 3 will produce a more accurate reconstruction than separate spatial and spectral reconstructions. Solving for ρ is a large scale, nonlinear problem due to the unknown field map; however, B0 maps can be readily obtained from 1H data and utilized to calculate 13C fields, ψ = γ(13C)B0 (23). With a known field map, Eq. 3 can be posed as lem in matrix notation:
| [4] |
where S is the signal matrix, E is the encoding matrix, which is a function of the field map, position in k-space and time (k-t space), ρ is the signal distribution for each metabolite, and ξ is the noise error. Given sufficient sampling in the k-t domain, the encoding matrix will be well conditioned and signal distributions will be accurately determined using least square minimization of the L2- norm of the differences:
| [5] |
In this reconstruction, both spectral and spatial reconstructions are performed jointly via least squares optimization (e.g., matrix inversion). While previous methods have accounted for phase accrual due to chemical shift, they either neglect field inhomogeneity (19) or demodulate the field map in image space (17), only correcting for field heterogeneities at the echo-time TE and not for each k-space point. In contrast, in this approach, phase accrual due to field heterogeneities is corrected at each k-space point, further reducing off-resonance blurring artifacts.
k-t Sampling
Traditional approaches to spiral CSI require repeated spectral interleaves (i.e., echoes), resulting in a single point in spatial k-space that is acquired at multiple echo times. Jointly solving for both spatial and spectral encoding removes this constraint. Subsequently, there is substantial freedom in the design of k-t sampling strategies and a vast array of sampling patterns which provide a sufficiently conditioned encoding matrix, including traditional approaches.
Leveraging the joint k-t reconstruction, we propose a highly efficient, modified spiral trajectory as shown in Figure 1. In this design, a spiral trajectory is designed with a δk between adjacent spiral rotations that is η times smaller than required for the nominal field of view (FOV). Ignoring the time domain, spatial k-space is over-sampled by a factor of η. For a given region in k-t space, time is also effectively sampled η times greater than a spiral designed for this nominal FOV. As a result, there is an effective δt between adjacent spiral rotations that can be utilized for chemical species separation. Data are then reconstructed at the nominal FOV, thereby enabling spectral reconstruction from the k-t spiral trajectory.
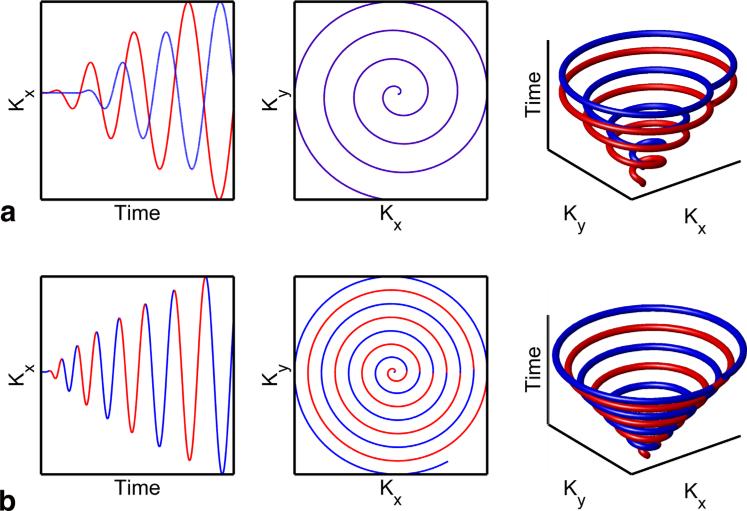
FIG. 1.
Comparison of k-t trajectories between echo time shifted spiral interleaves and a spiral trajectory prescribed for an extended FOV. (a) Two single-shot spirals are shifted in time by δTE, tracing out two conical patterns in k-t space separated by a fixed DTE. Fully sampled spatial k-space is explicitly acquired in successive echoes (red and blue). (b) A spiral with a prescribed FOV that is two times larger than the nominal FOV (η = 2) results in a more densely sampled kx-ky space. In k-t space, this covers a single conical surface, but densely samples time in a single shot. If rotations are split into odd (red) and even rotations (blue), spatial k-space will be fully sampled. This is functionally similar to the acquisition of k-space in two echoes but requires only a single shot. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
This is demonstrated in Figure 1 with a comparison of a spiral designed with η = 2 and a conventional two echo spiral with shifted echo times. For the oversampled spiral case, if rotations are split into odd (red) and even rotations (blue), spatial k-space will be fully sampled. This is functionally similar to the acquisition of k-space in two echoes, in which fully sampled k-space is explicitly acquired in successive (red and blue) echoes. These adjacent rotations are analogous to and can be modeled as additional echoes, with the temporal spacing determining the spectral noise performance. For constant linear velocity spirals designed for fixed gradient limits, the rotation time (i.e., angular velocity in k-space) is a function of the subject size, slew rate, k-space radius, and η. Similar to multi-echo fat/water separation techniques, the phase accrual between each rotation is critical for optimal performance. For clarity, we define δTE as the spacing between spectral interleaves (i.e., echoes) and δt as the spacing between adjacent spiral rotations.
Methods
All experiments were performed on or assumed the performance of a 4.7T small animal scanner (MRBR 4.7T/310, Agilent Technologies, Santa Clara, CA). The scanner was equipped with gradient coils with a maximum strength of 400 mT/m and a slew rate of 2580 T/m/s. A commercially available single-channel dual-tune 1H/13C volume coil (Doty Scientific, Columbia, SC) was used for all phantom and in vivo experiments. k-t reconstructions were performed using iterative conjugate gradient minimization implemented in C++ on a 64-bit Linux workstation (4 × 8 core AMD 6140, 128 GB ram). The encoding matrix and its transpose were evaluated by direct computation (Eq. 3).
Multi-Echo δTE Analysis
In some situations, single echo k-t sampling will be impractical and multiple echoes will be required to sufficiently sample k-t space. For a conventional IDEAL approach, the optimal δTE is chosen by maximizing the number of signal averages (NSA) for each chemical species (21,24). For joint k-t reconstruction, direct NSA calculations are computationally impractical due to the large size of the encoding matrix described in Eq. 4. In order to determine the optimal δTE for multiple spectral interleaves, digital simulations were performed with interspiral δTEs ranging from 0.1 ms to 3.0 ms in 0.1 ms increments using 2–5 echo times and FOV factors (η) of 1, 4, and 7. These FOV factors were chosen to correspond to conventional IDEAL, gradient limited, and high-performance gradient cases, respectively. Field inhomogeneities and T2* decay were not considered in the δTE analysis. Full details of the simulation are described below. Root mean square error (RMSE) was calculated by comparing the signal in each metabolite region of interest (ROI) with the ground truth (digital phantom). The optimal δTE was chosen as the smallest δTE which minimized RMSE for all metabolites.
Digital Simulations
Spiral 13C acquisitions were simulated in Matlab (R2009b, The MathWorks, Natick, MA) for [1-13C]pyruvate and downstream metabolites at 4.7T. Simulations utilized a high-resolution digital phantom comprised of four distinct circles with smooth edges, representing pyruvate, lactate (δf = 614 Hz), alanine (δf 272 Hz), and pyruvate-hydrate (δf = 433 Hz). Metabolite ratios were simulated as 5 for pyruvate/lactate and 10 for both pyruvate/alanine and pyruvate/pyruvate-hydrate. 13C bicarbonate was excluded from simulations because of very low signal levels in our principle test application of renal imaging (25). From the phantom, k-space data were simulated by direct evaluation of Eq. 4 under idealized and realistic conditions. In idealized conditions, noise, field map heterogeneity, and T2* were not considered. Under realistic conditions, independently realized complex noise was added to simulated k-space data to mimic the SNR obtained in vivo. Realistic conditions also considered a Gaussian field map with off-resonance range of ±25 Hz and a global T2* of 15 ms, corresponding to a linewidth of 21 Hz. This simulated field map corresponds to ±100 Hz for 1H imaging and is at the high end of what we typically observe on our small-bore, pre-clinical imaging system.
Constant linear velocity Archimedean spirals were designed with time-optimal gradients (26) for η factors ranging from 1 to 9, corresponding to designed FOVs ranging from 32 × 32 mm2 to 288 × 288 mm2. In-plane resolution was 2 × 2 mm2 for all simulations, while the slew rate was reduced for lower η in order to keep the acquisition time of 30 ms constant. A five echo acquisition was simulated, with TR/TE = 50 ms/0.6 ms, corresponding to a simulated scan time ranging from 50 to 250 ms. An echo spacing of 2.0 ms was chosen in order to provide good noise performance across all echo combinations. Data were reconstructed using 1–5 echoes in order to determine the minimum number of echoes required for accurate reconstruction. Ten independent noise realizations were performed for each simulation. Reconstruction fidelity was assessed by measuring the RMSE of each metabolite as well as pyruvate to metabolite signal ratios.
Phantom Validation
A thermally polarized multispectral phantom comprised of 13C urea (δf = –488 Hz), [1-13C]glycine (δf = 0 Hz), and [1,4-13C2]succinate (δf = 493 Hz) was imaged to experimentally validate the reconstruction technique. A [1-13C]pyruvate phantom was not used due to the development of impurities in the neutralized solution over time (27). Each compound was placed in a separate 1 mL syringe and doped with 4 mM Multihance (Bracco, Princeton, NJ) to shorten T1. The three syringes were placed in a triangular configuration and inserted into a 50 mL vial filled with water. Spiral data were initially obtained with an FOV of 32 × 32 mm2 and a 24 × 24 matrix resulting in an in-plane resolution of 1.33 × 1.33 mm2. Five echoes were obtained per dataset, with a 20° flip-angle and TR/TE = 45 ms/0.6 ms. A δTE of 0.68 ms was used based on NSA performance analysis for the three chemical species. A 10-mm-thick slice and 768 averages were acquired to increase SNR, for a total scan time of 173 s. Three subsequent experiments were performed, with the FOV and matrix size concomitantly increased by a factor of 2–4. The readout time of 30.7 ms was held constant by derating the gradients in order to provide equal noise levels in raw data across all experiments. To correct the 13C data for phase errors arising from field inhomogeneity, a field map was obtained on the 1H channel with a multi-echo gradient echo sequence. A spectral–spatial excitation was used to minimize contamination from off-resonance 1H chemical species. Reconstructions were performed using 1–5 echoes. Reconstruction accuracy was measured by calculating the RMSE of each chemical species with respect to the fully sampled, five echo acquisition.
In Vivo Experiments
HP experiments were performed in a healthy female ICR mouse (43 g; 1 year of age) to assess the efficacy of k-t spiral in dynamic HP experiments. 30 μL samples of [1-13C]pyruvic acid (Sigma-Aldrich, St. Louis, MO) and 15 mM trityl radical (OX63; GE Healthcare) were inserted into a Hypersense polarizer (Tubney Woods, Abingdon, Oxfordshire, UK), cooled to 1.4 K and irradiated with 94.1 GHz microwaves for 1 h. Samples were subsequently dissoluted with 4 mL 100 mM NaOH/Tris buffer and 100 mg/L EDTA, resulting in a solution of 100 mM pyruvate at physiologic temperature and pH. Pyruvate was rapidly injected via a tail-vein catheter at a dose of 9 μL/g into the mouse. The mouse was fasted for 4.5 h prior to imaging in order to achieve a reproducible metabolic state. Animal respiration was monitored and temperature was maintained at 36.5 ± 0.5° C with a hotair blower. All experiments were performed under protocols approved by our Institutional Animal Care and Use Committee.
Two sets of HP 13C imaging experiments were performed. First, data were acquired with an FOV of 224 × 224 mm2 and a 112 × 112 matrix, corresponding to η = 7 and resulting in an in-plane resolution of 2 × 2 mm2. Five echoes were acquired, with a 30.24 ms readout duration and TR/TE/δTE = 50/0.6/2.0 ms for a scan time of 250 ms per timeframe. A 10° , 300 ms sinc excitation was used to excite a 10-mm-thick slice while minimizing chemical shift artifacts in the slice-select direction. Prior to spiral imaging, a slice-selective spectrum with a 5 kHz spectral bandwidth was acquired to determine the metabolite frequencies relative to the center frequency for use in the reconstruction. A 5 s delay was inserted between imaging frames to allow time for metabolism. In a second experiment, data were acquired with a reduced FOV factor of η = 4 to measure performance in situations where larger FOV factors are impractical due to gradient hardware limitations. All other scan parameters were identical. Data were corrected for RF decay and separately reconstructed using 1–5 echoes. Whole kidney ROIs were drawn based on an anatomical 1H image in order to calculate the average metabolite signal at each time frame. Fidelity was assessed by comparing metabolite dynamics for each reconstruction to the fully sampled five echo reconstruction. The difference between the two curves across time was assessed by calculating RMSE and intraclass correlation coefficients (ICC) for pyruvate and lactate.
For all HP experiments, an estimate of the field inhomogeneity was obtained prior to 13C imaging with a 1H multi-echo gradient echo sequence. Optimal echo times determined from fat-water NSA analysis and the field map was calculated using image-space IDEAL (28). Following 13C experiments, the spiral trajectory was measured on the 1H channel using a thin-slice excitation technique (29).
Results
Multi-Echo δTE Analysis
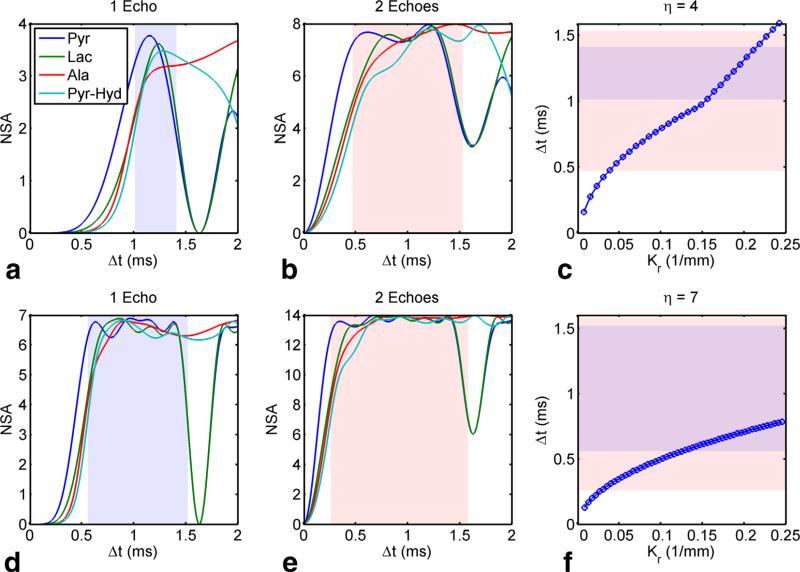
Figure 2 shows the NSA and temporal spacing for two different spatially oversampled trajectories (η 4 and 7), both with equal readout durations. For conventional spectroscopic imaging, NSA (21) describes the noise performance for individual metabolites and is, at maximum, equal to the number of echoes. Regions of acceptable noise performance were defined from the NSA plots as having ≥0.5 of the maximum for all metabolites and are indicated by the shaded regions. These NSA calculations assume constant temporal spacing (δt) between adjacent rotations, but approximate NSA dependence on k-space radius for the constant linear velocity spirals used in this work. For a single echo, the temporal spacing is insufficient in the center of k-space and thus some noise amplification is expected. For two echoes separated by 2 ms, the region of acceptable temporal spacing is substantially larger and comprises all but the center of k-space for both oversampling factors. Therefore, we expect variable noise performance throughout the readout, with higher noise for samples acquired at the center of k-space.
FIG. 2.
Number of signal averages (NSA) for η = 4 acquired with a single echo (a) and two echoes (b). Note that there are 4 and 8 equi-spaced δt's, respectively, between adjacent spiral rotations in a compared to b with δTE between echoes of 2 ms reflecting that used experimentally. For constant linear velocity spirals, temporal spacing varies as a function of k-space radius (c) making the noise performance vary with spatial frequency. The shaded regions indicate robust noise performance (empirically defined as NSA ≥ 0.5 of the maximum) for all four metabolites highlighting that the worst performance is at the center of k-space. The same analysis is repeated for η = 7, also with one echo (d) compared to two echoes (e), with similar dependence of noise performance as a function of k-space radius (f). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
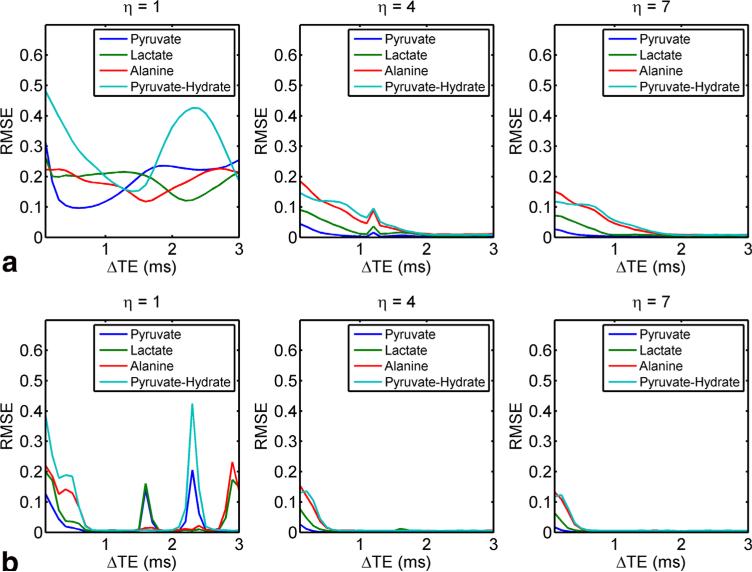
This is corroborated by Figure 3, which shows RMSE plots for pyruvate, lactate, alanine, and pyruvate-hydrate at 4.7T as a function of δTE. For η = 4 or 7, RMSE is less than 5% for δTE ≥ 1.1 ms using two echoes or δTE > 1.4 ms using five echoes. This broad RMSE minimum indicates that the choice of δTE is not critical to obtain optimal noise performance, so long as a sufficiently large echo spacing is prescribed to allow sufficient phase accrual between spectral interleaves. Forη = 1, which corresponds to conventional spiral IDEAL CSI, with only two echoes the system is under-determined, as reflected by the large RMSE for all δTE values. In contrast, using five echo times results in regions of local minima with low RMSE. Local maxima in RMSE for η = 1 correspond to in-phase images, resulting in poor noise performance at these δTE times. As aforementioned, δTE = 2 ms was chosen for all simulations and in vivo experiments to provide good noise performance for all values of η across all echo combinations.
FIG. 3.
Relative RMSE as a function of DTE for two echoes (a) and five echoes (b) for three FOV factors, corresponding to conventional IDEAL, gradient limited, and high-performance gradients, respectively. Poor noise performance is indicated by high RMSE and occurs when in-phase images are acquired. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Digital Simulations
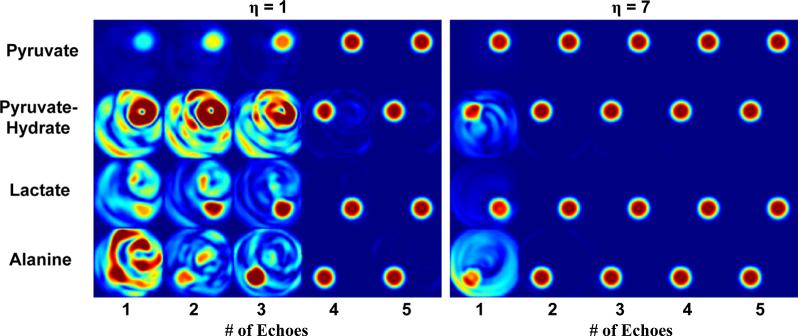
Representative metabolite images for an idealized case can be seen in Figure 4. For η = 1 (spiral designed for 32 mm FOV), metabolite maps reconstructed using fewer than four echoes are contaminated by spectral cross-talk. When four or more echoes are used, the system is fully determined and spectral contamination is minimal. In this case, the joint k-t reconstruction technique is similar to spiral IDEAL CSI but directly accounts for field map variations. When larger FOV factors are used, joint reconstruction reduces the number of echo images required, reducing both scan time and RF excitations. In simulations with η = 7, spectral cross-talk is reduced using a single echo and completely mitigated for two or more echoes (Fig. 4). This is confirmed by relative RMSE measurements, which are less than 25% for all metabolites using a single echo for η ≥ 7 and less than 10% using two echoes for η ≥ 3.
FIG. 4.
Digital simulation metabolite images reconstructed with k-t spiral. For a FOV factor of η = 1, at least four echoes are required to adequately separate off-resonant species. Larger FOV factors (η = 7) reduce the number of spectral interleaves (and therefore RF deposition) necessary to generate spectral images, enabling accelerated imaging with fewer than four echoes. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
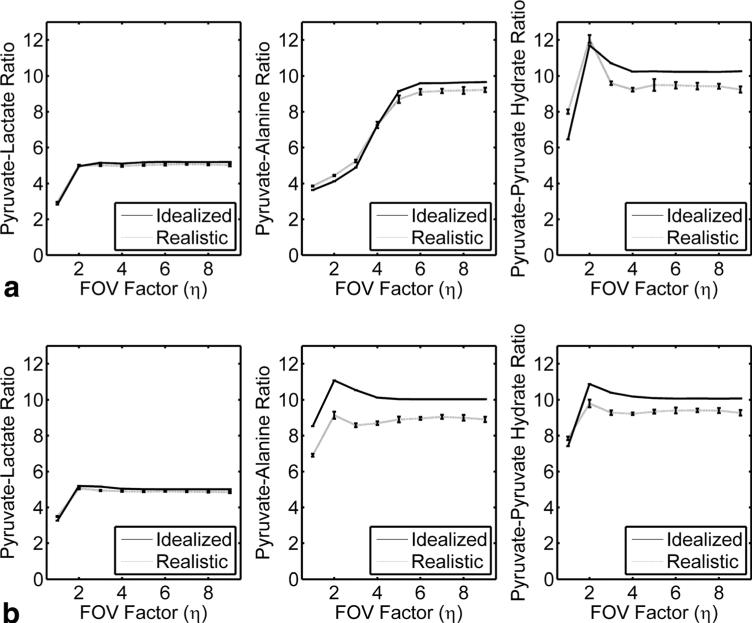
This can be observed in the measured signal ratios between metabolites across a range of FOV factors (Fig. 5). In the idealized case, which ignores field inhomogeneities, T2* decay and system noise, signal ratios using two echoes were within 2% for η ≥ 4. For only a single echo, all metabolite signal ratios were less than 5% for η ≥ 6. Even in the realistic case with field inhomogeneities and a short T2* , the k-t spiral reconstruction accurately measures metabolite ratios. Simulations incorporating realistic noise levels (5% independently realized complex noise in k-space) with field inhomogeneity (±25 Hz) and T2* (15 ms) resulted in metabolites signal ratios that were within 13% for all metabolites using two echo times for η ≥ 4. For a single shot, all metabolite signal ratios were less than 9% for η ≥ 6, indicating that high-fidelity single-shot imaging is attainable with a large FOV factor.
FIG. 5.
Signal ratios as a function of FOV factor (η) for simulations using one (a) or two (b)spectral interleaves. The idealized case was simulated in the absence of noise, T2* or field inhomogeneities. The realistic case was simulated with 5% noise, a 15 ms T2* and δB0 = ± 25 Hz. Under realistic conditions, signal ratios are within 9% for η ≥ 6 for all metabolites using only one echo and are within 13% for η ≥ 4 for all metabolites using two echoes.
Multispectral 13C Phantom
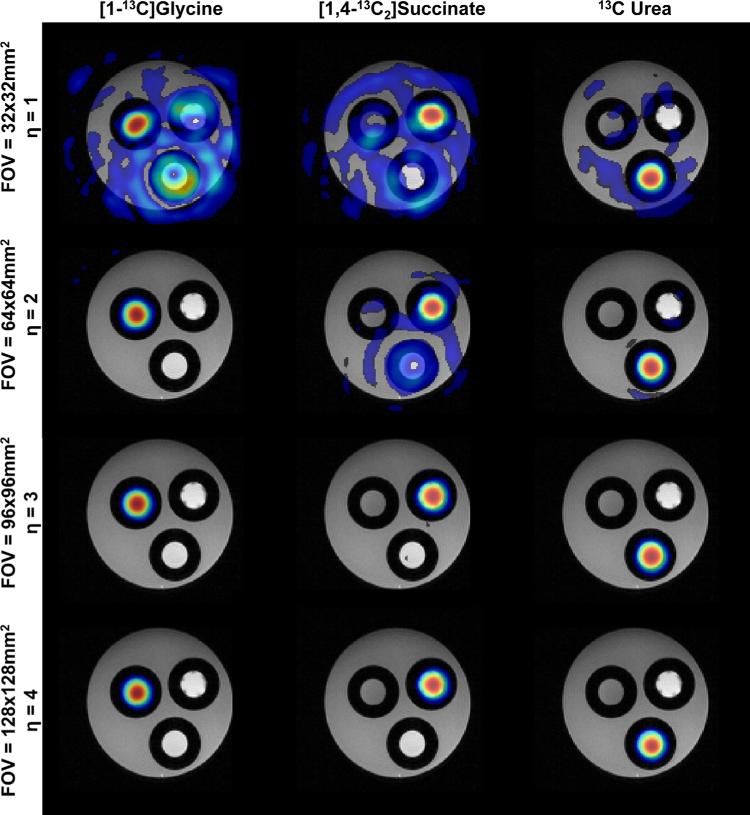
Data from the 13C multispectral phantom can be seen in Figure 6. Reconstruction with only one echo at a prescribed FOV of 32 × 32 mm2 (η = 1; Fig. 6, top row) results in images that are contaminated from spectral cross-talk. This is expected, as the system is underdeter-mined. As the FOV factor increases, adjacent spiral rotations can be modeled as intraspiral echoes, enabling reconstruction from data acquired with only a single echo and resulting in images that are free of spectral contamination. Qualitatively, this can be measured by calculating RMSE compared with a five echo acquisition as the reference standard. For η = 1, three or more echoes are required to reduce the RMSE to less than 20% for all three species, while increasing η to 4 reduces the RMSE to less than 5%with only a single RF excitation.
FIG. 6.
Single-echo multispectral 13C phantom data reconstructed with the k-t spiral algorithm. Chemical species images have been independently thresholded to 10% of the maximum signal intensity in each frame. Using only a single echo, data acquired with an FOV of 32 × 32 mm2 (corresponding to η = 1) show significant spectral cross-talk, indicating ill-posed conditions in the k-t reconstruction. When data are acquired with η = 3 or greater, correlation between adjacent spiral rotations can be modeled as intraspiral echo times, resulting in spectral images that can be reconstructed with a single shot. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
In vivo Feasibility
In vivo results from renal imaging using HP [1-13C]pyruvate are shown in Figure 7. Pyruvate and lactate images are in good agreement with underlying anatomy, with pyruvate images dominated by the vasculature and lac-tate predominantly confined to the kidneys and liver. Metabolite dynamics from the same experiment are shown in Figure 8. Average signal intensity from a whole kidney ROI reconstructed using all five echoes was considered as an internal reference standard. RMSE and ICC were calculated by comparing the metabolite dynamics for each reconstruction to the reconstruction using a fully sampled five echo dataset. The RMSE and ICC (Table 1) indicate strong signal fidelity for the single echo acquisition.
FIG. 7.
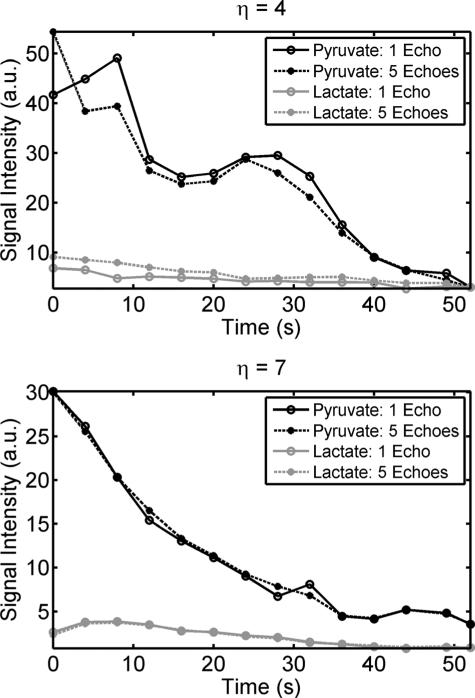
Single time point images of pyruvate and lactate as a function of number of echoes used for image reconstruction for separate experiments acquired with η = 4 and 7. With η = 4, single-shot imaging is not feasible and results in spectral contamination. However, metabolites can be separated with two or more echoes, half as many required for conventional IDEAL. Utilizing η = 7, images show limited contamination with as few as a single echo. This is due to the more complete sampling of k-t space. Each set of metabolite images has been equally displayed across the echo dimension and thresholded to 5% (pyruvate) and 20% (lactate) of the maximum signal intensity for each metabolite.
FIG. 8.
Metabolite time course for pyruvate and lactate for reconstructions using only a single echo or all five echoes. For an FOV factor η of 4, a single-shot acquisition poorly represents the true dynamics, as compared to the five echo reconstruction. In contrast, acquiring data with a larger η results in strong agreement, indicating that single-shot imaging with large η accurately measures metabolism.
Table 1.
Root Mean Square Error (RMSE) and Intraclass Correlation Coefficient (ICC) Between a Single Echo k-t Spiral Reconstruction and a Five Echo Reconstruction
|
η = 7 |
η = 4 |
|||
|---|---|---|---|---|
| RMSE | ICC | RMSE | ICC | |
| Pyruvate, single echo | 0.022 | 0.986 | 0.128 | 0.96 |
| Lactate, single echo | 0.036 | 0.912 | 0.11 | 0.84 |
| Pyruvate, two echoes | 0.015 | 0.994 | 0.123 | 0.98 |
| Lactate, two echoes | 0.02 | 0.973 | 0.033 | 0.95 |
| Pyruvate, three echoes | 0.013 | 0.997 | 0.043 | 0.985 |
| Lactate, three echoes | 0.017 | 0.994 | 0.041 | 0.954 |
| Pyruvate, four echoes | 0.006 | 0.998 | 0.009 | 0.993 |
| Lactate, four echoes | 0.009 | 0.999 | 0.040 | 0.968 |
With a large FOV factor (η = 7), both the RMSE and ICC indicate that a single-shot acquisition has high fidelity. Conversely, data acquired with a single shot using a smaller FOV factor (η = 4) result in metabolite dynamics that are poorly represented, especially for lactate. Using two echoes mitigates this error.
Data acquired with η = 4 present a more realistic FOV factor, representing image quality that can be obtained with gradients typical of a clinical system. Metabolite dynamics measured with only a single echo varies considerably compared with the five echo reconstruction (Fig. 8). Similarly, RMSE and ICC are substantially worse, with RMSE > 11% for both pyruvate and lactate using only a single echo (Table 1). Reconstruction with two echoes improves image quality and yields RMSE and ICC that agree well with the five echo dataset.
Discussion
In this work, we developed a novel method to generate both spectrally and spatially resolved images. This method jointly reconstructs spectral and spatial domains allowing flexible design of trajectories in k-t space and reduced sensitivity to B0 heterogeneity. Utilizing this approach, k-t spiral sampling was developed to leverage intraspiral rotation time to reduce the number of echoes required. Digital simulations of [1-13C]pyruvate metabolism and spectral 13C phantom scans were used to evaluate this method and demonstrated accurate reconstruction with a single-shot spiral acquisition with a large spatial oversampling factor, η. Finally, in vivo imaging experiments with HP [1-13C]pyruvate in a healthy mouse showed excellent fidelity with the single-shot technique when η was large. For a modest η, which represents the FOV factors that can be obtained with a clinical scanner platform, only two echoes were required to reconstruct data with acceptable RMSE and ICC.
Joint k-t reconstruction with modified spirals reduces the number of echoes required, resulting in images that can be acquired with fewer RF excitations. The remaining signal and imaging time can be utilized to enable imaging at a higher temporal resolution, improved spatial resolution or coverage, and/or with magnetization preparation to improve quantification. In the case of HP [1-13C]pyruvate, single echo k-t spiral offers the opportunity for fourfold faster imaging. This is without the considerable SNR penalty incurred when utilizing short readout multi-echo approaches which do not efficiently utilize the long T2* of 13C. This may allow the extension of 2D techniques to 3D approaches, where modeling is expected to be more accurate but scan times and RF requirements are presently prohibitive. For example, recently, Josan et al. (30) acquired a 3D spatial/1D spectral imaging volume in 4.5 s with spiral CSI. This approach required 36 excitations per volume. For single-shot k-t spiral, an equal number of slices would only require ~500 ms per volume and 12 excitations. This shorter scan time will allow the incorporation of physiological gating for cardiac applications and magnetization preparation for improved quantification.
In addition to the spectral imaging techniques discussed above, spectral-spatial (SPSP) RF pulses have also found wide use in HP 13C imaging. Unlike sinc pulses that have a broadband frequency response, SPSP pulses have a narrow excitation passband and provide strong off-resonance suppression by virtue of their design. However, they are highly sensitive to field inhomogeneities and are highly challenging for species with small relative chemical shifts. While off-resonance corrections to the trajectory are readily incorporated into reconstruction algorithms (31), these cannot correct for cross contamination of off-resonance chemical species. The effective flip-angle (and therefore SNR) will also decrease for high B0 heterogeneity because of the narrow SPSP passband. While this heterogeneity is mitigated by multiband spectral-spatial excitation (32) or higher-order shims and small bore sizes on pre-clinical scanners, it presents a significant challenge for large bore imaging on a clinical scanner and in areas of significant susceptibility (e.g., above the nasal fossa).
In our approach, 1H data provide a high-resolution dataset that can be averaged to improve estimates of field heterogeneity in regions of low signal. Errors in either the field map or assumed chemical shift, or unaccounted compounds will lead to off-resonance blurring and potential quantitation errors. Moreover, motion between the field map acquisition and the HP 13C experiment may lead to misregistration or alter the B0 field, hindering spectral separation. In practice, acquiring a spectrum prior to spiral imaging reduces the possibility that the frequency shifts are incorrect. However, a more elegant solution would be to simultaneously image 1H and 13C (33) using k-t spiral to provide 13C metabolite maps and a real time, inherently registered field map and FOV support derived from the 1H spiral data.
Errors in the B0 field map could be addressed with nonlinear reconstructions; however, the reconstruction time is, at the moment, impractical. Currently, joint k-t reconstruction is computationally intensive, requiring ~10 min to reconstruct a 128 × 128 dataset acquired with a single shot for four chemical species and 16 time points on a Linux workstation with a 4 × 8 core AMD 6140 and 128 GB memory. The potential exists to increase reconstruction speed by incorporating time segmentation of the k-space trajectory (34) or via implementing a GPU-based reconstruction.
In this work, we utilized 13C compounds with a relatively sparse spectrum. For more complex spectra, it is likely that multiple echoes may be needed. For example, bicarbonate production is routinely observed in HP brain (15) and heart (35) studies, requiring future simulations to determine the optimal echo spacing and η. Large FOV factors also require high-slew rates and strong gradients. Human in vivo use of k-t spiral is therefore likely to be more challenging than in the mouse model, and single-shot acquisitions may not be feasible for high-resolution scans.
Several other improvements to the k-t spiral method may improve performance but were not explored in this study. All data were acquired with a constant linear velocity spiral trajectory, resulting in short intraspiral temporal spacing at the center of spatial k-space and longer spacing at the edge. As shown in Figure 2, there is not sufficient phase accrual during the center of k-space, resulting in poor noise performance. This analysis does not evaluate the spatial component, however, and it would be of value to jointly analyze the spatial/spectral performance in future studies. Furthermore, the variation in noise performance with spatial frequency may depend on object sparsity.
Alternative spiral design strategies may allow for substantial improvements in performance. For example, a constant angular velocity spiral will by definition have uniform temporal spacing between adjacent spiral rotations (36). Trajectories could therefore be designed with optimal separation between adjacent spiral rotations based on NSA analysis, potentially providing increased noise performance in certain situations. However, one tradeoff for constant angular velocity spirals is the limit imposed on the obtainable η for a fixed readout duration.Alternatively, variable density or variable slew rate sampling could be utilized to provide a higher η or slower rotation at the center of k-space compared with the edge. Additionally, incorporating parallel imaging, which naturally increases the effective FOV, and compressed sensing may further improve noise performance. It should be noted that both parallel imaging and compressed sensing are highly compatible with the k-t reconstruction.
Conclusion
Jointly solving for both spatial and spectral encoding with a least-squares based optimization and reconstruction technique enables accelerated 2D spatial/1D spectral imaging of HP 13C metabolites. By reducing the RF requirements, k-t spiral improves flexibility in sampling strategies and reduces acquisition time, enabling higher temporal resolution.
Acknowledgments
Grant sponsors: NIH/NIMH 1 P50AG033514, GE Healthcare, Department of Radiology, University of Wisconsin-Madison.
REFERENCES
- 1.Ardenkjær-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc Natl Acad Sci USA. 2003;100:10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ardenkjaer-Larsen JH, Leach AM, Clarke N, Urbahn J, Anderson D, Skloss TW. Dynamic nuclear polarization polarizer for sterile use intent. NMR Biomed. 2011;24:927–932. doi: 10.1002/nbm.1682. [DOI] [PubMed] [Google Scholar]
- 3.Golman K, Rit Zandt, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic Imaging by Hyperpolarized 13C Magnetic Resonance Imaging for In vivo Tumor Diagnosis. Cancer Res. 2006;66:10855–10860. doi: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- 4.Gillies R, Gatenby R. Adaptive landscapes and emergent phenotypes: why do cancers have high glycolysis? J Bioenerg Biomemb. 2007;39:251–257. doi: 10.1007/s10863-007-9085-y. [DOI] [PubMed] [Google Scholar]
- 5.Kurhanewicz J, Vigneron DB, Brindle K, et al. Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia. 2011;13:81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallagher FA, Kettunen MI, Day SE, et al. Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature. 2008;453:940–943. doi: 10.1038/nature07017. [DOI] [PubMed] [Google Scholar]
- 7.Witney TH, Kettunen MI, Hu De, Gallagher FA, Bohndiek SE, Napolitano R, Brindle KM. Detecting treatment response in a model of human breast adenocarcinoma using hyperpolarised [1–13C] pyruvate and [1,4–13C2] fumarate. Br J Cancer. 2010;103:1400–1406. doi: 10.1038/sj.bjc.6605945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adalsteinsson E, Irarrazabal P, Topp S, Meyer C, Macovski A, Spielman DM. Volumetric spectroscopic imaging with spiral-based k-space trajectories. Mag Reson Med. 1998;39:889–898. doi: 10.1002/mrm.1910390606. [DOI] [PubMed] [Google Scholar]
- 9.Zhao L, Mulkern R, Tseng C-H, Williamson D, Patz S, Kraft R, Wals-worth RL, Jolesz FA, Albert MS. Gradient-Echo Imaging Considerations for Hyperpolarized129Xe MR. J Mag Reson, Series B. 1996;113:179–183. [PubMed] [Google Scholar]
- 10.Day SE, Kettunen MI, Gallagher FA, Hu DE, Lerche M, Wolber J, Golman K, Ardenkjaer-Larsen JH, Brindle KM. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med. 2007;13:1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- 11.Kohler SJ, Yen Y, Wolber J, et al. In vivo 13carbon metabolic imaging at 3T with hyperpolarized 13C-1-pyruvate. Magn Reson Med. 2007;58:65–69. doi: 10.1002/mrm.21253. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham CH, Chen AP, Albers MJ, Kurhanewicz J, Hurd RE, Yen Y-F, Pauly JM, Nelson SJ, Vigneron DB. Double spin-echo sequence for rapid spectroscopic imaging of hyperpolarized 13C. J Magn Reson. 2007;187:357–362. doi: 10.1016/j.jmr.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Larson PEZ, Bok R, Kerr AB, Lustig M, Hu S, Chen AP, Nelson SJ, Pauly JM, Kurhanewicz J, Vigneron DB. Investigation of tumor hyper-polarized [1–13C]-pyruvate dynamics using time-resolved multiband RF excitation echo-planar MRSI. Magn Reson Med. 2010;63:582–591. doi: 10.1002/mrm.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer D, Levin YS, Hurd RE, Glover GH, Spielman DM. Fast metabolic imaging of systems with sparse spectra: Application for hyper-polarized 13C imaging. Magn Reson Med. 2006;56:932–937. doi: 10.1002/mrm.21025. [DOI] [PubMed] [Google Scholar]
- 15.Mayer D, Yen Y-F, Takahashi A, Josan S, Tropp J, Rutt BK, Hurd RE, Spielman DM, Pfefferbaum A. Dynamic and high-resolution metabolic imaging of hyperpolarized [1–13C]-pyruvate in the rat brain using a high-performance gradient insert. Magn Reson Med. 2011;65:1228–1233. doi: 10.1002/mrm.22707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayer D, Yen Y-F, Tropp J, Pfefferbaum A, Hurd RE, Spielman DM. Application of subsecond spiral chemical shift imaging to real-time multislice metabolic imaging of the rat in vivo after injection of hyperpolarized 13C1-pyruvate. Magn Reson Med. 2009;62:557–564. doi: 10.1002/mrm.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brodsky EK, Holmes JH, Yu H, Reeder SB. Generalized k-space decomposition with chemical shift correction for non-cartesian water-fat imaging. Magn Reson Med. 2008;59:1151–1164. doi: 10.1002/mrm.21580. [DOI] [PubMed] [Google Scholar]
- 18.Schulte RF, Sperl JI, Haase A, Irkens M, Manglberger M, Weidl E, Kudielka G, Schwaiger M, Weisinger F. Advanced parallel imaging techniques for metabolic imaging with hyperpolarised 13C.. Proceedings of the 19th Annual Meeting of ISMRM; Montreal, Canada. 2011; Abstract 1522. [Google Scholar]
- 19.Wiesinger F, Weidl E, Menzel MI, Janich MA, Khegai O, Glaser SJ, Haase A, Schwaiger M, Schulte RF. IDEAL spiral CSI for dynamic metabolic MR imaging of hyperpolarized [1–13C]pyruvate. Magn Reson Med. 2012;68:8–16. doi: 10.1002/mrm.23212. [DOI] [PubMed] [Google Scholar]
- 20.Hu S, Lustig M, Chen AP, et al. Compressed sensing for resolution enhancement of hyperpolarized 13C flyback 3D-MRSI. J Magn Reson. 2008;192:258–264. doi: 10.1016/j.jmr.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reeder SB, Brittain JH, Grist TM, Yen Y-F. Least-squares chemical shift separation for 13C metabolic imaging. J Magn Reson Imag. 2007;26:1145–1152. doi: 10.1002/jmri.21089. [DOI] [PubMed] [Google Scholar]
- 22.Yen YF, Kohler SJ, Chen AP, et al. Imaging considerations for in vivo 13C metabolic mapping using hyperpolarized 13C-pyruvate. Magn Reson Med. 2009;62:1–10. doi: 10.1002/mrm.21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon JW, Fain SB, Johnson KM. Direct estimation of hyperpolarized metabolites with IDEAL spiral CSI.. In Proceedings of the 20th Annual Meeting of ISMRM; Melbourne, Australia. 2012; Abstract 4299. [Google Scholar]
- 24.Pineda AR, Reeder SB, Wen Z, Pelc NJ. Cramer–Rao bounds for three-point decomposition of water and fat. Magn Reson Med. 2005;54:625–635. doi: 10.1002/mrm.20623. [DOI] [PubMed] [Google Scholar]
- 25.Xu T, Mayer D, Gu M, Yen Y-F, Josan S, Tropp J, Pfefferbaum A, Hurd R, Spielman D. Quantification of in vivo metabolic kinetics of hyperpolarized pyruvate in rat kidneys using dynamic 13C MRSI. NMR Biomed. 2011;24:997–1005. doi: 10.1002/nbm.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lustig M, Seung-Jean K, Pauly JM. A fast method for designing time-optimal gradient waveforms for arbitrary k-space trajectories. Med Imag IEEE Trans. 2008;27:866–873. doi: 10.1109/TMI.2008.922699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margolis SA, Coxon B. Identification and quantitation of the impurities in sodium pyruvate. Anal Chem. 1986;58:2504–2510. [Google Scholar]
- 28.Hernando D, Kellman P, Haldar JP, Liang ZP. Robust water/fat separation in the presence of large field inhomogeneities using a graph cut algorithm. Magn Reson Med. 2010;63:79–90. doi: 10.1002/mrm.22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duyn JH, Yang Y, Frank JA, van der Veen JW. Simple correction method fork-space trajectory deviations in MRI. J Magn Reson. 1998;132:150–153. doi: 10.1006/jmre.1998.1396. [DOI] [PubMed] [Google Scholar]
- 30.Josan S, Spielman D, Yen Y-F, Hurd R, Pfefferbaum A, Mayer D. Fast volumetric imaging of ethanol metabolism in rat liver with hyperpolarized [1–13C]pyruvate. NMR Biomed. 2012;25:993–999. doi: 10.1002/nbm.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W, Meyer CH. Fast automatic linear off-resonance correction method for spiral imaging. Magn Reson Med. 2006;56:457–462. doi: 10.1002/mrm.20973. [DOI] [PubMed] [Google Scholar]
- 32.Larson PEZ, Kerr AB, Chen AP, Lustig MS, Zierhut ML, Hu S, Cunningham CH, Pauly JM, Kurhanewicz J, Vigneron DB. Multiband excitation pulses for hyperpolarized 13C dynamic chemical-shift imaging. J Magn Reson. 2008;194:121–127. doi: 10.1016/j.jmr.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson E, Wang K, Kurpad K, Erickson M, Rowland I, Fain S. Simultaneous proton and hyperpolarized carbon imaging.. Proceedings of the 18th Annual Meeting of ISMRM; Stockholm, Sweden. 2010; Abstract 1020. [Google Scholar]
- 34.Sutton BP, Noll DC, Fessler JA. Fast, iterative image reconstruction for MRI in the presence of field inhomogeneities. Med Imag IEEE Trans. 2003;22:178–188. doi: 10.1109/tmi.2002.808360. [DOI] [PubMed] [Google Scholar]
- 35.Chen AP, Hurd RE, Schroeder MA, Lau AZ, Gu Y-p, Lam WW, Barry J, Tropp J, Cunningham CH. Simultaneous investigation of cardiac pyruvate dehydrogenase flux, Krebs cycle metabolism and pH, using hyperpolarized [1,2–13C2]pyruvate in vivo. NMR Biomed. 2012;25:305–311. doi: 10.1002/nbm.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahn CB, Kim JH, Cho ZH. High-speed spiral-scan echo planar NMR imaging-I. Med Imag IEEE Trans. 1986;5:2–7. doi: 10.1109/TMI.1986.4307732. [DOI] [PubMed] [Google Scholar]